Abstract
The beneficial effects of Bifidobacterium are partly due to its immunostimulatory properties. These immunostimulatory properties may be linked to the presence of unmethylated CpG motifs specific to bacterial DNA, which may induce a TH1 response by activating Toll-like receptors (TLR). Using in silico analyses, PCR amplification, and dot blotting, we characterized the CpG content of various bifidobacterial strains and evaluated the immunostimulatory properties and genomic heterogeneity of these motifs in the genus. Our in silico study, based on entire genome sequences from five bifidobacterial strains, showed that Bifidobacterium genomes contain numerous CpG motifs, including 5′-purine-purine-CG-pyrimidine-pyrimidine-3′ and 5′-purine-TCG-pyrimidine-pyrimidine-3′ motifs, and biologically active sequences previously identified in lactic acid bacteria. We identified four CpG-rich sequences with Bifidobacterium longum NCC2705. Two sequences with a percent G+C of about 68% included 14 and 16 CpG motifs. Two sequences with a percent G+C of about 60% included 16 and 6 CpG motifs. These sequences induce the production of monocyte chemoattractant protein 1 (MCP-1) and tumor necrosis factor alpha (TNF-α) through a pattern of TLR9 stimulation on RAW 264.7 macrophages. No link could be established between their immunostimulatory properties, the number of CpG motifs, and percent G+C. We investigated inter- and intraspecies heterogeneity in 71 strains of various origins. These sequences were highly conserved in the genus. No link was found between the presence of the CpG-rich sequence and the origin of the strains (healthy, allergic, or preterm infants). The high frequency of CpG motifs in the DNA of Bifidobacterium may play an important role in the immunostimulatory properties of commensal or probiotic bifidobacterial strains.
Many studies have shown that initial intestinal colonization plays a crucial role in the development of the intestinal immune system (7, 12, 18, 46). Bifidobacteria are Gram-positive anaerobic bacteria accounting for up to 90% of the total gut microbiota in breast-fed babies and up to 75% in formula-fed babies (10). Their implantation takes place within the first 10 days of life. It has been suggested that these high levels of bifidobacteria in the human intestine contribute to human health, leading to their use as probiotics. Indeed, bifidobacteria have been shown to have beneficial effects against various diseases, including diarrhea associated with rotavirus or antibiotics, inflammatory bowel diseases (40), necrotizing enterocolitis (1), and allergy (16, 19, 37). The immunostimulatory properties of bifidobacteria may contribute to these beneficial effects. However, little is known about the contribution of these bacteria to immunity.
We previously characterized the effects of 10 strains of Bifidobacterium on TH1/TH2 balance in gnotobiotic mice. We showed that these strains had species-specific effects on the stimulation of immunity and strain-specific effects on the TH1/TH2 balance (32). Differences in the bacterial structures recognized by Toll-like receptors (TLR) could potentially account for these strain-specific effects (17, 36). DNA from Gram-positive and Gram-negative bacteria has been shown to induce a TH1 response (17, 36). These immunostimulatory properties may be linked to the presence of unmethylated CpG motifs, biologically active sequences with a high G+C content conserved in the bacterial genome. These motifs are known to exert immunostimulatory effects by activating Toll-like receptor 9, leading to the induction of TH1-type immune responses (55). These sequences are much more frequent in prokaryotes (95% of CpG motifs are unmethylated) than in eukaryotes (10 to 30%) (23). A few studies have shown the DNA of lactic acid bacteria to have immunostimulatory properties (9, 14, 27, 30, 31). Satokari et al. (42) hypothesized that bifidobacteria may have high frequencies of unmethylated CpG motifs due to their high G+C content, potentially accounting for their immunostimulatory effects.
We used in silico analyses, PCR amplification, and dot blotting of CpG-rich DNA fragments from various bifidobacterial strains to characterize the immunostimulatory properties of bacteria of the Bifidobacterium genus and to evaluate their genomic heterogeneity.
MATERIALS AND METHODS
Strains.
We carried out an in silico analysis of the five Bifidobacterium genomes for which entire genome sequences have been obtained and are publicly available: Bifidobacterium longum NCC2705 (NC_004307.2) (43), B. longum DJO10A (NC_010816) (28), B. longum subsp. infantis ATCC 15697 (NC_011593) (44), B. animalis subsp. lactis AD011 (NC_011835) (20), and B. adolescentis ATCC 15703 (NC_008618).
We also included 71 bifidobacterial strains isolated at our laboratory from the fecal microbiota of infants (culture collection of EA 4065 laboratory). Twenty-nine of these strains were isolated from the fecal microbiota of allergic infants, 32 were isolated from nonallergic infants, and 10 from preterm infants. These 71 strains comprised 17 B. longum group strains, 26 B. breve strains, 12 B. bifidum strains, 1 B. dentium strain, 10 B. catenulatum/pseudocatenulatum group strains, and 5 B. adolescentis strains.
All strains were grown on Wilkins Chalgren agar base (WCB) supplemented with 10 g/liter d-glucose, 0.5 g/liter l-cysteine, and Tween 80 (0.5% [vol/vol]) (4) and in trypticase-glucose-yeast-hemin (TGYH) broth. They were incubated at 37°C in an anaerobic chamber (du Scientific, Chemunex-AES Laboratoire, Bruz, France).
PCR was used to identify strains to the genus and species levels, as described by Kok et al. (24) and Mullié et al. (35), respectively. When necessary, we carried out 16S rRNA gene PCR amplification and sequencing using SD008 and SD1492 primers (47).
In silico analysis of Bifidobacterium genome.
In silico analysis was performed with the cgplot and cgreport programs of EMBOSS to identified CpG-rich sequences. We searched for 5′-purine-purine-CG-pyrimidine-pyrimidine-3′ and 5′-purine-TCG-pyrimidine-pyrimidine-3′ motifs, referred to here as type 1 and type 2 CpG motifs, respectively (25). We also searched for four immunostimulatory motifs previously identified in lactic acid bacteria: BL07 (5′-GCGTCGGTTTCGGTGCTCAC-3′) (52), OL-LB7 (5′-CGGCACGCTCACGATTCTTG-3′) (22), ID35 (5′-ACTTTCGTTTTCTGCGTCAA-3′) (14), and AT ODN (5′-ATTTTTAC-3′) (45).
The Blastn program of EMBOSS and the multiple sequence alignment software MultAlin, developed by Florence Corpet (6), were used for sequence alignments.
Preparation of genomic DNA.
Genomic DNA was prepared as described by Zhu et al. (57). Briefly, bacteria were collected after 48 h of incubation in 10 ml TGYH broth, by centrifugation at 13,000 rpm for 5 min. They were then lysed by incubation in Tris-HCl (pH 8, 100 μM), EDTA (pH 8, 1 mM), 1% SDS (Sigma-Aldrich, Lyon, France). Genomic DNA was extracted by adding one volume of benzoyl chlorate, incubating at 55°C for 30 min, adding a half volume of sodium acetate (3 M, pH 5.4), and incubating for a further 15 min on ice. The solution was centrifuged at 13,000 rpm for 10 min. DNA was precipitated with isopropanol.
PCR amplification of CpG-rich sequences.
