Abstract
The rdpA and sdpA genes encode two enantioselective α-ketoglutarate-dependent dioxygenases catalyzing the initial step of microbial degradation of the chiral herbicide (R,S)-2-(2,4-dichlorophenoxy)propionate (R,S-dichlorprop). Primers were designed to assess abundance and transcription dynamics of rdpA and sdpA genes in a natural agricultural soil. No indigenous rdpA genes were detected, but sdpA genes were present at levels of approximately 103 copies g of soil−1. Cloning and sequencing of partial sdpA genes revealed a high diversity within the natural sdpA gene pool that could be divided into four clusters by phylogenetic analysis. BLASTp analysis of deduced amino acids revealed that members of cluster I shared 68 to 69% identity, cluster II shared 78 to 85% identity, cluster III shared 58 to 64% identity, and cluster IV shared 55% identity to their closest SdpA relative in GenBank. Expression of rdpA and sdpA in Delftia acidovorans MC1 inoculated in soil was monitored by reverse transcription quantitative real-time PCR (qPCR) during in situ degradation of 2 and 50 mg kg−1 of (R,S)-dichlorprop. (R,S)-Dichlorprop amendment created a clear upregulation of both rdpA and sdpA gene expression during the active phase of 14C-labeled (R,S)-dichlorprop mineralization, particularly following the second dose of 50 mg kg−1 herbicide. Expression of both genes was maintained at a low constitutive level in nonamended soil microcosms. This study is the first to report the presence of indigenous sdpA genes recovered directly from natural soil and also comprises the first investigation into the transcription dynamics of two enantioselective dioxygenase genes during the in situ degradation of the herbicide (R,S)-dichlorprop in soil.
It has been reported that as many as 25% of all chemical pesticides exist as chiral molecules (12), otherwise known as enantiomers. (R,S)-2-(2,4-Dichlorophenoxy)propionate [(R,S)-dichlorprop] is a chiral herbicide used mostly in agriculture, lawn care, and industry for the control of broad-leaved weeds and belongs to the family of phenoxyalkanoic herbicides, which also includes other racemates such as the structurally related phenoxypropionate (R,S)-2-(4-chloro-2-methylphenoxy)propionate [(R,S)-mecoprop]. The R enantiomer of both dichlorprop and mecoprop is the only form providing herbicidal activity, and several European countries have now started implementing stricter regulations on the use of chiral herbicides, e.g., banning racemic dichlorprop in favor of R-enantiomer-specific formulations (10). Nevertheless, it is important to evaluate the impact of the racemic mixtures as not only are they still applied in some parts of the world, but instances of enantiomerization have been reported [e.g., (S)-dichlorprop converting to (R)-dichlorprop or vice versa] (26).
Degradation of (R,S)-dichlorprop in the environment is mainly mediated by biological activity (22, 26, 36). As in the case of other chiral pesticides (6, 35), preferential degradation of either the R or the S enantiomer of dichlorprop during microbial degradation has been reported (13, 22, 26). (R,S)-Dichlorprop enantioselectivity is largely dependent upon the behavior of the microbial community at hand and can be influenced by factors such as pH (3, 23), soil type (36), and the environment (e.g., soil, water, or sludge) (4, 13, 47). The enantioselectivity observed during the microbial transformation of chiral dichlorprop implies that enzymes involved in degradation of this compound are somehow able to differentiate between enantiomers. For phenoxyalkanoic acid herbicides, the first step in the catabolic pathway usually begins with etherolytic cleavage of the side chain that is catalyzed by an α-ketoglutarate-dependent dioxygenase, thus converting the herbicides into corresponding phenols. For phenoxyacetate herbicides such as achiral 2,4-dichlorophenoxyacetate (2,4-D) and 4-chloro-2-methylphenoxyacetate (MCPA), this dioxygenase is typically encoded by the tfdA gene (2, 7, 11). Although a few strains are capable of degrading at least one enantiomer of dichlorprop using TfdA, increasing amounts of data have recently revealed the existence of two α-ketoglutarate-dependent dioxygenases distinct from TfdA that exhibit opposite stereoselectivities toward degradation of (R,S)-dichlorprop and (R,S)-mecoprop. These dioxygenases, designated RdpA and SdpA, target each of the two R and S enantiomers, respectively (31, 45). Unfortunately, studies focusing on enantiospecific degradative enzymes are far more scarce than studies on the involvement of TfdA during 2,4-D and MCPA degradation.
