Abstract
Exposure to heat stress has been recognized as one of the major factors leading to the breakdown of the coral-alga symbiosis and coral bleaching. Here, we describe the presence of three new cytochrome P450 (CYP) genes from the reef-building coral endosymbiont Symbiodinium (type C3) and changes in their expression during exposure to severe and moderate heat stress conditions. Sequence analysis of the CYP C-terminal region and two conserved domains, the “PERF” and “heme-binding” domains, confirmed the separate identities of the CYP genes analyzed. In order to explore the effects of different heat stress scenarios, samples of the scleractinian coral Acropora millepora were exposed to elevated temperatures incrementally over an 18-h period (rapid thermal stress) and over a 120-h period (gradual thermal stress). After 18 h of gradual heating and incubation at 26°C, the Symbiodinium CYP mRNA pool was approximately 30% larger, while a further 6°C increase to a temperature above the average sea temperature (29°C after 72 h) resulted in a 2- to 4-fold increase in CYP expression. Both rapid heat stress and gradual heat stress at 32°C resulted in 50% to 90% decreases in CYP gene transcript abundance. Consequently, the initial upregulation of expression of CYP genes at moderately elevated temperatures (26°C and 29°C) was followed by a decrease in expression under the greater thermal stress conditions at 32°C. These findings indicate that in the coral-alga symbiosis under heat stress conditions there is production of chemical stressors and/or transcriptional factors that regulate the expression of genes, such as the genes encoding cytochrome P450 monooxygenases, that are involved in the first line of an organism's chemical defense.
The cytochrome P450 (CYP) enzymes are a superfamily of heme-thiolate proteins (43). These enzymes are include different forms and are found in all aerobic eukaryotes and some prokaryotes (42). Typically, CYPs oxidize a substrate by incorporation of a single oxygen atom, which defines them as monooxygenase-type enzymes. They catalyze hydroxylation, epoxidation, N, S, and O dealkylation, N oxidation, sulfoxidation, dehalogenation, and a number of other more complex reactions (9, 27, 40). Also, CYP enzymes play a major role in xenobiotic metabolism and in phase I drug metabolism due to the oxidation of exogenous and endogenous chemicals, including drugs and carcinogens (21). As a result of the versatile biosynthetic potential of CYPs, there has been growing interest in using these enzymes in drug development and discovery of novel valuable chemicals, as well as in bioremediation (24, 26, 33, 53, 68).
The role of cytochrome P450s in xenobiotic metabolism and steroidogenesis in marine organisms has been widely recognized. CYP activity has been detected in the tissues of marine Cnidaria, Mollusca, Arthropoda, Annelida, and Echinodermata, and higher levels have usually been observed in hepatic organs and tissues carrying out steroidogenesis, as described by Rewitz et al. (48). In the phylum Cnidaria, the presence of CYP genes has been demonstrated using expressed sequence tag (EST) sequences from freshwater Hydra sp. (48), and in the sea anemone Nematostella vectensis, CYPs have been recognized as a “chemical defensome” because of their role in protection against chemical stressors (25). In the scleractinian corals, CYP CO (carbon monoxide) difference spectra have been observed for the species Favia fragum, Siderastrea siderea, and Montastraea faveolata (22, 47). Environmentally induced changes in CYP activity were observed in the coral Stylophora pistillata after exposure to hyposaline conditions (18), as well as in Madracis mirabilis after exposure to the photosynthesis inhibitor Irgarol (17). Furthermore, inducible CYP expression has been demonstrated in the scleractinian coral Montastraea faveolata after exposure to benzo[a]pyrene and in Pocillopora damicornis after exposure to fuel oil (47, 54). The great substrate diversity of CYPs is also thought to be the result of evolutionary adaptation driven by environmental conditions (20). P450 activity involving specific xenobiotic biotransformation processes has also been reported for unicellular green algae (13, 65), while the function of CYP in the marine diatom Skeletonema costatum has not been determined yet (74).
Cytochrome P450 enzymes have not been found yet in unicellular photosynthetic dinoflagellates belonging to the genus Symbiodinium. Based on ribosomal and chloroplast DNA, symbiotic dinoflagellates are grouped into eight phylogenetic clades (clades A to H) that are further separated into distinct subclades (10, 46, 57, 58). These dinoflagellates live in mutualistic symbiosis with many marine organisms, including the reef-building corals. Rapid changes in the external environment are known to destabilize the symbiosis between corals and their dinoflagellate symbionts, leading to loss of the symbionts from the tissues of the coral hosts (29, 73). The breakdown of the symbiosis between corals and their dinoflagellates begins with photosynthetic dysfunction (11, 31) due to damage to photosystem II (2, 37, 62, 72). Overreduction of the light reactions leads to production of oxygen radicals (superoxide and singlet oxygen), which can result in damage to proteins and other cellular constituents, followed by breakdown of cell-cell signaling and removal of the symbiont (23). This is manifested at the colony level as a dramatic loss of the brown pigment of the symbiotic dinoflagellate from coral colonies (coral bleaching).
Due to the increase in carbon dioxide emission, climate change scenarios predict that global temperatures will rise at least 2°C by 2050 to 2100 (30). With a continuing global warming trend, mass coral bleaching events are predicted to increase in frequency and severity (29). Corals exposed to heat stress produce reactive oxygen species (ROS), mainly in symbiotic dinoflagellates, which destabilize the functionality of the coral-alga symbiosis (38, 73). The cytochrome P450 hemoproteins are involved in cell detoxification and protection from oxidative stress (25), and therefore we hypothesized that Symbiodinium CYP enzymes would be upregulated in response to thermal stress in the reef-building coral Acropora millepora. In the present study, we aimed to identify cytochrome P450 monooxygenase genes in the coral dinoflagellates and to evaluate the effect of a range of thermal stress scenarios on the expression of CYP genes and their role in the cnidarian-dinoflagellate symbiosis.
MATERIALS AND METHODS
Sequence identity and analysis.
EST libraries were created for Symbiodinium (type C3) isolated from the coral host Acropora aspera after exposure to thermal and nutrient stresses (36). In the Symbidoinium EST database, five putative P450 homologues were identified using BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences of three putative CYP genes included sequences encoding two conserved regions, a “PERF domain” and a “heme-binding” region (3), and were used for evaluation. The presence of cysteine in the conserved heme-binding region was necessary since cysteine thiolate binds the heme prosthetic group as the fifth ligand. The sequences corresponding to cytochrome P450 genes and accession numbers are shown in Table 1. Sequence analysis was performed using the BLAST search options tblastx and blastx, while translated sequences were analyzed using the tblastn option and blastp. The highest-scoring hits with E-values less than 10−5 were used. Sequence alignment was performed using web-based BioManager-ANGIS services (http://bioman5.angis.org.au) with ClustalW (66), and alignments were viewed using GeneDoc software.
