Abstract
New primers were designed for the amplification of dsrAB genes by nested PCR to investigate the diversity of sulfate-reducing prokaryotes (SRP) in environments with low bacterial cell density. The success of the nested PCR for the determination of SRP diversity was estimated by terminal-restriction fragment length polymorphism analysis in the Reigous, a small creek at an inactive mine (Carnoulès, France), which constitutes an extreme acidic arsenic-rich environment. Nested PCR limits were evaluated in dsrAB-rich sediments, and this technique was compared to direct PCR using either known primers (DSR1F/DSR4R) or new primers (dsr619AF/dsr1905BR). The comparison of clone libraries revealed that, even if the levels of diversity observed were not identical, nested PCR did not reduce the diversity compared to that of direct DSR1F/DSR4R PCR. Clone sequences were affiliated mainly with the Desulfobacteraceae and Desulfohalobiaceae families. Many sequences (∼30%) were related to a deeply branching lineage unaffiliated with any cultured SRP. Although this dsrAB cluster was found in all libraries, the new primers better amplified this lineage, providing more information on this unknown bacterial group. Thanks to these new primers in nested PCR, the SRP community from Carnoulès could be characterized. Specific SRP populations were obtained according to environmental characteristics. Desulfomicrobiaceae-related sequences were recovered in samples with low pH, low levels of dissolved oxygen, and high As content, while sequences belonging to the deeply branching group were found in a less extreme sample. Furthermore, for the first time, dsrAB sequences related to the latter group were recovered from freshwater.
Sulfate-reducing prokaryotes (SRP) constitute a functional group of physiologically diverse anaerobes sharing the ability to use sulfate as a terminal electron acceptor during the consumption of organic matter with the concomitant production of sulfide. They are ubiquitous in the environment and have crucial roles in the biogeochemical cycling of carbon and sulfur. Sulfate reduction could be responsible for up to 50% of organic matter degradation in high-sulfate environments such as estuarine and marine sediments (25). Active sulfate reduction also has been reported in low-sulfate environments such as soils and freshwater sediments (1, 19). SRP also are known to play a role in the biodegradation and biotransformation of a number of environmental pollutants (12, 41). Recent studies also have shown that SRP are present in mining environments (5) and that microbial sulfate reduction could be important in permanently acidic (pH 2 to 3) mine tailing sites, suggesting that SRP can be active under very acidic conditions (42). Due to their great ecological importance, SRP have been intensively studied during the last few decades.
Most of the molecular studies on the bacterial diversity in complex communities have been based on 16S rRNA gene analysis (5, 8, 55). However, retrieved 16S rRNA sequences frequently are not related to any cultivated organism, and thus it becomes impossible to infer a likely ecophysiology for the organism containing the gene. An alternative approach to infer physiology from environmental sequences is to retrieve functional gene sequences coding for enzymes that are essential to the target metabolisms. The dissimilatory sulfite reductase (Dsr) catalyzes the final steps in sulfate and sulfite reduction, and it is therefore an essential enzyme in SRP metabolism. Dsr proteins are multisubunit enzymes that catalyze the six-electron reduction of sulfite to sulfide in anaerobic sulfite/sulfate-reducing prokaryotes (56). Moreover, a reverse dsr has been described in sulfur-oxidizing prokaryotes, such as Thiobacillus denitrificans strain RT and Chromatium vinosum strain D, with a proposed function in sulfide oxidation (45, 46).
They all contain siroheme and [Fe4-S4] prosthetic centers and consist of at least two different polypeptides in an α2β2 structure (24). The ubiquity of Dsr and its high sequence conservation has made this enzyme ideal for assessing the biodiversity of SRP in anoxic environments (54) and provides a basis for culture-independent molecular diversity studies of natural sulfate-reducing assemblages using PCR primers broadly specific for a large fragment of all known dsrAB genes (6, 7, 37, 52). The DSR1F and DSR4R primers (54) have been used extensively in environmental studies to provide molecular profiles of SRP communities. This primer set amplified most of the α and β subunits of the dsr gene, allowing the detection of members of all known SRP groups. However, these studies used PCR techniques, which require a minimal number of target copies of the gene to retrieve sequences of interest, thus constituting an obstacle in the diversity analysis. Nested PCR is a modification of standard PCR that is aimed at increasing amplification and specificity. It is particularly useful in situations where the detection of low numbers of bacterial cells in complex environmental samples is required (9, 29).
In this paper, we describe new PCR primers for use in nested PCR to amplify dsrAB genes in low-cell-density water to provide baseline information on the occurrence and distribution of SRP. These primer sets were evaluated and validated using an SRP-rich sediment sample collected from a wastewater treatment plant of an oil refinery. For this purpose, the diversity obtained with DSR1F/4R primers was compared to that obtained with the new primer set used in both direct and nested PCRs. By means of diversity retrieval, our results support the development of a molecular methodology for the specific detection of SRP communities in low-cell-biomass environments.
MATERIALS AND METHODS
Reference strains and environmental samples.
The specificity of the new primers was tested in PCRs with template DNA from cultures of SRP type species: Desulfotomaculum nigrificans DSM771T, Desulfobulbus propionicus DSM2032T, Desulfobacterium autotrophicum DSM3382T, Desulfobacter curvatus DSM3379T, Desulfococcus multivorans DSM2059T, and Desulfovibrio africanus DSM2603T.
A total of four environmental samples were investigated for dsrAB amplification and diversity analysis. (i) A microbial mat sample from a wastewater treatment plant of an oil refinery located on the shore of the Etang de Berre (EB) on the French Mediterranean Sea (43°29′05″N; 5°11′17″E). (ii) Bacterial communities were recovered from 300 ml filtered water from groundwaters of a borehole, between 10 and 12 m deep, located in the center of the mine tailings (CARN-A) and from 30 m downstream in the Reigous creek (CARN-B) of the Carnoulès mine (Gard, France) (44°06′41″N; 04°01′27″E). Differences in the physical-chemical parameters were observed between water samples from CARN-A (low pH [∼3] and low dissolved oxygen content [∼1 mg liter−1]) and CARN-B (higher pH [∼4] and dissolved oxygen content [∼5 mg liter−1]) (5). Both samples have high concentrations of As(III) and As(V) (100 to 350 mg liter−1), Fe (750 to 2,700 mg liter−1), and SO42− (2,000 to 7,500 mg liter−1) (5). Samples from the Adour estuary sediments (southwest French coast) (iii) and oligotrophic waters from the Mediterranean Sea (iv) were tested only for dsrAB amplification. Samples were kept in liquid nitrogen and stored at −80°C until analysis.
