Abstract
Clostridium perfringens and Clostridium difficile are associated with scours in the neonatal piglet and are an economic concern in swine production. The objective of this study was to characterize the prevalence and diversity of C. perfringens and C. difficile isolates obtained from scouring neonatal piglets in a large integrated production system, as well as in smaller independently owned regional farms. Rectal swabs were collected from 333 pigs at 11 sites in an integrated swine production system and from an additional 180 pigs at 16 regional farms located throughout the Midwest. C. perfringens was isolated from 89.8% of the pigs swabbed at the integrated sites, and C. difficile was isolated from 57.7% of these pigs. Of the pigs from the regional farms sampled, 95.6% were positive for isolation of C. perfringens and 27.2% were positive for C. difficile. Toxigenic isolates were typed using random amplified polymorphic DNA (RAPD) PCR, and were placed in four dendrograms for C. perfringens and C. difficile populations isolated from the integrated sites and regional farms. Diversity indices showed that there was greater diversity in C. difficile populations and in populations isolated from the regional farms. A subset of isolates from the C. difficile dendrograms were further toxinotyped by amplification of the pathogenicity locus and subsequent digestion by HincII, AccI, and EcoRI. Of the 45 isolates typed, 44 were determined to be toxinotype V. The results of this study illustrate the diversity of C. perfringens and C. difficile isolates and the prevalence of these pathogens in swine production sites.
Enteric clostridial infections in swine occur predominantly in the neonatal period, and Clostridium perfringens type A and Clostridium difficile infections have been recognized with increasing frequency in the swine industry (42). C. perfringens and C. difficile are Gram-positive, anaerobic, spore-forming bacteria. Clostridial spores can persist in the fecal matter and environment of pigs, which can make the spread of these bacteria and passage from sow to piglet difficult to control. Clostridial infection usually occurs in piglets within the first 7 days after birth and may be associated with an underdeveloped normal microbiota and antibiotic administration (42). C. perfringens and C. difficile infection causes diarrhea in neonatal piglets, which can lead to low weaning weights, preweaning mortality, and economic impact on swine production.
The pathogenicity of C. perfringens is associated with several toxins, and this species is classified into types based on the production of the four major toxins: the alpha, beta, epsilon, and iota toxins. C. perfringens type A (alpha toxin) and type C (alpha and beta toxins) are the only types known to infect swine, although other types can infect a number of additional animal species and also humans (12, 34). Disease caused by C. perfringens type C has been controlled by vaccination in swine herds in recent years; however, the virulence mechanisms of C. perfringens type A are not well understood. C. perfringens type A strains are considered commensal microbes in the intestinal tract of healthy pigs, which can make diagnosis of the disease caused by C. perfringens difficult. There is strain-to-strain variation in the virulence of type A strains, and it is possible that several toxins and enzymes play a role in pathogenesis (25, 32, 36).
Enteric infection caused by C. difficile has emerged as a common diagnosis in neonatal pigs in recent years (35). Virulent strains of C. difficile are associated with two toxins: the enterotoxin TcdA (toxin A) and the cytotoxin TcdB (toxin B) (12, 39). This pathogen is known to cause disease in a variety of other animals, including calves, lambs, dogs, and horses (17, 34). C. difficile has also been associated with hospitalization and antibiotic use in humans, and recently there have been epidemic outbreaks of C. difficile-associated disease (CDAD) due to the emergence of a hypervirulent strain in hospitals worldwide (18, 38). This strain is a toxinotype III (ribotype 027) strain, contains the binary toxin CDT, and has an 18-bp deletion in the tcdC regulatory gene (9, 14).
Several techniques have been used to type C. perfringens and C. difficile in both humans and animal species. The common typing methods include multilocus sequence typing (MLST) (15, 18, 20), pulsed-field gel electrophoresis (PFGE) (7, 18, 21), random amplified polymorphic DNA (RAPD) typing (5, 9, 19), PCR ribotyping (3, 7, 13, 16, 18, 28), and toxinotyping (14, 28, 29, 30). Generally, these methods have been used to type C. perfringens in attempts to differentiate pathogenic strains from commensals and to type C. difficile as an epidemiology tool to identify clusters or strains that are associated with disease outbreaks.
Understanding the diversity of toxigenic strains in commercial swine herds may lead to a greater understanding of the pathogenesis of Clostridium in neonatal pigs and aid in the development of effective intervention methods for controlling clostridial disease outbreaks. Therefore, the focus of this study was to assess the prevalence and diversity of pathogenic clostridia in neonatal pigs showing clinical signs of clostridial disease at farm sites in a large integrated swine production system and in a sample of smaller independently owned regional swine farms located throughout the Midwest.
MATERIALS AND METHODS
Sample collection.