Although degradation of (R,S)-dichlorprop in various environments is well reported (for a review, see reference 32), very few bacterial strains able to oxidize both enantiomers of dichlorprop using the RdpA and SdpA enzymes have been isolated to date. The best described are Sphingomonas herbicidovorans MH (16, 48) and Delftia acidovorans MC1 (28, 29), from which rdpA and sdpA genes were first isolated and sequenced (30, 39). Enzyme activity from these strains has shown that RdpA dioxygenase is generally highly specific to the R enantiomer of 2-phenoxypropionates and shows very weak activity toward phenoxyacetates, whereas SdpA is enantioselective to the S enantiomer but can also convert certain phenoxyacetates such as 2,4-D and MCPA with various catalytic activities (27, 31, 44, 45). The genetic background has only recently become available and shows that rdpA sequences reported to date are nearly identical while sdpA sequences between S. herbicidovorans MH and D. acidovorans MC1 are far more variable (30, 39). In D. acidovorans MC1, plasmid-borne rdpA and sdpA genes were deduced to be constitutively expressed based on enzyme activity during induction studies with (R)- and (S)-dichlorprop in mineral salts solution (MSS) (29). Studies of S. herbicidovorans MH have looked specifically at mRNA and found that both genes were constitutively expressed (30), although earlier studies on enzyme activity reported constitutive activity only for SdpA (33). It is noteworthy to mention that such induction studies conducted on pure cultures grown and tested in laboratory media may not always reflect the reality observed in more complex soil environments.
Although RdpA and SdpA from a few dichlorprop-degrading strains have been biochemically characterized, there is scarcely any information available regarding the rdpA and sdpA genes when it comes to their presence, natural abundance, and activation in soil environments. In an effort to better understand the diversity of genes involved in mecoprop degradation, Zakaria et al. (46) reported the presence of rdpA and sdpA genes from soil enrichment cultures grown on either (R)-mecoprop or (R,S)-mecoprop but did not provide any quantitative data or time course analysis. This might be due to the difficulties often encountered while working with direct quantification of nucleic acids in soil. Recent advances in detection of mRNA in soil have helped overcome some of these problems and now make it possible to achieve reliable quantitative transcription analysis of functional genes in soil (34).
This work aims to provide an initial report on the natural abundance and the in situ transcription dynamics of rdpA and sdpA genes during degradation of (R,S)-dichlorprop in soil. New sets of specific primers for detection by SYBR green quantitative real-time PCR (qPCR) were designed and tested to target rdpA and sdpA genes. Soil DNA extracts were screened for the presence of these genes by qPCR followed by cloning and sequencing for phylogenetic analysis. To investigate regulation of rdpA and sdpA gene expression under in situ conditions, D. acidovorans MC1 cells were inoculated directly into soil microcosms amended with (R,S)-dichloprop and reverse transcription combined with qPCR was used to follow transcription dynamics over time. Herbicide degradation was followed by generating progress curves of accumulated [14C]CO2 evolved from (R,S)-[14C]dichlorprop mineralization.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
D. acidovorans MC1 and S. herbicidovorans MH (DSMZ, Braunschweig, Germany; DSM11019), both carrying rdpA and sdpA genes, were grown and maintained under selective conditions in mineral salts solution (MSS) (29) supplemented with 400 mg liter−1 of (R,S)-dichlorprop. Cupriavidus necator AEO106(pRO101) carrying the tfdA gene was grown in LB medium supplemented with 10 mg ml−1 tetracycline. Incubations for growth of all strains were performed at 28°C with shaking at 200 rpm.
