Abstract
A tetR family transcriptional regulatory gene (SCO1712) was identified as a global antibiotic regulatory gene from a Streptomyces interspecies DNA microarray analysis. SCO1712 disruption in Streptomyces coelicolor not only upregulated antibiotic biosynthesis through pathway-specific regulators when a previously identified pleiotropic downregulatory wblA was expressed but also further stimulated antibiotic production in a wblA deletion mutant, implying that SCO1712 might encode a novel antibiotic downregulator.
Streptomycetes are well-characterized Gram-positive filamentous soil bacteria with a complex life cycle involving morphological differentiation. They are widely used as natural sources of a variety of commercially valuable enzymes and secondary metabolites, including antibiotics, antitumor agents, immunosuppressants, and enzyme inhibitors (3, 6, 8, 15, 16). The regulation of secondary metabolite production in Streptomyces species involves multiple and parallel regulatory networks that are complicatedly intertwined and sensitive to both nutritional and environmental factors (2, 4, 19).
Although several pathway-specific antibiotic regulatory genes have been identified based on their typical location within the biosynthetic pathway gene cluster, global antibiotic regulatory genes are more difficult to identify among the more than 300 annotated putative regulatory open reading frames (ORFs) present in the Streptomyces coelicolor genome sequence and still remain largely unknown in most Streptomyces species (1, 2). Recently, so-called “-omics-guided reverse engineering” approaches, including comparative transcriptomics and proteomics, were successfully used to identify alterations in gene expression associated with the overproduction of secondary metabolites in industrial streptomyces strains (9, 10, 11, 12, 13). Especially, interspecies genome-wide screening using S. coelicolor cDNA microarrays together with antibiotic-overproducing industrial strains of related streptomycetes led to the discovery of putative global downregulator genes affected by unidentified mutations in the industrial strains (9, 14). Previously, we reported on the characterization of an unidentified novel downregulator gene via comparisons of gene transcription profiles using DNA microarrays (9). Overexpression of this gene, which was identified as wblA (18), inhibited the biosynthesis of doxorubicin (DXR) in S. peucetius as well as the production of antibiotics such as actinorhodin (ACT), undecylprodigiosin (RED), and calcium-dependent antibiotic (CDA) in S. coelicolor, suggesting that wblA and its orthologs act globally among streptomycetes as downregulators of antibiotic biosynthesis (9). In this brief communication, we report the identification of another antibiotic downregulator gene from S. coelicolor, a tetR family transcriptional regulator gene named SCO1712, from further analysis of the previous interspecies DNA microarray results. We show that SCO1712 overexpression led to a significant reduction of antibiotic production in both ACT-producing S. coelicolor and DXR-producing S. peucetius. In addition, SCO1712 disruption in S. coelicolor not only upregulated antibiotic biosynthesis through pathway-specific regulators in the presence of the wblA transcript but also further stimulated antibiotic production in a wblA deletion mutant, implying that SCO1712 might encode a wblA-independent antibiotic downregulator.
Although SCO1712 was not initially selected among the previously identified 160 S. coelicolor potential candidate genes affecting DXR production (9), its ortholog in S. peucetius exhibited more than a 4-fold decrease of transcript levels in the DXR-overproducing mutant strain in a repeated microarray analysis (see Fig. S1A in the supplemental material). In addition, the culture time-dependent comparative microarray analysis revealed that SCO1712 expression is considerably lower in S. coelicolor M145, which produces abundant ACT, than in S. coelicolor J1501, which produces relatively little ACT (see Fig. S1B). SCO1712 encodes a 205-amino-acid (aa) protein with an N-terminal TetR family helix-turn-helix (HTH) DNA binding domain (see Fig. S2A). To determine the in vivo biological significance of SCO1712, it was cloned next to an ermE* promoter in a Streptomyces-Escherichia coli shuttle expression vector, pSE34, followed by interspecies transformation into both S. coelicolor M145 and the S. peucetius industrial mutant. As shown in Fig. 1A, a noticeable decrease in the blue-pigment antibiotic ACT was observed in the SCO1712-expressing S. coelicolor in plate culture, though the previously identified SCO3579 (wblA) had a stronger inhibitory effect on antibiotic production. The SCO1712-expressing S. peucetius industrial mutant also displayed more than 3-fold less of the red DXR pigment during growth in liquid medium as well as in plate cultures (Fig. 1B), suggesting that SCO1712 may be another downregulator that broadly functions to inhibit antibiotic biosynthesis in streptomycetes. Since SCO1712 is the third ORF of a putative translationally coupled three-gene operon (http://streptomyces.org.uk/), the other two upstream ORFs, SCO1713 (encoding a 34-aa hypothetical protein) and/or SCO1714 (encoding a 189-aa possible secreted protein with unknown function), might also be involved in antibiotic regulation. However, overexpression of SCO1713 and/or SCO1714 failed to downregulate blue-pigment ACT biosynthesis in S. coelicolor (see Fig. S3), implying that the in vivo biological effect of SCO1712 as an antibiotic downregulator does not seem to be directly related to the functions of SCO1713 and/or SCO1714.