Rectal swabs were collected from neonatal pigs at 11 sites of an integrated commercial swine company with approximately 150,000 sows. One swab was obtained from each of 333 individual pigs exhibiting clinical signs of scours at the sites. Each of the sites was sampled a minimum of two times during a 9-month period (January to September 2006). To compare the clostridial diversity in a large integrated operation to the clostridial diversity at smaller farms, rectal swabs were collected from scouring neonatal pigs at swine farms containing less than 2,500 sows located throughout the Midwest. These farms had a history of scours in which clostridial species were determined to be the likely causative agents, as indicated by a veterinarian. During a 3-month period, 16 farm sites in Iowa, Indiana, South Dakota, Minnesota, Michigan, Ohio, Illinois, and Oklahoma were sampled once. A total of 180 swabs representing the 16 farm sites were collected from piglets showing clinical signs of clostridium-related diarrhea.
Clostridium isolation.
Bacteria were harvested from the swabs by vortexing each swab in a 10-ml tube containing 0.1% peptone. The samples were heated at 75°C for 30 min, and the spores were plated. Samples were plated on selective medium for isolation of C. perfringens by using perfringens agar base (Oxoid Ltd., Cambridge, United Kingdom) supplemented with d-cycloserine (TSC; Oxoid Ltd., Cambridge, United Kingdom) and 5% egg yolk emulsion, and samples were plated on selective medium for isolation of C. difficile by using difficile agar base (Oxoid Ltd., Cambridge, United Kingdom) supplemented with cysteine hydrochloride, norofloxacin, and moxalactam (CDMN; Oxoid Ltd., Cambridge, United Kingdom), as well as 7% defibrinated horse blood. The samples were plated using a Spiral Autoplate 4000 (Spiral Biotech, Bethesda, MD), and plates were incubated in an anaerobic chamber (Coy Laboratory, Grass Lake, MI) for 24 to 48 h at 37°C. A maximum of three isolated colonies of both C. perfringens and C. difficile with characteristic colony morphology were picked from the plates representing one swab sample and grown anaerobically at 37°C for 24 h in prereduced brain heart infusion (BHI) broth (Anaerobe Systems, Morgan Hill, CA). Isolates were harvested by centrifugation for 5 min at 6,000 × g, resuspended in 50 mM Tris-HCl supplemented with 15% sucrose, and frozen at −20°C for subsequent DNA isolation. DNA was isolated using a 96-well format and a DNeasy 96 tissue kit (Qiagen, Valencia, CA) according to the manufacturer's protocol.
Toxin screening.
Multiplex PCR (mPCR) was performed with the isolates to determine the toxin genes present and to confirm the species of the isolates. The C. perfringens mPCR targeted the four major toxins (alpha, beta, epsilon, and iota toxins) as described by Yoo et al. (43). The presumptive C. difficile isolates were screened for the presence of the tcdA and tcdB toxin genes (6). Briefly, the reaction mixture for each PCR (final volume, 50 μl) contained 5 μl 10× PCR buffer (without MgCl2), 1 μl 10 mM deoxynucleoside triphosphate (dNTP) mixture, 0.5 μl 5 U/μl Platinum Taq polymerase (Invitrogen, Carlsbad, CA), 3 μl 50 mM MgCl2, the appropriate forward and reverse primers (2.5 μl 10 pM primer), 25.5 μl double-distilled H2O (ddH2O), and 5 μl genomic DNA. Amplification for both mPCRs was performed with a GeneAmp PCR system thermal cycler (Applied Biosystems, Foster City, CA). The cycling conditions for the C. perfringens mPCR began with preincubation at 94°C for 5 min, which was followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min (43). The cycling conditions for the C. difficile mPCR began with preincubation at 94°C for 3 min, which was followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s (6). The PCR products were identified by electrophoresis using a 2% 96-well E-gel (Invitrogen, Carlsbad, CA) and were visualized using UV transillumination.
Genotyping.
Random amplified polymorphic DNA typing was used to characterize the diversity of the toxigenic C. perfringens and C. difficile isolates. PCR amplification was carried out using stable Ready-to-Go RAPD beads (GE Healthcare, United Kingdom) which contained two thermostable polymerases (AmpliTaq and the Stoffel fragment), dNTPs, and buffer. To each bead mixture (total volume, 25 μl), 5 μl of 5 pM primer GTTTCGCTCC, 4 μl of genomic DNA, and 16 μl of ddH2O were added. Amplification for RAPD PCR analysis was performed with a thermal cycler (Applied Biosystems, Foster City, CA) programmed for one cycle of 4 min at 95°C followed by 45 cycles of 94°C for 1 min, 36°C for 1 min, and 72°C for 2 min. DNA fragments were separated by gel electrophoresis using a 1% agarose gel run at 70 V for 120 min and were analyzed using Bio-Numerics software (Applied Maths Inc., Austin, TX). Four dendrograms were constructed, one each for the toxigenic integrated and regional C. perfringens and C. difficile isolates. The dendrograms were generated using the Dice coefficient and an unweighted-pair group method (unweighted-pair group method using average linkages [UPGMA]) to assess the clostridial diversity in each of the populations. Dendrograms were color coded by sampling site, and a similarity coefficient of 80% was used to define unique clusters. Shannon's diversity index (H′), maximum H′ (Hmax), and Pielou's evenness index (J′) were calculated for each of the four dendrograms (22, 31). Greater values for Shannon's index indicate greater diversity, while higher values for Pielou's index indicate a more even distribution among members of a population.