Soil microcosm setup and sampling.
Organic agricultural topsoil classified as Typic Argiudoll (USDA soil taxonomy) with 19% clay, 18% silt, 62% sand, and 1.2% carbon and pH 7.2 was obtained from Sjællands Odde, Zeeland, Denmark, and stored at 4°C until use. Prior to setup of microcosms, soil was passed through a 4-mm sieve and water content was determined by baking a subsample at 105°C for 24 h. To prepare for inoculation into soil, D. acidovorans MC1 cells were grown to late stationary phase as indicated above in MSS supplemented with (R,S)-dichlorprop and harvested by centrifugation at 8,000 × g for 4 min at 4°C. Cells were washed twice and resuspended in 1× phosphate-buffered saline (PBS; 25 mM phosphate buffer, 125 mM NaCl, pH 7.4), and 1 ml was transferred to nonselective PYE medium (29) for growth overnight. Cells were once again harvested by centrifugation, washed twice, and resuspended in 1× PBS. An optical density (OD) of 1 was estimated to correspond to 5 × 108 CFU ml−1, and cell suspensions were adjusted in order to apply 1 ml per soil microcosm to reach a cell density of 5 × 107 cells g of soil−1. After addition of cells, soil water content was adjusted to 18% [wet weight; including subsequent addition of (R,S)-dichlorprop solution] with sterile Milli-Q water and soil was mixed mechanically in plastic bags and divided into glass flasks each containing 30-g soil subsamples. A stock solution of unlabeled (R,S)-dichlorprop was prepared in dimethyl sulfoxide (DMSO; VWR, Fontenay-sous-Bois, France) whereas (R,S)-[ring-U-14C]dichlorprop (specific activity, 3.56 MBq mg−1; radiochemical purity, 97.41%; Izotop, Budapest, Hungary) stock solution was prepared in methanol (LabScan, Dublin, Ireland). From these, an (R,S)-dichlorprop solution was prepared with 0.1 M Na2HPO4 (pH 8.45) and applied to each microcosm as a top amendment in a fixed volume of 1.1 ml to obtain a final concentration of 2 or 50 mg kg−1 unlabeled (R,S)-dichlorprop containing trace amounts of labeled (R,S)-dichlorprop (28,000 dpm g soil−1). Control microcosms containing no herbicide and only 0.1 M Na2HPO4 were also prepared. Finally, a glass vial containing 1 ml of 1 M NaOH was placed on top of the soil in each flask to capture [14C]CO2 evolved during (R,S)-[14C]dichlorprop mineralization and bottles were closed airtight and incubated in the dark at 20°C. Soil sampling and replacement of [14C]CO2 traps were done at appropriate time intervals during the course of herbicide degradation following the method described by Nicolaisen et al. (34). Furthermore, triplicate soil samples were taken before addition of herbicide and degrader strain to evaluate the natural diversity of rdpA and sdpA genes. All soil samples (0.5 g) were immediately frozen in liquid nitrogen and stored at −70°C until nucleic acid extraction. A second amendment with (R,S)-dichlorprop was made at day 13 after mineralization of the first amendment had ceased.
Mineralization curve analysis.
Cumulative (R,S)-[14C]dichlorprop mineralization data were obtained as described previously (34) and corrected for the removal of 0.5-g soil samples taken over time during herbicide degradation in (R,S)-[14C]dichlorprop-amended soils.
Nucleic acid extraction.
DNA and RNA were coextracted from soil samples as previously described (34) with the following modification: 1 μl of glycogen (Roche, Basel, Switzerland) was added during incubation with polyethylene glycol (PEG 6000) to help nucleic acid precipitation. Samples were kept on ice during the whole procedure, and final extracts were resuspended in 50 μl of DNase- and RNase-free diethyl pyrocarbonate (DEPC)-treated water (Fluka; Sigma-Aldrich, Saint Louis, MO) and stored at −70°C.
