Abstract
Micro-Raman spectroscopy is a fast and sensitive tool for the detection, classification, and identification of biological organisms. The vibrational spectrum inherently serves as a fingerprint of the biochemical composition of each bacterium and thus makes identification at the species level, or even the subspecies level, possible. Therefore, microorganisms in areas susceptible to bacterial contamination, e.g., clinical environments or food-processing technology, can be sensed. Within the scope of point-of-care-testing also, detection of intentionally released biosafety level 3 (BSL-3) agents, such as Bacillus anthracis endospores, or their products is attainable. However, no Raman spectroscopy-compatible inactivation method for the notoriously resistant Bacillus endospores has been elaborated so far. In this work we present an inactivation protocol for endospores that permits, on the one hand, sufficient microbial inactivation and, on the other hand, the recording of Raman spectroscopic signatures of single endospores, making species-specific identification by means of highly sophisticated chemometrical methods possible. Several physical and chemical inactivation methods were assessed, and eventually treatment with 20% formaldehyde proved to be superior to the other methods in terms of sporicidal capacity and information conservation in the Raman spectra. The latter fact has been verified by successfully using self-learning machines (such as support vector machines or artificial neural networks) to identify inactivated B. anthracis-related endospores with adequate accuracies within the range of the limited model database employed.
The detection of biological warfare agents requires methods for detecting and rapidly identifying bacterial endospores—such as Bacillus anthracis, the etiological agent of the acute fatal zoonosis anthrax in mammals—that are released in buildings or distributed in the environment. A great number of different technologies and combinations, such as DNA detection by PCR or DNA sequencing (42), are applied for genetic analysis. In addition, different microscopic approaches, such as atomic force microscopy (64) or fluorescence microscopy (26, 27), mass spectroscopy, and infrared (23, 34, 50) and Raman spectroscopy (15), have been used. With these optical detection methods, preparation and analysis time can be considerably shortened relative to that required for currently established methods based on, e.g., microbiology, immunoassays, and genetic and molecularly based approaches for identification.
The feasibility of a variety of approaches for pathogen detection relying on Raman spectroscopy, including nonresonant Raman spectroscopy (10, 54), UV resonance Raman spectroscopy (11, 35), surface-enhanced Raman spectroscopy (SERS) (12, 62), nonlinear Raman spectroscopy (39, 40), and Raman imaging (18, 22, 46), has already been verified. In some of these studies, the possibility of performing detection and identification on a single-organism level via a micro-Raman setup was exploited.
Because Raman spectra provide a snapshot of the total molecular composition of single cells, they inherently contain all the information needed to accurately identify microorganisms to the subspecies level. Although many Raman marker bands for single biosubstances or classes of biosubstances are known, an overall understanding of the spectral fingerprint in the spectra of bacteria is usually neither attainable nor necessary, if the typing strategy is based on a pattern-matching algorithm with a comprehensive database of single-cell Raman reference spectra. The recognition algorithms applied rely mainly on unsupervised and supervised multivariate chemometrical methods (6, 13, 45, 47).
Some advantages over classical microbiological and other typing methods are apparent. First, time-consuming cultivation steps prior to the actual measurements are redundant, since detectability at the single-organism level is ensured through the use of high-numerical-aperture illumination and light-gathering optics. Second, sample preparation is limited to a minimum and generally consists only in the isolation of the microorganisms from their native surroundings. Third, there is no reliance on costly taxospecific consumables with limited shelf lives. Furthermore, the complete sequence of operations—starting with isolation, proceeding through the acquisition of Raman spectra, and ending with chemotaxonomic identification—can be performed automatically. Thus, with a micro-Raman-based sensor, rapid detection of highly pathogenic microorganisms at the very place of contamination is possible.
If biosafety level-3 (BSL-3) endospores, such as those of B. anthracis, are the pathogens to be identified, an appropriate inactivation procedure is essential. Unfortunately, dormant spores exhibit incredible hardiness against germicidal agents. Several parameters contributing to the spores' intrinsic resistance are discussed in the literature. The complex shell-like structure of endospores plays a key role in their resistance: the central core, which is the analogue of the protoplast or germ cell of a growing cell, is surrounded by several protective barriers containing the inner membrane, germ cell wall, cortex outer membrane, and coats. In some species a thin exosporium may also be present. Some exosporia contribute to spore resistance by their mere impermeability to exogenous chemicals or by their ability to react with or detoxify chemical agents. The other characteristics contributing to resistance can be found in the spore core: the low water content (25 to 50% of wet weight), the saturation of spore DNA with a group of unique proteins called α/β-SASP (small acid-soluble spore proteins), or the high mineral content in the form of divalent cations, in particular Ca2+. Most of these cations seem to be chelated by dipicolinic acid (DPA), since calcium dipicolinate (CaDPA), the calcium chelate of dipicolinic acid (2,6-pyridinedicarboxylic acid), accounts for 5 to 15% of the spores' molecular weight (52).
Actually, several inactivation techniques are available, and their efficiencies have been studied intensively. Reliable disintegration of endospores has been achieved by means of heat inactivation (boiling, moist heat, dry heat), radiation (microwave [57], UV [63], gamma, electron beam [16]), desiccation, high pressure (31), and the application of various liquid or gaseous antiseptics and disinfectants (1, 51). Classically, inactivation of endospores is quantified by measuring the (logarithmic) reduction factors in CFU. Less time-consuming are viability assays based on the detection of DPA via luminescence microscopy, where the germination of endospores is triggered and subsequently the released DPA is detected by means of terbium dipicolinate luminescence under UV excitation. The terbium dipicolinate fluorescence method has been described as the most sensitive technique, allowing the determination of nanomolar concentrations of DPA (8).
Unfortunately, none of the methods mentioned above have been tested for their applicability with micro-Raman spectroscopy so far. That fact is the motivation for this work, where we systematically assess a number of well-established methods for the inactivation of pathogenic endospores with regard to sufficient microbial inactivation and appropriate identification on a single-endospore level via micro-Raman spectroscopy.
MATERIALS AND METHODS
Bacillus strains.
The following strains were examined in this study: B. anthracis 4463, B. anthracis 13/38, Bacillus subtilis ATCC 6633, Bacillus mycoides DSM 299, Bacillus sphaericus DSM 1867, Bacillus thuringiensis ATCC 10792, and B. thuringiensis DSM 350. All but the B. anthracis strains, which were provided by the Federal Research Institute for Animal Health (FLI, Jena, Germany), were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). For the selection of the most suitable inactivation method, spores of the two Bacillus thuringiensis strains (DSM 350 and ATCC 10792) and the two Bacillus anthracis strains (13/38 and 4463) were used.
Sample preparation.
Bacterial cultures were prepared on blood agar plates. From each strain a spore suspension (107 spores per ml) was prepared. Briefly, one loop of bacterial mass was inoculated into tryptone glucose broth (TGB), consisting of 2.5 g yeast extract, 5.0 g tryptone, and 1.0 g glucose per 1,000 ml double-distilled water (pH 7.2 ± 0.2; autoclaved at 121°C for 20 min). After incubation at 37°C for 3 days, 1 ml of the inoculated TGB was inoculated onto yeast extract agar consisting of 10.00 g peptone, 2.0 g yeast extract, 0.04 g MnSO4 H2O, and 15 g agar per 1,000 ml double-distilled water (pH 7.0 ± 0.2; autoclaved at 121°C for 20 min). After cultivation at 37°C for 8 to 10 days, the spores were harvested by washing with 5 ml double-distilled water and centrifugation at 2,500 × g for 10 min. The sediment was washed four times using double-distilled water before the suspension was heated at 75°C for 10 min. The spore suspensions were stored at 4°C.
Inactivation.
For inactivation of spore suspensions, formaldehyde (F) solutions (Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany) at final concentrations of 10% and 20%, a 0.5% Wofasteril-Alcapur (CH3COOOH, H2O2, CH3COOH) mixture (Kesla, Bitterfeld-Wolfen, Germany), peracetic acid (PAA) at 1% and 2%, and the household hygiene detergent Danchlorix (NaOCl; Colgate-Palmolive GmbH, Hamburg, Germany), undiluted and at 5%, were tested. As a control, the procedure was conducted using double-distilled water. Furthermore, autoclaving was carried out at 134°C for 1 h.