TABLE 1.
GenBank accession numbers, designations, functions, and best BLAST (tblastn) hits (E < 1.0 × 10−5) for CYP genes
| Gene | GenBank accession no. | Annotationa | Most similar species | E value |
|---|---|---|---|---|
| CYP-1 | EH058016 | Cytochrome P450c17 (steroid 17-alpha-monooxygenase) | Anguilla japonica | 2.00E−20 |
| CYP-2 | EH036607 | Cytochrome P450 | Acidobacteria (“Candidatus Koribacter versatilis Ellin345”) | 2.00E−11 |
| CYP-3 | FE864302 | Cytochrome P450, family 20, subfamily A, polypeptide 1 | Danio rerio | 2.00E−34 |
| CYP-4 | EH036898 | Cytochrome P450 | Aedes aegypti | 1.00E−07 |
| CYP-5 | FE865804 | CYP356A1 | Crassostrea gigas | 1.00E−21 |
In general, P450 proteins have a role in secondary metabolite biosynthesis, transport, and catabolism.
Experimental design.
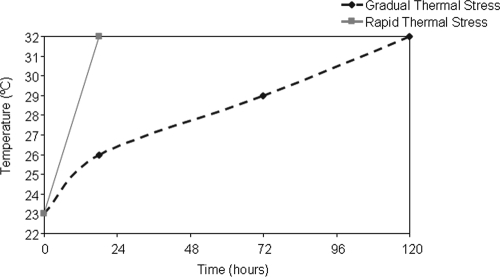
Coral nubbins (length, 4 to 5 cm) of A. millepora harboring the Symbiodinium C3 type (35) were collected in May 2009 from the reef flat surrounding Heron Island, Great Barrier Reef, Australia (23°25′S, 152°07′E). Coral nubbins were removed using wire cutters and immediately transported to a seawater facility. Corals nubbins were then attached to nylon threads and suspended in flowthrough seawater aquaria. Samples were acclimated for 48 h in 9 large experimental aquarium tanks (60 liters; 3 tank replicates per treatment) at the winter mean local ambient temperature (23 to 24°C) and at the ambient light levels, which were adjusted using shade cloth to mimic the conditions of the natural reef flat. To examine the influence of temperature stress, coral nubbins were distributed randomly in systems subjected to two experimental temperature regimens (Fig. 1), a gradual thermal stress regimen (temperature increased from 23°C to 32 ± 1°C at a rate of 1°C per ∼10 h over a 120-h period) and a rapid thermal stress regimen (temperature increased from 24°C to 32 ± 1°C at a rate of 1°C per ∼2.5 h over an 18-h period), as well as in a control group (temperature kept at 23 to 24 ± 1°C [ambient temperature]). The gradual thermal stress regimen was relevant to the natural bleaching conditions on the Great Barrier Reef, where the temperature was reported to be between 31 and 32°C over a 5-day period (5, 6, 15), while the rapid thermal stress regimen was more representative of the Heron Island reef flat dynamics observed in 2002, when the temperature oscillated between 25°C and 32°C over an approximately 24-h period and the maximum rate of increase was ∼2°C per h (15). The water temperature was recorded every 2 min using StowAway TidbiT loggers placed in each aquarium (Onset Computer Corporation, Bourne, MA). A subset of coral branches (n = 4) was removed from the treatment and control tanks at 0, 18, 72, and 120 h. Corals nubbins were immediately snap-frozen in liquid nitrogen and stored at −80°C before they were processed.
FIG. 1.
Stress regimens. Coral nubbins were exposed to two stress regimens, a gradual thermal stress regimen (temperature increased at a rate of 1°C per ∼10 h) and a rapid thermal stress regimen (temperature increased at a rate of 1°C per ∼2.5 h), as well as a control regimen (temperature stable at 23 to 24 ± 1°C [ambient temperature]). The nubbins were exposed to the rapid thermal stress regimen for 18 h and to the gradual thermal stress and control regimens for 120 h.
Extraction of total RNA.
RNA was separately extracted from 4 coral branches using small fragments (length, 0.5 to 1 cm) of coral nubbins, which were cut with a bone cutter and ground directly in liquid nitrogen. The resulting powder was put in 700 ml of RLT buffer with beta-mercaptoethanol (RNeasy kit; Invitrogen), homogenized with a hand homogenizer (Tissue-Tearor; Biospec Products, Inc.), and centrifuged for 3 min at 13,000 × g at 4°C. The aqueous phase was then used to extract total RNA with an RNeasy kit (Invitrogen) by following the manufacturer's instructions. RNA quantity and integrity were analyzed with an Agilent 2100 bioanalyzer, and 500 ng of high-quality total RNA (integrity number, >7) was subjected to reverse transcription (RT) for cDNA synthesis.
Synthesis of cDNA for qPCR.
Reverse transcription was carried out using the QuantiTect reverse transcription procedure (Qiagen, Australia). Briefly, 0.5 μg of purified total RNA was used per reaction mixture and incubated in genomic DNA wipeout buffer at 42°C for 2 min to eliminate any traces of genomic DNA, and this was followed by reverse transcription at the same temperature for 30 min. The cDNA obtained, diluted 1:10 prior to use, was used as a template in the quantitative PCR (qPCR) analysis. In parallel, RNA isolation (52) and cDNA synthesis using the QuantiTect reverse transcription procedure (Qiagen, Australia) were carried out for an A. millepora egg-sperm sample (free of algal symbionts).
Primer design.
The sequences corresponding to cytochrome P450 genes were identified by performing a BLAST search with the Symbiodinium (C3) EST database, as shown in Table 1. Specific primers were designed using Primer Express 2.0 software (Applied Biosystems, United States). The designations of the genes used for real-time RT-PCR analysis, GenBank accession numbers, primer sequences, amplicon lengths, and melting temperatures are shown in Tables 1 and 2. Standard PCR amplification was performed using selected primers (Table 2) to confirm the reproducibility of the primers used for Symbiodinium (C3) isolated from A. millepora and the lack of amplification for coral. The PCR conditions were as follows: an initial step consisting of 94°C for 1 min, followed by 35 cycles of 94°C 20 s, 60°C 20 s, and 72°C 1.5 min and then extension at 72°C for 10 min and storage of samples at 4°C.