DNase treatment and reverse transcription.
To eliminate contaminating DNA, 8-μl aliquots of each nucleic acid sample were treated with 2 units of RQ1 RNase-free DNase I (Promega, Madison, WI) according to the manufacturer's protocol. Immediately after, 4-μl subsamples of each DNase-treated extract were used as a template for reverse transcription with the Omniscript reverse transcription kit (Qiagen, Crawley, United Kingdom). Reaction mixtures were prepared using 400 ng of random hexamer primers (Promega), 4 U Super RNase inhibitor (Ambion, Austin, TX), and reagents provided in the kit for a final volume of 20 μl. Incubation for 90 min at 37°C was followed by inactivation of the transcriptase for 10 min at 95°C. Resulting cDNA samples were then stored at −20°C until use for quantitative real-time PCR. In parallel, DNase-treated control reaction mixtures were prepared for each RNA sample without addition of the reverse transcriptase to ensure that no DNA contamination was present.
Primer design.
PCR primers for SYBR green quantitative real-time PCR (qPCR) were designed using the BioEdit v.7.0.9.0 sequence alignment software (14) along with Primer-BLAST. Primer-BLAST is a combination of the Primer 3 primer design software (37) and BLAST (1) that searches the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html) in order to avoid nontarget sequence amplification. Design of rdpA primers was based on the alignment of conserved regions from the 16 available rdpA sequences retrieved from the GenBank database, while sdpA primer design was based on sequences of the only three available strains known to carry sdpA: D. acidovorans MC1 (AY327575), S. herbicidovorans MH (AJ628860), and Rhodoferax sp. P230 (DQ406818). The two sdpA reverse primers differ in composition by only 2 nucleotides (Table 1) and were used separately instead of degenerate primers to enable specific detection of the gene from D. acidovorans MC1 (sdpA_r1). An equal mixture of sdpA_r1 and sdpA_r2 reverse primers was used only when screening for the presence of naturally occurring sdpA sequences. Specificity of each primer pair was further tested by qPCR (conditions indicated below) using DNA extracted as described above from pure cultures of D. acidovorans MC1 and S. herbicidovorans MH for positive controls while DNA extracted from C. necator AEO106(pRO101) served as a negative control. PCR amplification products were separated by 1.5% agarose gels, and fragments were purified and sequenced.
TABLE 1.
Oligonucleotide primers designed and used in this study for SYBR green qPCR
| Target gene | Primer | Model gene(s) used for primer design | Sequence (5′→3′) | Product length (bp)a |
|---|---|---|---|---|
| rdpA | rdpA_f | 16 known rdpA sequences in GenBank | CACTTACCAGGTCATCTA | |
| rdpA_r | GTGTACATGCTCAGAAAAC | 285 | ||
| sdpA | sdpA_f | D. acidovorans MC1, S. herbicidovorans MH, Rhodoferax sp. P230 | ATGCCGACAGCACCTACAT | |
| sdpA_r1 | D. acidovorans MC1 | GGTCTCGGGATGCACCTT | 287 | |
| sdpA_r2 | S. herbicidovorans MH, Rhodoferax sp. P230 | GGTCTCGGGATGGATCTT | 290 |
PCR product lengths for different combinations of forward and reverse primers are indicated. Primer sdpA_f in combination with sdpA_r1 produces a 287-bp product while sdpA_f in combination with sdpA_r2 produces a 290-bp product.
Quantitative real-time PCR.