Design of internal primers for the dsrAB genes.
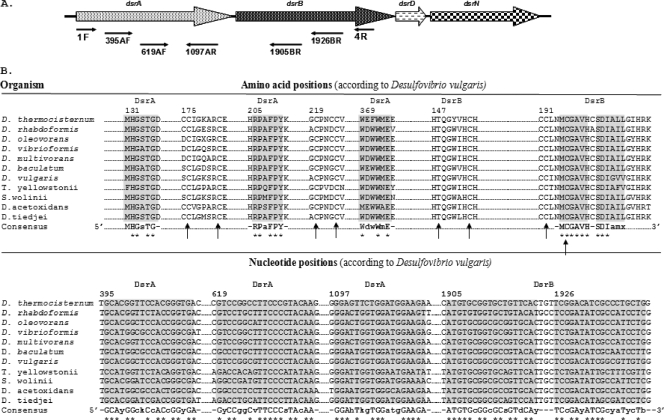
dsrAB gene sequences from the Functional Gene Pipeline/Repository (FGPR), available at http://fungene.cme.msu.edu/, were aligned using Clustal X version 1.83 (51), and primers for PCR amplification were designed manually based on conserved regions. Two forward primers (dsr395AF and dsr619AF) and one reverse primer (dsr1097AR) were designed within the dsrA gene, and two reverse primers (dsr1905BR and dsr1926BR) were designed within the dsrB gene (Fig. 1 for sequences). IDT software (available at http://www.idtdna.com) was used to evaluate the formation of hetero/homodimers and the stability of the primer match of novel primer sets. The setting parameters for primer characteristic determination were 250 nM primer and 50 mM Na+ salt, and the ΔG temperature was 25°C (43).
FIG. 1.
(A) dsrABDN operon of Desulfovibrio vulgaris. The locations of the primers designed in this study and that of the known primers DSR1F and DSR4R (54) are indicated. (B) Alignment fragments for the priming sites on DsrAB (∼750 amino acids) and the dsrA and dsrB genes (∼1,900 nucleotides). Uppercase letters in the consensus sequence indicate the unit fully conserved at the position among the SRP analyzed. Lowercase letters in the consensus correspond to designated ambiguities. The consensus sequence for the reverse primer was processed for proper orientation. The complete [Fe4S4]-seroheme binding site motif (Cys/Thr-X5-Cys)-Xn-(Cys-X3-Cys) is indicated by arrows. The dsrA and dsrB genes shown in the alignments are from Desulfotomaculum thermocisternum DSM10259T (AF074396), Desulfobulbus rhabdoformis DSM8777T (AJ250473), Desulfobacterium oleovorans (AF482464), Desulfobacter vibrioformis DSM8776T (AJ250472), Desulfococcus multivorans DSM2059T (U58126), Desulfomicrobium baculatum (AF4282460), Desulfovibrio vulgaris DSM644T (U16723), Thermodesulfovibrio yellowstonii DSM 11347, Syntrophobacter wolinii DSM 2805 (AF418192), Desulfobacca acetoxidans (AF482453), and Desulfomonile tiedjei DSM 6799 (AF334595).
The in silico coverage and specificity of the new primer sets were tested using the probeCheck free online software (31).
DNA extraction and PCR conditions.
DNA extractions were performed using the UltraClean Soil DNA Isolation kit according to the recommendation of the manufacturer (MoBio Laboratories, Inc.). For water samples, a volume of 300 ml was filtered through a sterile 0.22-μm-nucleopore filter, transferred to a cryotube, and frozen in liquid nitrogen. Filters then were cut with a sterile scalpel before processing. All extracted genomic DNA samples were stored at −20°C.
PCRs were performed with 1× PCR buffer, 0.2 mM each deoxynucleoside triphosphate (dNTP), 1.5 mM MgCl2, 0.2 μM each primer, 2.5 U of Taq DNA polymerase (Eurobio), and DNA template (between 0.5 to 5 ng for Carnoulès samples and 1 to 100 ng for Berre samples) in a final volume of 50 μl. DNA amplification was performed with a minicycler PTC 200 (MJ Research) starting with 5 min at 94°C, followed by 35 cycles consisting of denaturation (45 s at 94°C), annealing (45 s at either 55 or 48°C for DSR1F/4R and DSR1Fmix/4Rmix [56] or 54°C for the newly designed primers), extension (90 s at 72°C), and a final extension at 72°C for 10 min. The nested PCR was performed using DSR1F/4R or DSR1Fmix/4Rmix primers for the first run and with different combinations of the new sets for the second run, applying the same thermal profile and number of cycles and using as the template the first PCR product or a 100-fold dilution when necessary. All of the PCR products were checked in 1.0% (wt/vol) agarose electrophoresis in 1× Tris-borate-EDTA (TBE) buffer and stained with ethidium bromide (0.2 μg·ml−1). DNA bands were detected under UV light.
T-RFLP analyses.
Terminal-restriction fragment length polymorphism (T-RFLP) analyses were conducted with DNA extracted from the two water samples of the Carnoulès mine to compare the levels of SRP diversity obtained with the different combinations of internal primers used for nested PCR. The nested PCR was performed as described above, but the forward primer was fluorescently labeled with TET fluorochrome (5-tetrachloro-fluorescein). T-RFLP analyses were performed as previously described (4) using 3 U of Sau3AI, TaqαI, and RsaI (New England Biolabs) as restriction enzymes.