TABLE 2.
Primer sequences, amplicon lengths, and melting temperatures for the primers used for real-time RT-PCR analysis
| Gene | Forward primer |
Reverse primer |
Amplicon length (bp) | ||
|---|---|---|---|---|---|
| Sequence (5′-3′) | Melting temp (°C) | Sequence (5′-3′) | Melting temp (°C) | ||
| CYP-1 | CCCGAACGTTTCTTGGACTCT | 60 | TCAGTTTTGGCCAGGGACTCT | 60 | 120 |
| CYP-2 | GCATGCTGACGCACAGTGA | 59 | GCCAAAAGGCATGTGCATCT | 60 | 101 |
| CYP-3 | TGTGGCCAGAACCTCAAAGG | 60 | CTTTCCCGCAAAGCCAAAT | 59 | 101 |
Quantitative PCR and gene expression analysis.
Quantitative PCR assays were performed using SYBR green PCR master mixture (Applied Biosystems, Warrington, Cheshire, United Kingdom), 384-well plates, a 7900HT fast real-time PCR system (Applied Biosystems, United States), and an Eppendorf 5075 robot (Applied Biosystems, United States). The PCR conditions were as follows: initial denaturation for 10 min at 95°C, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. At the end of the procedure, a dissociation step was included, which consisted of 95°C for 2 min, 60°C for 15 s, and 95°C for 15 s. The final reaction volume was 10 μl, and the mixture contained 300 nM primers (Table 2). All reactions were carried out with three technical replicates. The levels of expression of targeted cytochrome P450 genes were quantified using geNorm software (http://medgen.ugent.be/∼jvdesomp/genorm/) (69). The relative quantification method was used for estimation of the expression of cytochrome P450 genes by using reference genes (Table 3) with the most stable expression patterns and specificity for Symbiodinium cultures under different experimental conditions (N. Rosic, M. Pernice, and O. Hoegh-Guldberg, submitted for publication). The expression of each gene was determined from the cycle threshold (CT) value, which corresponded to the number of cycles required for the fluorescent signal from PCR amplification to reach a fixed threshold (71). CT values were obtained for a specific threshold of 0.1 and were transformed into quantities using the maximal PCR efficiency for each gene (E = 2). A real-time dissociation curve was used to check primer specificity and the presence of a unique PCR product.
TABLE 3.
GenBank accession numbers, functions, and primer sequences for housekeeping genes used for real-time RT-PCR analysis
| Gene | Function | Forward primer (5′-3′) | Reverse primer (5′-3′) | GenBank accession no. |
|---|---|---|---|---|
| Cal | Calmodulin | GGCACCATCACCACCAAAG | TCTGCGTCAACCTCGTTGATC | EH037752 |
| Rp-S4 | Ribosomal protein S4 | CCGCACAAACTGCGTGAGT | CGCTGCATGACGATCATCTT | EH036413 |
| SAM | S-Adenosyl-l-methionine synthetase | GCCTACATTTGCCGACAGATG | AATGGCTTGGCAACACCAAT | EH036622 |
| Tub | Beta-tubulin | TGACGCAGCAGATGTTTGATG | CGACATACGTCCACGGAAGAG | EH037669 |
Statistical analyses were done using the Statistica 7.0 software (Statsoft Inc., Tulsa, OK). Data normality was confirmed using the Kolmogorov-Smirnov test, and the influence of time and treatment on gene expression was tested by performing a two-way analysis of variance (ANOVA). The levels of expression of the algal CYP genes isolated from thermally stressed corals were compared to control levels of expression at each time point (for coral nubbins not exposed to elevated temperatures) using a t test (n = 4). Values were considered significantly different if the P value was <0.05. In this paper data are expressed as means ± standard deviations.
RESULTS
Sequence analysis of the Symbiodinium CYP genes.
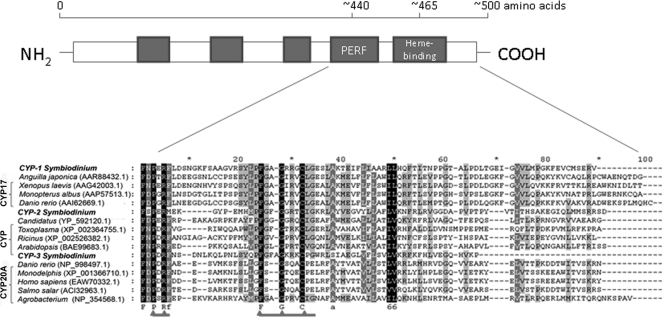
Five contigs corresponding to CYP genes with representative ESTs are shown in Table 1. Multiple-sequence alignment of the three CYP contigs with sequences corresponding to the C-terminal region including the conserved “PERF” domain (PXFX) and “heme-binding” region (FXXGXRXCXG) was performed with ClustalW (66). Three CYP genes were designated CYP-1, CYP-2, and CYP-3, and an alignment of the sequences encoded by these genes is shown in Fig. 2. The EST sequences of two other contigs, CYP-4 and CYP-5, corresponded to sequences encoding the N-terminal CYP protein region and therefore were not included in the evaluation described below. The sequence analysis of CYP-1, CYP-2, and CYP-3 confirmed the variability of the sequences of the CYP genes. At the protein level, despite considerable sequence differences, two well-conserved domains in the C-terminal region of CYP enzymes, the PERF domain (PXRX) and the absolutely conserved heme-binding region (FXXGXRXCXG), were recognized, and they are indicated in Fig. 2. Variants of the conserved PERF domain were found in the sequences encoded by all Symbiodinium CYP genes; there was a PERF region in the sequence encoded by CYP-1 and there was a PERM region in the sequence encoded by CYP-2, while a PDRF domain was found in the sequence encoded by the CYP-3 gene. The heme-binding domain was conserved in all translated CYP genes. However, a slight variation in the heme-binding region encoded by the CYP-3 gene was noticed; an extra amino acid was found between highly conserved F and G residues (Fig. 2).
FIG. 2.