The inactivation efficacy of each suspension was tested after treatment for 15 min, 30 min, 1 h, 2 h, and 4 h. For the inactivation experiment, 1 ml of the spore suspension was mixed with 9 ml of disinfectant in a 10-ml tube also containing 10 glass beads to achieve the respective final concentration. During the treatment time, the tubes were shaken continuously at 200 rpm in a laboratory shaker (Gerhardt Shaker RO 15). The treatment was stopped by centrifugation at 2,500 × g for 10 min. After three additional washing steps, including centrifugation, the sediment was resuspended in 1 ml double-distilled water. For the inactivation control, 100 μl of the suspension was inoculated into TGB, and after 24 h of incubation at 37°C, 100 μl of this broth was inoculated onto blood agar plates. These plates were incubated for 2 days at 37°C to determine if the inactivation of spores was successful.
Spectroscopic instrumentation.
The Raman spectroscopic measurements were carried out with two different micro-Raman setups. The Raman spectra for the explorative studies were measured with a micro-Raman spectrometer (LabRam HR system with inverse microscope; Horiba Jobin Yvon). A Compass frequency-doubled Nd:YAG laser (Coherent Inc.) provides the excitation light with a wavelength at 532 nm and a power of 1.5 mW incident on the sample. The integration time for single-endospore Raman spectra was 30 s. A Leica PL Fluotar 100× objective focuses the laser light onto the sample within a focus of less than 1 μm in diameter. The spectrometer has an entrance slit of 100 μm and a focal length of 800 mm and is equipped with a 300-line/mm grating. A charge-coupled device (CCD) camera operating at 220 K is utilized over a range of 298 to 3,373 cm−1. The resolution of the spectra is approximately 10 cm−1.
In order to establish a database of Raman spectra, a second micro-Raman device was employed (Bio Particle Explorer; rap.ID Particle Systems GmbH, Berlin, Germany), that allowed automated measurements of single-cell Raman spectra with an excitation light of 532 nm from a solid-state frequency-doubled Nd:YAG module (LCM-S-111-NNP25; Laser-export Co. Ltd.). An Olympus MPLFLN-BD 100× objective focuses the Raman excitation light onto the sample with a spot size of <1 μm laterally, so that approximately 3.5 mW hits the sample. The integration time per Raman spectrum (276 cm−1 to 3,204 cm−1) was 5 s. After removal of the Rayleigh scattering, the 180° back-scattered Raman light is diffracted with a single-stage monochromator (HE 532; Horiba Jobin Yvon) with a 300-line/mm grating and is collected with a thermoelectrically cooled CCD camera (DV401-BV; Andor Technology) with a spectral resolution of ca. 7 cm−1. All of the Raman spectra were collected under ambient conditions.
Electron microscopy.
Micrographs of differently treated endospores were obtained with a scanning electron microscope (JSM-6700F; JEOL) operated at 15.0 keV.
Multivariate analysis.
For the hierarchical cluster analysis (HCA), the Opus Ident software package, version 6.5 (Bruker Optik GmbH, Ettlingen, Germany), was utilized exclusively. The preprocessing of the spectra comprised the first derivative and vector normalization. The spectral regions between 684 and 1,775 cm−1, as well as those between 2,763 cm−1 and 3,194 cm−1, were used for cluster analysis using the squared Euclidian distance and Ward algorithm.
For more-complex calculations, such as self-learning machines, GNU R (41) was used.
The main structure of the chemometric procedure is explained below. Basically, there are three steps: preprocessing, training of the self-learning machines, and validation.
The preprocessing of every data set was always the same. First the cosmic spikes and the backgrounds of the spectra were removed. The latter is necessary because the spectral background differs for different measurements of one particular substance, so it would disturb the training process of the self-learning machines. For this purpose, the SNIP algorithm was adopted (48). Afterwards, vector normalization took place to make the spectra comparable. Here, the spectrum was divided by its 2-norm. With principal-component analysis (PCA) (37), the dimensionality of the problem was reduced and, in addition, white noise was removed by cutting off the scores after a particular channel in the new spectral space. The exact number of scores used depends on the size of the data set. A good choice for the number of scores is to use 2 to 5% of the number of data points. The reduction of the dimensionality of the problem is necessary to avoid overfitting (58).
If a new set of spectra had to be labeled, the preprocessing of this new data set was the same before PCA. To convert both sets in the same spectral space, we did not perform PCA with the combined data set but rotated the new set by the loadings of the PCA of the first data set into the spectral space of the first data set.
After the preprocessing was done, the classifiers were trained. The binary classifiers investigated are (i) artificial neural networks (ANN) with a feed-forward topology (44), (ii) a support vector machine (SVM) with a radial basis kernel (3), and linear discriminant analysis (LDA) (9).
These binary classifiers are not capable of distinguishing between more than two classes. There are several methods for combining the classifiers so that they can be used as multiclass classifiers. Here the “one-against-the-rest” (OATR) schema is adopted (55). The idea is that every class is separated from the rest by one binary classifier. Afterwards there are n binary classifiers for n classes. If a new spectrum now has to be labeled, it is designated by the respective classifier. If all classifiers put this new spectrum into the “rest” class, this spectrum is labeled as “unknown.” The same happens if more than one classifier labels this spectrum as its associated class. An ambiguity results in labeling as “unknown.”
If there is more than one classifier, it is possible to combine them by a majority-voting scheme (24), where the basic rule from Kittler (24) is adapted to the novelty detection scheme. If the majority votes for one class, it is labeled with this class; if not, it is labeled as “unknown”. This procedure might enhance accuracy, but in some cases it delivers the same or even worse accuracy. This combined classifier is investigated for the purpose of identifying bacterial endospores; the three classifiers (ANN, SVM, and LDA) are combined by this method.
Finally, validation of the chosen classifiers is needed. There are several methods for estimating the quality of a classifier, e.g., hold-out, cross-validation, and bootstrapping (25).
To obtain the best classifier, cross-validation was used, and this accuracy was taken as the accuracy of the classifier. To estimate the generalization error, the hold-out technique was applied. One set of bacteria from each of the Bacillus species analyzed is used for training, and another, completely independent batch of the particular strain, which has been cultured separately under exactly the same conditions, is used as the validation set. The hold-out method provides a way to estimate the accuracy of the procedure with the connected database for a real application.
RESULTS
Inactivation results.
It is not surprising that the analysis and handling of intentionally released BSL-3 level agents, or their products, require either BSL-3 facilities or complete inactivation if samples from contaminated areas are prepared for dispatch to standard laboratories. Within the scope of the work presented here, we have tested a number of inactivation methods: heat inactivation (autoclaving) and inactivation by formaldehyde (F), peracetic acid (PAA), or sodium hypochlorite. The disinfectants evaluated were chosen according to a selection of valid lists and guidelines with approved disinfectants for spores. For surface disinfection in laboratories and hospitals in Germany, no approved sporicidal disinfectant is available (43, 59). However, the WHO Guidelines for the Surveillance and Control of Anthrax in Humans and Animals recommend, for example, the use of formaldehyde and peracetic acid (56). The disadvantages of peracetic acid include its corrosive properties and pungent odor. By the combination of peracetic acid with aluminum hydroxide, e.g., Wofasteril-Alcapur (Kesla, Bitterfeld-Wolfen, Germany), the disinfectant becomes odorless. Because other authors have also shown the sporicidal potency of commercial household products containing sodium hypochlorite bleach against anthrax spores (2), we tested the product Danchlorix (NaOCl; Colgate-Palmolive GmbH, Hamburg, Germany). The most reliable disinfection procedure for spores so far has been autoclaving at 134°C.
The results of the inactivation experiments with spores of Bacillus thuringiensis and Bacillus anthracis strains are summarized in Table 1. Each entry in the table represents one independent inactivation experiment. It can be seen that the 20% formaldehyde solution, undiluted Danchlorix, and 2% peracetic acid were the most reliable and fastest spore-inactivating substances. Even after the shortest application time of 15 min, all four strains tested exhibited no viability after inactivation with these agents. In some cases, single strains showed higher resistance to the treatment; e.g., only B. thuringiensis strain DSM 350 survived parts of the 1% PA treatment. On the other hand, some disinfectants, such as the 10% formaldehyde or 5% Danchlorix treatment, showed time dependency.
TABLE 1.