Cloning and analysis of drsAB sequences.
dsrAB DNA fragments obtained by direct PCR amplification with DSR1F/4R or dsr619AF/1905BR or in nested PCR runs were ligated into the pCRTOPO2.1 TA cloning vector (Invitrogen, Netherlands) and transformed into competent Escherichia coli TOP 10F′ cells (Invitrogen, Netherlands) according to the manufacturer's instructions. Recombinant clones were analyzed by PCR using primers M13F (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′). Inserts of clones from Etang de Berre libraries were directly sequenced using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) with M13 primers by following the manufacturer's procedures. When necessary, the internal degenerate sequencing primers 1FI (5′-CAGGAYGARCTKCACCG-3′) and 1RI (5′-CCCTGGGTRTGRAYRAT-3′) (11) were used to obtain full-length dsr gene sequences. Inserts of clones obtained from water samples of Carnoulès mine libraries (CARN-A and CARN-B) were analyzed first by RFLP, performed using the same restriction enzyme as that in T-RFLP analyses. The digestion products were analyzed by 3% agarose gel electrophoresis. One clone of each RFLP pattern was sequenced as described above.
The dsrAB sequences were assembled by using the software Sequencher v4.1.4 and were compared to sequences deposited in the GenBank DNA database by using the BLAST algorithm (2). Alignments were achieved by using Clustal X v1.83 (51) and corrected with ProSeq v2.9 (13) before drawing the design of phylogenetic trees with Mega v4 (49). The tree was inferred using the neighbor-joining method (44). The evolutionary distances were computed using the Poisson correction method and all positions containing alignment gaps; missing data were eliminated only in pairwise sequence comparisons. The confidence level of the phylogenetic tree topology was evaluated by performing 100 bootstrap replications.
Diversity analyses.
To estimate the diversity coverage and determine if the number of clones from each library was sufficient, rarefaction analyses were performed using Paleontological Statistics version 1.19 software (18) as previously described (4). The homologous coverage of each DNA library was calculated using the equation CX = 1 − (NX/n), where NX is the number of unique sequences (singletons) and n is the total number of sequences (17). UniFrac, available at http://bmf.colorado.edu/unifrac, was used to determine whether communities from libraries were significantly different using the UniFrac significance test and lineage-specific analysis (33, 34).
Quantification of 16S rRNA gene copy numbers.
For the cell counting of Carnoulès mine samples, the 16S rRNA gene was targeted using primers 338F/518R (23). A cloned 16S rRNA from an environmental sample was chosen to create a standard curve (47). The amplification and detection of DNA were performed using the Stratagene Mx3005P system and the Brilliant SYBR Green QPCR Master Mix (Stratagene) by following the manufacturer's instructions. Eight replicates were performed for each sample. The detection limit was determined to be below 102 16S rRNA copies per ml using a template dilution series. Results were expressed as the number of 16S copies per ml of water.
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to the EMBL database and assigned accession no. FM212281 to FM212436 for Etang de Berre libraries and FM211674 to FM211687 for the Carnoulès library.
RESULTS
Primer design and T-RFLP analyses of low-species-richness environment.
To overcome the difficulty of detecting small numbers of SRP in complex microbial communities from natural and extreme environments, internal primers for the nested PCR amplification of dsrAB genes were designed. The alignment of dsrAB gene sequences from SRP, which were available in the specific database Functional Gene Pipeline/Repository, showed conserved regions allowing the design of degenerate primers for the amplification of most dsrAB genes. These dsrAB primer pairs lead to a single amplification of expected size (approximately 700, 1,300, or 1,500 bp), except for the combination dsr619AF and dsr1097AR (data not shown), using DNA from a representative of each of the six generic/suprageneric sulfate-reducing bacterial groups proposed by Daly and coworkers (8) as the template.
The primer sets giving positive amplification were used in nested PCR on low-cell-number environmental samples (CARN-A and CARN-B) (5) after a first round with the DSR1F/4R primer set. The estimation of bacterial abundance in CARN-A and CARN-B was carried out by the real-time PCR amplification of the 16S rRNA genes. The 16S rRNA genes were quantified to be 4.5 × 103 ± 1.3 × 103 and 2.1 × 104 ± 3.8 × 103 per ml of water for CARN-A and CARN-B, respectively.
This two-step PCR leads to good amplification. The success in diversity recovering was estimated by the number of operational taxonomic units (OTU) obtained by T-RFLP analysis. The dsr619AF/dsr1905BR combination revealed the highest OTU number (from 11 to 18) for both samples. Using this set of primers, Sau3AI and TaqαI as restriction enzymes revealed more OTU than RsaI (data not shown).
When the newly developed primers (dsr619AF and dsr1905BR) were manually aligned against dsrAB sequences obtained from FGPR databases, they targeted all important suprageneric SRP groups (Fig. 1; also see Fig. S1 in the supplemental material) except for those related to Archaeoglobus spp. The dsrAB gene amplification using this primer pair either in direct or nested PCR also was positive with (i) DNA from the Etang de Berre sediments, (ii) DNA from the water column of the Mediterranean Sea, and (iii) DNA from Adour estuary sediments (Table 1). Amplification was obtained with all templates tested in nested PCR, indicating that this couple could be used for different environmental samples. Moreover, in silico analysis showed that this primer set does not amplify reverse operating dissimilatory sulfite reductase (rDSR) even if dsr619AF shared some degree of nucleotide similarity with pure-culture representatives containing an rDSR gene (see Fig. S2 in the supplemental material).
TABLE 1.
Amplification of dsrAB genes with different primer pairs from environmental samplesa
| Origin | Sample type | Template | Primer pair |
Nested PCR | |
|---|---|---|---|---|---|
| DSR1F/DSR4R | dsr619AF/dsr1905BR | ||||
| Carnoulès | Water from acid mine drainage | DNA | − | − | + |
| Mediterranean Sea | Water | DNA | − | ND | + |
| Etang de Berre | Sediment from brackish lagoon | DNA | + | + | + |
| Adour estuary | Sediment | DNA | + | ND | + |
−, no amplification; +, amplification; ND, not done.
Influence of the first-round PCR on diversity coverage.