Diagram of the cytochrome P450 primary structure (48), showing the distribution of the major conserved domains (filled boxes), and multiple-sequence alignment of the Symbiodinium CYP C-terminal regions, including the conserved PERF domain (PXRF) and heme-binding region (FXXGXRXCXG), constructed with Clustal W. Alignment was performed using the following pairwise alignment options: Blossum30 protein weight matrix, a gap-opening penalty of 10, and a gap extension penalty of 0.1. Predicted amino acid sequences encoded by the Symbiodinium ESTs designated CYP-1, CYP-2, and CYP-3 were aligned with the selected sequences producing the best hits according to a BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The identical residues in all sequences are indicated; amino acids conserved in 100% of the sequences are indicated by white letters with a black background, amino acids conserved in >80% of the sequences are indicated by white letters with a gray background, and amino acids conserved in >60% of the sequences are indicated by black letters with a gray background.
The closest BLAST (tblastn) hits (E value, <1.0 × 10−5) for the deduced protein sequences are shown in Table 2. For the CYP-1 gene the C-terminal domain data indicated that there was a metazoan origin, and the closest relationship was the relationship with the cytochrome P450 steroid 17-alpha-hydroxylase/17,20-lyase found in the Japanese eel (Anguilla japonica), a frog (Xenopus laevis), and the zebrafish (Danio rerio). In contrast, the C-terminal domain of the CYP-2 gene product exhibited the highest levels of similarity with the C-terminal region of a putative cytochrome P450 in unicellular microorganisms (Acidobacteria and Toxoplasma gondii) and higher plants (Arabidopsis thaliana and Nicotiana tabacum). A metazoan origin was again indicated for the CYP-3 gene product, and the closest connection was the connection with the sequence encoding an “orphan” cytochrome P450 (family 20, subfamily A, polypeptide 1) found in Homo sapiens, zebrafish, Salmo salar, and a marsupial (Monodelphis domestica). Based on data for the C-terminal region, all three CYP genes found in symbiotic dinoflagellates contained novel sequences compared to the genes encoding other CYP hemoproteins. In addition, due to symbiosis and the pool of mixed algal and coral transcriptomes, we examined whether three CYP genes were present in a dinoflagellate-free coral host using cDNA obtained from an A. millepora egg-sperm specimen. Our results confirmed that the CYP genes discovered did not have a coral origin (PCR amplification was not successful).
Effect of elevated temperatures on CYP gene expression.
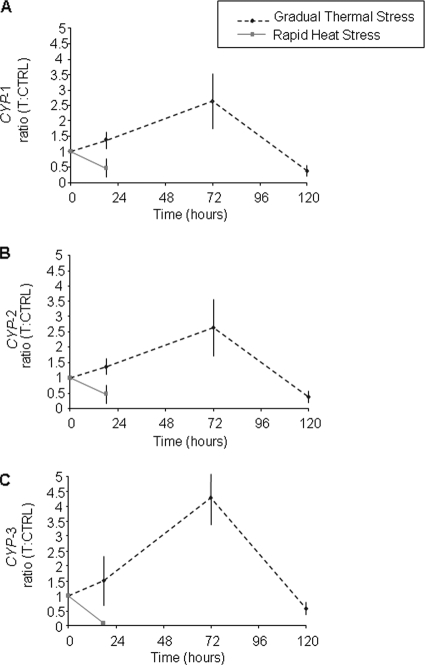
A real-time dissociation curve confirmed the presence of a specific PCR product in all amplification reactions. The relative quantities of CYP genes were normalized using the quantities of two reference genes with the most stable expression pattern as defined by the GeNorm analysis (69). The housekeeping genes used were the ribosomal protein S4 (Rp-S4) and S-adenosyl-l-methionine synthetase (SAM) genes, which had an average expression stability value (M) of 0.351 and a pairwise variation for V2/3 of 0.137. In Fig. 3, the ratios of the CYP gene expression for the treatment to the CYP gene expression for the control at different time points are shown for CYP-1 (Fig. 3A), CYP-2 (Fig. 3B), and CYP-3 (Fig. 3C). During gradual thermal stress there was a significant interaction between time and treatment for the levels of CYP-1, CYP-2, and CYP-3 expression (P < 0.001, two-way ANOVA). However, elevated temperatures affected the transcript abundance of each Symbiodinium CYP gene compared to the control at each time point. Increases in CYP gene expression were observed after 18 h of incubation at 26°C, and significant increases (P < 0.01) were observed for CYP-1 (30%) and CYP-2 (40%). In contrast, rapid heat stress with the temperature reaching 32°C after 18 h led to significant decreases in the CYP mRNA levels (P < 0.01) and nearly 50% reductions in CYP-1 and CYP-2 expression and a ∼90% reduction in CYP-3 transcript abundance (Fig. 3). Under more moderate heat stress conditions, the CYP gene expression significantly increased (P < 0.01) after 72 h of incubation at 29°C; the expression of CYP-1 and CYP-2 increased 2.4-fold (240%), whereas a 4.2-fold (420%) increase was observed for CYP-3. Again, gradual extended exposure to heat stress with the temperature reaching 32°C after 120 h resulted in reductions in the CYP transcript levels compared to the control levels of approximately 66% for CYP-1 and CYP-2 transcripts (P < 0.01) and of 41% for CYP-3 transcripts (P < 0.05).
FIG. 3.
Regulation of CYP gene expression in Symbiodinium symbiotic with a coral host (A. millepora) exposed to rapid and gradual thermal stress regimens with a maximum temperature of 32°C. The relative quantities were normalized using data for two of the most stable reference genes defined by the GeNorm analysis, the Rp-S4 and SAM genes (M = 0.351) with pairwise variations for V2/3 of 0.137. The ratio of the CYP gene expression for the treated sample to the CYP gene expression for the control at each time point (T:CTRL) is shown for CYP-1 (A), CYP-2 (B), and CYP-3 (C). All data are the means ± standard deviations of values obtained for four independent biological replicates.
DISCUSSION
Elevated sea temperatures have been recognized as one of the major factors leading to coral bleaching (29); consequently, the possible role of new Symbiodinium CYP enzymes in the response to heat stress in a coral host was examined in this study. Over the last decade the use of high-throughput sequencing technologies has led to the discovery of new genes, followed by identification of new functions and pathways. Existing EST databases have increased our knowledge about functional genomics and provided a baseline for further functional characterizations and discoveries (64). In these terms, EST sequences were also a helpful tool for obtaining information about enzyme identity and phylogeny (56). In this study, using the Symbiodinium (C3) EST database (36), we identified three CYP genes that were characterized by sequences encoding a unique C-terminal region and preserved conserved PERF and heme-binding domains. In all of the sequences encoded by the putative CYP genes, we recognized an absolutely conserved cysteine in the heme-binding region, confirming the heme-thiolate nature of the enzymes discovered (43). An overview of the CYPs' conserved domains typically found in the majority of membrane-bound CYPs (48) is shown in Fig. 2. The majority of eukaryotic P450 proteins are membrane-bound proteins with molecular masses of approximately 50 kDa and have a C terminus surrounding the cysteine residue and a hydrophobic N-terminal membrane domain (70). The molecular evolution of the CYP superfamily, in contrast to many other protein families, does not reflect its phylogeny due to numerous genetic changes, including horizontal gene transfer, gene duplications and fusions, and intron loss (12).