Results of inactivation experiments on spores of two Bacillus thuringiensis strains and two Bacillus anthracis strainsa
| Disinfectant | Growth of sporesb inactivated at the following time: |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 min |
30 min |
1 h |
2 h |
4 h |
||||||||||||||||
| A | B | C | D | A | B | C | D | A | B | C | D | A | B | C | D | A | B | C | D | |
| Formaldehyde | ||||||||||||||||||||
| 10% | + | − | + | + | + | − | + | + | + | − | − | − | − | − | − | − | + | − | − | − |
| 20% | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Danchlorix | ||||||||||||||||||||
| Undiluted | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 5% | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − |
| 0.5% Wofasteril-Alcapur mixture | + | + | + | + | + | + | − | − | + | + | − | − | + | + | − | − | + | + | − | − |
| Peracetic acid | ||||||||||||||||||||
| 1% | − | − | − | − | + | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − |
| 2% | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Autoclaving at 134°C | NT | NT | NT | NT | NT | NT | NT | NT | − | − | − | − | NT | NT | NT | NT | NT | NT | NT | NT |
| Distilled water (control) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
Capital letters represent strains as follows: A, B. thuringiensis DSM 350; B, B. thuringiensis ATCC 10792; C, B. anthracis strain 13/38; D, B. anthracis strain 4463.
−, no growth observed after inactivation; +, visible growth observed after inactivation; NT, not tested. The initial spore concentration was 1 × 107 spores per ml.
Analysis of endospore morphology via electron microscopy.
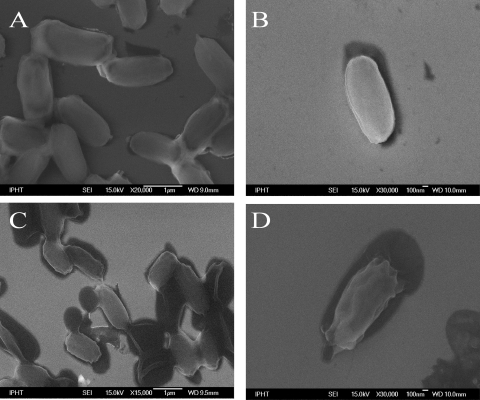
To assess spore morphology and integrity after inactivation via formaldehyde, peracetic acid, or autoclaving, scanning electron microscopic (SEM) images of single B. thuringiensis DSM 350 endospores were prepared. In Fig. 1A to D, representative images of spores treated by these methods (20% formaldehyde for 15 min [Fig. 1B], 2% peracetic acid for 15 min [Fig. 1C], or 134°C autoclaving for 15 min [Fig. 1D]) are shown in comparison to an image of untreated, viable endospores (Fig. 1A).
FIG. 1.
Electron micrographs of differently treated endospores of B. thuringiensis DSM 350. (A) Viable spores not subjected to inactivation treatment; (B) spores inactivated by 20% formaldehyde for 15 min; (C) spores inactivated by 1% peracetic acid for 15 min; (D) spores inactivated by autoclaving at 134°C for 15 min.
Aiming to minimize the ratio of the outer surface to the cell volume, the untreated spores (Fig. 1A) have a roundish shape and a smooth surface.
The most obvious physical changes occurred for the spores when they were exposed to moist heat and pressure through the autoclaving process. They exhibit a wrinkled shape and have obviously lost internal volume, as evidenced by their decreased outer form and the presence of a corona of undifferentiated solids around the spore perimeter. These morphological changes seem to arise from a swelling and splitting of the cells, followed by a subsequent collapsing into a wrinkled form—a behavior well known in the literature (38). During this process, intracellular material might be released, which explains the surrounding undefined material.
The formaldehyde (Fig. 1B)- and PAA (Fig. 1C)-treated cells exhibit greater similarity to native spores. But for the latter (Fig. 1C) it is noteworthy that next to the brighter cells with apparently good structural integrity, some endospores are in contact with dark, roundish, closely attached pools, which seem to originate from outflows from the spores, since the cell wall seems to be cracked open. Nothing like this can be seen in the images of the viable endospores (Fig. 1A). Nevertheless, the absence/presence of an exosporium cannot be verified by these pictures.
Analysis of single-endospore Raman spectra.
Apart from characterizing single cells according to their outward morphology by means of SEM, molecular assessment of cells on a single-endospore level can be achieved by means of micro-Raman spectroscopy. Here, the overall composition of the endospores is probed, and by the observation of a few Raman spectra, a first evaluation can easily be made. Furthermore, a more-sophisticated but fast analysis via unsupervised hierarchical cluster analysis (HCA), which pools the spectra by spectral similarity, is performed. A maximum of correspondence between the Raman profiles of inactivated spores and those of viable endospores is desirable. Furthermore, the slight spectral differences, on which the subsequent phenotypic identification is based, should still remain after the treatment with disinfectants.
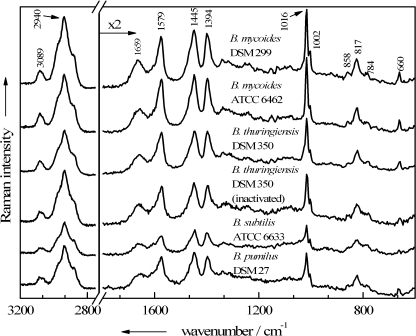
Since single Raman spectra of viable cells serve as a standard, spectra of native Bacillus sp. endospores are shown in Fig. 2. Independently of the species, all spore spectra are dominated by signals that can be assigned to CaDPA: the C—COO− stretching vibration at 817 cm−1, the symmetric and asymmetric COO− stretching vibrations at 1,394 cm−1 and 1,579 cm−1, three pyridine ring vibrations at 660 cm−1, 1,016 cm−1, and 1,445 cm−1, and the aromatic C—H stretching mode at 3,089 cm−1. Small alterations in the CaDPA band intensities within these species may arise from differences in the spore composition of the particular culture, emerging, e.g., from fluctuating CaDPA contents in single spores among a population (19). The band positions, on the other hand, remain stable among the spectra; this is not necessarily the case, since the use of divalent manganese as a sporulation accelerator can cause some minor CaDPA band shifts (53).
FIG. 2.
Single-endospore Raman spectra of untreated viable spores of five different Bacillus strains plus one spectrum of a formaldehyde-inactivated B. thuringiensis endospore.
Except for the CH2/3-stretching vibrations at 2,940 cm−1, all bands from the other major biological constituents of the cells exhibit lower intensities but are still clearly recognizable. Some contributions arise exclusively due to proteins (1,002 cm−1, ring breath vibration of phenylalanine; 1,659 cm−1, amide I vibration; 858 cm−1, one of the ring breathing modes of tyrosine) or DNA/RNA moieties (784 cm−1, O—P—O stretching of cytosine/uracil). In addition, some bands of those biomolecules overlap with CaDPA signals, e.g., the second ring breathing mode of tyrosine at 824 cm−1 with the carboxylate stretching mode and the deformation mode of CH2/3, as well as the ring vibrations of guanine and adenine, which are expected at around 1,450 cm−1 and at 1,570 cm−1 with the pyridine ring vibrations of the DPA.
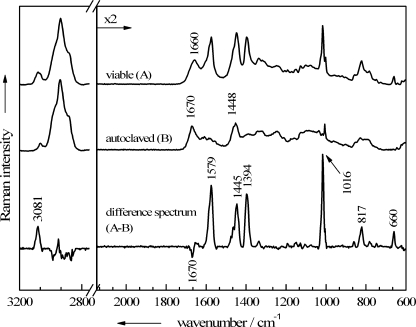
All endospores that have been sterilized by autoclaving exhibit the same changes in their Raman spectra. As examples, two mean spectra of Bacillus mycoides DSM 299 are shown in Fig. 3: the top spectrum was calculated from 20 single viable endospore spectra, and the middle spectrum from the spectra of 20 single endospores inactivated by autoclaving. The third spectrum represents the subtraction of the inactivated-cell spectrum from the spectrum of one of the viable spores. It can be seen clearly that the spectra of autoclaved endospores are basically devoid of any CaDPA bands, which normally dominate endospore spectra. However, bands from the other major biological constituents of the cells still appear in the spectra of autoclaved cells. Only the amide I vibration at 1,660 cm−1 is shifted significantly, to 1,670 cm−1, after autoclaving. This Raman band is highly sensitive to changes in the secondary structures of proteins (60), which most likely occurred during the denaturation and subsequent aggregation due to autoclaving. UV-resonance Raman spectroscopy correlated with X-ray data revealed that the amide I band shows the highest wave numbers in proteins with unordered secondary structures, followed by those with β-sheets and α-helices, respectively (4).
FIG. 3.