To check the influence of the primer sets (DSR1Fmix/4Rmix or the original DSR1F/4R) and the annealing temperature in the first PCR round on diversity coverage, T-RFLP analysis was carried out on samples from Etang de Berre sediments (EB) and from Carnoulès mine waters (CARN-A and CARN-B). The levels of richness of species retrieved were similar whatever the primer pair used. The richness highly depended on annealing temperature and sample: for EB samples, higher diversity was found at 48°C than at 55°C, whereas for Carnoulès samples the diversity was higher at 55°C (see Fig. S3 in the supplemental material). Qualitatively, the levels of diversity were similar for all samples whatever the primer and annealing temperature used except for the CARN-B sample, for which the diversity coverage with DSR1F/4R at 55°C was higher than those obtained with the other PCR conditions.
Efficiency in determination of diversity using the new primers and the nested PCR: analysis of Etang de Berre sediments.
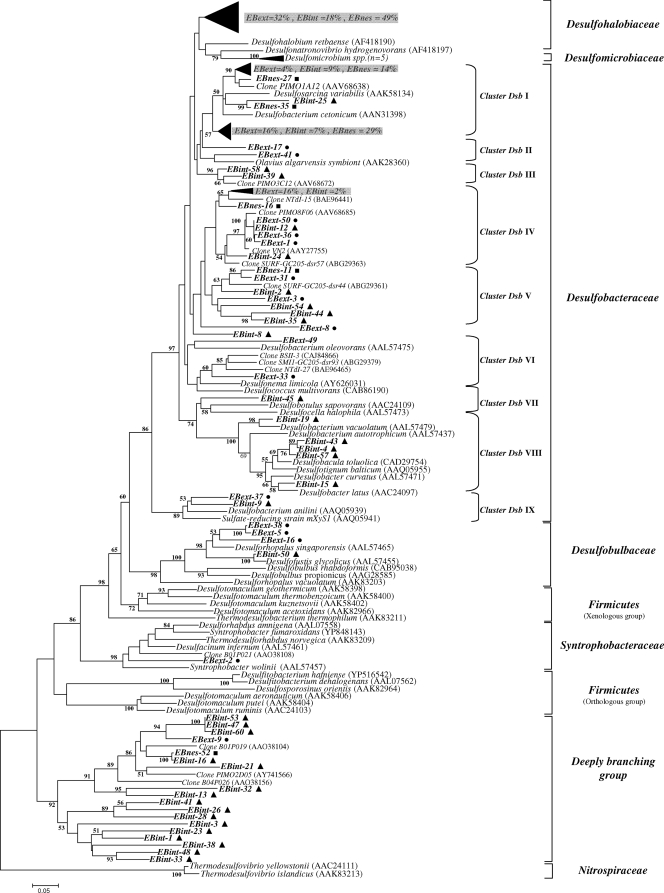
The diversity of the SRP recovered in the Etang de Berre sediment, an SRP-rich environment, was analyzed by constructing three dsrAB gene libraries; (i) library EBext was obtained with the primer set DSR1F/DSR4R, (ii) library EBint was obtained with the primer set dsr619AF/1905BR, and (iii) library EBnes was obtained by nested PCR (first-round PCR, DSR1F/DSR4R; second-round PCR, dsr619AF/1905BR). A total of 156 clones were sequenced (50, 54, and 52 for EBext, EBint, and EBnes libraries, respectively). Two phylotypes were considered different when they shared sequence identities lower than 90% (21) to be conceptually consistent with the 97% threshold for grouping 16S rRNA gene sequences (48). The rarefaction analysis of the libraries indicated that the saturation plateau was not reached for each library (data not shown). The EBint library (dsr619AF/dsr1905BR primers) showed greater species richness than EBext (DSR1F/DSR4R primers) and EBnes (nested PCR) libraries for the number of clones analyzed.
Sequence analysis showed that sequences affiliated with the Desulfobacteraceae and Desulfohalobiaceae families were found to be dominant whatever the library, representing 90, 70, and 98% of sequences found in the EBext, EBint, and EBnes libraries, respectively (data not shown). Sequences affiliated with a deeply branching lineage were found in the three libraries, representing 28% of the sequences in the EBint library, 2% for Ebext, and 2% for EBnes. These dsrAB sequences, which form a deep-branching lineage in the phylogenetic tree, were first described in 2003 by Dhillon et al. (11). Few sequences related to the Desulfobulbaceae family were found in both the EBext and EBint libraries (6 and 2% of the sequences, respectively). Only one sequence from EBext was affiliated with members of the Syntrophobacteraceae cluster. UniFrac significance tests made on phylogenetic trees based on translated dsrA sequences (α subunit) showed no significant difference (P = 0.35) between EBext and EBnes, but differences were marginally significant (P = 0.02) between EBext and EBint.
The main phylotype, whatever the clone library, was affiliated with DsrA sequences of Desulfohalobium retbaense and Desulfonatronovibrio hydrogenovorans (see Fig. 3). However, most of the sequences of the three libraries fell within the Desulfobacteraceae family. The sequences affiliated with this family were grouped in nine clusters (clusters Dsb I to IX) as shown in Fig. 2. Most of the analyzed sequences (54, 30, and 46% for EBext, EBint, and EBnes libraries, respectively) were found in clusters Dsb I, IV, and V. Except for cluster Dsb I, in which sequences were associated with the Desulfosarcina and Desulfobacterium genera, clusters Dsb IV and V could not be directly related to known cultured SRP DsrA sequences, but they were found to be related to environmental sequences that have been retrieved from freshwater mudflats of the Seine Estuary (clone VN2) (26), from Nankai Trough deep-sea sediment cores (clone NTDI-15) (20), from the Plum Island Salt Marsh (clone PIMO8F06) (3), and from subsurface sediments of Green Canyon in the Gulf of Mexico (clones SURF-GC205-dsr57 and -dsr44) (28). Sequences related to clusters Dsb III, VII, and VIII were exclusively found in the EBint library, whereas clusters Dsb II and VI were found in EBext. The dominant groups were detected by T-RFLP analysis (Fig. 3A and B) mainly affiliated with Desulfohalobiaceae and Dsb I clusters, confirming the predominance of these phylotypes at this site.
FIG. 3.