C. difficile toxinotyping.
A subset of strains representing the majority (>70%) of the diversity in both the commercial and regional farm C. difficile dendrograms were toxinotyped using the method of Rupnik et al. (27, 28, 29, 30). Briefly, the A3 fragment of the tcdA gene and the B1 fragment of the tcdB gene were amplified by PCR (29) and subsequently digested with restriction enzymes. The A3 fragment was digested with EcoRI, and the B1 fragment was digested with both HincII and HindIII (New England Biolabs, Ipswich, MA). The banding patterns were then compared to those of previously described reference strains to determine the toxinotype of each isolate (27, 28, 30).
RESULTS
Prevalence of Clostridium.
The results of an analysis of the prevalence of toxigenic C. perfringens and C. difficile at the integrated and regional farm sites are shown in Table 1. C. perfringens type A was isolated from 299 of the 333 pigs sampled (89.8%) in the large integrated production system. There were 794 isolates that contained the alpha toxin and were confirmed to be C. perfringens type A. No other type of C. perfringens was isolated. Altogether, C. difficile was cultured from 192 of the 333 pigs in the integrated farm sites (57.7%). The presence of tcdA and/or tcdB confirmed that 476 isolates were C. difficile isolates. All of the sites in the integrated production system were positive for C. perfringens and C. difficile.
TABLE 1.
Prevalence of C. perfringens and C. difficile at the integrated sites and regional farms
| Species | Location | No. of pigs positivea | % of pigs positive | Total no. of toxigenic isolatesb | No. of sites sampled | No. of sites positive | % of sites positive |
|---|---|---|---|---|---|---|---|
| C. perfringens | Integrated | 299 | 89.8 | 794 | 11 | 11 | 100.0 |
| Regional | 172 | 95.6 | 472 | 16 | 16 | 100.0 | |
| C. difficile | Integrated | 192 | 57.7 | 476 | 11 | 11 | 100.0 |
| Regional | 49 | 27.2 | 102 | 16 | 10 | 62.5 |
A total of 333 pigs from 11 integrated sites and 180 pigs from 16 regional farms were sampled.
Isolates were considered toxigenic if they contained any of the toxin genes screened by mPCR (alpha, beta, epsilon, or iota toxin gene for C. perfringens or toxin A and/or toxin B gene for C. difficile).
C. perfringens was isolated from 172 of the 180 pigs swabbed (95.6%) at all 16 of the regional Midwest farms (Table 1). Of the 472 C. perfringens isolates recovered, 469 were C. perfringens type A. The remaining three isolates were C. perfringens type C and were isolated from pigs at the same regional farm. Forty-nine of the 180 pigs (27.2%) at 62.5% of the farms were positive for the presence of C. difficile. A total of 102 toxigenic isolates acquired from pigs at the regional farms were confirmed to be C. difficile by mPCR.
Diversity of Clostridium isolates.
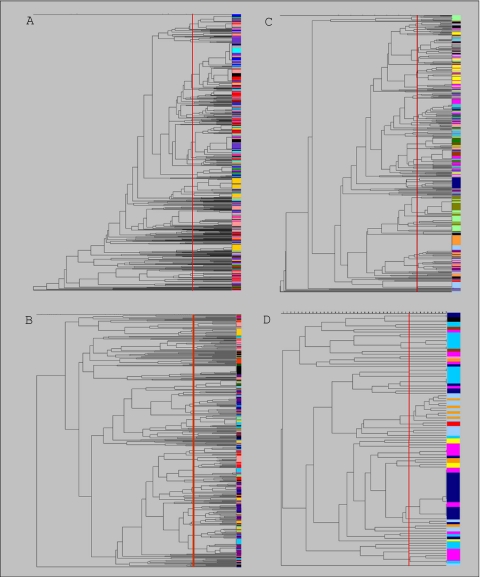
Banding patterns from RAPD typing were used to create dendrograms characterizing the genetic relatedness of the confirmed C. perfringens and C. difficile isolates. Dendrograms were constructed and used to identify clusters of closely related isolates and to compare the diversity of the four population subsets. The dendrogram of C. perfringens isolates recovered from the integrated production system consisted of 138 clusters representing 794 isolates when a similarity coefficient of 80% was used (Fig. 1A). Isolates connected to the same branch with similarity coefficients of ≥80% were considered members of the same cluster, and isolates connected at 100% similarity had identical RAPD banding patterns. The largest cluster in the dendrogram contained 131 isolates (16.5% of the population), 47 of which had identical RAPD types. The C. difficile dendrogram was comprised of 476 isolates grouped into 126 clusters (Fig. 1B). The largest cluster in the C. difficile dendrogram consisted of 30 isolates (6.3% of the population). Thirteen of the 30 isolates in the largest C. difficile cluster had identical RAPD types. Of the 126 clusters in the C. difficile dendrogram, 53 consisted of a single isolate.