The sequence analyses at the protein level showed that the product of the putative CYP-1 gene had the greatest similarity to a P450 steroid 17-alpha-monooxygenase (Table 1). This CYP is well known for its role in steroid metabolism, catalyzing both steroid 17-hydroxylation and C17,20-lyase reactions (34, 75). In mammals, cytochrome P450 17α-hydroxylase (CYP17) is a membrane-bound hemoprotein necessary for maintenance of sexual characteristics and is also involved in the stress response (34). The implications of the steroid metabolism in symbiotic dinoflagellates for the steroid signaling pathways between algal symbionts and their coral hosts are not clear and require further study. Identification of the CYP-2 gene as a gene encoding a P450 monooxygenase was based on the presence of conserved domains, but there was great sequence variability in the nonconserved regions compared to other P450 family members, suggesting that it may belong to a new CYP family (Fig. 2). Conversely, the Symbiodinium CYP-3 gene product showed a high level of sequence similarity with the known cytochrome P450 CYP20A1. Because its biological function is not known, this CYP20A1 protein is the so-called “orphan” P450 (28, 60, 61), and its possible role during heat stress in the coral-alga symbiosis has not been elucidated yet. The high level of similarity of the CYP-1 and CYP-3 sequences to the sequences of metazoan P450 genes suggests that there might have been host contamination, while our results for aposymbiotic A. millepora coral egg-sperm specimens indicated that CYP genes discovered did not originate from coral. Further molecular information is needed to explore the possibility of horizontal gene transfer, which has been found, for example, in the Nematostella genome for microbial genes involved in the shikimic acid pathway (59).
For all three CYP genes, similar expression patterns were induced by heat stress. Upregulation of CYP mRNA synthesis by moderately elevated temperatures (26°C and 29°C) was followed by a decrease in CYP mRNA abundance after the highest temperature (32°C) was reached (Fig. 3). An initial increase in CYP gene abundance under moderate heat stress conditions might occur due to the involvement of CYP enzymes in protection of cells from oxidative stress. Together with other oxidative detoxification proteins, members of the cytochrome P450 protein superfamily are involved in an organism's defense against chemical stressors (25). It has been shown that changes in the expression profile of oxidative stress enzymes are likely to occur first in algal symbionts (51). Also, oxidative stress and ROS production that occur first in symbiotic dinoflagellates may lead to initial upregulation of CYP mRNA synthesis. However, the negative effects of a very high temperature (32°C) on CYP transcript abundance under both rapid and gradual heat stress conditions may be an indication of irreversible cellular changes. Similarly, photosynthesis was greatly impaired in cultured Symbiodinium exposed to temperatures above 30°C (32). In addition, a decrease in the Symbiodinium cell density in the coral tissue was noted during heat stress treatments, whereas a significant decrease in the number of algal cells was observed after 72 and 120 h (data not shown), indicating that there was coral bleaching. During heat stress-induced bleaching, the rate of cell death increased for both the cnidarian host and the endosymbiotic algae (19), and the role of the host was proposed to be protection of its algal symbionts from oxidative stress (49).
In other organisms, P450 activity has also been affected by temperature conditions. A positive effect on P450 expression has been observed in mice exposed to heat stress while they are consuming infected food (7), whereas downregulation of the P450 17-alpha hydroxylase and P450 aromatase mRNA levels was observed in hens subjected to hyperthermal conditions (55). Additional changes in the environment, such as hyposalinity conditions (18) and exposure to toxic substances (47, 54), also affected CYP activity in scleractinian corals and other marine organisms. In two unicellular green algae, cytochrome P450 monooxygenase activity was reported during biotransformation of herbicides (65). In plants, the cytochrome P450 enzymes play a major role in one of the nitric oxide (NO) biosynthetic pathways, catalyzing the oxidation of N-hydroxyguanidines and the formation of nitrogen oxides, including NO (39). NO signaling has been found to be involved in the induction of cell death and in interactions with ROS (44), and several studies have reported links between NO synthesis and the breakdown of a cnidarian-alga symbiosis (8, 45, 67). Also, ROS generated during thermal stress in symbiotic dinoflagellates (16, 41) has been related to coral bleaching (38, 50, 73).
Similarities in the changes in expression between CYP genes suggest that there are comparable regulation mechanisms and that CYP enzymes may be involved in related oxidative stress metabolic pathways. Previous studies have reported that increases in temperature induced oxidative stress which was related to the first signs of breakdown in the functionality of the coral-alga symbiosis, leading to the coral bleaching phenomenon (38, 73). As thermal stress is one of the major environmental stressors challenging the well-being of the coral reef ecosystem and the stability of the coral-alga symbiosis, it has been predicted that a further increase in the average ocean temperature will lead to mass coral bleaching, followed by a decline in coral cover and changes in the composition of coral species due to various levels of susceptibility and in other coral reef organisms (14). On the other hand, the diversity of symbiotic algae and their ability to change their composition might affect the ability of coral to respond to future increases in temperatures (1, 4, 63). Understanding the mechanisms that regulate the genes encoding widespread enzymes, such as cytochrome P450s, which are involved in the first line of defense in the organisms exposed to various stressors, should bring us a step closer to obtaining a more complete picture of the stress responses in symbiotic dinoflagellates exposed to elevated temperature and to understanding the susceptibility of Symbiodinium and the reef-building corals to global climate change.
Acknowledgments
We thank all members of the Dove-Hoegh-Guldberg lab and especially Olga Pantos and Charlotte Kvennefors for their comments on the manuscript. We thank Ruth Reef for the coral egg-sperm sample.
This project was supported by the ARC Centre for Excellence in Reef Studies and the Coral Reef Targeted Research Project (www.gefcoral.org).
Footnotes
Published ahead of print on 12 March 2010.