PCR amplifications of selected sequences on the basis of in silico analyses and of dot blot probings were performed with GeneAmp PCR system 2700 (Applied Biosystems, Courtaboeuf, France), with primers designed on the basis of the B. longum NCC2705 sequence (Table 1). The PCR mix (50 μl) contained 1 U of Taq DNA polymerase (Invitrogen, Fisher-Bioblock, Illkirch, France), 5 μl of 10 μM primers, 4 μl of 10 mM dNTP, and various quantities of 50 mM MgCl2. It was subjected to thermal cycling as follows: 10 min at 96°C followed by 35 cycles of 30 s of denaturation at 97°C, 30 s at hybridization temperature, and 2 min of extension at 72°C, and then a final extension phase for 7 min at 72°C (Table 1).
TABLE 1.
Primers used for amplification of the different CpG-rich sequences or fragments of these sequences
| Primer | Forward primer 5′-3′ | Reverse primer 5′-3′ | Quantity of 50 mM MgCl2 in PCR mix (μl) | Hybridizing temp (°C) | Fragment size (bp) |
|---|---|---|---|---|---|
| Primers used for amplification of the different sequences | |||||
| H2 | AGACCCGTGAGGTGATTGTC | GAACTGCAGGTTCCACAGGT | 2 | 60 | 1,555 |
| H3 | CACTTCATCTTCCGCACCG | TTGATGTCGAACTGCTGGAG | 2 | 60 | 1,074 |
| N2 | ACATGAAGGAGCACGACTGC | CGAGACACACCTTGTCTGGA | 2 | 55 | 1,431 |
| N3 | AACGGCCCATATGTCTGAGG | AGGGCATGGTTCATTTGC | 6 | 50 | 771 |
| Primers used for fabrication of the dot blot probes | |||||
| H2 Dot | GCGAACCTGTCACTGGAAAT | AAAAATCAGAGGGGCTGGTT | 2 | 60 | 152 |
| H3 Dot | GATTTGGCGGAGAAGATCAA | TTGATGTCGAACTGCTGGAG | 4 | 194 | |
| N2 Dot | AGCTTTCGGGTGGAGAATTT | CAACCAGACCGTCATGTCAC | 4 | 159 | |
| N3 Dot | GGCCCATATGTCTGAGGA | GGCGGTTCTGTCATCGTATT | 6 | 214 |
These CpG-rich PCR products (amplified fragments containing unmethylated CpG-rich sequences) were inserted into the pCR2.1 TOPO vector (Invitrogen, Fisher-Bioblock) according to the manufacturer's instructions. The transformation and storage buffer (TSB) method was used to transform Escherichia coli TOP10 (5). Automatic DNA sequencing was performed by Genome Express (Grenoble, France).
Dot blotting.
We transferred 2 μg of Bifidobacterium DNA onto a nylon membrane with a Minifold 1 dot blotter (Schleicher & Schuell) and using the alkaline method. Membranes were then blocked by exposure to UV for 3 min and probed with peroxidase-conjugated PCR-amplified fragments, which were detected with the ECL direct nucleic acid-labeling and detection system (Amersham Biosciences, GE Healthcare, Bukinghamshire, England) according to the manufacturer's instructions. Hybridizations were performed overnight at 42°C, and the membranes were subjected to high-stringency washes before incubation with chemoluminescent substrate for ECL detection.
Biological activity of CpG-rich sequences. (i) Preparation of the sequences.
The CpG-rich PCR products obtained from plasmid amplification were purified using the endo-free plasmid maxi kit (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions. Endotoxin content was assessed with the Limulus amebocyte lysate kinetic-QCL kit (Cambrex, Lonza, Saint Beauzire, France), according to the manufacturer's instructions, in a Cambrex Elx 808CSE. For all CpG-rich PCR products and DNA, endotoxin concentrations were below 0.01 EU/ml.
(ii) Macrophage stimulation.
RAW 264.7 murine macrophages were seeded in 48-well plates at a density of 5 × 105 cells/well. The plates were incubated overnight at 37°C under an atmosphere containing 5% CO2 and 95% air. Cells were stimulated with CpG-rich PCR product (10 μg/ml) and genomic DNA (250 ng/ml) in fresh Dulbecco modified Eagle medium (DMEM) (containing 4.5 g/liter glucose and l-glutamine) supplemented with 1% pyruvate and 10% heat-inactivated fetal bovine serum (Gibco, Fisher Bioblock). Lipopolysaccharide (LPS) (1 μg/ml) and the oligodeoxynucleotide (ODN) CpG 1826 (10 μg/ml) were used as positive controls. Three independent assays were performed. Supernatants were collected after 6, 12, and 24 h.
Cytokine determinations were carried out with the mouse inflammation cytometric bead array (CBA) kit (BD Biosciences, le Pont de Claix, France) according to the manufacturer's instructions. Detection limits for the CBA were as follows: interleukin-6 (IL-6), 5 pg/ml; IL-10, 17.5 pg/ml; monocyte chemoattractant protein 1 (MCP-1), 52.7 pg/ml; gamma interferon (IFN-γ), 2.5 pg/ml; tumor necrosis factor alpha (TNF-α), 7.3 pg/ml; and IL-12p70, 10.7 pg/ml.
(iii) Involvement of TLR9.
Macrophages were stimulated, as described above, in the presence of ODN 2088, an oligonucleotide that inhibits TLR9 signaling by disruption of the colocalization of CpG ODNs with TLR9 in endosomal vesicles without affecting cellular binding and uptake (InvivoGen, France). This ODN was used at the same concentration as that of the CpG-rich PCR products. Supernatants were collected after 6 h, as recommended by the manufacturer's instructions.
Statistical analysis.
Mann-Whitney U tests were used to assess the significance of differences in cytokine concentrations between samples. We evaluated differences in the numbers of sequence motifs present in the genome of a strain as a function of the origin of that strain, by carrying out Student's t tests. P values below 0.05 were considered to be statistically significant. Data were analyzed using SPSS software (version 12.0).
RESULTS
CG-rich sequences and CpG motifs in Bifidobacterium genomes.
We carried out in silico analyses of the five Bifidobacterium strains for which entire genome sequences are publicly available. These genomes were between 1,933,695 bp and 2,832,748 bp in size and contained between 84 and 86% coding sequences. The G+C content of the three B. longum strains was 60%, and that of the B. animalis subsp. lactis AD011 and B. adolescentis ATCC 15703 strains was 59%.
