Abstract
To ascertain whether on animal farms there reside extended-spectrum β-lactamase (ESBL) and plasmidic class C β-lactamase-producing Escherichia coli isolates potentially pathogenic for humans, phylogenetic analyses, pulsed-field gel electrophoresis (PFGE) typing, serotyping, and virulence genotyping were performed for 86 isolates from poultry (57 isolates) and pig (29 isolates) farms. E. coli isolates from poultry farms carried genes encoding enzymes of the CTX-M-9 group as well as CMY-2, whereas those from pig farms mainly carried genes encoding CTX-M-1 enzymes. Poultry and pig isolates differed significantly in their phylogenetic group assignments, with phylogroup A predominating in pig isolates and phylogroup D predominating in avian isolates. Among the 86 farm isolates, 23 (26.7%) carried two or more virulence genes typical of extraintestinal pathogenic E. coli (ExPEC). Of these, 20 were isolated from poultry farms and only 3 from pig farms. Ten of the 23 isolates belonged to the classic human ExPEC serotypes O2:H6, O2:HNM, O2:H7, O15:H1, and O25:H4. Despite the high diversity of serotypes and pulsotypes detected among the 86 farm isolates, 13 PFGE clusters were identified. Four of these clusters contained isolates with two or more virulence genes, and two clusters exhibited the classic human ExPEC serotypes O2:HNM (ST10) and O2:H6 (ST115). Although O2:HNM and O2:H6 isolates of human and animal origins differed with respect to their virulence genes and PFGE pulsotypes, the O2:HNM isolates from pigs showed the same sequence type (ST10) as those from humans. The single avian O15:H1 isolate was compared with human clinical isolates of this serotype. Although all were found to belong to phylogroup D and shared the same virulence gene profile, they differed in their sequence types (ST362-avian and ST393-human) and PFGE pulsotypes. Noteworthy was the detection, for the first time, in poultry farms of the clonal groups O25b:H4-ST131-B2, producing CTX-M-9, and O25a-ST648-D, producing CTX-M-32. The virulence genes and PFGE profiles of these two groups were very similar to those of clinical human isolates. While further studies are required to determine the true zoonotic potential of these clonal groups, our results emphasize the zoonotic risk posed especially by poultry farms, but also by pig farms, as reservoirs of ESBL- and CMY-2-encoding E. coli.
Extended-spectrum β-lactamases (ESBL) and plasmidic class C β-lactamases have emerged as clinically relevant antimicrobial resistance mechanisms. Enterobacteriaceae harboring these enzymes include pathogens of animals and humans and commensals of their intestinal tract, such as Escherichia coli, a potential reservoir of these resistances. Recently, we demonstrated that farms are an important source of E. coli isolates carrying the genes encoding both types of lactamases (2, 22) and identified the conjugative plasmids harboring these resistances (3).
Some pathotypes of E. coli are capable of causing intestinal diseases, while others, referred to as extraintestinal pathogenic E. coli (ExPEC), are responsible for extraintestinal infections. Usually, commensal E. coli isolates harbor no or only very few virulence factors (VFs), while ExPEC isolates have specialized VFs enabling them to colonize host surfaces, injure host tissues, and avoid or subvert host defense systems (15). ExPEC isolates have been implicated in a wide range of human and animal infections. Human ExPEC, avian pathogenic E. coli (APEC), and other animal ExPEC isolates significantly overlap with respect to their O antigens, phylogenetic groups, and virulence genotypes (1, 5, 6, 8, 17, 26). Furthermore, some APEC strains appear to belong to the same clonal groups as human ExPEC isolates (18, 23, 24). Studies of E. coli isolates from humans have shown that commensal isolates are characteristically derived from E. coli phylogenetic group A or B1, while intestinal pathogenic E. coli types belong to phylogenetic group A, B1, or D. In contrast, ExPEC isolates typically are assigned to E. coli phylogenetic group B2 and, to a lesser extent, group D (16).
There are many reports in the literature describing the molecular characteristics of E. coli isolates from human clinical samples and from sick animals, and several studies have examined the degree of molecular similarities between pathogenic isolates of animal versus human origin. However, little is known about the characteristics of E. coli isolates from food-producing animal farms, although isolates from these sources may well pose a risk for human health. A recent study suggested that not only the intestine but also the external environment of healthy chickens acts as a reservoir for ExPEC with zoonotic potential (13). In previous work, we showed that farms are indeed reservoirs of isolates expressing ESBL and plasmidic class C β-lactamases (2, 3, 22). Our findings were based on analyses of ESBL- and CMY-2-expressing E. coli isolates present in fecal material obtained from poultry and pig farms where the animals did not show sign of E. coli infection. We have now expanded this study to ascertain whether E. coli farm isolates carrying these mechanisms of resistance are potentially pathogenic for humans. Accordingly, poultry and pig isolates were compared with respect to phylogenetic group, ESBL and CMY-2 types, virulence profile, sequence type (ST), and pulsed-field gel electrophoresis (PFGE) pulsotypes. Finally, animal isolates belonging to classical ExPEC serotypes were compared with E. coli strains known to cause extraintestinal infections in humans.