All qPCRs were performed in the Mx3000P qPCR system (Stratagene, La Jolla, CA) using the Brilliant SYBR Green QPCR Master Mix (Stratagene). Reaction mixtures were prepared with 10 μl 2× Brilliant QPCR Master Mix (including SYBR green 1 dye and PCR buffer, deoxynucleoside triphosphates [dNTPs], SureStart Taq DNA polymerase, and 5 mM MgCl2); 0.4 μM (each) primer (DNA Technology A/S, Risskov, Denmark); 20 μg of bovine serum albumin (New England Biolabs, Ipswich, MA); 2 μl of template DNA (dilution 1/10), cDNA, or DNase-treated RNA; and DNase/RNase-free water (Sigma-Aldrich) for a final volume of 20 μl. The qPCR thermal cycling program was the same for all primer sets and consisted of an initial 15-min incubation at 95°C for enzyme activation, 40 cycles of 45 s at 95°C for denaturation, 45 s at 55°C for annealing, and 1 min at 72°C for extension with a final elongation for 2 min at 72°C. A specific dissociation curve with melting temperature (Tm) of the amplified product was generated at the end of the qPCR program by including a cycle of 95°C for 1 min and 55°C for 30 s, finally reaching 95°C by increments of 0.5°C s−1, each followed by a fluorescence acquisition step. These curves were used to ensure PCR product specificity by observation of a single melting peak, and results were further confirmed by the presence of single PCR fragments of the expected size in 1.5% agarose gels.
Standard curves generated with DNA extracted from soils spiked with D. acidovorans MC1.
To enable quantitative detection of both rdpA and sdpA genes, a standard curve was generated by adding 100 μl of D. acidovorans MC1 cells (following the same method as for inoculation into soil microcosms) to 7 Bio101 Lysing Matrix E tubes (MP Biomedicals, Solon, OH) each containing 500 mg of soil to reach cell densities of 108, 107, 106, 105, 104, 103, and 102 cells g of soil−1. Nucleic acids were extracted as described above and used as templates for qPCR. The resulting DNA standard curve was used to quantify both DNA and mRNA from rdpA and sdpA with gene expression data being presented as ratios of mRNA normalized to DNA as first described by Nicolaisen et al. (34). Recovery rates of nucleic acids were regarded as equal for each extraction based on previously obtained extraction efficiencies from this soil where mRNA/DNA ratios remained unaffected over a wide dynamic range despite a low recovery of mRNA (34). qPCR runs always included a triplicate standard curve, and amplification efficiency (E) was calculated from the slope of the standard curve using the equation E = 10(−1/slope) −1.
qPCR analysis of all samples collected during the soil microcosm experiments was done on the DNA, cDNA, and DNase-treated RNA fractions using the newly designed rdpA and sdpA primer sets shown in Table 1. In addition, qPCR screening for the presence of naturally occurring rdpA and sdpA genes in soil was done prior to (R,S)-dichlorprop exposure and degrader strain inoculation. In these cases, reaction mixtures were prepared using a mix of 0.2 μM (each) sdpA reverse primer in order to target all known sequences.
Cloning and sequencing.
No PCR product for rdpA could be detected in soil samples prior to herbicide exposure. However, PCR products obtained by the sdpA primers were used to construct clone libraries. Briefly, PCR was performed using the sdpA_f and sdpA_r1/sdpA_r2 primers and reaction mixtures were prepared containing 4 μl of 5× Phusion GC buffer (Finnzymes, Helsinki, Finland), 800 μM dNTPs (Fermentas, Burlington, Canada), 0.4 μM forward sdpA primer, 0.2 μM (each) reverse sdpA primer, 20 μg of bovine serum albumin (New England Biolabs), 1.25 U of Phusion DNA polymerase (Finnzymes), and 2 μl of template DNA and DNase/RNase-free water (Sigma-Aldrich) for a final volume of 20 μl. The thermal cycling program was as follows: 98°C for 30 s followed by 35 cycles of 10 s at 98°C, 30 s at 62°C, and 30 s at 72°C and a final extension of 10 min at 72°C. PCR products were run through a SeaKem GTG 1% agarose gel, and fragments corresponding to sdpA were excised and purified with the Qiaquick gel extraction kit according to the manufacturer's protocol (Qiagen). Cleaned PCR products were ligated into the pCR2.1-TOPO vector and transformed into TOP10 chemically competent Escherichia coli cells following the manufacturer's protocol (Invitrogen, Carlsbad, CA). Forty clones were randomly selected, and both DNA strands were sequenced (Uppsala Genome Center, Uppsala, Sweden) using the M13 universal primers (Invitrogen). Resulting forward and reverse sequences were assembled into contigs and submitted to a BLASTn and BLASTp query of the GenBank database for nucleic acid and putative amino acid sequences, respectively, in order to find most homologous sequences.