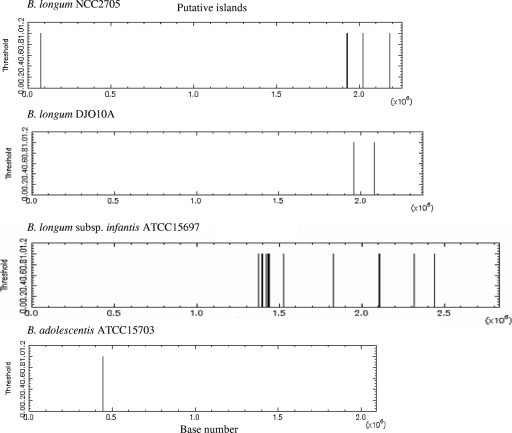
We searched for sequences of more than 200 bp with a C+G content exceeding 80%. No such sequences were found with B. animalis subsp. lactis AD011, whereas B. adolescentis ATCC 15703 contained one such sequence (212 bp, 76% G+C), B. longum DJO10A contained two such sequences (238 bp, 76%; 261 bp, 78%), B. longum NCC2705 contained four such sequences (265 bp, 78%; 291 bp, 77%; 213 bp, 76%; and 238 bp, 76%), and B. longum subsp. infantis ATCC 15697 contained 19 such sequences (between 202 bp and 527 bp and between 74 and 80% CG) (Fig. 1). Many sequences of more than 200 bp, with the percent C+G exceeding 70%, were identified: 543 in B. animalis subsp. lactis AD011, 421 in B. adolescentis ATCC 15703, 661 in B. longum NCC2705, 709 in B. longum DJO10A, and 948 in B. longum subsp. infantis ATCC 15697. Some of these sequences were longer than 500 bp: 27 sequences for B. animalis subsp. lactis AD011, 24 for B. adolescentis ATCC 15703, 60 for B. longum NCC2705, 70 for B. longum DJO10A, and 141 for B. longum subsp. infantis ATCC 15697.
FIG. 1.
Location of putative CpG islands more than 200 bp in length and with a G+C content exceeding 80% within the Bifidobacterium genome. No sequences were found with B. animalis subsp. lactis AD011. Data were obtained with the cpgplot program.
The numbers of type 1 and type 2 CpG motifs on the 5′-to-3′strand were similar for B. longum NCC2705, B. longum DJO10A, and B. adolescentis ATCC 15703: a mean of 21,723 (± 4%). These motifs were 11% less frequent in B. animalis subsp. lactis AD011 (18,699 motifs) and 28% more frequent in B. longum subsp. infantis ATCC 15697 (26,915 motifs) (Table 2). Type 1 CpG motifs, which are palindromic, were present at similar frequencies on both strands. This was not the case for type 2 CpG motifs. However, the number of repeats on the 3′-to-5′ strand was close to the number on the 5′-to-3′ strand (Table 2).
TABLE 2.
In silico search of immunostimulatory motifs on sense and antisense strands of Bifidobacterium genomesa
| CpG motif | Sequence | B. longum NCC2705 | B. longum DJO10A | B. longum subsp. infantis ATCC 15697 | B. adolescentis ATCC 15703 | B. animalis subsp. lactis AD011 |
|---|---|---|---|---|---|---|
| Type 1b | AACGTT | 296 | 313 | 375 | 433 | 151 |
| AACGTC | 541 | 611 | 698 | 703 | 386 | |
| AACGCT | 424 | 414 | 560 | 589 | 285 | |
| AACGCC | 1,203 | 1,253 | 1,546 | 1,191 | 894 | |
| AGCGTT | 485 | 492 | 501 | 476 | 320 | |
| AGCGTC | 629 | 661 | 817 | 705 | 572 | |
| AGCGCT | 469 | 486 | 485 | 370 | 361 | |
| AGCGCC | 995 | 1,054 | 1,174 | 928 | 838 | |
| GACGTTc | 528 | 571 | 729 | 730 | 385 | |
| GACGTC | 648 | 668 | 865 | 844 | 778 | |
| GACGCT | 527 | 660 | 859 | 744 | 541 | |
| GACGCC | 1,516 | 1,651 | 2,097 | 1,371 | 1,294 | |
| GGCGTT | 1,186 | 1,300 | 1,540 | 1,144 | 894 | |
| GGCGTC | 1,538 | 1,661 | 2,046 | 1,460 | 1,353 | |
| GGCGCT | 975 | 1,070 | 1,179 | 933 | 921 | |
| GGCGCC | 1,014 | 1,054 | 1,490 | 1,112 | 1,433 | |
| Type 1 total | 12,974 | 13,919 | 16,961 | 13,733 | 11,406 | |
| Type 2 | ATCGTT | 553 (596) | 564 (640) | 666 (718) | 573 (593) | 405 (341) |
| ATCGTC | 1,250 (1,236) | 1,312 (1,312) | 1,591 (1,600) | 1,408 (1,406) | 1,204 (1,152) | |
| ATCGCT | 501 (435) | 516 (455) | 545 (508) | 425 (434) | 350 (295) | |
| ATCGCC | 2,079 (2,044) | 2,171 (2,196) | 2,460 (2,513) | 1,951 (1,863) | 1,642 (1,715) | |
| GTCGTTd | 663 (632) | 693 (692) | 898 (834) | 661 (681) | 672 (594) | |
| GTCGTC | 960 (931) | 1,076 (999) | 1,275 (1,286) | 1,011 (988) | 1,100 (1,138) | |
| GTCGCT | 526 (508) | 576 (514) | 570 (628) | 537 (550) | 536 (507) | |
| GTCGCC | 1,562 (1,496) | 1,638 (1,597) | 1,949 (1,964) | 1,338 (1,290) | 1,384 (181) | |
| Type 2 total | 8,094 (7,878) | 8,546 (8,405) | 9,954 (10,051) | 7,904 (7,805) | 7,293 (7,123) | |
| Total | 21,068 | 22,465 | 26,915 | 21,637 | 18,699 | |
| Published motifse | ||||||
| BL07 | GCGTCGGTTTCGGTGCTCAC | 1 (0) | 1 (0) | 1 (0) | 0 (0) | 0 (0) |
| OL-LB7 | CGGCACGCTCACGATTCTTG | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| ID35 | ACTTTCGTTTTCTGCGTCAA | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Active motif of ID35 | TTTCGTTT | 16 (22) | 20 (21) | 26 (28) | 42 (46) | 13 (13) |
| AT ODN | ATTTTTAC | 6 (4) | 8 (3) | 8 (13) | 11 (9) | 2 (1) |
Values indicate the number of the times that each sequence appears in the respective genome. The values in parentheses indicate the results for the antisense strand.
Palindromic sequences, found equally on antisense strand.
Sequence for optimal immunostimulatory motif of murine cells.
Sequence for optimal immunostimulatory motif of human cells.
BL07, a sequence from a B. longum strain, was found only once on the sense strand of the three B. longum strain genomes. OL-LB7, originally identified in a Lactobacillus strain, was not detected in any of the strains analyzed here. ID35 ODN, a sequence from Lactobacillus rhamnosus GG, was absent from these Bifidobacterium genomes, but its active region (TTTCGTTT) was present on both strands of the B. animalis subsp. lactis AD011 (26 repeats), B. longum NCC2705 (38 repeats), B. longum DJO10A (41 repeats), B. longum subsp. infantis ATCC 15697 (54 repeats), and B. adolescentis (88 repeats) genomes. AT ODN, originally identified in another Lactobacillus strain, was also present on both strands of all the Bifidobacterium genomes, with three copies for B. animalis subsp. lactis AD011, 10 for B. longum NCC2705, 11 for B. longum DJO10A, 21 for B. longum subsp. infantis ATCC 15697, and 20 for B. adolescentis ATCC 15703 (Table 2).
Selection of CpG-rich sequences in B. longum NCC2705 genome.