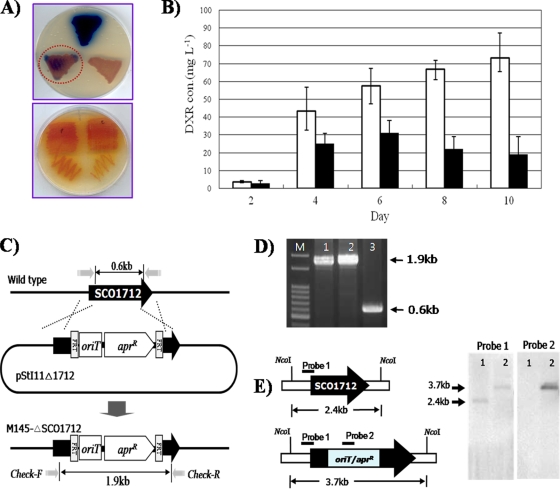
FIG. 1.
(A) Upper plate, R2 yeast extract (R2YE) plate cultures of S. coelicolor transformants harboring some of the previously identified putative regulatory genes (clockwise from top): empty expression vector pSE34, SCO3579, and SCO1712 (highlighted by circle). Lower plate, nitrogen-defined yeast extract (NDYE) plate cultures of the S. peucetius overproducing mutant harboring pSE34 (bottom right) and SCO1712 (bottom left). (B) Average of triplicates of doxorubicin (DXR) volumetric productivities from NDYE liquid cultures of recombinant S. peucetius overproducing mutant strains. Open bars, S. peucetius mutant containing pSE34; filled bars, S. peucetius mutant containing overexpressed SCO1712. (C) Schematic representation of PCR-targeted gene replacement disruption of SCO1712 and apramycin resistance (Aprr)/oriT. (D) Confirmation of constructed ΔSCO1712 mutant by PCR with a check primer pair. Lanes: M, 100-bp DNA ladder; 1, S. coelicolor M145-Δ1712 mutant genomic DNA; 2, StI11Δ1712 disrupted cosmid; 3, S. coelicolor M145 wild-type genomic DNA. (E) Confirmation of constructed ΔSCO1712 mutant by Southern hybridization with probe 1 (portion of SCO1712) and probe 2 (portion of oriT). Lanes: 1, S. coelicolor M145 wild-type genomic DNA; 2, S. coelicolor M145-Δ1712 mutant genomic DNA.
Although in silico sequence analysis of SCO1712 as well as its overexpression in both S. coelicolor and the S. peucetius industrial mutant was consistent with its downregulatory role in antibiotic biosynthesis, we sought to confirm the in vivo function of SCO1712 using a gene disruption approach (5). SCO1712 carried by the StI11 cosmid was replaced with an apramycin resistance/oriT cassette, generating pStI11Δ1712 (http://streptomyces.org.uk/), which was introduced into S. coelicolor M145 by conjugative gene transfer (Fig. 1C). Construction of the SCO1712 mutant (named S. coelicolor M145-ΔSCO1712) was confirmed by PCR analysis. The expected size of 0.6 kb for the PCR-amplified bands was observed in genomic DNA samples isolated from S. coelicolor M145, while a band of the expected size (1.9 kb) was observed in genomic DNA samples isolated from S. coelicolor M145-ΔSCO1712 (Fig. 1D). Moreover, the two different theoretically calculated NcoI digestion patterns were observed in Southern hybridization with S. coelicolor M145 and S. coelicolor M145-ΔSCO1712 (Fig. 1E), implying that SCO1712 was specifically disrupted as expected. Both S. coelicolor M145 and S. coelicolor M145-ΔSCO1712 mutant strains were cultured on modified R5 plates as well as liquid medium for production of both ACT and RED, followed by visual observation of the plates and antibiotic measurements from the liquid cultures using UV spectrophotometric quantification (7).