Terminal-restriction fragment length polymorphism electropherograms for the 5′-end dsrAB fragments digested with Sau3AI. (A) Fragments obtained from direct amplification with DSR1F/4R of Etang de Berre sediment sample (EBext). (B) Fragments obtained from direct amplification with dsr619AF/1905BR and nested PCR of an Etang de Berre sediment sample (EBint and EBnes). (C) Fragments obtained from nested PCR of Carnoulès acid mine drainage samples (CARN-A and CARN-B).
FIG. 2.
Phylogenetic tree based on the translated (α subunit) amino acid sequences of PCR-amplified dsrAB genes from the Etang de Berre with selected sequences from other different environmental sources and formally described species. The tree was inferred using the neighbor-joining method, and evolutionary distances were computed using the Poisson correction method. There was a total of 195 positions in the final data set. The scale bar corresponds to 0.05 substitution per site. Percentages of 100 bootstrap resamplings that supported the branching orders in each analysis are shown above or near the relevant nodes. Bootstrap values are shown for branches with more than 50% bootstrap support. Circles, EBext clone sequences; triangles, EBint clone sequences; squares, EBnes clone sequences.
A deeply branching dsrAB lineage also was common to the three libraries but appeared to be dominant in the EBint library (28%); only one sequence was found in both the EBext and EBnes libraries (Fig. 2). Indeed, T-RFLP analysis allowed us to detect these groups only for the EBint sample (Fig. 3B). This dsrAB cluster, unaffiliated with any cultured SRP, was preferentially amplified by primers dsr619AF/dsr1905BR. The topology of the tree constructed by the ML (maximum-likelihood) and the MP (maximum-parsimony) methods was not different from that of the neighbor-joining tree presented here (data not shown), supporting the postulate of this deep-branching lineage. Lineage-specific analysis (UniFrac) showed no significant difference (P > 1) between libraries for all of the lineages except for the deeply branching lineage (P < 0.001).
Nested PCR for low-cell-biomass ecosystem studies: analysis of Carnoulès acid mine drainage.
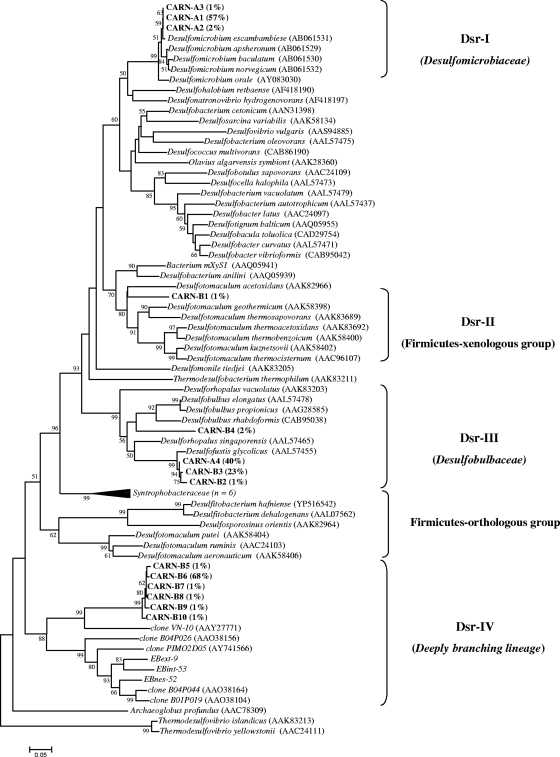
Two dsrAB clone libraries were constructed from CARN-A and CARN-B sites using dsr619AF/1905BR in nested PCR, since the direct amplification with DSR1F/DSR4R was not possible. Ninety-six clones were analyzed for each library. Sau3AI and TaqαI RFLP analysis of clones revealed 4 different patterns for CARN-A and 10 different patterns for CARN-B. A sufficient number of clones analyzed in CARN-A and CARN-B libraries was confirmed by the high-diversity-coverage values (0.98 and 0.94 for CARN-A and -B, respectively) and the rarefaction curves reaching a plateau (data not shown).
Phylogenetic analysis revealed four distinct clusters (Dsr-I to Dsr-IV) (Fig. 4); only cluster Dsr-III was observed in both samples. Cluster Dsr-I includes clones only from CARN-A samples (clones CARN-A1 to CARN-A3, with 60% of the total clones analyzed) and are closely related to the Desulfomicrobiaceae family (supported by high bootstrap values). Clone CARN-B1 fell within the Dsr-II cluster, and it is closely related to the laterally acquired dsrAB Firmicutes group (22, 56). Cluster Dsr-III includes clones CARN-B4 (2%), CARN-A4 (40%), and CARN-B3 (23%) and is closely related to the glycolate oxidizer Desulfofustis glycolicus (16), belonging to the Desulfobulbaceae family. Cluster Dsr-IV encompasses clones CARN-B5 to -B10 (73% of the total clones analyzed from the CARN-B library). These sequences could not be directly related to known cultured SRP. They were found to be related to environmental sequences of the deeply branching lineage of dsrAB sequences also retrieved from EB libraries. Clones related to these sequences have been retrieved from a hydrothermal site (Guaymas Basin, Gulf of California) (11) and a mudflat of the Seine estuary (26). As observed by library analysis, terminal restriction fragments corresponding to sequences affiliated with cluster Dsr-I also were found to be dominant by T-RFLP analysis. The main clusters (Dsr-III and -IV) were detected by T-RFLP analysis on CARN-B samples, showing that the deeply branching group (cluster Dsr-IV) was dominant in these waters (Fig. 3C).
FIG. 4.
Phylogenetic tree based on the translated (α subunit) amino acid sequences of PCR-amplified dsrAB genes from CARN-A and CARN-B with selected sequences recovered from other different environmental sources and formally described species. The tree was inferred using the neighbor-joining method, and evolutionary distances were computed using the Poisson correction method. There were a total of 224 positions in the final data set. The scale bar corresponds to 0.05 substitution per site. Percentages of 100 bootstrap resamplings that supported the branching orders in each analysis are shown above or near the relevant nodes. Bootstrap values are shown for branches with more than 50% bootstrap support. The relative abundance of each RFLP pattern is indicated in brackets.