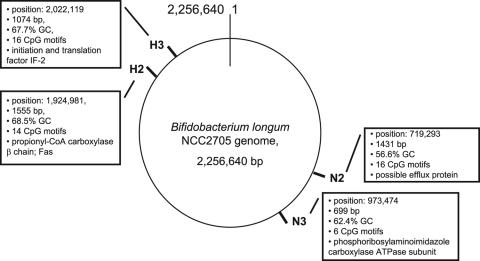
Based on our in silico analysis of the B. longum NCC2705 genome, we chose four CpG-rich sequences with various G+C contents (Fig. 2). Two sequences with a percent G+C of about 68%—H2 (1,555 bp) and H3 (1,074 bp)—included 200-bp islets with high G+C contents (76% and 78%, respectively). These two sequences were located close together. Two sequences with a percent G+C similar to that for the whole genome were selected and named N2 (1,431 bp) and N3 (699 bp). The number of type 1 and type 2 CpG motifs identified in these sequences was 14 for H2, 16 for H3 and N2, and 6 for N3.
FIG. 2.
Characteristics of CpG-rich sequences of Bifidobacterium longum NCC2705, including position, length, percent G+C, number of types 1 and 2 CpG motifs, and associated coding region. CoA, coenzyme A; Fas, fatty acid synthase.
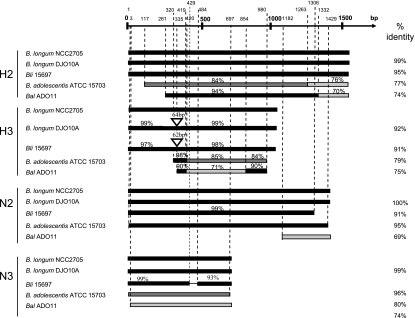
The variability of these four CpG-rich sequences in the various Bifidobacterium spp. studied here is illustrated in Fig. 3. H2 and H3 were highly conserved in the genomes of B. longum DJO10A and B. longum subsp. infantis ATCC 15697 compared to those of B. longum NCC2705, but parts of their 5′ end were missing in the B. adolescentis ATCC 15703 and B. animalis subsp. lactis AD011 genomes. A 341-bp fragment at the 3′ end of H3, with a G+C content of 67.4% and containing six CpG motifs, was duplicated in the genomes of B. longum DJO10A and B. longum subsp. infantis ATCC 15697. N2 was strongly conserved in the B. longum DJO10A and B. adolescentis ATCC 15703 genomes, but its 3′ end was partly deleted in the B. longum subsp. infantis ATCC 15697 genome, and this sequence was not conserved in the B. animalis subsp. lactis AD011 genome, which contained only 228 bp of the 3′ end. N3 was present in all the genomes studied but was highly conserved only in the B. longum genomes.
FIG. 3.
Variability of H2, H3, N2, and N3 sequences from the Bifidobacterium strains for which entire genome sequences have been published. Comparisons with B. longum NCC2705 were performed with MultAlin by Florence Corpet (6). The overall percentage of identity to B. longum NCC2705 is shown in the last column. Percent identity for a specific fragment: ▪, >90%;  , > 80%;
, > 80%;  , < 80%.
, < 80%.  , insertion into sequence;
, insertion into sequence;  , deletion from sequence. Bli 15697, B. longum subsp. infantis ATCC 15697; Bal AD011, B. animalis subsp. lactis AD011.
, deletion from sequence. Bli 15697, B. longum subsp. infantis ATCC 15697; Bal AD011, B. animalis subsp. lactis AD011.
The sequences used for probing in the dot blot assay were highly conserved in the five strains.
Immunostimulant activity of DNA and CpG-rich PCR products.
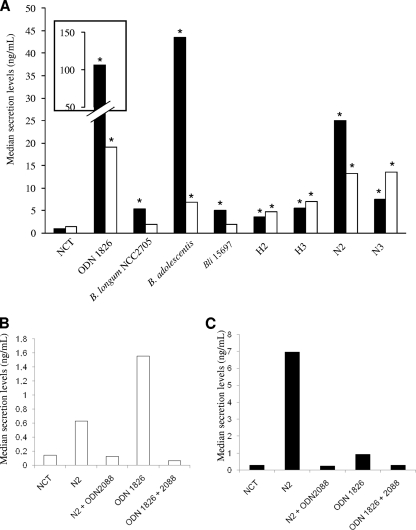
RAW 264.7 macrophages were stimulated with CpG-rich DNA fragments from B. longum NCC2705 and genomic DNA (250 ng/ml) from B. longum NCC2705, B. adolescentis ATCC 15703, and B. longum subsp. infantis ATCC 15697. The viability of RAW macrophages after 24 h of culture with CpG-rich PCR products and DNA was confirmed by trypan blue exclusion (data not shown). CpG-rich PCR products displayed immunostimulatory activity from 6 h of stimulation onwards, peaking at 12 h and remaining stable thereafter until 24 h (data not shown). The 12-h time point was selected to ensure that we were measuring the direct impact of CpG-rich PCR products, rather than the indirect effects of TNF-α. Our positive control, ODN 1826, a TLR9 agonist known to induce a pro-TH1 response, gave levels of TNF-α and MCP-1 production significantly higher (124 and 14 times, respectively) than those obtained for unstimulated macrophages (the negative control). No induction of IL-6 was observed. LPS, a TLR4 agonist, induced high levels of TNF-α and MCP-1 and a 91-fold increase of IL-6 compared to the negative control (data not shown). The four CpG-rich PCR products had cytokine induction profiles similar to that of ODN 1826. They induced significant increases in TNF-α (levels more than six times those of the negative control) and MCP-1 (more than three times those of the negative control) but did not induce IL-6, IL-10, IFN-γ, or IL-12 (Fig. 4A). N2 was the most active, increasing TNF-α and MCP-1 levels by factors of 29 and 10, respectively. The three genomic DNAs that were used significantly induced TNF-α production by factors of at least 6 up to 51 for B. adolescentis ATCC 15703. Only B. adolescentis ATCC 15703 DNA significantly stimulated MCP-1 production, to levels five times those of the negative control (Fig. 4A).
FIG. 4.
(A) Secretion of cytokines and chemokines by RAW 264.7 macrophages after 12 h of stimulation with 10 μg/ml CpG-rich PCR products (H2, H3, N2, and N3), 250 ng/ml DNA from B. longum NCC2705, B. longum subsp. infantis ATCC 15697, and B. adolescentis ATCC 15703, 10 μg/ml CpG-ODN 1826 as a positive control, 100 ng/ml LPS as a positive control, or no stimulation as a negative control (NCT). ▪, TNF-α; □, MCP-1. (B and C) MCP-1 (B) and TNF-α (C) levels after 6 h of stimulation by N2 CpG-rich PCR product (10 μg/ml) with or without ODN 2088 (10 μg/ml), an inhibitor of TLR9 signaling. Cytokines and chemokines were dosed by the mouse inflammation cytometric bead array (CBA) kit (BD Biosciences, France). Data are presented as medians. *, P < 0.05.