REFERENCES
- 1.Abrego, D., K. E. Ulstrup, B. L. Willis, and M. J. van Oppen. 2008. Species-specific interactions between algal endosymbionts and coral hosts define their bleaching response to heat and light stress. Proc. Biol. Sci. 275:2273-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aro, E. M., I. Virgin, and B. Andersson. 1993. Photoinhibition of photosystem. II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143:113-134. [DOI] [PubMed] [Google Scholar]
- 3.Bak, S., R. A. Kahn, H. L. Nielsen, B. L. Moller, and B. A. Halkier. 1998. Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol. Biol. 36:393-405. [DOI] [PubMed] [Google Scholar]
- 4.Baskett, M. L., S. D. Gaines, and R. M. Nisbet. 2009. Symbiont diversity may help coral reefs survive moderate climate change. Ecol. Appl. 19:3-17. [DOI] [PubMed] [Google Scholar]
- 5.Berkelmans, R. 2002. Time-integrated thermal bleaching thresholds of reefs and their variation on the Great Barrier Reef. Mar. Ecol. Prog. Ser. 229:73-82. [Google Scholar]
- 6.Berkelmans, R., and B. L. Willis. 1999. Seasonal and local spatial patterns in the upper thermal limits of corals on the inshore Central Great Barrier Reef. Coral Reefs 18:219-228. [Google Scholar]
- 7.Bhusari, S., Z. Liu, L. B. Hearne, D. E. Spiers, W. R. Lamberson, and E. Antoniou. 2007. Expression profiling of heat stress effects on mice fed ergot alkaloids. Toxicol. Sci. 95:89-97. [DOI] [PubMed] [Google Scholar]
- 8.Bouchard, J. N., and H. Yamasaki. 2008. Heat stress stimulates nitric oxide production in Symbiodinium microadriaticum: a possible linkage between nitric oxide and the coral bleaching phenomenon. Plant Cell Physiol. 49:641-652. [DOI] [PubMed] [Google Scholar]
- 9.Brown, C. M., B. Reisfeld, and A. N. Mayeno. 2008. Cytochromes P450: a structure-based summary of biotransformations using representative substrates. Drug Metab. Rev. 40:1-100. [DOI] [PubMed] [Google Scholar]
- 10.Coffroth, M. A., and S. R. Santos. 2005. Genetic diversity of symbiotic dinoflagellates in the genus Symbiodinium. Protist 156:19-34. [DOI] [PubMed] [Google Scholar]
- 11.Coles, S. L., and P. L. Jokiel. 1977. Effects of temperature on photosynthesis and respiration in hermatypic corals. Mar. Biol. 43:209-216. [Google Scholar]
- 12.Degtyarenko, K. N., and A. I. Archakov. 1993. Molecular evolution of P450 superfamily and P450-containing monooxygenase systems. FEBS Lett. 332:1-8. [DOI] [PubMed] [Google Scholar]
- 13.DellaGreca, M., A. Fiorentino, M. R. Iesce, M. Isidori, A. Nardelli, L. Previtera, and F. Temussi. 2003. Identification of phototransformation products of prednisone by sunlight: toxicity of the drug and its derivatives on aquatic organisms. Environ. Toxicol. Chem. 22:534-539. [PubMed] [Google Scholar]
- 14.Donner, S. D. 2009. Coping with commitment: projected thermal stress on coral reefs under different future scenarios. PLoS One 4:e5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dove, S. 2004. Scleractinian corals with photoprotective host pigments are hypersensitive to thermal bleaching. Mar. Ecol. Prog. Ser. 272:99-116. [Google Scholar]
- 16.Dove, S., J. C. Ortiz, S. Enriquez, M. Fine, P. Fisher, R. Iglesias-Prieto, D. Thornhill, and O. Hoegh-Guldberg. 2006. Response of holosymbiont pigments from the scleractinian coral Montipora monasteriata to short-term heat stress. Limnol. Oceanogr. 51:1149-1158. [Google Scholar]
- 17.Downs, C., and A. Downs. 2007. Preliminary examination of short-term cellular toxicological responses of the coral Madracis mirabilis to acute Irgarol 1051 exposure. Arch. Environ. Contam. Toxicol. 52:47-57. [DOI] [PubMed] [Google Scholar]
- 18.Downs, C. A., E. Kramarsky-Winter, C. M. Woodley, A. Downs, G. Winters, Y. Loya, and G. K. Ostrander. 2009. Cellular pathology and histopathology of hypo-salinity exposure on the coral Stylophora pistillata. Sci. Total Environ. 407:4838-4851. [DOI] [PubMed] [Google Scholar]
- 19.Dunn, S. R., J. C. Thomason, M. D. Le Tissier, and J. C. Bythell. 2004. Heat stress induces different forms of cell death in sea anemones and their endosymbiotic algae depending on temperature and duration. Cell Death Differ. 11:1213-1222. [DOI] [PubMed] [Google Scholar]
- 20.Fu, C., J. Xiong, and W. Miao. 2009. Genome-wide identification and characterization of cytochrome P450 monooxygenase genes in the ciliate Tetrahymena thermophila. BMC Genomics 10:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujita, K., and T. Kamataki. 2002. Genetically engineered bacterial cells co-expressing human cytochrome P450 with NADPH-cytochrome P450 reductase: prediction of metabolism and toxicity of drugs in humans. Drug Metab. Pharmacokinet. 17:1-22. [DOI] [PubMed] [Google Scholar]
- 22.Garcia, E., R. Ramos, and C. Bastidas. 2005. Presence of cytochrome P450 in the Caribbean corals Siderastrea siderea and Montastraea faveolata. Presencia del citocromo P450 en las especies de coral Siderastrea siderea y Montastraea faveolata. Caribe 31:23-30. [Google Scholar]
- 23.Gates, R. D., G. Baghdasarian, and L. Muscatine. 1992. Temperature stress causes host cell detachment in symbiotic cnidarians: implications for coral bleaching. Biol. Bull. 182:324-332. [DOI] [PubMed] [Google Scholar]
- 24.Gillam, E. M. 2005. Exploring the potential of xenobiotic-metabolising enzymes as biocatalysts: evolving designer catalysts from polyfunctional cytochrome P450 enzymes. Clin. Exp. Pharmacol. Physiol. 32:147-152. [DOI] [PubMed] [Google Scholar]