Baseline-corrected mean Raman spectra, each calculated from 20 single-endospore spectra of B. mycoides DSM 299. Shown are spectra for viable, untreated spores (A) and spores inactivated via autoclaving (B), as well as a difference spectrum (A-B).
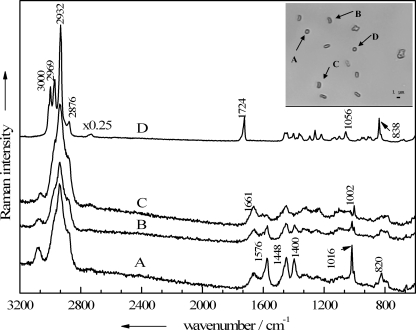
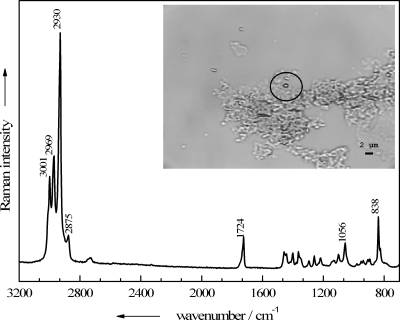
In the bright-field image in the Fig. 4 inset, a small grouping of B. thuringiensis endospores, inactivated with 1% peracetic acid for 15 min, is shown. The micro-Raman spectra of the four labeled particles, which exhibit rather similar morphologies, were recorded and are shown in Fig. 4. Raman spectrum A in Fig. 4 represents a typical endospore spectrum, displaying the CaDPA bands mentioned above at 1,016, 1,400, 1,448, and 1,576 cm−1, as well as signals typical of vegetative cells, e.g., the amide I band at 1,661 cm−1 and a sharp signal due to the ring vibration of phenylalanine at 1,002 cm−1. To a lesser extent this is also true for spectrum B in Fig. 4, where again the DPA bands occur, but compared to the signals of the other bacterial components, their intensities are considerably decreased. The Raman spectrum of particle C (Fig. 4) shows almost no CaDPA bands any more, suggesting that a leakage of intracellular components, including CaDPA, took place. Although particle D is not phenotypically different from the others, its Raman spectrum does not resemble the other Raman spectra: none of the CaDPA or protein marker bands mentioned above for either vegetative cells or endospores appears. Instead, the Raman spectrum is representative of poly(3-hydroxybutyrate) (PHB), a biodegradable polymer that can be produced by certain bacteria in large amounts in response to physiological stress (21). Recent work dealing with the analysis of microbial PHB by Raman spectroscopy has been published (7, 18) and allows for a rather complete band assignment. The bands at 3,000, 2,969, 2,932, and 2,876 cm−1 all arise from different CH2/3-streching vibrations of the polymer; the 1,724-cm−1 band is assignable to the C=O stretching vibration of both crystalline and amorphous PHB. In the lower-wave-number region, the most intense signals, at 1,056 and 838 cm−1, are also important hallmarks for this polyester and can be assigned to the C—O and the C—COO stretching vibrations, respectively.
FIG. 4.
Single-particle Raman spectra of four particles present in a B. thuringiensis DSM 350 endospore suspension treated with 1% peracetic acid for 15 min. (Inset) Corresponding bright-field image of the analyzed particles A to D.
This effect of finding PHB particles beside intact spores is also observed when the spores are treated with a higher concentration of PAA for longer periods. This has been verified by analyzing two B. thuringiensis and B. anthracis endospore suspensions after treatment with 1% or 2% PAA for 15, 30, 60, 120, or 240 min. For each sample, 30 single cells were measured, and roughly 20% were free of CaDPA after the suspensions had been treated for 15 or 30 min. After a 1-h exposure to PAA, nearly half of the cells seemed to be depleted of the DPA salt, and after 2 and 4 h, not a single intact endospore could be detected, and all cells gave spectra like spectrum C in Fig. 4. There is no difference between suspensions exposed to different concentrations of PAA (1% or 2%); even after treatment for 2 h with 1% PAA, all cells were depleted of the DPA salt. Therefore, this biocide not only affects the cells unevenly but also differs strongly in its effect according to the treatment time.
The sodium hypochlorite-containing bleaching agent Danchlorix presents a more uniform but also undesirable effect on the endospores. Even when Danchlorix was diluted 1:20, and after a short exposure time, not a single cell was left for Raman measurements according to the bright-field image. As shown in the Fig. 5 inset, only broad patterns of unidentifiable material could be found next to just a few single particulate entities, which turned out, again, to be PHB accumulations. The Raman spectrum of one of these deposits (circled) is shown in Fig. 5.
FIG. 5.
Raman spectrum of a single particle found in a B. thuringiensis DSM 350 endospore suspension treated with 1:20-diluted Danchlorix. (Inset) Bright-field image with the corresponding particle circled.
In contrast to the results with these two biocides, the interaction of formaldehyde with the endospores yields inactivated cells whose Raman spectra correspond well with those of native endospores without exception—no matter how long (15 to 240 min) the treatment and how high the concentration of formaldehyde (10% or 20%). In Fig. 2, a Raman spectrum of a single formaldehyde-inactivated B. thuringiensis endospore is shown in order to demonstrate its striking similarity with the spectra of untreated cells. It shows no remarkable changes, such as relative intensity variations or even the appearance of new signals, in the spectral profile.
Evaluation of the inactivation methods.
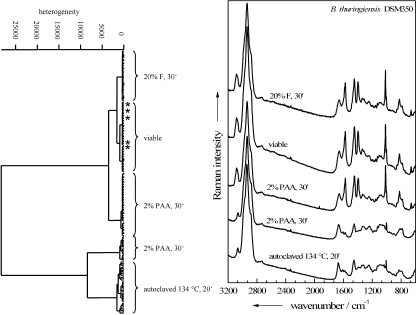
The Raman spectra were analyzed more thoroughly by performing HCA on an ensemble of differently treated endospores of B. thuringiensis DSM 350. This ensemble included cells inactivated by autoclaving or by treatment with 2% PAA or 20% formaldehyde. These inactivated species were chosen because, as Table 1 shows, only 2% PAA and 20% formaldehyde treatments guaranteed consistent inactivation of B. thuringiensis DSM 350 endospores after a 30-min exposure. Additionally, some spectra of viable endospores were added in order to determine which—if any—of the inactivation methods results in endospores whose Raman spectra are most similar to those of viable endospores. The resultant dendrogram is shown in Fig. 6. For most of the samples, all 40 Raman spectra of the same sample are grouped in the same cluster. The five outliers (asterisked) all belong to formaldehyde-treated spectra, which are wrongly classified into the cluster of untreated endospores. All in all, of 160 spectra, 155 (96.7%) are correctly classified.
FIG. 6.
Dendrogram obtained from a hierarchical cluster analysis performed on Raman spectra of differently treated single endospores of B. thuringiensis DSM 350. Shown are the Raman spectra (right) and dendrogram (left) for viable, untreated cells and for endospores inactivated either by 20% formaldehyde for 30 min (20% F, 30′), by 2% peracetic acid for 30 min (2% PAA, 30′), or by autoclaving at 134°C for 20 min (134°C, 20′). Asterisks mark wrongly classified spectra.
Two major groups, subdivided into three and two subclusters, respectively, can be identified in the dendrogram. One main cluster comprises spectra of formaldehyde- or PAA-treated endospores or viable endospores. The second main cluster is composed of spectra of endospores inactivated by means of autoclaving or PAA. Each of the five distinguishable subclusters can be explained according to the mean spectra shown in Fig. 6. All spectra devoid of CaDPA signals are assigned to the same main cluster. This is true for all of the autoclaved samples and a fraction of the PAA-treated samples. Interestingly, the remaining spectral information for these obviously severely altered endospores is prominent enough to produce a clear partition according to the inactivation procedure.
In the other main cluster, the second partition of the PAA-treated endospores can be located next to the joint cluster of the formaldehyde-inactivated and native endospores, suggesting that among the biocide treatments examined, inactivation with formaldehyde causes the fewest spectral changes from the spectra of viable endospores. Thus, the occurrence in the dendrogram of the five formaldehyde-inactivated endospores falsely classified as native endospores is tolerable and emphasizes the high similarity of the spectra of the two kinds of samples.