FIG. 1.
Dendrograms of toxigenic C. perfringens (A) and C. difficile (B) isolates from the 11 integrated sites and toxigenic C. perfringens (C) and C. difficile (D) isolates from the 16 regional farms. Isolates from each of the sites studied are indicated by a different color. The dendrograms were constructed using the Dice coefficient and UPGMA. The vertical red line in each dendrogram indicates a similarity coefficient of 80% and is used to indicate unique clusters.
A total of 92 unique clusters were obtained for the 469 toxigenic C. perfringens strains isolated from the regional Midwest farms (Fig. 1C). Five clusters in the dendrogram contained more than 25 isolates each, and the largest cluster consisted of 38 isolates (7.9% of the population), 29 of which had identical RAPD types. Fifty-one unique clusters were identified for the 102 toxigenic C. difficile isolates (Fig. 1D). Nineteen of these clusters contained only one isolate. The largest cluster in the regional C. difficile dendrogram contained 20 isolates (19.6% of the population), and all but one isolate belonged to two RAPD type groups of isolates with identical banding patterns.
In general, the clostridial isolates from the integrated production sites and the regional farms did not cluster solely by farm, site, or sampling time. C. difficile isolates appeared to be more genetically diverse than C. perfringens isolates, as indicated by fewer C. difficile isolates per cluster in the dendrograms on average (Table 2). Shannon's index data also indicated that there was a greater diversity among C. difficile isolates than among C. perfringens isolates from the integrated production sites. The evenness index (J′), which indicates even distribution of isolates in clusters, was lowest for the dendrogram comprised of C. perfringens isolates from the integrated sites.
TABLE 2.
Diversity and evenness indices calculated from RAPD dendrograms for isolates from the integrated sites and regional farmsa
| Species | Location | No. of clusters | Avg no. of isolates/ cluster | Shannon's index (H′) | Hmax | Pielou's evenness index (J′) |
|---|---|---|---|---|---|---|
| C. perfringens | Integrated | 138 | 5.75 | 3.71 | 4.92 | 0.75 |
| Regional | 92 | 5.09 | 3.83 | 4.52 | 0.85 | |
| C. difficile | Integrated | 126 | 3.77 | 4.23 | 4.84 | 0.87 |
| Regional | 51 | 2.04 | 3.47 | 3.93 | 0.88 |
The integrated and regional C. perfringens dendrograms contained 794 and 469 isolates, respectively. The integrated C. difficile dendrogram contained 476 isolates, and the regional C. difficile dendrogram contained 102 isolates.
C. difficile toxinotyping.
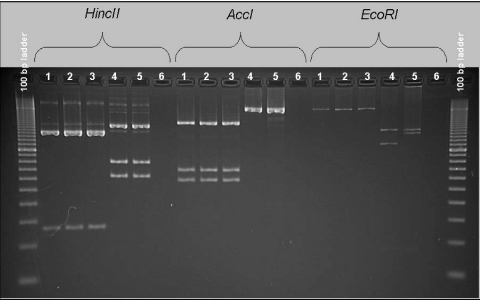
A subset of the C. difficile strains from the integrated production system and the regional farms representing the majority of the diversity from both dendrograms was characterized further by toxinotyping. Figure 2 shows the results of toxinotyping of representative C. difficile isolates based on restriction enzyme banding patterns of the A3 and B1 PCR fragments of the pathogenicity locus. All 18 of the isolates from the integrated sites typed were toxinotype V (ribotype 078 or 066) (Table 3). In addition, 26 of the 27 C. difficile isolates from the regional farms were determined to be toxinotype V; the exception was an isolate that was toxinotype I (ribotype 003, 012, or 102) (Table 3).
FIG. 2.
Restriction enzyme patterns of PCR fragments A3 and B1 from four representative C. difficile isolates examined in this study. Toxinotypes of the isolates were determined by comparison to previously described reference strains (27, 28, 30). Lanes 1 to 3, toxinotype V; lane 4, toxinotype I; lane 5, toxinotype 0 (positive-control strain ATCC 9689); lane 6, negative control.
TABLE 3.