MATERIALS AND METHODS
E. coli isolates.
The study material consisted of 86 ESBL- and CMY-2-producing E. coli isolates obtained during 2003 from the floors of poultry (59 isolates) and pig (27 isolates) farms located in different geographic areas of northeast Spain (Catalonia) (2, 22). None of the animals was suspected of harboring an E. coli infection. Also, 80 human isolates causing extraintestinal infections (urinary, sepsis, or meningitis) were included in the study for purposes of comparison. These were obtained from patients with extraintestinal infections admitted to two hospitals in Galicia (northwest Spain).
The E. coli reference strains ECOR 1 (A), ECOR 30 (B1), ECOR 39 (D), and ECOR 65 (B2) were used as positive controls for phylogenetic grouping by PCR. The following E. coli strains were used as positive controls for ExPEC characterization and virulence gene detection: J96 (sfa/focDE, papA, and papC), CCUG31249 (kpsM II); FV35 (afa/draBC); CFT073 (iucD and tsh), FV7561 (afa/draBC, iucD, and kpsM II); Ec9192 (sfa/focDE, iucD, kpsM II, ibeA, neuC-K1, tsh, and fimAvMT78), Ec9180 (papC, papG II, sfa/focDE, hlyA, cnf1, iucD, and kpsM II), and Ec9182 (papC, papG III, sfa/focDE, hlyA, cnf1, iucD, and kpsM II) (26, 27).
Antimicrobial susceptibility test.
Susceptibility to β-lactam antibiotics and characterization of β-lactamase were determined as previously described (2). Susceptibility to other antimicrobials was tested by the disc diffusion method, following CLSI recommendations (12). For statistical studies, the median number of resistance markers to non-β-lactam antibiotics for each isolate was calculated.
Phylogenetic analysis.
All isolates were assigned to one of four phylogenetic groups (A, B1, B2, or D) based on the results of multiplex PCR, as previously described (9).
Virulence genotyping.
Virulence gene carriage was analyzed as described elsewhere (15) using primers specific for 12 genes and operons encoding virulence factors (VFs) characteristic of ExPEC. These consisted of adhesins: pyelonephritis-associated pili (papA and papC), S and F1C fimbriae (sfa/focDE), Dr-binding adhesins (afa/draBC), and the fimA variant MT78 allele of type 1 fimbriae (fimAvMT78); the three toxins α-hemolysin (hlyA), cytotoxic necrotizing factor (cnf1), and temperature-sensitive hemagglutinin serine protease (tsh); one ferric aerobactin receptor (iucD); and three protection/invasion-encoding genes corresponding to the group II capsule gene (kpsM II), the K1 kps variant (neuC-K1), and the invasion of brain endothelium gene (ibeA). The papC-positive strains were further tested for papG I, papG II, and papG III alleles. All assays were carried out in duplicate using independently prepared boiled lysates of each isolate. For further studies, the median of the VFs per isolate was calculated.
Conventional serotyping and molecular subtyping of the O25 serogroup.
The presence of O and H antigens was determined by a previously described method (14), in which all available O (O1 to O185) and H (H1 to H56) antisera were used. The antisera were absorbed with the corresponding cross-reacting antigens to remove nonspecific agglutinins. The molecular subtypes O25a and O25b were identified based on a recently described molecular approach involving allele-specific PCR (10).
PFGE analysis.
XbaI PFGE analysis was carried out as previously described (23). Profiles were analyzed with the BioNumerics fingerprinting software (Applied Maths, St.-Martens-Latem, Belgium). Cluster analysis of the Dice similarity indices based on the unweighted pair group method using average linkages (UPGMA) was done to generate a dendrogram describing the relationship among PFGE profiles. Isolates were considered to be related and to belong to the same PFGE cluster if their Dice similarity index was >85%, according to Tenover's criteria (≤6 bands of difference) (31).
ST determination.
Multilocus sequence typing (MLST) was carried out as previously described (23). The seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) were amplified and sequenced using the primers and protocol specified on the E. coli MLST website (http://mlst.ucc.ie/mlst/dbs/Ecoli). Sequences were reviewed by visual inspection with BioEdit Sequence Alignment Editor (version 7.0.9; Ibis Biosciences). The ClustalW2 program was used for sequence alignment. The allelic profile of the seven gene sequences and the STs were obtained via the electronic database at the E. coli MLST website.
Statistical methods.
Comparisons of proportions were analyzed using chi-square statistics (Fisher's exact test for 2-by-2 tables). Nonparametric statistics (Mann-Whitney U test) were used to compare VFs and the non-β-lactam-resistance medians. The significance level was fixed at 5%.
RESULTS
Phylogenetic groups.
PCR analysis of the 86 isolates showed that phylogroups A (22.8%), B1 (38.6%), and D (31.6%) were predominant among isolates from poultry farms, whereas phylogroups A (55.2%) and B1 (34.5%) were the most frequently detected among isolates from pig farms. Poultry and pig isolates differed significantly in their phylogenetic group assignments, with phylogroup A more prevalent among pig isolates (P < 0.05) and phylogroup D associated with isolates of avian origin (P < 0.05) (Table 1).
TABLE 1.