ODN 2088, a TLR9 signaling inhibitor, inhibited the immunostimulatory effect of N2, resulting in lower TNF-α and MCP-1 levels not significantly different from those for the negative control (Fig. 4B and C).
Variability of the CpG-rich sequences in Bifidobacterium spp.
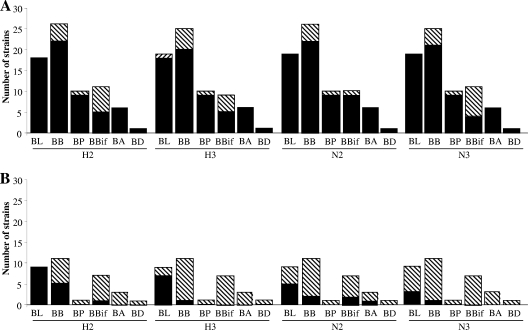
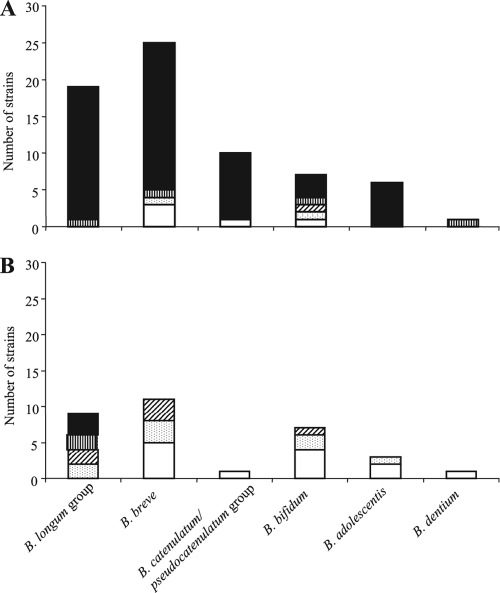
We assessed the variability of CpG-rich sequences in B. longum NCC2705, B. adolescentis ATCC 15703, B. longum subsp. infantis ATCC 15697, and 71 bifidobacterial strains, isolated at our laboratory, from the fecal microbiota of infants, by dot blotting (Fig. 5A and 6A). H2 and H3 were found to be present in 85% of the strains tested, N2 in 90%, and N3 in 84%. All 19 B. longum strains contained H2, N2, and N3, and 18 of these strains (95%) contained H3. All four CpG-rich sequences were present in 95% of B. longum strains; 85% of B. breve strains possessed H2, N2, and N3, and 80% of these strains contained H3. Three B. breve strains contained none of the sequences studied. All six B. adolescentis strains and nine of the 10 B. catenulatum/pseudocatenulatum group strains contained all four sequences. Greater variability was observed for the 12 B. bifidum strains, 90% of which contained N2, 56% H3, 45% H2, and 36% N3. The B. dentium strain tested contained H2, H3, and N3.
FIG. 5.
Interspecies variability of CpG-rich sequences in the genus Bifidobacterium. The presence of each sequence was evaluated by dot blotting (A) or PCR (B). Bars indicate the number of strains in which the sequence is present (▪) or absent (▧). BL, B. longum group; BB; B. breve; BP, B. catenulatum/pseudocatenulatum group; BIF, B. bifidum; BA, B. adolescentis; BD, B. dentium.
FIG. 6.
Intraspecies variability of CpG-rich sequences in the genus Bifidobacterium. The presence of sequences was evaluated by dot blotting (A) or by PCR (B). Bars represent the number of strains containing 4 sequences (▪), 3 sequences (▥), 2 sequences (▨), 1 sequence (░⃞), or no sequence (□).
These results were confirmed by PCR amplification for 32 strains (Fig. 5B and 6B). PCR confirmed the absence of the sequence in the genomes of strains for which no sequences were detected in dot blots, with the exception of two strains testing positive for H2 and H3 by PCR but without detection of the corresponding spot in dot blots. A total of 10 of the 24 strains testing positive for H2 by dot blotting (42%) tested negative by PCR; 18 of the 25 strains (72%) testing positive for H3 by dot blotting tested negative by PCR. For N2, 14 of the 28 strains (50%) testing positive by dot blotting tested negative by PCR, and for N3, 20 of the 25 (80%) strains testing positive by dot blotting tested negative by PCR.
There were no significant differences in the presence of the CpG-rich sequences, whatever the allergic status of the infant.
DISCUSSION
The immunostimulatory properties of lactic acid bacteria seem to result partly from the promotion of a TH1 response by their DNA (9, 22, 31). This effect may be linked to the presence in bacterial DNA of unmethylated CpG motifs, which are known to trigger TH1-type immune responses by activating Toll-like receptor 9 (55). Using in silico and molecular methods, we showed that Bifidobacterium genomes contain conserved CpG motifs with immunostimulatory activity but that there are differences in the distributions of these motifs between and within species.
Our in silico study showed that Bifidobacterium genomes contain numerous CpG motifs, with between 18,000 and 27,000 such motifs per DNA strand. Type 1 and 2 CpG motifs, as described in the literature, were found to be present and included the motifs known to stimulate murine and human cells, such as GACGTT and GTCGTT (25), respectively, and some of the immunostimulatory motifs identified in lactic acid bacteria (14, 45, 52). This may account for the biological activity observed with our various CpG-rich PCR products. We investigated the effect of these products on a macrophage cell line, as macrophages play a key role in the priming and activation of the immune system by commensal bacteria. Moreover, many studies (3, 54) have used the RAW 264.7 cell line for characterization of the mechanism of TLR9 activation by CpG-ODN and CpG-DNA. We found that the stimulation of these cells with Bifidobacterium DNA induced a pattern of cytokine production typical of TLR9 signaling in murine macrophages (8, 53), with the production of high levels of TNF-α and secretion of MCP-1 (significant only for B. adolescentis ATCC 15703) but no secretion of IL-6, IL-10, or IL-12. The active part of ID35, a sequence with a protective effect in a mouse model of allergic disease (15), was present in all five complete genomes studied, together with the AT ODN, a non-CpG immunostimulatory sequence (45). BL07, a sequence demonstrated to be protective against allergy in a mouse model (50, 51), seems conserved in the B. longum group. Indeed, Takahashi et al. (52) described this sequence for two B. longum strains (B. longum subsp. longum BB536 and B. longum subsp. infantis M-63) and for the B. breve M-16V genome (52), and we detected this sequence in B. longum NCC2705, B. longum DJO10A, and B. longum subsp. infantis ATCC 15697. This sequence was absent from B. adolescentis ATCC 15703 and B. animalis subsp. lactis AD011. These variations and differences in the number of CpG motifs pointed out heterogeneity within the genus. However, no significant differences in the immunostimulatory properties of the three DNAs studied were observed. We were, therefore, unable to establish a link between the number and the type of CpG motifs and the immunostimulatory properties of the strains. The importance to immunostimulation of the genotypic variations and of specific motifs remains to be demonstrated. The environment surrounding CpG motifs in DNA, which may include inhibitory G-rich sequences, may also affect their biological activity (26, 29, 39).