Toxinotypes of C. difficile isolates representative of populations isolated from the integrated sites and regional farms
| Location | No. of C. difficile isolatesa | No. of toxinotype V isolates | No. of toxinotype I isolates |
|---|---|---|---|
| Integrated | 18 | 18 | 0 |
| Regional | 27 | 26 | 1 |
Samples were obtained from a total of 333 pigs from 11 integrated sites and 180 pigs from 16 regional farms. The C. difficile isolates toxinotyped accounted for more than 70% of the diversity in the integrated and regional dendrograms containing 476 and 102 isolates, respectively.
DISCUSSION
The aims of this study were to assess the prevalence and diversity of C. perfringens and C. difficile isolates obtained from neonatal piglets in the United States swine industry and to compare the clostridial diversity of a large integrated swine production system with that of smaller regional farms. RAPD PCR was chosen as the initial typing method in this study because it allowed us to type a large number of isolates. This method facilitates study of changes in diversity over time and also provides a basis for subsampling in large populations. Previous studies have shown that this method is a rapid and reproducible way to distinguish closely related species of bacteria (5, 19, 24, 41).
RAPD typing of the toxigenic C. perfringens and C. difficile isolates revealed that both species appeared to be genetically diverse. However, on average, the C. difficile clusters contained fewer isolates for both the integrated sites and regional farms, and the diversity was higher for these isolates than for the C. perfringens isolates. This was in agreement with Shannon's index (H′) data except in the case of C. difficile isolates from the regional farms, for which the diversity index was lower than expected based on the average number of isolates per cluster. This was due to a bias in diversity indices that do not easily allow comparison of populations when sample sizes are unequal (1). Although Shannon's index (H′) was lowest for C. difficile isolates from regional farms, the calculated Hmax was also lower than that for any of the other clostridial populations. The evenness index (J′), which expresses H′ relative to Hmax, calculated for the C. difficile isolates from the regional farms was near the value calculated for the isolates at the integrated sites, indicating that there were similar distributions of isolates in the populations. This assessment of diversity is consistent with microarray data reported previously, which showed that a pool of 75 human and animal C. difficile isolates shared only 19.7% of their genes with the fully sequenced strain C. difficile 630 (38).
There were isolates of both C. perfringens and C. difficile that were present at more than one of the integrated sites or individual regional farms, as well as isolates that were unique. Overall, the dendrograms can be visualized as mosaics in which isolates did not cluster by site or sampling date. For the integrated production sites which were sampled multiple times, new isolates were added to the dendrogram at each sampling time. On average, there were more clostridial isolates per cluster for the integrated sites than for the regional farms, indicating that there was more clostridial diversity in the regional farms sampled. The lowest evenness index (J′) was calculated for the C. perfringens isolates obtained at the integrated sites. Several large clusters dominated the integrated site C. perfringens dendrogram, including a cluster containing 131 isolates (16.5% of the population). This may have been due to the fact that the integrated sites were sampled multiple times, as well as to the multitude of variations in farm management practices at the regional farms that could have influenced the diversity of microbial populations.
The apparent genetic diversity of the organisms further complicates identification of virulent strains associated with disease. The data suggest that the diversity of the populations even within a site or commercial production facility should be considered in development of effective treatments for the control of clostridial disease. Information about the genetic diversity of the Clostridium pathogens may result in greater understanding of methods used for disease control in neonatal piglets. Numerous prophylaxis and treatment methods, including antibiotics, vaccines, prebiotics, and probiotics, are now used in the swine industry with various degrees of success. In addition, control of C. difficile in swine herds is increasingly challenging due to the resistance of this organism to antibiotics and the general absence of other treatment options for producers.
Producers have faced challenges in finding effective means of controlling outbreaks in production systems, and diagnosis of neonatal scours due to Clostridium has become increasingly common (42). In this study, C. perfringens type A was isolated from all of the farm sites sampled for both the integrated production system and the regional farms, as well as from a majority of the pigs sampled at the sites. C. perfringens is a ubiquitous bacterium that is commonly found in the environment and intestinal tracts of piglets, and thus high isolation rates were expected. The pathogenesis of C. perfringens type A disease is not well understood, which makes the diagnosis of disease due to C. perfringens type A difficult. Commensal and pathogenic strains cannot be distinguished, and a C. perfringens strain isolated from a scouring piglet may or may not be the cause of the disease (33). Previous studies in our lab showed that isolates acquired from scouring piglets and isolates acquired from nonscouring piglets could not be differentiated by the presence of several virulence genes, toxins, and enzymes or by RAPD typing (4). It is possible that rapid bacterial proliferation, increased toxin production, gene transfer, environmental variables, or other toxins and regulatory systems may influence visual manifestation of C. perfringens type A disease.