Phylogenetic group distribution, antimicrobial susceptibility, and virulence factors of isolates from poultry and pig farms
| Trait | No. (%) of isolates from farms raisinga: |
|
|---|---|---|
| Poultry (n = 57) | Pigs (n = 29) | |
| Phylogenetic groups | ||
| A | 13 (22.8) | 16 (55.2)† |
| B1 | 22 (38.6) | 10 (34.5) |
| B2 | 4 (7.0) | 1 (3.4) |
| D | 18 (31.6)† | 2 (6.9) |
| β-Lactamase types | ||
| CTX-M-9 group | 37 (64.9)* | 0 |
| CTX-M-1 group | 3 (5.3) | 20 (69.0)* |
| SHV | 5 (8.8) | 9 (20.7) |
| TEM | 2 (3.5) | 0 |
| CMY-2 | 10 (17.5)† | 0 |
| Non-β-lactam resistanceb | ||
| Nalr | 53 (93.0)* | 15 (51.7) |
| Cipr | 20 (35.1) | 7 (24.1) |
| Median | 2 | 3 |
| VFs | ||
| papA | 2 (3.5) | 1 (3.4) |
| papC | 8 (14.0)† | 0 |
| sfa/focDE | 1 (1.8) | 0 |
| afa/draBC | 0 | 0 |
| iucD | 25 (43.9)* | 1 (3.4) |
| kpsM II | 8 (14.0) | 1 (3.4) |
| hlyA | 0 | 0 |
| cnf1 | 0 | 0 |
| ibeA | 1 (1.8) | 0 |
| neuC-K1 | 0 | 0 |
| fimAvMT78 | 7 (12.3) | 5 (17.2) |
| tsh | 12 (21.1) | 7 (24.1) |
| Median | 1 | 0 |
| Isolates with ≥2 virulence genes | 20 (35.1)† | 3 (1.0) |
*, P < 0.001; †, P < 0.05.
Nalr, nalidixic acid resistance; Cipr, ciprofloxacin resistance.
Antibiotic resistance.
As shown in Table 1, β-lactamases of group CTX-M-9 (64.9%) and type CMY-2 (17.5%) were significantly associated with poultry isolates (P < 0.001 and P < 0.015, respectively), while CTX-M-1 group enzymes (69.0%) were significantly associated only with isolates from pig farms (P < 0.001). In addition, resistance to nalidixic acid (93.0%) was significantly (P < 0.001) present in poultry farms. No significant difference was noted with respect to the median of non-β-lactam resistance.
Virulence genes.
The isolates were also analyzed for the presence of 12 virulence genes (Table 1). In 56% of the isolates from poultry farms, at least one virulence gene was detected. The frequencies with respect to the total poultry isolates were as follows, in descending order: iucD, 43.9%; tsh, 21.1%; papA/papC, 17.5%; kpsM II, 14.0%; fimAvMT78, 12.3%; and ibeA and sfa/focDE, 1.8% each. The virulence genes afa/draBC, hlyA, cnf1, and neuC-K1 were not detected. Furthermore, 41% of the isolates from pig farms also carried at least one virulence gene: tsh, 24.1%; fimAvMT78, 17.2%; and papA/papC, iucD, and kpsM II, 3.4% each. The genes afa/draBC, hlyA, cnf1, ibeA, neuC-K1, and sfa/focDE were not detected. Poultry and pig isolates differed significantly in two virulence genes, papC (P < 0.05) and iucD (P < 0.001), which were more frequently detected in poultry. Among the 86 farm isolates, 23 (26.7%) carried two or more virulence genes typical of ExPEC; of these, 20 were isolated from poultry farms and only 3 were from pig farms (P < 0.05).
Serotypes.
The high diversity of serotypes was reflected in the 34 O serogroups and 52 O:H serotypes identified (see Table S1 in the supplemental material). The most frequent O serogroups were O2 (8 isolates); O8 (5 isolates); O25 and O64 (4 isolates each); and O16, O20, O29, O103, and O112 (3 isolates each). Classic human ExPEC serotypes were only detected in 11 isolates belonging to serotypes O2:H6, O2:H7, O2:HNM, O15:H1, and O25:H4.
PFGE profiles.
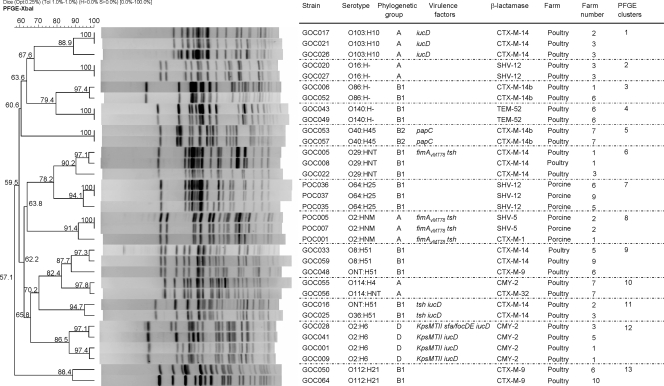
In accordance with the broad diversity established by serotyping, XbaI macrorestriction profiles obtained by PFGE showed a high heterogeneity among the 86 E. coli isolates. However, PFGE also revealed 13 clusters showing ≥85% similarity (11 clusters from poultry farms and 2 from pig farms), which included 33 of the 86 isolates. (Fig. 1) The pulsotypes of the remaining 53 isolates were unrelated. Isolates of each cluster shared the same serotype, phylogenetic group, and β-lactamase, with the exception of members of clusters 8, 9, and 10. Four clusters (no. 6, 8, 11, and 12) included isolates with two or more virulence genes isolated from different farms. Notably, two of these clusters (no. 8 and 12) included isolates belonging to classic human ExPEC serotypes.