The four CpG-rich sequences selected from the B. longum NCC2705 genome are double stranded and larger than the ODN used in the literature, such as BL07, OL-LB7, and ID35, which are generally about 20 bp long, and commercialized CpG ODN, such as ODN 1826 (14, 21, 22, 45, 52). These sequences include several CpG motifs and mimic the fragments likely to be released during bacterial lysis. The internalization pathway of large DNA fragments remains unknown, but our CpG-rich PCR products induced cytokine secretion in a dose-dependent manner: 1-μg/ml and 10-μg/ml concentrations of these sequences induced the pattern of cytokine, except that larger amounts of cytokines were produced in response to the 10-μg/ml dose (data not shown). This higher concentration, which has been used in several other studies (9, 52), was therefore used in all assays. Unlike LPS, which induces the production of large amounts of IL-6, these sequences induced the production of only TNF-α and MCP-1 in macrophages. The use of an inhibitor of TLR9 signaling confirms that these sequences induced their effects through the stimulation of TLR9 rather than TLR4. This observed activity was similar to that obtained with CpG-ODN T3, which stimulates expression of the genes encoding MCP-1 and TNF-α but not that of the gene encoding IL-10 (8). TNF-α secretion by macrophages was stimulated significantly more strongly by N2 than by H2 and H3 (N2 stimulation was five times stronger) or N3 (N2 stimulation was four times stronger).
N2 stimulated MCP-1 secretion by macrophages twice as strongly as H2 and H3. However, N2 contains a large number of type 1 and 2 CpG motifs, like H3, and has a percent G+C similar to that of the entire genome, like N3. We were therefore unable to establish a link between the immunostimulatory properties of these sequences, the number of CpG motifs they contain, and their percent G+C. The environment of CpG motifs in these sequences, as in genomic DNA, may also affect their immunostimulatory properties (26, 29, 39).
We assessed the variability of these CpG-rich sequences in the genus Bifidobacterium by two complementary methods, i.e., dot blot and PCR analyses.
Dot blotting showed the four sequences to be highly conserved throughout the genus (84% of the strains). The combination of a positive signal on dot blots with negative results for PCR indicated the occurrence of modifications or deletions at one or both ends of the sequence. Such a combination was observed for 71% (20/28) of the strains. For example, in silico analysis of the B. adolescentis ATCC 15703 genome showed N3 to be conserved, but a small number of modifications at the 5′ extremity of the sequence resulted in a mismatch with the primers used for PCR. Similarly, a large deletion in the 5′ ends of H2 and H3 led to negative PCR results. In contrast, the combination of a negative signal on dot blots with a positive result for PCR indicated modifications to the hybridization spot. Such a combination was obtained only for two strains. The combination of dot blot and PCR techniques showed that the frequency of sequence modifications was high for the genus (75%), with the exception of B. longum. This species predominantly displayed a high degree of conservation of the four sequences.
The B. breve, B. catenulatum/pseudocatenulatum group, and B. adolescentis genomes generally possess all four sequences, with or without modifications. The B. dentium strain has modified H2, H3, and N3 but does not have N2. B. bifidum strains have between zero and four of these sequences, which are frequently modified when present. Variations in the presence and abundance of CpG rich motifs depended on the bifidobacterial species considered. Modification in bifidobacterial establishment could lead to an inadequate maturation of the immune system (see reference 41 for discussion). Factors influencing intestinal microbiota composition, such as breastfeeding, may then affect the immunomodulatory effect of the microbiota.
However, the impact of these sequences on the immunity of the entire living strain needs to be clarified. Indeed, four of the B. longum strains containing the four sequences displayed immunostimulatory properties in a nonpathological monoxenic mouse model (32). They showed a strain-specific effect on TH1/TH2 balance which could be linked to modifications of N3 in two of them and of N2 in one of them. However, other structures, e.g., cell wall, cytoplasm, and secreted molecules, may interfere with the immune system (2, 13, 31, 33, 34).
Some authors have reported dysbiosis in allergic infants. Allergic status was associated with the presence of B. adolescentis, B. longum, or the B. catenulatum/pseudocatenulatum group or with a decrease in bifidobacterial diversity (see reference 38 for a review). Few in vitro studies have demonstrated a link between the immunostimulatory properties of Bifidobacterium and the characteristics of infants from whom the strains were isolated (country and allergic status) (11, 56). In our study, we did not observe any differences in the prevalence of the CpG-rich sequences according to the origin of the strain.
In conclusion, this study improves our understanding of the heterogeneity of the immunostimulatory properties of the genus Bifidobacterium at the genomic level. We identified immunostimulatory sequences within the Bifidobacterium genome that were able to induce TLR9 activation, which is known to trigger a TH1 orientation of the immune system. CpG-ODNs have been shown to counterbalance the TH2-dominant response when used as a vaccine adjuvant (49) and to have a protective effect against pro-TH2 diseases when added to the diet of monoxenic mice (48). Further studies of the ability of our sequences to induce a shift in the host immune response toward a TH1 orientation are required, including investigations of the protective properties of our sequences in an in vivo model of allergy. A high frequency of CpG motifs in the DNA of the bacterium may be important for the immunostimulatory properties of probiotic bifidobacteria.
Acknowledgments
The work of O. Ménard was supported by Danone Institute and the Société Française de Nutrition.
We particularly thank Gérard Corthier (UEPSD, INRA, Jouy en Josas, France) for his useful advice and Jacques Christian Darbord (Agence Générale des Equipements et Produits de Santé, Paris, France) for determining endotoxin activity.
Footnotes
Published ahead of print on 5 March 2010.