Phylogenetic analysis of the sdpA gene sequences.
The cloned gene sequences obtained with the sdpA primers were aligned with related sequences retrieved from GenBank (June 2009) using DIALIGN (25). Sites where nucleotide alignments were not resolved were excluded, and a final 190-bp alignment was used for phylogenetic analysis. The phylogenetic tree was constructed with the PAUP 4.0b10 software (40) using a neighbor-joining (NJ) analysis with 1,000 iterations to calculate bootstrap values. Tree topology was confirmed by maximum likelihood and parsimony analysis.
Nucleotide sequence accession numbers.
The nucleotide sequences of sdpA determined in this study have been deposited in the GenBank database under the accession numbers GQ922849 to GQ922883.
RESULTS
Primer design and evaluation.
Multiple alignment of the 16 rdpA sequences available in GenBank revealed highly conserved sequences. Aside from rdpA in D. acidovorans MC1, S. herbicidovorans MH, and Rhodoferax sp. P230 [all previously described for their involvement in (R,S)-dichlorprop degradation] (9, 16, 28), sequences belonged to Burkholderia sp. and uncultured bacterial clones. The rdpA primers designed from this alignment amplified a 285-bp region (Table 1). No product was detected when testing with C. necator AEO106(pRO101), which carries the tfdA gene instead of rdpA, whereas D. acidovorans MC1 and S. herbicidovorans MH, known to harbor rdpA, yielded amplicons of the desired length and sequence composition (data not shown). When tested by qPCR, dissociation curves produced a single melting peak with a Tm of approximately 89°C, thus indicating the presence of a single amplicon.
The sdpA primers were designed based on the alignment of three sequences. S. herbicidovorans MH and Rhodoferax sp. P230 have identical sdpA nucleic acid sequences while D. acidovorans MC1 shares only 75% identity to this sequence, thus restraining primer design options. A 19-mer forward primer was designed from a conserved region while two 18-mer reverse primers targeting the same area (differing only by 2 bp) had to be created for lack of a suitable consensus region (Table 1). The sdpA primers designed here were found suitable for detection of sdpA from all known strains, while this was not the case for previously published primers due to the presence of several mismatches at both primer hybridization sites for the S. herbicidovorans MH sequence (Fig. 1). The basis for using two separate reverse primers instead of one mix of degenerate primers was the intention to use sdpA_r1 to enable detection of the gene in D. acidovorans MC1 during transcription analysis while a combination of sdpA_r1/sdpA_r2 enabled us to better study the natural diversity of sdpA in soil. The strains tested for rdpA primer specificity were also used to test sdpA primer specificity and generated amplicon lengths as indicated in Table 1. Dissociation curves following qPCR produced a single melting peak at approximately 91.5°C for D. acidovorans MC1 and 90°C for S. herbicidovorans MH. For both rdpA and sdpA, primer specificity was further confirmed by the presence of a single band during agarose gel electrophoresis of qPCR products and sequencing of the amplified product (data not shown).
FIG. 1.
Alignment of partial sdpA nucleic acid sequences from D. acidovorans MC1 (sdpA_MC1) and S. herbicidovorans MH (sdpA_MH) showing primer hybridization sites. Underlined sequences indicate primer sites used by Schleinitz et al. (39) based on the D. acidovorans MC1 SdpA peptide sequence. Primer sites designed in this study are boxed and based on all available sdpA nucleic acid sequences.