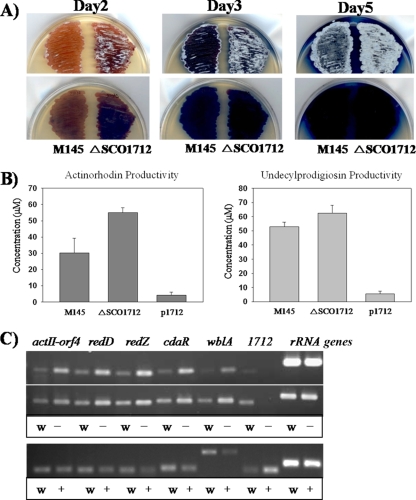
SCO1712 seems to affect morphogenesis, with aerial hypha formation initiation earlier in the SCO1712-disrupted strain than in the wild type (Fig. 2A). Although a slight reduction and delay in pigment production were observed in the plates of the wild type compared with the SCO1712 deletion mutant (Fig. 2A), approximately 62% more ACT and 22% more RED were observed in the S. coelicolor M145-ΔSCO1712 mutant strain than in the wild type under the same liquid culture conditions (Fig. 2B), providing strong evidence that SCO1712 is indeed another global downregulator of antibiotic biosynthesis in Streptomyces species. Since SCO1712 was cloned under the control of the constitutive ermE* promoter in an integrative pSET152 vector for complementation of the mutant 1712 strain, the expression levels were likely much higher than would have been seen in the wild type. That was probably the main reason that much lower levels of actinorhodin were observed in the S. coelicolor M145-ΔSCO171 strain containing the p1712 construct than in the wild type (Fig. 2B). The mutant 1712 strain was also functionally complemented to show reduced ACT production on the plate culture (see Fig. S2B in the supplemental material). To determine whether the noticeable change in antibiotic production upon gene disruption of SCO1712 from the S. coelicolor chromosome was controlled by pathway-specific activators, total RNA samples were prepared from S. coelicolor M145 and M145-ΔSCO1712 after 39 h (RED production period) and 78 h (ACT production period) of growth and used as a template for gene expression analysis by reverse transcription (RT)-PCR. Primers for RT-PCR were specific to sequences within the pathway-specific activator genes (see Table S1) and were designed to produce cDNAs of approximately 200 bp. A primer pair designed to amplify a cDNA from rRNA was used as an internal control. Transcripts were analyzed from the pathway-specific activator genes of the three major S. coelicolor antibiotics (i.e., actII-ORF4 for ACT, redD/Z for RED, and cdaR for CDA) and from the previously identified wblA. This analysis was carried out at least three times for each primer pair. Transcripts encoded by pathway-specific activator genes such as actII-ORF4, redD/Z, and cdaR were all significantly increased at both time points from S. coelicolor M145-ΔSCO1712 (Fig. 2C). As expected, an opposite transcription pattern was observed in the SCO1712-overexpressing S. coelicolor M145 strain (Fig. 2C). Taken together, these results strongly suggest that SCO1712 has a global inhibitory effect on antibiotic biosynthesis and that this may be due to direct or indirect control of the pathway-specific activators. Interestingly, the transcript profile of wblA was increased in M145-ΔSCO1712 (Fig. 2C), which is somewhat contradictory to the previous finding that wblA overexpression also causes a significant decrease in antibiotic production (9). Although this is complicated by the fact that wblA and SCO1712 genes appear to have similar antibiotic downregulating functions, the tetR-like SCO1712 protein might be able to repress the expression of wblA in a yet-unknown regulatory cascade. Since the transcripts of pathway-specific regulator genes were increased due to the SCO1712 gene disruption even in the presence of the wblA transcript, however, these RT-PCR results also strongly suggest that the effect of SCO1712 on actII-ORF4, redD/Z, and cdaR in S. coelicolor is believed to be indirectly related to, yet does not require, the presence of wblA.
FIG. 2.
(A) Time-dependent antibiotic production in modified R5 agar plate cultures of S. coelicolor M145 (left) and M145-Δ1712 (right). (B) Volumetric productivities (average of triplicates) of ACT (left) and RED (right) by wild-type (S. coelicolor M145), ΔSCO1712 (S. coelicolor M145-ΔSCO1712), and p1712 (S. coelicolor M145-ΔSCO1712 with pSET152 containing SCO1712) strains cultured in modified R5 for 4 days. (C) RT-PCR analysis of pathway-specific genes and the pleiotropic regulatory genes wblA and SCO1712. The amounts of total RNA samples from the two strains were measured to be equally present for comparable RT-PCR analyses. Assays in top panels were performed using samples isolated at 39 h (upper) and 78 h (lower). Lanes 1 and 2, actII-ORF4; lanes 3 and 4, redD; lanes 5 and 6, redZ; lanes 7 and 8, cdaR; lanes 9 and 10, wblA; lanes 11 and 12, SCO1712; lanes 13 and 14, rRNA genes. Odd-numbered lanes labeled w, RT-PCR with total RNA from S. coelicolor M145; even-numbered lanes labeled −, RT-PCR with total RNA from S. coelicolor M145-Δ1712 mutant. The assay in the bottom panel was performed using samples isolated at 78 h. Odd-numbered lanes labeled w, RT-PCR with total RNA from S. coelicolor M145; even-numbered lanes labeled +, RT-PCR with total RNA from the SCO1712-overexpressing S. coelicolor M145 mutant.