C. perfringens type C was a predominant clostridial problem in swine herds in the past; however, vaccination now seems to control populations of this organism (36). C. perfringens type C was not prevalent in the clostridium populations isolated at any of the farm sites in this study, and no isolates were recovered from samples obtained at the integrated production system sites, indicating that this organism was not a problem in the system. Only 3 C. perfringens type C isolates were found among the 502 isolates screened from the regional farms, and all of them were obtained from the same farm. Similar isolation rates were reported by Yoo et al. (43), who found that 85.7% of the isolates that they recovered from piglets showing clinical symptoms of enterotoxemia or necrotic enteritis were C. perfringens type A isolates and 14.3% of the isolates were C. perfringens type C isolates.
Published data for industry-wide rates of isolation of C. difficile from swine herds in the United States are limited. However, one study that reported that as many as 47.6% of litters and 90% of herds were infected with C. difficile in one commercial production system (33, 36). A survey of isolation of C. difficile from swine herds in Spain found that the isolation rates were between 0% and 64% for various farms (2). Other reports have also suggested that the incidence of scours due to C. difficile is underdiagnosed, mainly due to the difficulty of culturing this bacterium and the instability of the toxins during transport for enzyme-linked immunosorbent assay (ELISA) testing (35, 36, 42). Based on the isolation rates that we found, the presence of C. difficile seemed to be farm specific (present at some but not all farm sites). All of the sites in the integrated production system were culture positive for C. difficile, compared to 62.5% of the regional sites. The overall percentage of pigs harboring C. difficile was also higher for the integrated production system than for the regional farms. These differences may be due to a number of individual farm variables, including cleanliness, diet, genetic line, health status, season, and overall management practices. Further studies to document the percentages of C. difficile carriers in swine herds throughout the United States are warranted, as this organism appears to be an increasingly diagnosed cause of neonatal enteritis.
Due to interest in recent outbreaks of CDAD in hospitals worldwide and genetic typing of C. difficile isolates from human and animal sources, a subset of isolates from our data set were further toxinotyped to determine whether the toxinotypes of the swine isolates in this study were toxinotypes commonly isolated from humans. Of the isolates that were typed, all but one were determined to be toxinotype V. Toxinotype V isolates contain both tcdA and tcdB genes and the binary toxin CDT and have a 39-bp deletion in the tcdC regulation gene (11, 27). This is consistent with other reports indicating that toxinotype V is prevalent among pigs and calves (11, 16, 23). However, toxinotyping is based solely on variation in the pathogenicity locus and not on total genome diversity. The RAPD analysis indicated there is a high degree of diversity in the C. difficile genome even within the toxinotype V subgroup, as demonstrated by the results for the large number of isolates from various swine farms that were RAPD typed in this study.
Several other studies have raised concerns about a possible association of toxinotype V (ribotype 078) isolates in food animals and CDAD in humans (8, 10, 11, 14, 23). C. difficile, including toxinotype V isolates, has recently been cultured from retail pork and beef, and workers worldwide have reported finding toxinotype V isolates associated with community-acquired CDAD in humans (10, 11, 14, 26, 37, 40). Our data reaffirm that toxinotype V is a predominant toxinotype in swine herds in the United States; however, further studies to determine the clinical importance of toxinotype V isolates and the potential for interspecies transmission are warranted.
We determined that C. perfringens and C. difficile are prevalent throughout the swine industry and that C. difficile may be a leading underdiagnosed cause of neonatal enteritis. C. perfringens was commonly isolated from piglets at all of the farm sites sampled in this study; however, the percentage of piglets infected with C. difficile was different at different sites, and outbreaks may vary based on management practices and other variables. In this study, nearly 2,000 isolates from over 500 piglets were typed for multiple sites in a large integrated swine production system over time and for several smaller regional farms. To our knowledge, this is the most extensive assessment of clostridial diversity in scouring neonatal piglets representative of the swine industry in the United States. We also illustrated the significant genomic diversity of Clostridium isolates associated with neonatal diarrhea in piglets, especially isolates in the C. difficile toxinotype V subgroup.
Acknowledgments
We thank Ardean Veldkamp, Mike Engelhardt, and Ron White for their assistance with sample collection and Alexandra Smith for a critical review of the manuscript.
Footnotes
Published ahead of print on 5 March 2010.