Although the mean spectrum of the PAA-treated endospores that still contain CaDPA in Fig. 6 shows high similarity to the spectra of viable cells, slight distinctions that appear reproducibly among all spectra of the sample seem to be more pronounced than those for formaldehyde-treated cells. Additionally, roughly one-third of the PAA-treated cells were sorted out into the other main cluster together with the autoclaved cells. According to the cluster spectrum that is second from the bottom in Fig. 6, these Raman spectra emerge from CaDPA-depleted endospores; thus, due to their weak resemblance to the spectra of viable endospores, they are not suitable for identification purposes and have to be discarded.
Single-cell identification.
To assess whether the use of formaldehyde on endospores has a great impact on the possibility of distinguishing and identifying Bacillus endospores to the species level, the following data set was created: from each of four Bacillus strains (Bacillus mycoides DSM 299, Bacillus subtilis ATCC 6633, Bacillus sphaericus DSM 1867, and Bacillus thuringiensis DSM 350), four independent batches were prepared under the same conditions. Afterwards, two of the four batches per strain were inactivated with 20% formaldehyde for 2 h, while the other two remained viable. With one batch of viable endospores per strain, a reference or training database for viable spores was established by measuring approximately 100 single-endospore Raman spectra per strain. The same was done with four batches of inactivated endospores, one per strain. Thus, each of the databases, one for the viable and one for the inactivated spores, comprised roughly 400 Raman spectra. Based on these, the other four batches of viable spores and the four samples with the inactivated endospores were identified according to their taxa. Therefore, nearly 30 endospore Raman spectra were recorded for each sample, and each of them was identified to the species level by comparing the spectra to the appropriate database: if they had been inactivated, they were compared to the training database of inactivated spores, and if they had not been treated, the database of viable spores was used.
Tables 2 to 5 summarize the results obtained by using different classifiers for untreated native endospores versus formaldehyde-inactivated endospores. For the untreated-spore data set the first 9 scores were used, and for the inactivated-spore data set the first 10 scores were used, whereas the data set was not centered or scaled by channel before the PCA.
TABLE 2.
Confusion table showing the accuracy each classifiera after the classification of four untreated native Bacillus species in the training data setb
| Identification predicted by classifier | No. of spectra actually classified as indicated, grouped by classifier |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANN |
SVM |
LDA |
||||||||||
| B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | |
| Unknown | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 3 |
| B. mycoides | 137 | 0 | 0 | 0 | 139 | 0 | 0 | 1 | 138 | 0 | 0 | 0 |
| B. sphaericus | 0 | 87 | 0 | 0 | 0 | 86 | 0 | 0 | 0 | 86 | 0 | 0 |
| B. subtilis | 0 | 0 | 70 | 0 | 0 | 0 | 72 | 0 | 0 | 0 | 71 | 0 |
| B. thuringiensis | 2 | 0 | 0 | 121 | 0 | 0 | 0 | 120 | 0 | 0 | 0 | 121 |
The accuracy of each classifier for viable endospores was 99%.
The species used were B. mycoides DSM 299, B. sphaericus DSM 1867, B. subtilis ATCC 6633, and B. thuringiensis DSM 350.
TABLE 5.
Confusion table showing the accuracy of each classifier and of the combination of all classifiers after the identification of four 20% formaldehyde-inactivated Bacillus species in the hold-out data set
| Identification predicted by classifier | No. of spectra actually identified as indicated, grouped by classifier (% accuracy) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANN (83) |
SVM (80) |
LDA (80) |
All classifiers combined (82%) |
|||||||||||||
| B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | |
| Unknown | 4 | 1 | 5 | 3 | 5 | 0 | 1 | 5 | 6 | 0 | 7 | 6 | 5 | 1 | 4 | 7 |
| B. mycoides | 53 | 0 | 0 | 15 | 47 | 2 | 1 | 10 | 52 | 0 | 0 | 13 | 52 | 0 | 0 | 11 |
| B. sphaericus | 0 | 74 | 1 | 0 | 2 | 73 | 4 | 1 | 1 | 75 | 3 | 0 | 1 | 74 | 3 | 0 |
| B. subtilis | 0 | 0 | 32 | 0 | 0 | 0 | 32 | 1 | 0 | 0 | 28 | 0 | 0 | 0 | 31 | 0 |
| B. thuringiensis | 6 | 0 | 0 | 8 | 9 | 0 | 0 | 9 | 4 | 0 | 0 | 7 | 5 | 0 | 0 | 8 |
The results of the cross-validation with the associated confusion table for the viable endospores are listed in Table 2. The rows of the tables show the species identifications predicted by each classifier investigated, and the columns show the actual species identified. As can be seen, all three classifiers achieved accuracies in comparable dimensions, around 99%, for viable endospores. Table 3 shows analogous data for endospores inactivated by 20% formaldehyde. Although the accuracies of the classifiers are decreased to 81 to 89%, none of the classifiers is superior to the other two with respect to the classification rate. Most of the false species identifications were mix-ups between the B. mycoides and B. thuringiensis spectra, especially for the ANN results. However, spectra classified as unknown are not necessarily false-positive results.
TABLE 3.
Confusion table showing the accuracy of each classifier after the classification of four 20% formaldehyde-inactivated Bacillus species in the training data set
| Identification predicted by classifier | No. of spectra actually classified as indicated, grouped by classifier (% accuracy) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANN (83) |
SVM (89) |
LDA (84) |
||||||||||
| B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | |
| Unknown | 16 | 2 | 0 | 21 | 11 | 2 | 1 | 11 | 17 | 2 | 2 | 21 |
| B. mycoides | 153 | 0 | 1 | 21 | 166 | 0 | 0 | 16 | 157 | 0 | 0 | 25 |
| B. sphaericus | 2 | 111 | 0 | 1 | 0 | 110 | 0 | 0 | 1 | 110 | 0 | 0 |
| B. subtilis | 2 | 0 | 97 | 1 | 0 | 0 | 96 | 0 | 0 | 0 | 92 | 0 |
| B. thuringiensis | 13 | 0 | 0 | 39 | 9 | 1 | 0 | 57 | 12 | 1 | 1 | 39 |
After the classifiers were trained with the training data set, the spectra of the second batch were labeled. The accuracy of identification and the associated confusion table for untreated endospores are presented in Table 4, and the analogous data for formaldehyde-inactivated endospores are shown in Table 5. The last columns list the combined results of all three classifiers. Here, the majority voting scheme, discussed above, was employed: in case of a majority vote for one class, the spectrum is assigned to this class; otherwise, it is put into the “unknown” class.
TABLE 4.
Confusion table showing the accuracy of each classifier and of the combination of all classifiers after the identification of four untreated native Bacillus species in the hold-out data set
| Identification predicted by classifier | No. of spectra actually identified as indicated, grouped by classifier (% accuracy) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANN (82) |
SVM (81) |
LDA (81) |
All classifiers combined (83) |
|||||||||||||
| B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | B. mycoides | B. sphaericus | B. subtilis | B. thuringiensis | |
| Unknown | 8 | 2 | 3 | 9 | 9 | 0 | 0 | 1 | 16 | 0 | 2 | 10 | 13 | 0 | 3 | 7 |
| B. mycoides | 48 | 0 | 0 | 7 | 43 | 0 | 0 | 6 | 39 | 0 | 0 | 1 | 43 | 0 | 0 | 4 |
| B. sphaericus | 0 | 73 | 0 | 0 | 0 | 75 | 1 | 0 | 1 | 75 | 1 | 0 | 0 | 75 | 1 | 0 |
| B. subtilis | 0 | 0 | 35 | 0 | 1 | 0 | 27 | 0 | 0 | 0 | 35 | 0 | 0 | 0 | 34 | 0 |
| B. thuringiensis | 7 | 0 | 0 | 10 | 10 | 0 | 10 | 19 | 7 | 0 | 0 | 15 | 7 | 0 | 0 | 15 |
It is obvious that for both native and inactivated endospores, the identification rate lies in the same range, around 81 to 83%, for all of the classifiers. The formaldehyde treatment does not seem to have a negative impact on the identification results. Nevertheless, most of the spectra that were incorrectly identified or were not identified at all were those of B. thuringiensis, which were frequently mixed up with B. mycoides spectra. It is known from the nucleotide sequences of the 150-bp 3′-end 16S rRNA genes, as well as from those of the 70-bp 5′ 16S-23S internal transcribed spacer (ITS) region, that the two species are phylogenetically very similar (61). Indeed, their phenotypic and genotypic similarities recently led to a proposal to regroup them, together with B. anthracis and Bacillus cereus, into a single species (17).