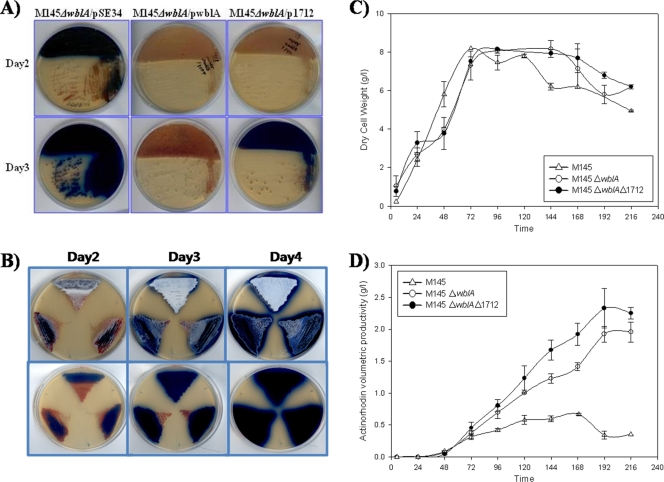
Expression of not only wblA but also SCO1712 complemented the ΔwblA mutant phenotype to delay ACT biosynthesis in the early culture stage (Fig. 3A), suggesting that wblA is not necessarily required for SCO1712 to function as an antibiotic downregulator. To further verify the wblA-independent phenotype, SCO1712 was additionally disrupted in the S. coelicolor M145ΔwblA mutant strain, resulting in the S. coelicolor M145ΔwblAΔSCO1712 double knockout mutant strain. There was no significant phenotypic difference observed between S. coelicolor M145ΔwblA and M145ΔwblAΔSCO1712 mutant strains, and both seemed to exhibit less sporulation but higher ACT production than did the wild type in the plate culture (Fig. 3B). While all three strains exhibited comparable growth patterns except for some extended lag phases observed in both mutants, in 9-day liquid fermentation cultures (Fig. 3C), an S. coelicolor M145ΔwblAΔSCO1712 mutant strain exhibited the highest ACT volumetric productivity (2.32 g/liter at 192 h), which is 6.8-fold and 1.2-fold higher than those of S. coelicolor M145 wild type (0.34 g/liter at 192 h) and S. coelicolor M145ΔwblA (1.92 g/liter at 192 h), respectively (Fig. 3D), implying that SCO1712 is not directly related to wblA function and is more likely to encode a wblA-independent antibiotic downregulator. Nonetheless, we cannot yet completely rule out the possibility that SCO1712 protein might also control wblA itself directly or indirectly. Once again, our results suggest that genome-wide screening using cDNA microarrays containing sequences from the S. coelicolor genome together with antibiotic-overproducing industrial strains of related streptomycetes may be an efficient approach to discover regulatory genes affected by unidentified mutations in the industrial strains. Moreover, sequential targeted gene disruptions of independently working downregulatory genes could provide an efficient and rational strategy for Streptomyces industrial strain improvement.
FIG. 3.
(A) Time-dependent antibiotic production in modified R5 plate cultures of S. coelicolor ΔwblA mutant (kindly provided by K. Chater) strains harboring pSE34 alone (left), pSE34 containing wblA (middle), and pSE34 containing SCO1712 (right). (B) Time-dependent antibiotic production in modified R5 plate cultures of wild-type S. coelicolor M145 (top), S. coelicolor ΔwblA (lower left), and S. coelicolor ΔwblAΔSCO1712 (lower right). The method for a PCR-targeted gene replacement disruption of SCO1712 in S. coelicolor ΔwblA was identical to those shown in Fig. 1 (data not shown). (C and D) Time-dependent growth curve (C) and ACT volumetric productivities (D) of wild-type S. coelicolor M145, S. coelicolor ΔwblA, and S. coelicolor ΔwblAΔSCO1712 cultured in modified R5 medium for 9 days in a 2-liter bioreactor (17). The averages of two independent fermentation experiments are shown with error bars.