REFERENCES
- 1.Alfaleh, K., and D. Bassler. 2008. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst. Rev. 23:CD005496. [DOI] [PubMed] [Google Scholar]
- 2.Amrouche, T., Y. Boutin, G. Prioult, and I. Fliss. 2006. Effects of bifidobacterial cytoplasm, cell wall and exopolysaccharide on mouse lymphocyte proliferation and cytokine production. Int. Dairy J. 16:70-80. [Google Scholar]
- 3.Ashkar, A. A., and K. L. Rosenthal. 2002. Toll-like receptor 9, CpG DNA and innate immunity. Curr. Mol. Med. 2:545-556. [DOI] [PubMed] [Google Scholar]
- 4.Butel, M. J., N. Roland, A. Hibert, F. Popot, A. Favre, A. C. Tessèdre, M. Bensaada, A. Rimbault, and O. Szylit. 1998. Clostridial pathogenicity in experimental necrotising enterocolitis in gnotobiotic quails and protective role of bifidobacteria. J. Med. Microbiol. 47:391-399. [DOI] [PubMed] [Google Scholar]
- 5.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U. S. A. 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forchielli, M. L., and W. A. Walker. 2005. The role of gut-associated lymphoid tissues and mucosal defence. Br. J. Nutr. 93(Suppl. 1):S41-S48. [DOI] [PubMed] [Google Scholar]
- 8.Gao, J. J., V. Diesl, T. Wittmann, D. C. Morrison, J. L. Ryan, S. N. Vogel, and M. T. Follettie. 2002. Regulation of gene expression in mouse macrophages stimulated with bacterial CpG-DNA and lipopolysaccharide. J. Leukoc. Biol. 72:1234-1245. [PubMed] [Google Scholar]
- 9.Ghadimi, D., R. Folster-Holst, M. de Vrese, P. Winkler, K. J. Heller, and J. Schrezenmeir. 2008. Effects of probiotic bacteria and their genomic DNA on TH1/TH2-cytokine production by peripheral blood mononuclear cells (PBMCs) of healthy and allergic subjects. Immunobiology 213:677-692. [DOI] [PubMed] [Google Scholar]
- 10.Harmsen, H. J., A. C. Wildeboer-Veloo, G. C. Raangs, A. A. Wagendorp, N. Klijn, J. G. Bindels, and G. W. Welling. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61-67. [DOI] [PubMed] [Google Scholar]
- 11.He, F., H. Morita, A. C. Ouwehand, M. Hosoda, M. Hiramatsu, J. Kurisaki, E. Isolauri, Y. Benno, and S. Salminen. 2002. Stimulation of the secretion of pro-inflammatory cytokines by Bifidobacterium strains. Microbiol. Immunol. 46:781-785. [DOI] [PubMed] [Google Scholar]
- 12.Hessle, C., B. Andersson, and A. E. Wold. 2000. Gram-positive bacteria are potent inducers of monocytic interleukin-12 (IL-12) while gram-negative bacteria preferentially stimulate IL-10 production. Infect. Immun. 68:3581-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoarau, C., C. Lagaraine, L. Martin, F. Velge-Roussel, and Y. Lebranchu. 2006. Supernatant of Bifidobacterium breve induces dendritic cell maturation, activation, and survival through a Toll-like receptor 2 pathway. J. Allergy Clin. Immunol. 117:696-702. [DOI] [PubMed] [Google Scholar]
- 14.Iliev, I. D., H. Kitazawa, T. Shimosato, S. Katoh, H. Morita, F. He, M. Hosoda, and T. Saito. 2005. Strong immunostimulation in murine immune cells by Lactobacillus rhamnosus GG DNA containing novel oligodeoxynucleotide pattern. Cell. Microbiol. 7:403-414. [DOI] [PubMed] [Google Scholar]
- 15.Iliev, I. D., M. Tohno, D. Kurosaki, T. Shimosato, F. He, M. Hosoda, T. Saito, and H. Kitazawa. 2008. Immunostimulatory oligodeoxynucleotide containing TTTCGTTT motif from Lactobacillus rhamnosus GG DNA potentially suppresses OVA-specific IgE production in mice. Scand. J. Immunol. 67:370-376. [DOI] [PubMed] [Google Scholar]
- 16.Inoue, Y., N. Iwabuchi, J. Z. Xiao, T. Yaeshima, and K. Iwatsuki. 2009. Suppressive effects of Bifidobacterium breve strain M-16V on T-helper type 2 immune responses in a murine model. Biol. Pharm. Bull. 32:760-763. [DOI] [PubMed] [Google Scholar]
- 17.Karlin, S., J. Mrazek, and A. M. Campbell. 1997. Compositional biases of bacterial genomes and evolutionary implications. J. Bacteriol. 179:3899-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson, H., C. Hessle, and A. Rudin. 2002. Innate immune responses of human neonatal cells to bacteria from the normal gastrointestinal flora. Infect. Immun. 70:6688-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, H., S. Y. Lee, and G. E. Ji. 2005. Timing of Bifidobacterium administration influences the development of allergy to ovalbumin in mice. Biotechnol. Lett. 27:1361-1367. [DOI] [PubMed] [Google Scholar]
- 20.Kim, J. F., H. Jeong, D. S. Yu, S. H. Choi, C. G. Hur, M. S. Park, S. H. Yoon, D. W. Kim, G. E. Ji, H. S. Park, and T. K. Oh. 2009. Genome sequence of the probiotic bacterium Bifidobacterium animalis subsp. lactis AD011. J. Bacteriol. 191:678-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitagaki, K., V. V. Jain, T. R. Businga, I. Hussain, and J. N. Kline. 2002. Immunomodulatory effects of CpG oligodeoxynucleotides on established Th2 responses. Clin. Diagn. Lab. Immunol. 9:1260-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitazawa, H., H. Watanabe, T. Shimosato, Y. Kawai, T. Itoh, and T. Saito. 2003. Immunostimulatory oligonucleotide, CpG-like motif exists in Lactobacillus delbrueckii ssp. bulgaricus NIAI B6. Int. J. Food Microbiol. 85:11-21. [DOI] [PubMed] [Google Scholar]
- 23.Klinman, D. M. 2004. Immunotherapeutic uses of CpG oligodeoxynucleotides. Nat. Rev. Immunol. 4:249-258. [DOI] [PubMed] [Google Scholar]
- 24.Kok, R. G., A. de Waal, F. Schut, G. W. Welling, G. Weenk, and K. J. Hellingwerf. 1996. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl. Environ. Microbiol. 62:3668-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krieg, A. M. 2000. The role of CpG motifs in innate immunity. Curr. Opin. Immunol. 12:35-43. [DOI] [PubMed] [Google Scholar]
- 26.Krieg, A. M., A. K. Yi, S. Matson, T. J. Waldschmidt, G. A. Bishop, R. Teasdale, G. A. Koretzky, and D. M. Klinman. 1995. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374:546-549. [DOI] [PubMed] [Google Scholar]
- 27.Lammers, K. M., P. Brigidi, B. Vitali, P. Gionchetti, F. Rizzello, E. Caramelli, D. Matteuzzi, and M. Campieri. 2003. Immunomodulatory effects of probiotic bacteria DNA: IL-1 and IL-10 response in human peripheral blood mononuclear cells. FEMS Immunol. Med. Microbiol. 38:165-172. [DOI] [PubMed] [Google Scholar]
- 28.Lee, J. H., V. N. Karamychev, S. A. Kozyavkin, D. Mills, A. R. Pavlov, N. V. Pavlova, N. N. Polouchine, P. M. Richardson, V. V. Shakhova, A. I. Slesarev, B. Weimer, and D. J. O'Sullivan. 2008. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenert, P., A. K. Yi, A. M. Krieg, L. L. Stunz, and R. F. Ashman. 2003. Inhibitory oligonucleotides block the induction of AP-1 transcription factor by stimulatory CpG oligonucleotides in B cells. Antisense Nucleic Acid Drug Dev. 13:143-150. [DOI] [PubMed] [Google Scholar]
- 30.Li, Y., X. Qu, H. Yang, L. Kang, Y. Xu, B. Bai, and W. Song. 2005. Bifidobacteria DNA induces murine macrophages activation in vitro. Cell. Mol. Immunol. 2:473-478. [PubMed] [Google Scholar]