DISCUSSION
To investigate SRP diversity in environments with low bacterial cell density, a nested PCR approach was developed. The detected signal was enhanced in the nested PCR thanks to both sufficient amounts of DNA amplified during the first-round PCR and the dilution of inhibitors during the second PCR round. This technique also can be useful to detect low dsrAB gene copy numbers in high-cell-biomass samples. To have a deeper characterization of SRP in extreme environments, we developed new primers encompassing the longer sequence of the dsrAB genes. Previous studies developed primers for a nested PCR-denaturing gradient gel electrophoresis (DGGE) strategy (36) based on the partial amplification of the dsrB gene (approximately 350 bp). Applying these primers to techniques other than DGGE should result in sequences that are too short. We also took into account that dsrAB genes are not restricted to sulfate-reducing bacteria, and some specialized sulfur-oxidizing bacteria employ a reversely operating dissimilatory sulfite reductase (rDSR) (32). This enzyme is homologous to, but phylogenetically clearly distinguishable from, the DSR present in anaerobic SRP. To avoid misinterpretation in the analysis of dsrAB sequences retrieved from environmental libraries with respect to the origin and the potential function of the organisms carrying these genes (56), the new primer sets were designed for no matching with rdsr sequences.
The primer pair DSR1F/DSR4R and the variant DSR1Fmix/4Rmix targeting conserved regions within dsrAB genes amplify a 1.9-kb fragment, encompassing most of the dsrA and dsrB subunit genes (54). This primer set has been used successfully to amplify dsrAB from almost 100 pure cultures representing all known SRP lineages (56), even if they are known to produce many unspecific amplifications when they are used at a low annealing temperature (56). Since these primers are suitable for studies of dsrAB gene diversity in environmental samples (7, 26, 38, 52), we used DSR1F/DSR4R as a diversity reference to evaluate the efficiency of the nested PCR in recovering the SRP diversity. Nevertheless, we amplified at 55°C to avoid unspecific amplification. The direct PCR and nested PCR libraries (EBext and EBnes, respectively) showed similar rarefaction patterns and no significant differences, suggesting that the second round of PCR with primers dsr619AF/dsr1905BR does not bias the diversity analysis using nested PCR.
Phylogeny based on translated dsrA sequences retrieved from Etang de Berre sediments using both direct (EBext and EBint libraries) and nested PCR (EBnes library) approaches revealed affiliation essentially with members of the Desulfohalobiaceae and Desulfobacteraceae families. The predominance of phylotypes of Desulfobacteraceae is consistent with other surveys of surface marine sediments (10, 27, 40, 53) and previous 16S rRNA gene library analysis of Etang de Berre sediments (39). The persistent and notable pattern of Desulfobacteraceae in sediments may be explained by their nutritional diversity, growth characteristics, and ecophysiological flexibility (41). Whatever the library, Desulfohalobium and Desulfonatronovibrio-like sequences were the dominant sequences, even if they are known to be alkaliphiles, incomplete oxidizers, and able to use hydrogen and a few organic compounds as the electron donor, which are not the conditions of the Berre lagoon sediment.
The main difference of diversity retrieval between the three libraries is the affiliation of sequences with the deeply branching group. The direct PCR using new primers (EBint library) encompasses many more sequences than the two other libraries. Sequences of this lineage have been obtained previously from salt marshes (3), hydrocarbon-rich hydrothermal vent sediments (11), shallow brackish water, and marine sediments (52). This new, deeply branching dsrAB lineage is well supported by a 92% bootstrap value. As this deeply branching lineage is not related to any cultured SRP, the physiology of this lineage is unknown. The so-far strictly marine or estuarine environmental occurrence of this SRP group and its preference for sediments that are rich in complex organic compounds may suggest strategies for bringing representatives into culture.
Thanks to the nested PCR approach, we obtained information on SRP diversity in environments with low cell biomass, such as the acid mine drainage from the abandoned Carnoulès mine. The presence of SRP in this environment has been described previously based on the 16S rRNA diversity approach (5), but clones related to SRP were affiliated only with uncultured bacteria. Because of the important role of SRP communities in acid mine drainage functioning (14, 15), the new approach described in this study enabled a more detailed description of the SRP diversity in Carnoulès acid mine drainage. The two stations were chosen because they differed in their physicochemical parameters (5). CARN-A waters (the center of the mine tailings), with low pH and dissolved oxygen content, were colonized by SRP related to the Desulfomicrobiaceae family (cluster Dsr-I), members of which are known to be strictly anaerobic. Moreover, Desulfomicrobium strain Ben-RB has been isolated from an arsenic-contaminated site in Australia (35) and has the ability to respire arsenate and release As(III). These waters also were colonized by bacteria belonging to the Desulfobulbaceae family, representatives of which can use alternatives to sulfate as electron acceptors, and some also can grow chemolithoautotrophically by the disproportionation of thiosulfate, sulfite, or elemental sulfur in the presence of a sulfide scavenger such as Fe(III) (30). Sequences related to this family have been found in the second station of the Carnoulès mine (CARN-B), which originated from the creek and showed higher pH and dissolved oxygen contents. These microorganisms are known to be physiologically diverse, and their physiological flexibility may enable this group to exploit the varied conditions existing with Carnoulès acid mine drainage.
CARN-B waters also were inhabited by bacteria related to the Gram-positive group and to the deeply branching lineage. The Desulfotomaculum spp. belonging to the Gram-positive sulfate-reducing group, which laterally acquired the dsrAB genes (56), have been reported to thrive under a variety of harsh conditions, including mine tailings and heavy-metal-contaminated estuarine sediments (7, 50). The extensive physiological capabilities, namely, spore production and the utilization of many different electron donors and acceptors, of Desulfotomaculum spp. can favor the existence of these bacteria in anthropogenically impacted or otherwise challenging environmental conditions. Only sequences from the CARN-B library fell within the deeply branching group described previously, and to the best of our knowledge this is the first study that reports on the presence of the deeply branching lineage in freshwaters.