Supplementary Material
Acknowledgments
The S. peucetius industrial mutant strain and the StI11 cosmid were kindly provided by Boryung Pharmaceutical Co. in South Korea and the John Innes Centre in the United Kingdom, respectively. In particular, we greatly appreciate the S. coelicolor ΔwblA strain as well as some critical comments kindly provided by Keith Chater at the John Innes Centre.
This work was supported by the Korean Systems Biology Program (MEST 2009-0065571) as well as a KOSEF grant (MEST 2009-0078663).
Footnotes
Published ahead of print on 26 February 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 2.Bibb, M. J. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208-215. [DOI] [PubMed] [Google Scholar]
- 3.Chater, K. F. 1989. Multilevel regulation of Streptomyces differentiation. Trends Genet. 5:372-377. [DOI] [PubMed] [Google Scholar]
- 4.Chng, C., A. M. Lum, J. A. Vroom, and C. M. Kao. 2008. A key developmental regulator controls the synthesis of the antibiotic erythromycin in Saccharopolyspora erythraea. Proc. Natl. Acad. Sci. U. S. A. 105:11346-11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hopwood, D. A. 2007. Therapeutic treasures from the deep. Nat. Chem. Biol. 3:457-458. [DOI] [PubMed] [Google Scholar]
- 7.Hopwood, D. A., T. Kieser, M. J. Bibb, M. J. Buttner, and K. F. Chater. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 8.Hranueli, D., J. Cullum, B. Basrak, P. Goldstein, and P. F. Long. 2005. Plasticity of the streptomyces genome-evolution and engineering of new antibiotics. Curr. Med. Chem. 12:1697-1704. [DOI] [PubMed] [Google Scholar]
- 9.Kang, S.-H., J. Huang, H.-N. Lee, Y.-A. Hur, S. N. Cohen, and E.-S. Kim. 2007. Interspecies DNA microarray analysis identifies WblA as a pleiotropic down-regulator of antibiotic biosynthesis in Streptomyces. J. Bacteriol. 189:4315-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, Y. J., J. Y. Song, M. H. Moon, C. P. Smith, S.-K. Hong, and Y. K. Chang. 2007. pH shock induces overexpression of regulatory and biosynthetic genes for actinorhodin production in Streptomyces coelicolor A3(2). Appl. Microbiol. Biotechnol. 76:1119-1130. [DOI] [PubMed] [Google Scholar]
- 11.Lee, H.-N., J.-H. Im, M.-J. Lee, S. Y. Lee, and E.-S. Kim. 2009. A putative secreted solute binding protein, SCO6569 is a possible AfsR2-dependent down-regulator of actinorhodin biosynthesis in Streptomyces coelicolor. Process Biochem. 44:373-377. [Google Scholar]
- 12.Lian, W., K. P. Jayapal, S. Charaniya, S. Mehra, F. Glod, Y.-S. Kyung, D. H. Sherman, and W.-S. Hu. 2008. Genome-wide transcriptome analysis reveals that a pleiotropic antibiotic regulator, AfsS, modulates nutritional stress response in Streptomyces coelicolor A3(2). BMC Genomics 9:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu, Y., W. Wang, D. Shu, W. Zhang, L. Chen, Z. Qin, S. Yang, and W. Jiang. 2007. Characterization of a novel two-component regulatory system involved in the regulation of both actinorhodin and a type I polyketide in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 77:625-635. [DOI] [PubMed] [Google Scholar]
- 14.Lum, A. M., J. Huang, C. R. Hutchinson, and C. M. Kao. 2004. Reverse engineering of industrial pharmaceutical-producing actinomycete strains using DNA microarrays. Metab. Eng. 6:186-196. [DOI] [PubMed] [Google Scholar]
- 15.Myles, D. C. 2003. Novel biologically active natural and unnatural products. Curr. Opin. Biotechnol. 14:627-633. [DOI] [PubMed] [Google Scholar]
- 16.Nodwell, J. R. 2007. Novel links between antibiotic resistance and antibiotic production. J. Bacteriol. 189:3683-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryu, Y.-G., M. J. Butler, K. F. Chater, and K. J. Lee. 2006. Engineering of primary carbohydrate metabolism for increased production of actinorhodin in Streptomyces coelicolor. Appl. Environ. Microbiol. 72:7132-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soliveri, J. A., J. Gomez, W. R. Bishai, and K. F. Chater. 2000. Multiple paralogous genes related to the Streptomyces coelicolor development regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology 146:333-343. [DOI] [PubMed] [Google Scholar]
- 19.Strauch, E., E. Takano, H. A. Baylis, and M. J. Bibb. 1991. The stringent response in Streptomyces coelicolor A3(2). Mol. Microbiol. 5:289-298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.