Standard curve performance in qPCR.
Extensive testing of primer pairs is essential when generating a suitable standard curve for use in quantitative PCR since factors such as secondary structures, PCR inhibitors, or amplicon length can affect primer amplification efficiency. Both standard curves in this work were established from serial dilutions of D. acidovorans MC1 cells inoculated into soil anticipated to represent DNA and mRNA copy numbers, as rdpA and sdpA are presumably single-copy genes in this strain (39). The rdpA standard curve was linear over 6 orders of magnitude (r2 = 1.00) from 108 to 103 copies g of soil−1, while the standard curve for sdpA was linear over 4 orders of magnitude (r2 = 0.999), ranging from 108 to 105 copies g of soil−1. The slope of the standard curve remained consistent between different runs and indicated qPCR efficiencies of approximately 82.5% and 102% for rdpA and sdpA, respectively. The 10-fold dilutions ranging from 104 to 102 copies g of soil−1 for sdpA did not follow a linear relationship and could not be used for accurate quantification. This was caused by the presence of indigenous sdpA genes (see below), which in turn created two melting peaks during the dissociation curve of qPCR products with Tm values of 91.5°C and 90°C, respectively. Due to the inevitable background of sdpA genes in soil, linear extrapolation of the standard curve was necessary for quantification of sdpA genes at levels below 105 copies g of soil−1.
Natural abundance of the rdpA and sdpA genes in agricultural soil.
We first determined the natural prevalence of the rdpA and sdpA genes in the Sjællands Odde agricultural soil. No qPCR product with the rdpA primers could be detected in DNA extracts recovered from agricultural soil prior to (R,S)-dichlorprop exposure (data not shown), indicating that indigenous rdpA genes are present at a level below the detection limit of the qPCR or they are simply absent in this environment. In contrast, sdpA was consistently detected at levels below 105 copies g of soil−1, corresponding approximately to 103 copies g of soil−1 according to the extrapolated standard curve, when soil DNA extracts were subjected to qPCR with a Tm value around 90°C.
Diversity of natural sdpA gene pool.
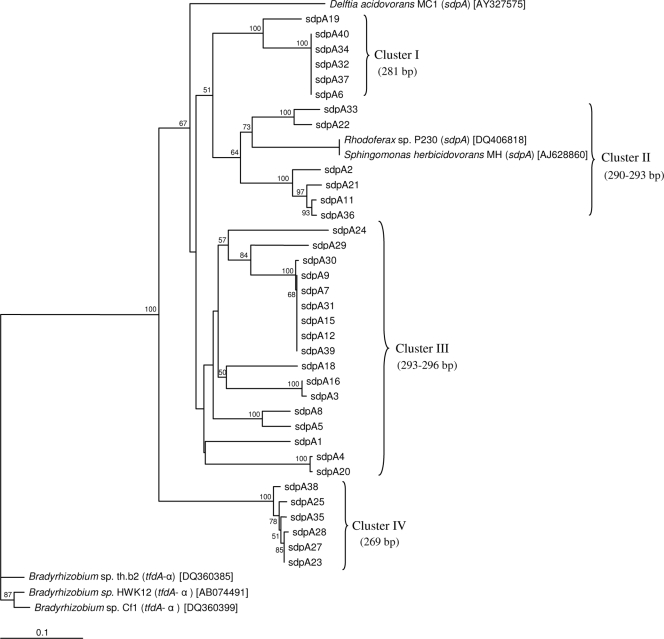
To confirm that products amplified by nonquantitative PCR were indeed sdpA, 35 clones were recovered from soil DNA extracts and successfully sequenced and assembled into contiguous consensus sequences. Analysis revealed differences in both base composition and sequence length. None of the partial gene sequences shared more than 82% identity to other known sdpA sequences as verified by BLASTn analysis of GenBank. Sequence lengths varied between 269 bp and 296 bp and were shortened to an unambiguous 190-bp region obtained by multiple alignment with DIALIGN. Although phylogenetic tree construction was based on this fixed 190-bp region, NJ analysis was able to separate sdpA clones into four major clusters (Fig. 2). Cluster I was composed exclusively of 281-bp sequences, cluster II included sequences closest to sdpA from S. herbicidovorans MH and Rhodoferax sp. P230 with lengths between 290 and 293 bp, and cluster III had lengths ranging from 293 to 296 bp and showed the largest diversity in base composition between sequences, while cluster IV had the shortest sequence of 269 bp. This clustering was reiterated by the tree topology obtained with maximum likelihood and parsimony analysis methods.