REFERENCES
- 1.Alatalo, R. 1981. Problems in the measurement of evenness in ecology. Oikos 37:199-204. [Google Scholar]
- 2.Alvarez-Perez, S., J. L. Blanco, E. Bouza, P. Alba, X. Gibert, J. Maldonado, and M. E. Garcia. 2009. Prevalence of Clostridium difficile in diarrhoeic and non-diarrhoeic piglets. Vet. Microbiol. 137:203-205. [DOI] [PubMed] [Google Scholar]
- 3.Arroyo, L. G., S. A. Kruth, B. M. Willey, H. R. Staempfli, D. E. Low, and J. S. Weese. 2005. PCR ribotyping of Clostridium difficile isolates originating from human and animal sources. J. Med. Microbiol. 54:163-166. [DOI] [PubMed] [Google Scholar]
- 4.Baker, A., E. Davis, T. Rehberger, T. Neumann, and D. Rosener. 2007. Prevalence of virulence factors contributing to pathogenesis of Clostridium perfringens among neonatal pigs in the Midwest, abstr. Z-015, p. 92. Abstr. 107th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 5.Barbut, F., N. Mario, M. Delmée, J. Gozian, and J. C. Petit. 1993. Genomic fingerprinting of Clostridium difficile isolates by using a random amplified polymorphic DNA (RAPD) assay. FEMS Microbiol. Lett. 114:161-166. [DOI] [PubMed] [Google Scholar]
- 6.Bélanger, S. D., M. Boissinot, N. Clairoux, F. J. Picard, and M. G. Bergeron. 2003. Rapid detection of Clostridium difficile in feces by real-time PCR. J. Clin. Microbiol. 41:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bidet, P., V. Lalande, B. Salauze, B. Burghoffer, V. Avesani, M. Delmée, A. Rossier, F. Barbut, and J. Petit. 2000. Comparison of PCR-ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for typing Clostridium difficile. J. Clin. Microbiol. 38:2484-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Debast, S. B., L. A. van Leengoed, A. Goorhuis, C. Harmanus, E. J. Kuijper, and A. A. Bergwerff. 2009. Clostridium difficile PCR ribotype 078 toxinotype V found in diarrhoeal pigs identical to isolates from affected humans. Environ. Microbiol. 11:505-511. [DOI] [PubMed] [Google Scholar]
- 9.Fawley, W. N., and M. H. Wilcox. 2001. Molecular epidemiology of endemic Clostridium difficile infection. Epidemiol. Infect. 126:343-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goorhuis, A., D. Bakker, J. Corver, S. B. Debast, C. Harmanus, D. W. Notermans, A. A. Bergwerff, F. W. Dekker, and E. J. Kuijper. 2008. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin. Infect. Dis. 47:1162-1170. [DOI] [PubMed] [Google Scholar]
- 11.Goorhuis, A., S. Debast, L. A. M. G. van Leengoed, C. Harmanus, D. W. Notermans, A. A. Bergwerff, and E. J. Kuijper. 2008. Clostridium difficile PCR ribotype 078: an emerging strain in humans and in pigs? J. Clin. Microbiol. 46:1157-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatheway, C. L. 1990. Toxigenic clostridia. Clin. Microbiol. Rev. 3:66-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Indra, A., D. Schmid, S. Huhulescu, M. Hell, R. Gattringer, P. Hasenberger, A. Fielder, G. Wewalka, and F. Allerberger. 2008. Characterization of clinical Clostridium difficile isolates by PCR ribotyping and detection of toxin genes in Austria, 2006-2007. J. Med. Microbiol. 57:702-708. [DOI] [PubMed] [Google Scholar]
- 14.Jhung, M. A., A. D. Thompson, G. E. Killgore, W. E. Zukowski, G. Songer, M. Warny, S. Johnson, D. N. Gerding, L. C. McDonald, and B. M. Limbago. 2008. Toxinotype V Clostridium difficile in humans and food animals. Emerg. Infect. Dis. 14:1039-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jost, B. H., H. T. Trinh, and J. G. Songer. 2006. Clonal relationships among Clostridium perfringens of porcine origin as determined by multilocus sequence typing. Vet. Microbiol. 116:158-165. [DOI] [PubMed] [Google Scholar]
- 16.Keel, K., J. S. Brazier, K. W. Post, S. Weese, and J. G. Songer. 2007. Prevalence of PCR ribotypes among Clostridium difficile isolates from pigs, calves, and other species. J. Clin. Microbiol. 45:1963-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keel, M. K., and J. G. Songer. 2006. The comparative pathology of Clostridium difficile-associated disease. Vet. Pathol. 43:225-240. [DOI] [PubMed] [Google Scholar]
- 18.Killgore, G., A. Thompson, S. Johnson, J. Brazier, E. Kuijper, J. Pepin, E. H. Frost, P. Savelkoul, B. Nicholson, R. J. van den Berg, H. Kato, S. P. Sambol, W. Zukowski, C. Woods, B. Limbago, D. N. Gerding, and L. C. McDonald. 2008. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J. Clin. Microbiol. 46:431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leflon-Guibout, V., J. L. Pons, B. Heym, and M. H. Nicolas-Chanoine. 1997. Typing of Clostridium perfringens strains by use of random amplified polymorphic DNA (RAPD) system in comparison with zymotyping. Anaerobe 3:245-250. [DOI] [PubMed] [Google Scholar]
- 20.Lemee, L., A. Dhalluin, M. Pestel-Caron, J. Lemeland, and J. Pons. 2004. Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types. J. Clin. Microbiol. 42:2609-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nauerby, B., K. Pedersen, and M. Madsen. 2003. Analysis by pulsed-field gel electrophoresis of the genetic diversity among Clostridium perfringens isolates from chickens. Vet. Microbiol. 94:257-266. [DOI] [PubMed] [Google Scholar]
- 22.Pielou, E. C. 1975. Ecological diversity. Wiley, New York, NY.