- 31.Medina, M., E. Izquierdo, S. Ennahar, and Y. Sanz. 2007. Differential immunomodulatory properties of Bifidobacterium longum strains: relevance to probiotic selection and clinical applications. Clin. Exp. Immunol. 150:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ménard, O., M. J. Butel, V. Gaboriau-Routhiau, and A. J. Waligora-Dupriet. 2008. Gnotobiotic mouse immune response induced by Bifidobacterium sp. strains isolated from infants. Appl. Environ. Microbiol. 74:660-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ménard, S., C. Candalh, J. C. Bambou, K. Terpend, N. Cerf-Bensussan, and M. Heyman. 2004. Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport. Gut 53:821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menard, S., D. Laharie, C. Asensio, T. Vidal-Martinez, C. Candalh, A. Rullier, F. Zerbib, F. Megraud, T. Matysiak-Budnik, and M. Heyman. 2005. Bifidobacterium breve and Streptococcus thermophilus secretion products enhance T helper 1 immune response and intestinal barrier in mice. Exp. Biol. Med. (Maywood) 230:749-756. [DOI] [PubMed] [Google Scholar]
- 35.Mullié, C., M. F. Odou, E. Singer, M. B. Romond, and D. Izard. 2003. Multiplex PCR using 16S rRNA gene-targeted primers for the identification of bifidobacteria from human origin. FEMS Microbiol. Lett. 222:129-136. [DOI] [PubMed] [Google Scholar]
- 36.Neujahr, D. C., C. F. Reich, and D. S. Pisetsky. 1999. Immunostimulatory properties of genomic DNA from different bacterial species. Immunobiology 200:106-119. [DOI] [PubMed] [Google Scholar]
- 37.Ohno, H., S. Tsunemine, Y. Isa, M. Shimakawa, and H. Yamamura. 2005. Oral administration of Bifidobacterium bifidum G9-1 suppresses total and antigen specific immunoglobulin E production in mice. Biol. Pharm. Bull. 28:1462-1466. [DOI] [PubMed] [Google Scholar]
- 38.Penders, J., E. E. Stobberingh, P. A. van den Brandt, and C. Thijs. 2007. The role of the intestinal microbiota in the development of atopic disorders. Allergy 62:1223-1236. [DOI] [PubMed] [Google Scholar]
- 39.Peter, M., K. Bode, G. B. Lipford, F. Eberle, K. Heeg, and A. H. Dalpke. 2008. Characterization of suppressive oligodeoxynucleotides that inhibit Toll-like receptor-9-mediated activation of innate immunity. Immunology 123:118-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Picard, C., J. Fioramonti, A. Francois, T. Robinson, F. Neant, and C. Matuchansky. 2005. Review article: bifidobacteria as probiotic agents—physiological effects and clinical benefits. Aliment. Pharmacol. Ther. 22:495-512. [DOI] [PubMed] [Google Scholar]
- 41.Round, J. L., R. M. O'Connell, and S. K. Mazmanian. 6 December 2009. Coordination of tolerogenic immune responses by the commensal microbiota. J. Autoimmun. 34:J220-J225. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satokari, R., T. Gronroos, K. Laitinen, S. Salminen, and E. Isolauri. 2009. Bifidobacterium and Lactobacillus DNA in the human placenta. Lett. Appl. Microbiol. 48:8-12. [DOI] [PubMed] [Google Scholar]
- 43.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U. S. A. 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sela, D. A., J. Chapman, A. Adeuya, J. H. Kim, F. Chen, T. R. Whitehead, A. Lapidus, D. S. Rokhsar, C. B. Lebrilla, J. B. German, N. P. Price, P. M. Richardson, and D. A. Mills. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U. S. A. 105:18964-18969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimosato, T., H. Kitazawa, S. Katoh, M. Tohno, I. D. Iliev, C. Nagasawa, T. Kimura, Y. Kawai, and T. Saito. 2005. Augmentation of T(H)-1 type response by immunoactive AT oligonucleotide from lactic acid bacteria via Toll-like receptor 9 signaling. Biochem. Biophys. Res. Commun. 326:782-787. [DOI] [PubMed] [Google Scholar]
- 46.Smits, H. H., A. J. van Beelen, C. Hessle, R. Westland, E. de Jong, E. Soeteman, A. Wold, E. A. Wierenga, and M. L. Kapsenberg. 2004. Commensal Gram-negative bacteria prime human dendritic cells for enhanced IL-23 and IL-27 expression and enhanced Th1 development. Eur. J. Immunol. 34:1371-1380. [DOI] [PubMed] [Google Scholar]
- 47.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sudo, N., Y. Aiba, N. Oyama, X. N. Yu, M. Matsunaga, Y. Koga, and C. Kubo. 2004. Dietary nucleic acid and intestinal microbiota synergistically promote a shift in the Th1/Th2 balance toward Th1-skewed immunity. Int. Arch. Allergy Immunol. 135:132-135. [DOI] [PubMed] [Google Scholar]
- 49.Sugai, T., M. Mori, M. Nakazawa, M. Ichino, T. Naruto, N. Kobayashi, Y. Kobayashi, M. Minami, and S. Yokota. 2005. A CpG-containing oligodeoxynucleotide as an efficient adjuvant counterbalancing the Th1/Th2 immune response in diphtheria-tetanus-pertussis vaccine. Vaccine 23:5450-5456. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi, N., H. Kitazawa, N. Iwabuchi, J. Z. Xiao, K. Miyaji, K. Iwatsuki, and T. Saito. 2006. Immunostimulatory oligodeoxynucleotide from Bifidobacterium longum suppresses Th2 immune responses in a murine model. Clin. Exp. Immunol. 145:130-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi, N., H. Kitazawa, N. Iwabuchi, J. Z. Xiao, K. Miyaji, K. Iwatsuki, and T. Saito. 2006. Oral administration of an immunostimulatory DNA sequence from Bifidobacterium longum improves Th1/Th2 balance in a murine model. Biosci. Biotechnol. Biochem. 70:2013-2017. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi, N., H. Kitazawa, T. Shimosato, N. Iwabuchi, J. Z. Xiao, K. Iwatsuki, S. Kokubo, and T. Saito. 2006. An immunostimulatory DNA sequence from a probiotic strain of Bifidobacterium longum inhibits IgE production in vitro. FEMS Immunol. Med. Microbiol. 46:461-469. [DOI] [PubMed] [Google Scholar]
- 53.Talati, A. J., H. J. Kim, Y. I. Kim, A. K. Yi, and B. K. English. 2008. Role of bacterial DNA in macrophage activation by group B streptococci. Microbes Infect. 10:1106-1113. [DOI] [PubMed] [Google Scholar]
- 54.Tross, D., L. Petrenko, S. Klaschik, Q. Zhu, and D. M. Klinman. 2009. Global changes in gene expression and synergistic interactions induced by TLR9 and TLR3. Mol. Immunol. 46:2557-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watson, J. L., and D. M. McKay. 2006. The immunophysiological impact of bacterial CpG DNA on the gut. Clin. Chim. Acta 364:1-11. [DOI] [PubMed] [Google Scholar]
- 56.Young, S. L., M. A. Simon, M. A. Baird, G. W. Tannock, R. Bibiloni, K. Spencely, J. M. Lane, P. Fitzharris, J. Crane, I. Town, E. Addo-Yobo, C. S. Murray, and A. Woodcock. 2004. Bifidobacterial species differentially affect expression of cell surface markers and cytokines of dendritic cells harvested from cord blood. Clin. Diagn. Lab. Immunol. 11:686-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, H., F. Qu, and L. H. Zhu. 1993. Isolation of genomic DNAs from plants, fungi and bacteria using benzyl chloride. Nucleic Acids Res. 21:5279-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]