DISCUSSION
Fast and reliable inactivation of bacterial endospores can be achieved with a great variety of different antiseptics and disinfectants. Most of the sporicidal agents provoke lysis and leakage of intracellular constituents by degrading most of the permeability barriers, which play different roles in the endospore's resistance. The various layers of proteinaceous spore coats, which surround the spore cortex, protect the spore from attacks by a large number of chemicals, particularly oxidizing agents such as hydrogen peroxide, sodium hypochlorite, chlorine dioxide, or ozone (52). Smaller hydrophobic and hydrophilic molecules are hindered on their way to the core by the extremely low permeability of the spore's inner membrane (36).
Most of the oxidizing agents, strong acids, and ethanol kill spores by causing some type of damage to the spore's permeability barriers, such that when the treated spores germinate, these damaged membranes rupture, resulting in spore death (36, 51). For some of the oxidizing agents, the location of damage could be pinpointed to the endospore's inner membrane (5); for others, only a general disruption of spore permeability barriers is reported. Interestingly, Cortezzo et al. found out that spore inactivation via a variety of oxidizing agents is not accompanied by loss of DPA (5), whereas acid-inactivated spores extrude DPA and other fibrillar material, including DNA (51). In addition, pretreatment with oxidizing agents makes the surviving spores more sensitive to inactivation by normally nonlethal heat and osmotic stresses in the presence of high salt concentrations in plating media.
These findings are in good agreement with the results we obtained by using PAA as a disinfectant. Among other peroxides, peracetic acid is frequently used in common disinfection procedures and is considered to be a more potent biocide than hydrogen peroxide. As with hydrogen peroxide, the hydroxyl radical appears to be of prime importance, and its inactivation mechanism is based not on DNA damage but rather on a weakening of the endospore's inner membrane by disrupting sulfhydryl (—SH) and sulfur (S—S) bonds of proteins (32, 33). As shown in Fig. 4 and in the dendrogram in Fig. 6, this internal damage only partially forces the spores to extrude core material, since most of the endospores remained intact and retained intracellular DPA after a short treatment (15 min) with highly diluted PAA (1%). The leakage is more pronounced at higher PAA concentrations (2%) and with longer exposure times (>1 h). Furthermore, the probability of measuring not only intact or harmed endospores but also metabolites of the vegetative cells increased with the inactivation time and PAA concentration, as shown in Fig. 4. According to these results, not a microbial cell but a residue of formerly intracellular granules of PHB has been measured. When PHB-producing cells die, PHB is released into the environment, where it is transformed into a denatured semicrystalline state (21).
In case of the chlorine-releasing product Danchlorix, the complete spore lysis we observed was also achieved in other studies with sodium hypochlorite (28). In those studies, loss of refractivity, separation of the spore coats from the cortex, extensive discharge of Ca2+, dipicolinic acid, and DNA/RNA, and finally lysis occurred.
Inactivation of endospores by wet heat had the same depleting effect. Here again, not the DNA but proteins are assumed to be the target (36), explaining the shift of the amide III band in the observed Raman spectra of Fig. 3. This is confirmed by previous work (38), where alterations of amide I and amide II bands due to autoclaving could be monitored by means of Fourier transform infrared spectroscopy (FT-IR), and where the absorption band at 1,570 cm−1, a diagnostic band for DPA, was lost, as shown in Fig. 3. Since the major part of this salt is deposited in the core of endospores, these ruptures due to autoclaving-induced cleavage of endospores seem to reach into the inner part of the cell, where most spore enzymes, as well as DNA, ribosomes, and tRNA, are located. Thus, not only denaturation of macromolecules and subcellular structures, including proteins, cytoplasmic membranes, and nucleic acids, occurs during autoclaving, but also a major loss of those biomolecules, as well as the ubiquitous DPA salt, takes place.
This method of extracting soluble microbial proteins can be beneficial for some analysis methods that rely on those proteins as biomarkers, e.g., for the matrix-assisted laser desorption-time-of-flight (MALDI-TOF)/intact-cell mass spectrometry (ICMS) methodology (29). But unlike these approaches, Raman spectroscopy does not analyze bulk samples or fractions thereof but intact single cells.
That is why all the types of chemical agents discussed above are inappropriate for the purpose of identifying bacterial spores by means of Raman spectroscopy, since retaining the structural integrity and most of the biochemical composition of single cells is a necessity for micro-Raman-based identification of inactivated pathogenic endospores. The considerable alterations of spore integrity by PAA, Danchlorix, and autoclaving obviously have a nonnegligible impact on the Raman spectra of the endospores, which coincide with a loss of information in the Raman spectra and thus with decreased identification accuracy. Additionally, the homogeneity of the inactivation treatment has to be guaranteed. The achievement of a maximum of uniformity among the sterilized cells is another important objective, since chemotaxonomic classification relies mainly on constantly recurring spectral patterns among an ensemble of single-cell spectra. If the inactivation treatment alters cells of the same species differently and thus decreases the uniformity of the respective spectra, these Raman data are less useful for building up a database for supervised identification routines. Alternatively, the database might comprise all the inactivation-induced spectral variances, but then it would probably gain a dimension of enormous extent.
Formaldehyde is also employed for a variety of decontamination processes, since it is sporicidal (49). Loshon et al. achieved a 99% killing rate for B. subtilis endospores in 40 min at 30°C with 25 g/liter formaldehyde (30). Endospores are inactivated by formaldehyde due to some unique features of this molecule. This small molecule can pass through all the protective layers to advance directly into the core of the endospore. There the genotoxic properties of formaldehyde take effect, causing spore inactivation at least in part by DNA damage; protein-DNA cross-linking is proposed to be one mutagenic mechanism of formaldehyde (30). However, the precise nature of the DNA damage is as yet unknown. The major resistance mechanisms of spores against formaldehyde are the saturation of spore DNA with α/β-SASPs and the recA-encoded repair pathway, which can repair at least some of the formaldehyde-induced lesions in DNA (36). The interaction of formaldehyde with proteins might also give rise to degradation of the spore integuments, but apparently not to such an extent that spore core material is released.
For the purposes of Raman spectroscopy, the use of formaldehyde to inactivate endospores is superior to any of the other treatments analyzed in this work. The inactivation experiments suggest the usage of this agent as a sporicide, since reliable, strain-independent, and fast inactivation of the Bacillus strains analyzed was achieved. It was shown by Raman spectroscopy that the formaldehyde inactivation technique is suitable for obtaining reproducible Raman spectra.
Possible reasons for competing intraspecies varieties in the Raman spectra are manifold, e.g., the preparation/inactivation process might induce spectral variances on the single-cell level, as can be seen in the case of PAA-inactivated endospores; cultivation parameters, such as growth time, temperature, and nutritional conditions, which have an impact on whole batches, are also factors to be reckoned with (14, 20). According to our results shown in Tables 4 and 5, at least the formaldehyde-induced inhomogeneities can be neglected, since all four different Bacillus strains were chemotaxonomically identified with the help of other, independently cultivated batches of the same strains grown under the same cultivation conditions. The interspecies distinctions are obviously still high enough to obtain satisfactory identification rates for different Bacillus species, which serve as B. anthracis models. This is noteworthy insofar as only a very limited model database was used, comprising just four different Bacillus species. The comparison between the accuracies of the cross-validation and the hold-out techniques for the bacterial database shows that the main problem of our current evaluation method is overfitting. Accuracy decreases if a trained classifier is adopted to an independent data set. This can be avoided by building a database up out of more than one batch and more data. But even with a larger and more diverse database, preprocessing and, most importantly, dimension reduction is necessary.
For anthrax detection, evaluation of as many different B. anthracis strains as possible, as well as genetic near neighbors, is mandatory. Currently, we are elaborating a micro-Raman-based procedure for the identification of B. anthracis strains inactivated with 20% formaldehyde according to the procedure described here.
Taken together, the results of this study showed that detection of the anthrax agent, isolated from real-world samples, via micro-Raman spectroscopy can be a reliable alternative for fast point-of-care testing. On-site diagnosis is ensured by inactivating the samples with the formaldehyde treatment described in this study, which is compatible with the micro-Raman identification approach. Further investigations should aim at analyzing possible interference due to, e.g., the influence of the native surrounding matrices or the isolation procedure. Additionally, efforts should be focused on the question of whether the inactivation efficacy of formaldehyde is reduced due to matrix effects.