In this study, we designed and validated primers to assess their potentials for investigating sulfate reducer diversity from low-cell-density samples using the nested PCR technique. Diversity retrieval was validated by the comparison of the levels of diversity observed in clone libraries constructed from DNA extracted from SRP-rich sediments with DSR1F/DSR4R, dsr619AF/dsr1905BR, and the combination of both previous primer pairs in nested PCR. Obviously, nested PCR did not reduce the diversity, as we could expect due to biases of the technique. The main levels of observed diversity were similar irrespective of the primer sets used except for the deeply branching dsrAB lineage, which is better targeted with the new primers. With those primers used in nested PCR, SRP communities from an extreme acidic environment containing a high arsenic concentration could be characterized, and for the first time we describe sequences belonging to a deeply branching lineage originating from freshwaters.
Supplementary Material
Acknowledgments
We acknowledge the financial support of the Aquitaine Regional Government Council (France), the Institut National des Sciences de l'Univers (INSU-CNRS-Cytrix—EC2CO3BIO project no. 075), and the ANR (RARE project no. 07-BLAN-0108). L.G. was supported partly by a doctoral grant from the Ministère de l'Enseignement Supérieur et de la Recherche (France).
We also thank O. Bruneel, C. Casiot, and F. Elbaz-Poulichet for access to the sampling site and S. Païssé and M. Dias for the amplification of dsrAB genes in samples from Mediterranean waters and from the Adour estuary sediments.
Footnotes
Published ahead of print on 12 March 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aeckersberg, F., F. Bak, and F. Widdel. 1991. Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch. Microbiol. 156:5-14. [Google Scholar]
- 2.Altschul, S., T. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahr, M., B. C. Crump, V. Klepac-Ceraj, A. Teske, M. L. Sogin, and J. E. Hobbie. 2005. Molecular characterization of sulfate-reducing bacteria in a New England salt marsh. Environ. Microbiol. 7:1175-1185. [DOI] [PubMed] [Google Scholar]
- 4.Bordenave, S., M. S. Goñi-Urriza, P. Caumette, and R. Duran. 2007. Effects of heavy fuel oil on the bacterial community structure of a pristine microbial mat. Appl. Environ. Microbiol. 73:6089-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruneel, O., R. Duran, K. Koffi, C. Casiot, A. Fourçans, F. Elbaz-Poulichet, and J. C. Personne. 2005. Microbial diversity in a pyrite-rich tailings impoundment (Carnoulès, France). Geomicrobiol. J. 22:249-257. [Google Scholar]
- 6.Castro, H., K. R. Reddy, and A. Ogram. 2002. Composition and function of sulfate-reducing prokaryotes in eutrophic and pristine areas of the Florida Everglades. Appl. Environ. Microbiol. 68:6129-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y. J., A. D. Peacock, P. E. Long, J. R. Stephen, J. P. McKinley, S. J. Macnaughton, A. K. M. Anwar Hussain, A. M. Saxton, and D. C. White. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium Mill tailings site. Appl. Environ. Microbiol. 67:3149-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly, K., R. J. Sharp, and A. J. McCarthy. 2000. Development of oligonucleotide probes and PCR primers for detecting phylogenetic subgroups of sulfate-reducing bacteria. Microbiology 146:1693-1705. [DOI] [PubMed] [Google Scholar]
- 9.Dar, S. A., J. G. Kuenen, and G. Muyzer. 2005. Nested PCR-denaturing gradient gel electrophoresis approach to determine the diversity of sulfate-reducing bacteria in complex microbial communities. Appl. Environ. Microbiol. 71:2325-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devereux, R., and G. W. Mundfrom. 1994. A phylogenetic tree of 16S rRNA sequences from sulfate-reducing bacteria in a sandy marine sediment. Appl. Environ. Microbiol. 60:3437-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhillon, A., A. Teske, J. Dillon, D. A. Stahl, and M. L. Sogin. 2003. Molecular characterization of sulfate-reducing bacteria in the Guaymas basin. Appl. Environ. Microbiol. 69:2765-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensley, B. D., and J. M. Suflita. 1995. Metabolism of environmental contaminants by mixed and pure cultures of sulfate-reducing bacteria, p. 293-332. In L. L. Barton (ed.), Sulfate-reducing bacteria, vol. 8. Peplum Press, New York, NY. [Google Scholar]
- 13.Filatov, D. A. 2002. PROSEQ: a software for preparation and evolutionary analysis of DNA sequence data sets. Mol. Ecol. Notes 2:621-624. [Google Scholar]
- 14.Fortin, D., and T. J. Beveridge. 1997. Microbial sulfate reduction within sulfidic mine tailings: formation of diagenetic Fe sulfides. Geomicrobiol. J. 14:1-21. [Google Scholar]
- 15.Fortin, D., R. Goulet, and M. Roy. 2000. Seasonal cycling of Fe and S in a constructed wetland: the role of sulfate-reducing bacteria. Geomicrobiol. J. 17:221-235. [Google Scholar]
- 16.Friedrich, M., and B. Schink. 1995. Isolation and characterization of a desulforubidin-containing sulfate-reducing bacterium growing with glycolate. Arch. Microbiol. 164:271-279. [Google Scholar]
- 17.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 18.Hammer, Ø., D. A. T. Harper, and P. D. Ryan. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4:9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm. [Google Scholar]
- 19.Jones, C. M., and J. E. Thies. 2007. Soil microbial community analysis using two-dimensional polyacrylamide gel electrophoresis of the bacterial ribosomal internal transcribed spacer regions. J. Microbiol. Methods 69:256-267. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko, R., T. Hayashi, M. Tanahashi, and T. Naganuma. 2007. Phylogenetic diversity and distribution of dissimilatory sulfite reductase genes from deep-sea sediment cores. Mar. Biotechnol. 9:429-436. [DOI] [PubMed] [Google Scholar]
- 21.Kjeldsen, K. U., A. Loy, T. F. Jakobsen, T. R. Thomsen, M. Wagner, and K. Ingvorsen. 2007. Diversity of sulfate-reducing bacteria from an extreme hypersaline sediment, Great Salt Lake (Utah). FEMS Microbiol. Ecol. 60:287-298. [DOI] [PubMed] [Google Scholar]
- 22.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane, D. J. 1991. rDNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 24.LeGall, J., and G. Fauque. 1988. Dissimilatory reduction of sulfur compounds, p. 587-639. In A. J. B. Zehnder (ed.), Biology of anaerobic organisms. John Wiley, New York, NY.