FIG. 1.
XbaI PFGE dendrogram of 13 clusters (similarity, ≥85%) including 33 isolates of poultry and porcine origins. The dendrogram was produced by the UPGMA algorithm based on a Dice similarity coefficient with a 1.0% band position tolerance (Tol).
Characteristics of isolates with two or more virulence genes.
Analysis of the 23 isolates carrying two or more virulence genes with respect to β-lactamases showed that they carried enzymes of the CTX-M-9 group (43.4%), CMY-2 (30.4%), CTX-M-1 group (13%), and SHV type (8.7%). The majority of the isolates (48%) belonged to phylogenetic group D and the remainder to groups A (26%) and B1 (22%). Only one isolate belonged to phylogenetic group B2 (4%). With reference to serotyping, 10 isolates (43.5%) belonged to classic human ExPEC serotypes O2:H6 (n = 4), O2:HNM (n = 3), O2:H7 (n = 1), O15:H1 (n = 1), and O25:H4 (n = 1). The serotypes of the other 13 farm isolates were highly heterogeneous.
Comparison of farm isolates of classic ExPEC serotypes with human clinical isolates.
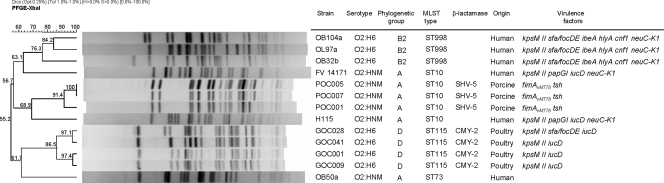
With the purpose of comparing those animal isolates of serotypes O2:HNM and O2:H6 with human isolates of the same serotypes, 61 clinical O2 isolates causing extraintestinal infections (urinary, sepsis, and meningitis) in humans were analyzed. The most prevalent O:H serotypes observed in human isolates were O2:HNM (24 isolates), O2:H4 (16 isolates), O2:H6 (14 isolates), and O2:H1 (6 isolates). Thus, 62.3% of these human isolates belonged to O2:HNM and O2:H6 detected among isolates from poultry and pig farms. Most (73.7%) of the human isolates belonged to phylogroup B2, none of the O2:H6 isolates belonged to phylogroup D, and only three of the O2:HNM isolates belonged to phylogroup A. The phylogroups, sequence types, PFGE pulsotypes and virulence genes of six of these clinical isolates of the O2:HNM and O2:H6 serotypes were compared with three pig O2:HNM and four poultry O2:H6 isolates (Fig. 2). Avian farm isolates of serotype O2:H6 belonged to phylogroup D and ST115, while human clinical isolates of this serotype belonged to phylogroup B2 and ST998. Furthermore, they differed in their background of virulence genes and PFGE pulsotypes. The three pig isolates and two human clinical isolates of serotype O2:HNM belonged to phylogroup A and ST10. No similarities were found with respect to virulence genes and PFGE pulsotypes. The remaining O2:HNM human isolate of phylogroup A showed a different sequence type (ST73). Human O2:H6 and O2:HNM isolates were negative for ESBL and CMY-2 enzymes, in contrast to isolates from poultry farms (positive for CMY-2) and pig farms (positive for SHV-5).
FIG. 2.
XbaI PFGE dendrogram of 13 isolates belonging to serotypes O2:HNM and O2:H6 of human and farm origins. The dendrogram was produced by the UPGMA algorithm based on a Dice similarity coefficient with a 1.0% band position tolerance.
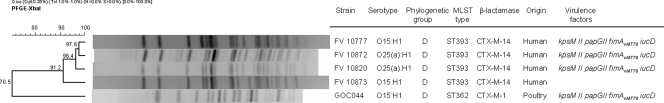
In the same way, the only O15:H1 avian isolate was compared with two human clinical isolates of this serotype. Although all shared the phylogroup D and the virulence gene profile, they differed in their sequence types (ST362-avian and ST393-human) and PFGE pulsotypes (Fig. 3). Whereas the poultry isolate expressed CTX-M-1, the two O15:H1 human clinical isolates were positive for CTX-M-14.
FIG. 3.
XbaI PFGE dendrogram of five isolates belonging to serotypes O15:H1 and O25a:H1 of human and farm origins. The dendrogram was produced by the UPGMA algorithm based on a Dice similarity coefficient with a 1.0% band position tolerance.