Acknowledgments
Funding of the research projects “Pathosafe” FKZ 13N9547 and FKZ 13N9549 from the Federal Ministry of Education and Research, Germany (BMBF), is gratefully acknowledged.
We also thank Franka Jahn (Institute of Photonic Technology, Germany) for performing the EM experiment and Katja Fischer (Friedrich Loeffler Institute, Germany) for doing the inactivation experiments.
Footnotes
Published ahead of print on 5 March 2010.
REFERENCES
- 1.Baron, P. A., C. F. Estill, J. K. Beard, M. J. Hein, and L. Larsen. 2007. Bacterial endospore inactivation caused by outgassing of vaporous hydrogen peroxide from polymethyl methacrylate (Plexiglas). Lett. Appl. Microbiol. 45:485-490. [DOI] [PubMed] [Google Scholar]
- 2.Black, D. G., T. M. Taylor, H. J. Kerr, S. Padhi, T. J. Montville, and P. M. Davidson. 2008. Decontamination of fluid milk containing Bacillus spores using commercial household products. J. Food Prot. 71:473-478. [DOI] [PubMed] [Google Scholar]
- 3.Burges, C. J. C. 1998. A tutorial on support vector machines for pattern recognition. Data Mining Knowledge Discov. 2:121-167. [Google Scholar]
- 4.Chi, Z., X. G. Chen, J. S. W. Holtz, and S. A. Asher. 1998. UV resonance Raman-selective amide vibrational enhancement: quantitative methodology for determining protein secondary structure. Biochemistry 37:2854-2864. [DOI] [PubMed] [Google Scholar]
- 5.Cortezzo, D. E., K. Koziol-Dube, B. Setlow, and P. Setlow. 2004. Treatment with oxidizing agents damages the inner membrane of spores of Bacillus subtilis and sensitizes spores to subsequent stress. J. Appl. Microbiol. 97:838-852. [DOI] [PubMed] [Google Scholar]
- 6.De Gelder, J., P. Scheldeman, K. Leus, M. Heyndrickx, P. Vandenabeele, L. Moens, and P. De Vos. 2007. Raman spectroscopic study of bacterial endospores. Anal. Bioanal. Chem. 389:2143-2151. [DOI] [PubMed] [Google Scholar]
- 7.De Gelder, J., D. Willemse-Erix, M. J. Scholtes, J. I. Sanchez, K. Maquelin, P. Vandenabeele, P. De Boever, G. J. Puppels, L. Moens, and P. De Vos. 2008. Monitoring poly(3-hydroxybutyrate) production in Cupriavidus necator DSM 428 (H16) with Raman spectroscopy. Anal. Chem. (Washington, DC) 80:2155-2160. [DOI] [PubMed] [Google Scholar]
- 8.Fichtel, J., J. Koester, J. Rullkoetter, and H. Sass. 2007. Spore dipicolinic acid contents used for estimating the number of endospores in sediments. FEMS Microbiol. Ecol. 61:522-532. [DOI] [PubMed] [Google Scholar]
- 9.Fisher, R. 1936. The use of multiple measurements in taxonomic problems. Ann. Eugenics 7:179-188. [Google Scholar]
- 10.Gaus, K., P. Rösch, R. Petry, K.-D. Peschke, O. Ronneberger, H. Burkhardt, K. Baumann, and J. Popp. 2006. Classification of lactic acid bacteria with UV-resonance Raman spectroscopy. Biopolymers 82:286-290. [DOI] [PubMed] [Google Scholar]
- 11.Ghiamati, E., R. Manoharan, W. H. Nelson, and J. F. Sperry. 1992. UV resonance Raman spectra of Bacillus spores. Appl. Spectrosc. 46:357-364. [Google Scholar]
- 12.Guicheteau, J., L. Argue, D. Emge, A. Hyre, M. Jacobson, and S. Christesen. 2008. Bacillus spore classification via surface-enhanced Raman spectroscopy and principal component analysis. Appl. Spectrosc. 62:267-272. [DOI] [PubMed] [Google Scholar]
- 13.Harz, M., M. Kiehntopf, S. Stöckel, P. Rösch, E. Straube, T. Deufel, and J. Popp. 2009. Direct analysis of clinical relevant single bacterial cells from cerebrospinal fluid during bacterial meningitis by means of micro-Raman spectroscopy. J. Biophotonics 2:70-80. [DOI] [PubMed] [Google Scholar]
- 14.Harz, M., P. Rösch, K. D. Peschke, O. Ronneberger, H. Burkhardt, and J. Popp. 2005. Micro-Raman spectroscopic identification of bacterial cells of the genus Staphylococcus and dependence on their cultivation conditions. Analyst 130:1543-1550. [DOI] [PubMed] [Google Scholar]
- 15.Harz, M., P. Rösch, and J. Popp. 2009. Vibrational spectroscopy—a powerful tool for the rapid identification of microbial cells at the single-cell level. Cytometry A 75:104-113. [DOI] [PubMed] [Google Scholar]
- 16.Helfinstine, S. L., C. Vargas-Aburto, R. M. Uribe, and C. J. Woolverton. 2005. Inactivation of bacillus endospores in envelopes by electron beam irradiation. Appl. Environ. Microbiol. 71:7029-7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helgason, E., O. A. Okstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A.-B. Kolsto. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermelink, A., A. Brauer, P. Lasch, and D. Naumann. 2009. Phenotypic heterogeneity within microbial populations at the single-cell level investigated by confocal Raman microspectroscopy. Analyst 134:1149-1153. [DOI] [PubMed] [Google Scholar]
- 19.Huang, S.-S., D. Chen, P. L. Pelczar, V. R. Vepachedu, P. Setlow, and Y.-Q. Li. 2007. Levels of Ca2+-dipicolinic acid in individual Bacillus spores determined using microfluidic Raman tweezers. J. Bacteriol. 189:4681-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutsebaut, D., K. Maquelin, P. De Vos, P. Vandenabeele, L. Moens, and G. J. Puppels. 2004. Effect of culture conditions on the achievable taxonomic resolution of Raman spectroscopy disclosed by three Bacillus species. Anal. Chem. 76:6274-6281. [DOI] [PubMed] [Google Scholar]
- 21.Jendrossek, D., and R. Handrick. 2002. Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 56:403-432. [DOI] [PubMed] [Google Scholar]
- 22.Kalasinsky, K. S., T. Hadfield, A. A. Shea, V. F. Kalasinsky, M. P. Nelson, J. Neiss, A. J. Drauch, G. S. Vanni, and P. J. Treado. 2007. Raman chemical imaging spectroscopy reagentless detection and identification of pathogens: signature development and evaluation. Anal. Chem. 79:2658-2673. [DOI] [PubMed] [Google Scholar]
- 23.Kirschner, C., K. Maquelin, P. Pina, N. A. N. Thi, L. P. Choo-Smith, G. D. Sockalingum, C. Sandt, D. Ami, F. Orsini, S. M. Doglia, P. Allouch, M. Mainfait, G. J. Puppels, and D. Naumann. 2001. Classification and identification of enterococci: a comparative phenotypic, genotypic, and vibrational spectroscopic study. J. Clin. Microbiol. 39:1763-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kittler, J. 1998. Combining classifiers: a theoretical framework. Pattern Anal. Appl. 1:18-27. [Google Scholar]
- 25.Kohavi, R. 1995. A study of cross-validation and bootstrap for accuracy estimation and model selection, p. 1137-1143. In Proceedings of the 14th International Joint Conference on Artificial Intelligence, vol. 2. Morgan Kaufmann Publishers Inc., San Francisco, CA. [Google Scholar]
- 26.Krause, M., B. Radt, P. Rösch, and J. Popp. 2007. The investigation of single bacteria by means of fluorescence staining and Raman spectroscopy. J. Raman Spectrosc. 38:369-372. [Google Scholar]
- 27.Krause, M., P. Rösch, B. Radt, and J. Popp. 2008. Localizing and identifying living bacteria in an abiotic environment by a combination of Raman and fluorescence microscopy. Anal. Chem. 80:8568-8575. [DOI] [PubMed] [Google Scholar]
- 28.Kulikovsky, A., H. S. Pankratz, and H. L. Sadoff. 1975. Ultrastructural and chemical changes in spores of Bacillus cereus after action of disinfectants. J. Appl. Bacteriol. 38:39-46. [DOI] [PubMed] [Google Scholar]
- 29.Lasch, P., H. Nattermann, M. Erhard, M. Staemmler, R. Grunow, N. Bannert, B. Appel, and D. Naumann. 2008. MALDI-TOF mass spectrometry compatible inactivation method for highly pathogenic microbial cells and spores. Anal. Chem. 80:2026-2034. [DOI] [PubMed] [Google Scholar]
- 30.Loshon, C. A., P. C. Genest, B. Setlow, and P. Setlow. 1999. Formaldehyde kills spores of Bacillus subtilis by DNA damage and small, acid-soluble spore proteins of the alpha/beta-type protect spores against this DNA damage. J. Appl. Microbiol. 87:8-14. [DOI] [PubMed] [Google Scholar]
- 31.Margosch, D., M. G. Gaenzle, M. A. Ehrmann, and R. F. Vogel. 2004. Pressure inactivation of Bacillus endospores. Appl. Environ. Microbiol. 70:7321-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marquis, R. E., G. C. Rutherford, M. M. Faraci, and S. Y. Shin. 1995. Sporicidal action of peracetic acid and protective effects of transition metal ions. J. Ind. Microbiol. 15:486-492. [DOI] [PubMed] [Google Scholar]
- 33.McDonnell, G., and A. D. Russell. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naumann, D. 2000. Infrared spectroscopy in microbiology, p. 102-131. In R. A. Meyers (ed.), Encyclopedia of analytical chemistry. John Wiley & Sons, Chichester, United Kingdom.