- 25.Goldstone, J. V. 2008. Environmental sensing and response genes in cnidaria: the chemical defensome in the sea anemone Nematostella vectensis. Cell Biol. Toxicol. 24:483-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guengerich, F. P. 2002. Cytochrome P450 enzymes in the generation of commercial products. Nat. Rev. Drug Discov. 1:359-366. [DOI] [PubMed] [Google Scholar]
- 27.Guengerich, F. P. 2007. Mechanisms of cytochrome P450 substrate oxidation: minireview. J. Biochem. Mol. Toxicol. 21:163-168. [DOI] [PubMed] [Google Scholar]
- 28.Guengerich, F. P., Z. L. Wu, and C. J. Bartleson. 2005. Function of human cytochrome P450s: characterization of the orphans. Biochem. Biophys. Res. Commun. 338:465-469. [DOI] [PubMed] [Google Scholar]
- 29.Hoegh-Guldberg, O. 1999. Climate change, coral bleaching and the future of the world's coral reefs. Mar. Freshw. Res. 50:839-866. [Google Scholar]
- 30.Hoegh-Guldberg, O., P. J. Mumby, A. J. Hooten, R. S. Steneck, P. Greenfield, E. Gomez, C. D. Harvell, P. F. Sale, A. J. Edwards, K. Caldeira, N. Knowlton, C. M. Eakin, R. Iglesias-Prieto, N. Muthiga, R. H. Bradbury, A. Dubi, and M. E. Hatziolos. 2007. Coral reefs under rapid climate change and ocean acidification. Science 318:1737-1742. [DOI] [PubMed] [Google Scholar]
- 31.Hoegh-Guldberg, O., and G. J. Smith. 1989. The effect of sudden changes in temperature, light and salinity on the population density and export of zooxanthellae from the reef corals Stylophora pistillata Esper and Seriatopora hystrix Dana. J. Exp. Mar. Biol. Ecol. 129:279-303. [Google Scholar]
- 32.Iglesias-Prieto, R., J. L. Matta, W. A. Robins, and R. K. Trench. 1992. Photosynthetic response to elevated temperature in the symbiotic dinoflagellate Symbiodinium microadriaticum in culture. Proc. Natl. Acad. Sci. U. S. A. 89:10302-10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kellner, D. G., S. A. Maves, and S. G. Sligar. 1997. Engineering cytochrome P450s for bioremediation. Curr. Opin. Biotechnol. 8:274-278. [DOI] [PubMed] [Google Scholar]
- 34.Kolar, N. W., A. C. Swart, J. I. Mason, and P. Swart. 2007. Functional expression and characterisation of human cytochrome P45017alpha in Pichia pastoris. J. Biotechnol. 129:635-644. [DOI] [PubMed] [Google Scholar]
- 35.LaJeunesse, T. C., W. K. W. Loh, R. Van Woesik, O. Hoegh-Guldberg, G. W. Schmidt, and W. K. Fitt. 2003. Low symbiont diversity in southern Great Barrier Reef corals, relative to those of the Caribbean. Limnol. Oceanogr. 48:2046-2054. [Google Scholar]
- 36.Leggat, W., O. Hoegh-Guldberg, S. Dove, and D. Yellowlees. 2007. Analysis of an EST library from the dinoflagellate (Symbiodinium sp.) symbiont of reef-building corals. J. Phycol. 43:1010-1021. [Google Scholar]
- 37.Lesser, M. P. 2004. Experimental biology of coral reef ecosystems. J. Exp. Mar. Biol. Ecol. 300:217-252. [Google Scholar]
- 38.Lesser, M. P. 1997. Oxidative stress causes coral bleaching during exposure to elevated temperatures. Coral Reefs 16:187-192. [Google Scholar]
- 39.Mansuy, D., and J. L. Boucher. 2002. Oxidation of N-hydroxyguanidines by cytochromes P450 and NO-synthases and formation of nitric oxide. Drug Metab. Rev. 34:593-606. [DOI] [PubMed] [Google Scholar]
- 40.Nelson, D. R., T. Kamataki, D. J. Waxman, F. P. Guengerich, R. W. Estabrook, R. Feyereisen, F. J. Gonzalez, M. J. Coon, I. C. Gunsalus, O. Gotoh, K. Okuda, and D. W. Nebert. 1993. The P450 superfamily—update on new sequences, gene-mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 12:1-51. [DOI] [PubMed] [Google Scholar]
- 41.Nesa, B., and M. Hidaka. 2009. High zooxanthella density shortens the survival time of coral cell aggregates under thermal stress. J. Exp. Mar. Biol. Ecol. 368:81-87. [Google Scholar]
- 42.Omura, T. 1999. Forty years of cytochrome P450. Biochem. Biophys. Res. Commun. 266:690-698. [DOI] [PubMed] [Google Scholar]
- 43.Omura, T. 2005. Heme-thiolate proteins. Biochem. Biophys. Res. Commun. 338:404-409. [DOI] [PubMed] [Google Scholar]
- 44.Palavan-Unsal, N., and D. Arisan. 2009. Nitric oxide signalling in plants. Bot. Rev. 75:203-209. [Google Scholar]
- 45.Perez, S., and V. Weis. 2006. Nitric oxide and cnidarian bleaching: an eviction notice mediates breakdown of a symbiosis. J. Exp. Biol. 209:2804-2810. [DOI] [PubMed] [Google Scholar]
- 46.Pochon, X., J. I. Montoya-Burgos, B. Stadelmann, and J. Pawlowski. 2006. Molecular phylogeny, evolutionary rates, and divergence timing of the symbiotic dinoflagellate genus Symbiodinium. Mol. Phylogenet. Evol. 38:20-30. [DOI] [PubMed] [Google Scholar]
- 47.Ramos, R., and E. Garcaa. 2007. Induction of mixed-function oxygenase system and antioxidant enzymes in the coral Montastraea faveolata on acute exposure to benzo(a)pyrene. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 144:348-355. [DOI] [PubMed] [Google Scholar]
- 48.Rewitz, K. F., B. Styrishave, A. Lobner-Olsen, and O. Andersen. 2006. Marine invertebrate cytochrome P450: emerging insights from vertebrate and insects analogies. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 143:363-381. [DOI] [PubMed] [Google Scholar]
- 49.Richier, S., P. Furla, A. Plantivaux, P.-L. Merle, and D. Allemand. 2005. Symbiosis-induced adaptation to oxidative stress. J. Exp. Biol. 208:277-285. [DOI] [PubMed] [Google Scholar]
- 50.Richier, S., C. Sabourault, J. Courtiade, N. Zucchini, D. Allemand, and P. Furla. 2006. Oxidative stress and apoptotic events during thermal stress in the symbiotic sea anemone, Anemonia viridis. FEBS J. 273:4186-4198. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Lanetty, M., S. Harii, and O. Hoegh-Guldberg. 2009. Early molecular responses of coral larvae to hyperthermal stress. Mol. Ecol. 18:5101-5114. [DOI] [PubMed] [Google Scholar]