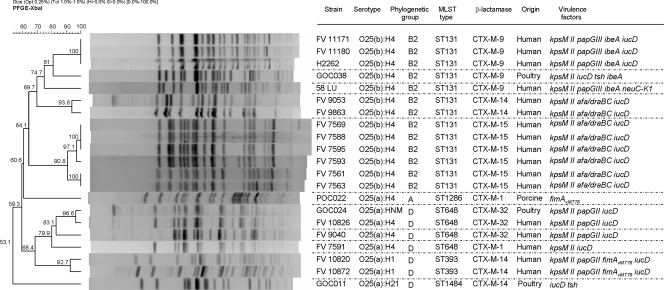
Finally, the phylogroups, sequence types, PFGE pulsotypes, and virulence genes of the four animal farm isolates belonging to serogroup O25 (O25a and O25b subtypes) were compared with 17 human clinical isolates of the same serogroup (Fig. 4). It is noteworthy that the PFGE profile of one poultry isolate (GOC024) showed a 96.6% similarity to that of a human clinical isolate (FV10826). Moreover, it is also remarkable that both isolates belonged to phylogenetic group D, molecular subtype O25a; they also had a shared sequence type (ST648). In addition, they carried both the gene encoding the CTX-M-32 enzyme and the virulence genes kpsM II, papG II, and iucD. Of particular interest was the presence of a cluster that comprised four O25b:H4-ST131 isolates but with a similarity of 81%. All of them (three human clinical isolates and one isolate from a poultry farm), which also shared expression of the CTX-M-9 enzyme, belonged to phylogenetic group B2 and carried three virulence genes in common (kpsM II, iucD, and ibeA).
FIG. 4.
XbaI-PFGE dendrogram of 21 isolates belonging to O25 serogroup of human and farm origin. The dendrogram was produced by the UPGMA algorithm based on a Dice similarity coefficient with a 1.0% band position tolerance.
Interestingly, we detected two human isolates of serotype O25a:H1 that had the molecular subtype ST393 typical of human O15:H1 isolates. In addition, these O25a:H1 isolates carried the same background of virulence genes (kpsM II, papG II, fimAvMT78, and iucD). Consequently, we compared the PFGE profiles of these two O25a:H1 human isolates with those of the two O15:H1 human isolates. The two O25a:H1 isolates were found to cluster with the two O15:H1 isolates, showing 91.2% similarity (Fig. 3).
DISCUSSION
The aim of this work was to investigate the potential zoonotic risk of ESBL- and CMY-2-producing E. coli isolates from poultry and pig farms, due to the relevance of these mechanisms of resistance in human clinical. To this end, the phylogenetic origin, antimicrobial resistance to non-β-lactam antibiotics, serotype, PFGE profile, and presence of several VFs were studied in 86 ESBL- and CMY-2-producing E. coli isolates obtained from poultry and pig farms. To our knowledge, this is, remarkably, the first study to analyze the serotypes and sequence types of ESBL- and CMY-2-producing E. coli isolates from farms, allowing the identification of new clonal groups, some of which show classical ExPEC serotypes associated with extraintestinal infections in humans.
Our results statistically corroborated previous findings (2) showing that E. coli isolates from poultry farms carried genes encoding enzymes of the CTX-M-9 group as well as the enzyme CMY-2, whereas isolates from pig farms were found to carry mainly genes encoding enzymes of the CTX-M-1 group. In addition, resistance to nalidixic acid was significant in poultry farms. The possible reasons for this have been discussed elsewhere (2). Poultry and pig isolates differed significantly in their assignments to phylogenetic groups, with phylogroup A being more prevalent among pig isolates than among poultry isolates, whereas phylogroup D was associated with isolates of avian origin. Several studies have suggested that virulent clonal groups of human ExPEC isolates are derived primarily from phylogroup B2 and to a lesser extent from phylogroup D (19). In the present study, only 7.0% of the poultry isolates and 3.4% of the pig isolates belonged to phylogroup B2, while significantly, 31.6% of poultry isolates versus 6.9% of pig isolates belonged to phylogroup D. Based on the phylogroup distribution, isolates from poultry origin are more likely to be pathogenic than those of pig origin. This possibility is supported by the virulence gene profiles, as 20 of 23 isolates carrying two or more virulence genes were of avian origin. Furthermore, most of those isolates belonged to phylogroup D.
It has been shown that APEC, uropathogenic E. coli (UPEC), and neonatal meningitis E. coli (NMEC) strains are genetically different from one another, such that a classification of ExPEC strains into subpathotypes has been proposed (19). According to this classification, the majority of APEC strains belong to phylogenetic groups A and D, while strains of the two human pathotypes are mostly from group B2 (19, 20). In the present study, most of the isolates carrying two or more virulence factors from poultry farms belonged to group A (15%) or D (55%), with characteristics similar to those of the APEC subpathotype. Other authors have reported different findings. For example, 42% of UPEC isolates (33) and 44.5% of APEC isolates (13) were shown to belong to groups D and B2, respectively. Our data, however, corroborate the hypothesis of Johnson et al. (17): i.e., that E. coli detected in retail meats derives primarily from the microbiota present in the poultry environment, as the phylogenetic group distribution and virulence gene profiles in the poultry isolates of our study were similar to those of E. coli isolate from retail chicken products in the United States (17).