- 35.Nelson, W. H., R. Dasari, M. Feld, and J. F. Sperry. 2004. Intensities of calcium dipicolinate and Bacillus subtilis spore Raman spectra excited with 244 nm light. Appl. Spectrosc. 58:1408-1412. [DOI] [PubMed] [Google Scholar]
- 36.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson, K. 1901. On lines and planes of closest fit to systems of points in space. Philos. Mag. 2:559-572. [Google Scholar]
- 38.Perkins, D. L., C. R. Lovell, B. V. Bronk, B. Setlow, P. Setlow, and M. L. Myrick. 2004. Effects of autoclaving on bacterial endospores studied by Fourier transform infrared microspectroscopy. Appl. Spectrosc. 58:749-753. [DOI] [PubMed] [Google Scholar]
- 39.Pestov, D., X. Wang, G. O. Ariunbold, R. K. Murawski, V. A. Sautenkov, A. Dogariu, A. Sokolov, and M. O. Scully. 2008. Single-shot detection of bacterial endospores via coherent Raman spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 105:422-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrov, G. I., R. Arora, V. V. Yakovlev, X. Wang, A. V. Sokolov, and M. O. Scully. 2007. Comparison of coherent and spontaneous Raman microspectroscopies for noninvasive detection of single bacteria endospores. Proc. Natl. Acad. Sci. U. S. A. 104:7776-7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Development Core Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 42.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 43.Robert Koch-Institut. 2007. Liste der vom Robert Koch-Institut geprüften und anerkannten Desinfektionsmittel und -verfahren Stand vom 31.5.2007 (15. Ausgabe). Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 50:1335-1356. [DOI] [PubMed] [Google Scholar]
- 44.Rojas, R. 1996. Neural networks: a systematic introduction. Springer, Berlin, Germany.
- 45.Rösch, P., M. Harz, K.-D. Peschke, O. Ronneberger, H. Burkhardt, and J. Popp. 2006. Identification of single eukaryotic cells with micro-Raman spectroscopy. Biopolymers 82:312-316. [DOI] [PubMed] [Google Scholar]
- 46.Rösch, P., M. Harz, K.-D. Peschke, O. Ronneberger, H. Burkhardt, A. Schuele, G. Schmauz, M. Lankers, S. Hofer, H. Thiele, H.-W. Motzkus, and J. Popp. 2006. On-line monitoring and identification of bioaerosols. Anal. Chem. 78:2163-2170. [DOI] [PubMed] [Google Scholar]
- 47.Rösch, P., M. Harz, M. Schmitt, K.-D. Peschke, O. Ronneberger, H. Burkhardt, H.-W. Motzkus, M. Lankers, S. Hofer, H. Thiele, and J. Popp. 2005. Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations. Appl. Environ. Microbiol. 71:1626-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryan, C. G., E. Clayton, W. L. Griffin, S. H. Sie, and D. R. Cousens. 1988. SNIP, a statistics-sensitive background treatment for the quantitative analysis of PIXE spectra in geoscience applications. Nucl. Instrum. Methods Phys. Res. B 34:396-402. [Google Scholar]
- 49.Sagripanti, J. L., and A. Bonifacino. 1996. Comparative sporicidal effects of liquid chemical agents. Appl. Environ. Microbiol. 62:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samuels, A. C., A. P. Snyder, D. K. Emge, D. St. Amant, J. Minter, M. Campbell, and A. Tripathi. 2009. Classification of select category A and B bacteria by Fourier transform infrared spectroscopy. Appl. Spectrosc. 63:14-24. [DOI] [PubMed] [Google Scholar]
- 51.Setlow, B., C. A. Loshon, P. C. Genest, A. E. Cowan, C. Setlow, and P. Setlow. 2002. Mechanisms of killing spores of Bacillus subtilis by acid, alkali and ethanol. J. Appl. Microbiol. 92:362-375. [DOI] [PubMed] [Google Scholar]
- 52.Setlow, P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101:514-525. [DOI] [PubMed] [Google Scholar]
- 53.Stöckel, S., S. Meisel, R. Böhme, M. Elschner, P. Rösch, and J. Popp. 2009. Effect of supplementary manganese on the sporulation of Bacillus endospores analysed by Raman spectroscopy. J. Raman Spectrosc. 40:1469-1477. [Google Scholar]
- 54.Tarcea, N., M. Harz, P. Rösch, T. Frosch, M. Schmitt, H. Thiele, R. Hochleitner, and J. Popp. 2007. UV Raman spectroscopy—a technique for biological and mineralogical in situ planetary studies. Spectrochim. Acta A 68:1029-1035. [DOI] [PubMed] [Google Scholar]
- 55.Tax, D. M. J., and R. P. W. Duin. 2002. Using two-class classifiers for multiclass classification, p. 124-127. In R. Kasturi et al. (ed.), 16th International Conference on Pattern Recognition: Proceedings, vol. 2. IEEE Computer Society Press, Los Alamitos, CA. [Google Scholar]
- 56.Turnbull, P. C. B. 2008. Guidelines for the surveillance and control of anthrax in humans and animals. WHO/EMC/ZDI/98/6. World Health Organization, Geneva, Switzerland. [PubMed]
- 57.Vaid, A., and A. H. Bishop. 1998. The destruction by microwave radiation of bacterial endospores and amplification of the released DNA. J. Appl. Microbiol. 85:115-122. [Google Scholar]
- 58.Vapnik, V. N. 2000. The nature of statistical learning theory. Springer, New York, NY.
- 59.Verbund für Angewandte Hygiene. 31 July 2006, accession date. Desinfektionsmittel-Liste. Flächendesinfektion. Desinfektionsmittel-Kommission im Verbund für Angewandte Hygiene (VAH) e.V., Bonn, Germany.
- 60.Williams, R. W. 1983. Estimation of protein secondary structure from the laser Raman amide I spectrum. J. Mol. Biol. 166:581-603. [DOI] [PubMed] [Google Scholar]
- 61.Xu, D., and J.-C. Cote. 2003. Phylogenetic relationships between Bacillus species and related genera inferred from comparison of 3′ end 16S rDNA and 5′ end 16S-23S ITS nucleotide sequences. Int. J. Syst. Evol. Microbiol. 53:695-704. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, X., M. A. Young, O. Lyandres, and R. P. Van Duyne. 2005. Rapid detection of an anthrax biomarker by surface-enhanced Raman spectroscopy. J. Am. Chem. Soc. 127:4484-4489. [DOI] [PubMed] [Google Scholar]
- 63.Zhao, J., V. Krishna, B. Hua, B. Moudgil, and B. Koopman. 2009. Effect of UVA irradiance on photocatalytic and UVA inactivation of Bacillus cereus spores. J. Photochem. Photobiol. B 94:96-100. [DOI] [PubMed] [Google Scholar]
- 64.Zolock, R. A., G. Li, C. Bleckmann, L. Burggraf, and D. C. Fuller. 2006. Atomic force microscopy of Bacillus spore surface morphology. Micron 37:363-369. [DOI] [PubMed] [Google Scholar]