- 23.Pirs, T., M. Ocepek, and M. Rupnik. 2008. Isolation of Clostridium difficile from food animals in Slovenia. J. Med. Microbiol. 57:790-792. [DOI] [PubMed] [Google Scholar]
- 24.Power, E. G. 1996. RAPD typing in microbiology—a technical review. J. Hosp. Infect. 34:247-265. [DOI] [PubMed] [Google Scholar]
- 25.Reggentin, P., and R. Schauer. 1997. Clostridial sialidases, p. 423-437. In J. I. Rood, B. A. McClane, J. G. Songer, and R. W. Titball (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, Inc., San Diego, CA.
- 26.Rodriguez-Palacios, A., H. R. Staempfli, T. Duffield, and J. S. Weese. 2007. Clostridium difficile in retail ground meat, Canada. Emerg. Infect. Dis. 13:485-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rupnik, M. 2008. Heterogeneity of large clostridial toxins: importance of Clostridium difficile toxinotypes. FEMS Microbiol. Rev. 32:541-555. [DOI] [PubMed] [Google Scholar]
- 28.Rupnik, M., J. S. Brazier, B. I. Duerden, M. Grabnar, and S. L. J. Stubbs. 2001. Comparison of toxinotyping and PCR ribotyping of Clostridium difficile strains and description of novel toxinotypes. Microbiology 147:439-447. [DOI] [PubMed] [Google Scholar]
- 29.Rupnik, M., V. Braun, F. Soehn, M. Janc, M. Hofstetter, R. Laufenberg-Feldmann, and C. von Eichel-Streiber. 1997. Characterization of polymorphisms in the toxin A and B genes of Clostridium difficile. FEMS Microbiol. Lett. 148:197-202. [DOI] [PubMed] [Google Scholar]
- 30.Rupnik, M., V. Avesani, M. Janc, C. von Eichel-Streiber, and M. Delmée. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J. Clin. Microbiol. 36:2240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon, C. E., and W. Weaver. 1949. The mathematical theory of communication. University Illinois Press, Urbana, IL.
- 32.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Songer, J. G., and D. J. Taylor. 2006. Clostridial infections, p. 613-628. In B. E. Straw, J. J. Zimmerman, S. D'Allaire, and D. J. Taylor (ed.), Diseases of swine, 9th ed. Blackwell Publishing, Ames, IA.
- 34.Songer, J. G. 1996. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9:216-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Songer, J. G., and M. A. Anderson. 2006. Clostridium difficile: an important pathogen of food animals. Anaerobe 12:1-4. [DOI] [PubMed] [Google Scholar]
- 36.Songer, J. G., and F. A. Uzal. 2005. Clostridial enteric infections in pigs. J. Vet. Diagn. Invest. 17:528-536. [DOI] [PubMed] [Google Scholar]
- 37.Songer, J. G., H. T. Trinh, G. E. Killgore, A. D. Thompson, L. C. McDonald, and B. M. Limbago. 2009. Clostridium difficile in retail meat products, U.S. A., 2007. Emerg. Infect. Dis. 15:819-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stabler, R. A., D. N. Gerding, J. G. Songer, D. Drudy, J. S. Brazier, H. T. Trinh, A. A. Witney, J. Hinds, and B. W. Wren. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 188:7297-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voth, D. E., and J. D. Ballard. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18:247-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weese, J. S., B. P. Avery, J. Rousseau, and R. J. Reid-Smith. 2009. Detection and enumeration of Clostridium difficile spores in retail pork and beef. Appl. Environ. Microbiol. 75:5009-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams, J. G. K., A. R. Kubelik, K. J. Livak, J. A. Rafalski, and S. V. Tingey. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yaeger, M., N. Funk, and L. Hoffmann. 2002. A survey of agents associated with neonatal diarrhea in Iowa swine including Clostridium difficile and porcine reproductive and respiratory syndrome virus. J. Vet. Diagn. Invest. 14:281-287. [DOI] [PubMed] [Google Scholar]
- 43.Yoo, H. S., S. U. Lee, K. Y. Park, and Y. H. Park. 1997. Molecular typing and epidemiological survey of prevalence of Clostridium perfringens types by multiplex PCR. J. Clin. Microbiol. 35:228-232. [DOI] [PMC free article] [PubMed] [Google Scholar]