The serotyping of E. coli O and H antigens is an established method to identify major bacterial clonal types, trace outbreaks of infection, detect disease reservoirs, and follow the emergence and spread of new pathogenic and/or resistant strains (7). To our knowledge, ours is the first study analyzing the serotypes of poultry and pig E. coli isolates producers of different ESBL types and CMY-2. Among the high diversity of serotypes that were found, with a total of 52 O:H serotypes, 5 (O2:H6, O2:H7, O2:HNM, O15:H1, and O25:H4) were considered as classic human ExPEC serotypes (4, 15, 20). Interestingly, 10 (90.9%) of the 11 animal isolates belonging to the cited ExPEC serotypes carried two or more virulence genes, in contrast to only 13 (17.3%) of the remaining 75 isolates (P < 0.005).
In accordance with the high diversity identified by serotyping, PFGE molecular analysis revealed highly heterogeneous profiles. Nonetheless, 13 PFGE clusters (similarity >85%) comprised isolates expressing similar traits regarding serotype, phylogenetic origin, and VF and β-lactamase expression. Moreover, the majority of clusters included E. coli isolates obtained from different farms, suggesting the successful expansion of these bacteria among some of the farms studied. Four of these clusters contained isolates with two or more virulence genes, and two of them exhibited the classic human ExPEC serotypes O2:HNM and O2:H6. Nevertheless, a comparison of these animal isolates with human isolates of the same serotypes showed differences in the background of virulence genes, sequence types, and PFGE pulsotypes. Interestingly, two O2:HNM human isolates showed the same sequence type (ST10) as the three O2:HNM pig isolates. Our avian farm CMY-2-producing isolates of serotype O2:H6 belonged to phylogroup D and ST115 and thus were of a different ST from the human clinical CMY-2 isolates from Norway that were reported by Naseer et al. (25).
Presently, the most important worldwide drug-resistant E. coli clonal groups causing extraintestinal infections in humans are O15:H1 ST393 and O25b:H4 ST131 (7, 28). In this study, only one O15:H1 isolate of avian origin was detected and compared with two human clinical isolates of this serotype. Although all of them shared phylogroup D and the virulence gene profile, they differed in sequence type (ST362-avian and ST393-human) and PFGE pulsotype. In contrast to the CTX-M-1-producing poultry isolate, the two O15:H1 human clinical isolates were positive for CTX-M-14. We have recently detected the emergence of O15:H1 CTX-M-14-producing isolates among E. coli strains causing extraintestinal infections in humans (unpublished data).
The intercontinental emergence of clonal group O25b:H4-ST131 producing CTX-M-15 and characterized by an extensive virulence profile has been reported in hospital and community settings of several countries (27). Human clinical isolates of this clonal group are known to produce numerous types of CTX-M enzymes (7, 11, 29) and even CMY-2 (25). Thus, in this context, the detection, for the first time, of the O25b:H4-ST131 clonal group producing CTX-M-9 on poultry farms is noteworthy. A comparison of the unique poultry isolate with human isolates of the same clonal group showed that they possessed a similar virulence profile and PFGE pulsotypes (similarity 81%). Indeed, the virulence profile of these CTX-M-9 isolates is very similar to that of an adherent-invasive human E. coli isolate (21). Recently, Vincent et al. (32) detected one isolate of this clonal group (albeit negative for ESBL production) in retail chicken. Our own recent findings suggest that the O25b:H4-ST131 clonal group producing CTX-M-9 group enzymes is frequently present in retail chicken products (unpublished data). In the present study, this clonal group was not identified among isolates from pig farms, although it was previously detected by Schierack et al. (30) in healthy piglets in Germany and by our group in piglets with diarrhea in Spain (unpublished data). To date, the porcine O25b:H4-ST131 isolates identified have not been shown to produce ESBL enzymes.
Remarkably, ours is the first study demonstrating the presence of the O25a-ST648-D clonal group producing CTX-M-32 in poultry farms and in human clinical strains. Furthermore, between the poultry ST648 isolate and one clinical human isolate a similarity of 96.6% was determined by PFGE analysis; identical backgrounds of virulence genes were determined as well.
Interestingly, the two human isolates of serotype O25a:H1 included in this study were of ST393, which is typical of O15:H1 isolates. A comparison of the two isolates with human O15:H1 ST393 isolates revealed that they shared the same background of virulence genes and had very similar PFGE profiles. As far as we know, this is the first report of an association between ST393 and the O25a:H1 serotype. A similar phenomenon was described in clonal human uropathogenic isolates of ST69 (serotypes O17:H18, O15:H18, O17:HNM, O25:H18, O73:H18, O77/17:H18, ONT:H18, ONT:HNM) and ST95 (O1:H7, O2:H7, O18:H7) (20). Thus, there seems to be a better association between ST and H antigen than between O serogroup and sequence type.
In conclusion, our study provides clear evidence that isolates from poultry and pig farms differ with respect to ESBL and CMY-2 enzymes, phylogenetic group, virulence genes, and serotype. Phylogroup distribution and virulence profiles suggest that isolates from poultry are potentially more pathogenic for humans than those from pigs. Particular attention should be paid to the detection in poultry farms of the O25b:H4-ST131-B2 clonal group, producing CTX-M-9, and the O25a-ST648-D clonal group, producing CTX-M-32, as their backgrounds of virulence genes and PFGE profiles are very similar to those of clinical human isolates. However, further studies are required to determine the true zoonotic potential of these clonal groups.