- 52.Rosic, N., and O. Hoegh-Guldberg. 2010. A method for extracting a high-quality RNA from the symbiotic dinoflagellate applicable in gene expression studies. J. Appl. Phycol. 22:139-146. [Google Scholar]
- 53.Rosic, N. N. 2009. Versatile capacity of shuffled cytochrome P450s for dye production. Appl. Microbiol. Biotechnol. 82:203-210. [DOI] [PubMed] [Google Scholar]
- 54.Rougee, L., C. A. Downs, R. H. Richmond, and G. K. Ostrander. 2006. Alteration of normal cellular profiles in the scleractinian coral (Pocillopora damicornis) following laboratory exposure to fuel oil. Environ. Toxicol. Chem. 25:3181-3187. [DOI] [PubMed] [Google Scholar]
- 55.Rozenboim, I., E. Tako, O. Gal-Garber, J. A. Proudman, and Z. Uni. 2007. The effect of heat stress on ovarian function of laying hens. Poult. Sci. 86:1760-1765. [DOI] [PubMed] [Google Scholar]
- 56.Sanderson, M. J., and M. M. McMahon. 2007. Inferring angiosperm phylogeny from EST data with widespread gene duplication. BMC Evol. Biol. 7(Suppl. 1):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santos, S. R., R. A. Kinzie III, K. Sakai, and M. A. Coffroth. 2003. Molecular characterization of nuclear small subunit (18S)-rDNA pseudogenes in a symbiotic dinoflagellate (Symbiodinium, Dinophyta). J. Eukaryot. Microbiol. 50:417-421. [DOI] [PubMed] [Google Scholar]
- 58.Santos, S. R., D. J. Taylor, R. A. Kinzie III, M. Hidaka, K. Sakai, and M. A. Coffroth. 2002. Molecular phylogeny of symbiotic dinoflagellates inferred from partial chloroplast large subunit (23S)-rDNA sequences. Mol. Phylogenet. Evol. 23:97-111. [DOI] [PubMed] [Google Scholar]
- 59.Starcevic, A., S. Akthar, W. C. Dunlap, J. M. Shick, D. Hranueli, J. Cullum, and P. F. Long. 2008. Enzymes of the shikimic acid pathway encoded in the genome of a basal metazoan, Nematostella vectensis, have microbial origins. Proc. Natl. Acad. Sci. U. S. A. 105:2533-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stark, K., and F. P. Guengerich. 2007. Characterization of orphan human cytochromes P450. Drug Metab. Rev. 39:627-637. [DOI] [PubMed] [Google Scholar]
- 61.Stark, K., Z. L. Wu, C. J. Bartleson, and F. P. Guengerich. 2008. mRNA distribution and heterologous expression of orphan cytochrome P450 20A1. Drug Metab. Dispos. 36:1930-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takahashi, S., and N. Murata. 2008. How do environmental stresses accelerate photoinhibition? Trends Plant Sci. 13:178-182. [DOI] [PubMed] [Google Scholar]
- 63.Takahashi, S., S. M. Whitney, and M. R. Badger. 2009. Different thermal sensitivity of the repair of photodamaged photosynthetic machinery in cultured Symbiodinium species. Proc. Natl. Acad. Sci. U. S. A. 106:3237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanguy, A., N. Bierne, C. Saavedra, B. Pina, E. Bachare, M. Kube, E. Bazin, F. Bonhomme, P. Boudry, V. Boulo, I. Boutet, L. Cancela, C. Dossat, P. Favrel, A. Huvet, S. Jarque, D. Jollivet, S. Klages, S. Lapègue, R. Leite, J. Moal, D. Moraga, R. Reinhardt, J. F. Samain, E. Zouros, and A. Canario. 2008. Increasing genomic information in bivalves through new EST collections in four species: development of new genetic markers for environmental studies and genome evolution. Gene 408:27-36. [DOI] [PubMed] [Google Scholar]
- 65.Thies, F., T. Backhaus, B. Bossmann, and L. H. Grimme. 1996. Xenobiotic biotransformation in unicellular green algae. Involvement of cytochrome P450 in the activation and selectivity of the pyridazinone pro-herbicide metflurazon. Plant Physiol. 112:361-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trapido-Rosenthal, H., S. Zielke, R. Owen, L. Buxton, B. Boeing, R. Bhagooli, and J. Archer. 2005. Increased zooxanthellae nitric oxide synthase activity is associated with coral bleaching. Biol. Bull. 208:3-6. [DOI] [PubMed] [Google Scholar]
- 68.Turner, N. J. 2009. Directed evolution drives the next generation of biocatalysts. Nat. Chem. Biol. 5:567-573. [DOI] [PubMed] [Google Scholar]
- 69.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.VonWachenfeldt, C., and E. F. Johnson. 1995. Structures of eukaryotic cytochrome P450 enzymes, p. 183-223. In P. R. Ortiz de Montellano (ed.), Cytochrome P450: structure, mechanism and biochemistry, 2nd ed. Plenum Press, New York, NY.
- 71.Walker, N. J. 2002. Tech.Sight. A technique whose time has come. Science 296:557-559. [DOI] [PubMed] [Google Scholar]
- 72.Warner, M. E., W. K. Fitt, and G. W. Schmidt. 1999. Damage to photosystem II in symbiotic dinoflagellates: a determinant of coral bleaching. Proc. Natl. Acad. Sci. U. S. A. 96:8007-8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weis, V. M. 2008. Cellular mechanisms of cnidarian bleaching: stress causes the collapse of symbiosis. J. Exp. Biol. 211:3059-3066. [DOI] [PubMed] [Google Scholar]
- 74.Yang, S., R. S. S. Wu, H. O. L. Mok, Z. P. Zhang, and R. Y. C. Kong. 2003. Identification of a novel cytochrome P450 cDNA, CYP97E1, from the marine diatom Skeletonema costatum bacillariophyceae. J. Phycol. 39:555-560. [Google Scholar]
- 75.Yang, W. H., L. B. Lutz, and S. R. Hammes. 2003. Xenopus laevis ovarian CYP17 is a highly potent enzyme expressed exclusively in oocytes: evidence that oocytes play a critical role in Xenopus ovarian androgen production. J. Biol. Chem. 278:9552-9559. [DOI] [PubMed] [Google Scholar]