- 25.Leloup, J., A. Loy, N. J. Knab, C. Borowski, M. Wagner, and B. B. Jørgensen. 2007. Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ. Microbiol. 9:131-142. [DOI] [PubMed] [Google Scholar]
- 26.Leloup, J., F. Petit, D. Boust, J. Deloffre, G. Bally, O. Clarisse, and L. Quillet. 2005. Dynamics of sulfate-reducing microorganisms (dsrAB genes) in two contrasting mudflats of the Seine estuary (France). Microb. Ecol. 50:307-314. [DOI] [PubMed] [Google Scholar]
- 27.Liu, X., C. E. Bagwell, L. Wu, A. H. Devol, and J. Zhou. 2003. Molecular diversity of sulfate-reducing bacteria from two different continental margin habitats. Appl. Environ. Microbiol. 69:6073-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd, K. G., L. Lapham, and A. Teske. 2006. An anaerobic methane-oxidizing community of ANME-1b archaea in hypersaline gulf of Mexico sediments. Appl. Environ. Microbiol. 72:7218-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Löffler, F. E., Q. Sun, J. Li, and J. M. Tiedje. 2000. 16S rRNA gene-based detection of tetrachloroethene-dechlorinating Desulfuromonas and Dehalococcoides species. Appl. Environ. Microbiol. 66:1369-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovley, D. R., and E. J. P. Phillips. 1994. Novel processes for anaerobic sulfate production from elemental sulfur by sulfate-reducing bacteria. Appl. Environ. Microbiol. 60:2394-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loy, A., R. Arnold, P. Tischler, T. Rattei, M. Wagner, and M. Horn. 2008. probeCheck—a central resource for evaluating oligonucleotide probe coverage and specificity. Environ. Microbiol. 10:2894-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loy, A., S. Duller, C. Baranyi, M. Mussmann, J. Ott, I. Sharon, O. Beja, D. Le Paslier, C. Dahl, and M. Wagner. 2009. Reverse dissimilatory sulfite reductase as phylogenetic marker for a subgroup of sulfur-oxidizing prokaryotes. Environ. Microbiol. 11:289-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macy, J. M., J. M. Santini, B. V. Pauling, A. H. O'Neill, and L. I. Sly. 2000. Two new arsenate/sulfate-reducing bacteria: mechanisms of arsenate reduction. Arch. Microbiol. 173:49-57. [DOI] [PubMed] [Google Scholar]
- 36.Miletto, M., P. L. E. Bodelier, and H. J. Laanbroek. 2007. Improved PCR-DGGE for high resolution diversity screening of complex sulfate-reducing prokaryotic communities in soils and sediments. J. Microbiol. Methods 70:103-111. [DOI] [PubMed] [Google Scholar]
- 37.Minz, D., S. Fishbain, S. J. Green, G. Muyzer, Y. Cohen, B. E. Rittmann, and D. A. Stahl. 1999. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryotic preference for anoxia. Appl. Environ. Microbiol. 65:4659-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paissé, S., F. Coulon, M. Goñi-Urriza, L. Peperzak, T. J. McGenity, and R. Duran. 2008. Structure of bacterial communities along a hydrocarbon contamination gradient in a coastal sediment. FEMS Microbiol. Ecol. 66:295-305. [DOI] [PubMed] [Google Scholar]
- 40.Pérez-Jiménez, J. R., and L. J. Kerkhof. 2005. Phylogeography of sulfate-reducing bacteria among disturbed sediments, disclosed by analysis of the dissimilatory sulfite reductase genes (dsrAB). Appl. Environ. Microbiol. 71:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Postgate, J. R. 1984. The sulphate reducing bacteria, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 42.Praharaj, T., and D. Fortin. 2004. Indicators of microbial sulfate reduction in acidic sulfide-rich mine tailings. Geomicrobiol. J. 21:457-467. [Google Scholar]
- 43.Rahmann, S., and C. Gräfe. 2004. Mean and variance of the Gibbs free energy of olionucleotides in the nearest neighbor model under varying conditions. Bioinformatics 20:2928-2933. [DOI] [PubMed] [Google Scholar]
- 44.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 45.Schedel, M., and H. G. Trueper. 1979. Purification of Thiobacillus denitrificans siroheme sulfite reductase and investigation of some molecular and catalytic properties. Biochim. Biophys. Acta 568:454-467. [DOI] [PubMed] [Google Scholar]
- 46.Schedel, M., M. Vanselow, and H. G. Trüper. 1979. Siroheme sulfite reductase isolated from Chromatium vinosum. Purification and investigation of some of its molecular and catalytic properties. Arch. Microbiol. 121:29-36. [Google Scholar]
- 47.Smith, C. J., D. B. Nedwell, L. F. Dong, and A. M. Osborn. 2006. Evaluation of quantitative polymerase chain reaction-based approaches for determining gene copy and gene transcript numbers in environmental samples. Environ. Microbiol. 8:804-815. [DOI] [PubMed] [Google Scholar]
- 48.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 49.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 50.Tebo, B. M., and A. Y. Obraztsova. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 162:193-198. [Google Scholar]
- 51.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomsen, T. R., K. Finster, and N. B. Ramsing. 2001. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl. Environ. Microbiol. 67:1646-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tiquia, S. M. 2008. Diversity of sulfate-reducing genes (dsrAB) in sediments from Puget Sound. Environ. Technol. 29:1095-1108. [DOI] [PubMed] [Google Scholar]
- 54.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zverlov, V., M. Klein, S. Lücker, M. W. Friedrich, J. Kellermann, D. A. Stahl, A. Loy, and M. Wagner. 2005. Lateral gene transfer of dissimilatory (bi)sulfite reductase revisited. J. Bacteriol. 187:2203-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.