Supplementary Material
Acknowledgments
We are deeply indebted to G. Prats for providing us with several of the strains used in this study.
This work was partially supported by grants PI020918, PI070971, REIPI RD06/0008/1018-0025-101, and PS09/01273 (Ministerio de Ciencia e Innovación Español, Instituto de Salud Carlos III, Fondo de Investigación Sanitaria); 2009SGR1106 (Generalitat de Catalunya); 09TAL007261PR and 2007/000044-0 (Xunta de Galicia); and AGL-2008-02129 (Ministerio de Educación y Ciencia Español). V. Blanc was a recipient of a predoctoral fellowship from the Universitat Autònoma de Barcelona (Spain). A. Mora acknowledges the Ramón y Cajal program from the Ministerio de Educación y Ciencia Español.
Footnotes
Published ahead of print on 12 March 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bidet, P., F. Mahjoub-Messai, J. Blanco, J. Blanco, M. Dehem, Y. Aujard, E. Bingen, and S. Bonacorsi. 2007. Combined multilocus sequence typing and O serogrouping distinguishes Escherichia coli subtypes associated with infant urosepsis and/or meningitis. J. Infect. Dis. 196:297-303. [DOI] [PubMed] [Google Scholar]
- 2.Blanc, V., R. Mesa, M. Saco, S. Lavilla, G. Prats, E. Miró, F. Navarro, P. Cortés, and M. Llagostera. 2006. ESBL- and plasmidic class C β-lactamase-producing E. coli strains isolated from poultry, pig and rabbit farms. Vet. Microbiol. 118:299-304. [DOI] [PubMed] [Google Scholar]
- 3.Blanc, V., P. Cortés, R. J. Mesa, E. Miró, F. Navarro, and M. Llagostera. 2008. Characterisation of plasmids encoding extended-spectrum β-lactamase and CMY-2 in Escherichia coli isolated from animal farms. Int. J. Antimicrob. Agents 31:76-78. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, J. E., J. Blanco, M. Blanco, M. P. Alonso, and W. H. Jansen. 1994. Serotypes of CNF1-producing Escherichia coli strains that cause extraintestinal infections in humans. Eur. J. Epidemiol. 10:707-711. [DOI] [PubMed] [Google Scholar]
- 5.Blanco, J. E., M. Blanco, A. Mora, and J. Blanco. 1997. Production of toxins (enterotoxins, verotoxins, and necrotoxins) and colicins by Escherichia coli strains isolated fom septicemic and healthy chickens: relationship with in vivo pathogenicity. J. Clin. Microbiol. 11:2953-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanco, M., J. E. Blanco, M. P. Alonso, A. Mora, C. Balsalobre, F. Munoa, A. Juárez, and J. Blanco. 1997. Detection of pap, sfa and afa adhesin-encoding operons in uropathogenic Escherichia coli strains: relationship with expression of adhesins and production of toxins. Res. Microbiol. 148:745-755. [DOI] [PubMed] [Google Scholar]
- 7.Blanco, M., M. P. Alonso, M. H. Nicolas-Chanoine, G. Dahbi, A. Mora, J. E. Blanco, C. López, P. Cortés, M. Llagostera, V. Leflon-Guibout, B. Puentes, R. Mamani, A. Herrera, M. A. Coira, F. García-Garrote, J. M. Pita, and J. Blanco. 2009. Molecular epidemiology of Escherichia coli producing extended-spectrum β-lactamases in Lugo (Spain): dissemination of clone O25b:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 63:1135-1141. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y. M. M., P. J. Wright, C. S. Lee, and G. F. Browning. 2003. Uropathogenic virulence factors in isolates of Escherichia coli from clinical cases of canine pyometra and feces of healthy bitches. Vet. Microbiol. 94:57-69. [DOI] [PubMed] [Google Scholar]
- 9.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clermont, O., M. Lavollay, S. Vimont, C. Deschamps, C. Forestier, C. Branger, E. Denamur, and G. Arlet. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024-1028. [DOI] [PubMed] [Google Scholar]
- 11.Clermont, O., H. Dhanji, M. Upton, T. Gibreel, A. Fox, D. Boyd, M. R. Mulvey, P. Nordmann, E. Ruppé, J. L. Sarthou, T. Frank, S. Vimont, G. Arlet, C. Branger, N. Woodford, and E. Denamur. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother. 64:274-277. [DOI] [PubMed] [Google Scholar]
- 12.CLSI. 2003. Performance standards for antimicrobial disk susceptibility testing. Approved standard M02-A10, 10th ed. CLSI, Wayne, PA.
- 13.Ewers, C., E. M. Antão, I. Diehl, H. C. Philipp, and L. H. Wieler. 2009. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl. Environ. Microbiol. 75:184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guinée, P. A. M., W. H. Jansen, T. Wadström, and R. Sellwood. 1981. Escherichia coli associated with neonatal diarrhoea in piglets and calves. Curr. Top. Vet. Med. Anim. Sci. 13:126-162. [Google Scholar]
- 15.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, J. R., and T. A. Russo. 2002. Extraintestinal pathogenic Escherichia coli: “the other bad E. coli.” J. Lab. Clin. Med. 139:155-162. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, J. R., A. C. Murray, A. Gajewski, M. Sullivan, P. Snippes, M. A. Kuskowski, and K. E. Smith. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47:2161-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, T. J., S. Kariyawasam, Y. Wannemuehler, P. Mangiamele, S. J. Johnson, C. Doetkott, J. A. Skyberg, A. M. Lynne, J. R. Johnson, and L. K. Nolan. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 189:3228-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, T. J., Y. Wannemuehler, S. J. Johnson, A. L. Stell, C. Doetkott, J. R. Johnson, K. S. Kim, L. Spanjaard, and L. K. Nolan. 2008. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl. Environ. Microbiol. 74:7043-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manges, A. R., H. Tabor, P. Tellis, C. Vincent, and P. P Tellier. 2008. Endemic and epidemic lineages of Escherichia coli that cause urinary tract infections. Emerg. Infect. Dis. 14:1575-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Medina, M., X. Aldeguer, M. Lopez-Siles, F. González-Huix, C. López-Oliu, G. Dahbi, J. E. Blanco, J. Blanco, L. J. Garcia-Gil, and A. Darfeuille-Michaud. 2009. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm. Bowel Dis. 15:872-882. [DOI] [PubMed] [Google Scholar]
- 22.Mesa, R. J., V. Blanc, A. R. Blanch, P. Cortés, J. J. González, S. Lavilla, E. Miró, M. Muniesa, M. Saco, M. T. Tórtola, B. Mirelis, P. Coll, M. Llagostera, G. Prats, and F. Navarro. 2006. Extended-spectrum β-lactamase-producing Enterobacteriaceae in different environments (humans, food, animal farms and sewage). J. Antimicrob. Chemother. 58:211-215. [DOI] [PubMed] [Google Scholar]
- 23.Mora, A., C. López, G. Dabhi, M. Blanco, J. E. Blanco, M. P. Alonso, A. Herrera, R. Mamani, S. Bonacorsi, M. Moulin-Schouleur, and J. Blanco. 2009. Extraintestinal pathogenic Escherichia coli O1:K1:H7/NM from human and avian origin: detection of clonal groups B2 ST95 and D ST59 with different host distribution. BMC Microbiol. 9:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moulin-Schouleur, M., C. Schouler, P. Tailliez, M. R. Kao, A. Bree, P. Germon, E. Oswald, J. Mainil, M. Blanco, and J. Blanco. 2006. Common virulence factors and genetic relationships between O18:K1:H7 Escherichia coli isolates of human and avian origin. J. Clin. Microbiol. 44:3484-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naseer, U., B. Haldorsen, G. S. Simonsen, and A. Sundsfjord. 2010. Sporadic occurrence of CMY-2-producing multidrug-resistant Escherichia coli of ST-complexes 38 and 448, and ST131 in Norway. Clin. Microbiol. Infect. 16:171-178. [DOI] [PubMed] [Google Scholar]
- 26.Naves, P., G. del Prado, L. Huelves, M. Gracia, V. Ruiz, J. Blanco, G. Dahbi, M. Blanco, M. D. C. Ponte, and F. Soriano. 2008. Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microb. Pathog. 45:86-91. [DOI] [PubMed] [Google Scholar]
- 27.Nicolas-Chanoine, M. H., J. Blanco, V. Leflon-Guibout, R. Demarty, M. P. Alonso, M. M. Caniça, Y. J. Park, J. P. Lavigne, J. Pitout, and J. R. Johnson. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273-281. [DOI] [PubMed] [Google Scholar]
- 28.Olesen, B., F. Scheutz, M. Menard, M. N. Skov, H. J. Kolmos, M. A. Kuskowski, and J. R. Johnson. 2009. Three-decade epidemiological analysis of Escherichia coli O15:K52:H1. J. Clin. Microbiol. 47:1857-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oteo, J., K. Diestra, C. Juan, V. Bautista, A. Novais, M. Pérez-Vázquez, B. Moyác, E. Miró, T. M. Coque, A. Oliver, R. Cantón, F. Navarro, J. Campos, and Spanish Network in Infectious Pathology Project (REIPI). 2009. Extended-spectrum β-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int. J. Antimicrob. Agents 34:173-176. [DOI] [PubMed] [Google Scholar]
- 30.Schierack, P., N. Walk, C. Ewers, H. Wilking, H. Steinrück, M. Filter, and L. H. Wieler. 2008. ExPEC-typical virulence-associated genes correlate with successful colonization by intestinal E. coli in a small piglet group. Environ. Microbiol. 10:1742-1751. [DOI] [PubMed] [Google Scholar]
- 31.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent, C., P. Boerlin, D. Daignault, C. M. Dozois, L. Dutil, C. Galanakis, R. J. Reid-Smith, P. P. Tellier, P. A. Tellis, K. Ziebell, and A. R. Manges. 2010. Food reservoir for Escherichia coli causing urinary tract infections. Emerg. Infect. Dis. 16:88-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao, L., S. Gao, H. Huan, X. Xu, X. Zhu, W. Yang, Q. Gao, and X. Liu. 2009. Comparison of virulence factors and expression of specific genes between uropathogenic Escherichia coli and avian pathogenic E. coli in a murine urinary tract infection model and a chicken challenge model. Microbiology 155:1634-1644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.