FIG. 2.
Phylogenetic tree of partial sdpA sequences recovered from soil prior to (R,S)-dichlorprop exposure. Analysis was based on 190-bp unambiguously aligned sdpA sequences using the NJ method. Bootstrap values of >50% from 1,000 replicate samplings are displayed at the nodes. The scale bar indicates 0.1 nucleotide substitution per site. Values in parentheses below cluster numbers indicate lengths in base pairs obtained from sequencing of PCR products amplified with the sdpA primers. Strains carrying the tfdA-α gene were used as an outgroup. Aside from sdpA, tfdA-α genes are the most closely related to the sequences recovered in this study as determined by BLASTn analysis of the GenBank database.
The phylogenetic diversity observed in the nucleic acid sequences was maintained when translated into putative amino acid sequences. BLASTp analysis indicated that all SdpA amino acid sequences shared the highest identity to both S. herbicidovorans and Rhodoferax sp. P230 (identical sequence composition) or to D. acidovorans MC1 (Table 2). Percent identities of partial SdpA sequences to their closest relatives in GenBank revealed a clear separation that corresponded well to each of the four clusters created by the sdpA phylogenetic tree (Fig. 2). The sdpA sequences ascribed to cluster I shared 68 to 69% identity, those ascribed to cluster II shared 78 to 85% identity, those ascribed to cluster III shared 58 to 64% identity, and those ascribed to cluster IV shared 55% identity to their closest match.
TABLE 2.
Identities of deduced partial amino acid sequences of SdpA recovered from agricultural soil, obtained by BLASTp analysis of the GenBank database
| Uncultured bacterium clone(s) | Clustera | Amino acid lengthb | % Identityc | Closest relative(s) in GenBankd |
|---|---|---|---|---|
| sdpA19 | I | 93 | 69 (67/97) | S. herbicidovorans MH, Rhodoferax sp. P230 |
| sdpA40, sdpA34, sdpA32, sdpA37, sdpA6 | I | 93 | 68 (66/97) | S. herbicidovorans MH, Rhodoferax sp. P230 |
| sdpA33, sdpA22 | II | 97 | 78 (76/97) | S. herbicidovorans MH, Rhodoferax sp. P230 |
| sdpA2, sdpA21, sdpA11, sdpA36 | II | 96 | 85 (82/96) | S. herbicidovorans MH, Rhodoferax sp. P230 |
| sdpA24 | III | 98 | 63 (62/98) | S. herbicidovorans MH, Rhodoferax sp. P230 |
| sdpA29 | III | 98 | 58 (57/98) | D. acidovorans MC1 |
| sdpA30, sdpA9, sdpA7, sdpA31, sdpA15, sdpA12, sdpA39 | III | 98 | 60 (59/98) | D. acidovorans MC1 |
| sdpA18 | III | 97 | 64 (63/97) | S. herbicidovorans MH, Rhodoferax sp. P230 |
| sdpA16 | III | 98 | 60 (59/98) | S. herbicidovorans MH, Rhodoferax sp. P230 |
| sdpA3 | III | 98 | 61 (60/98) | S. herbicidovorans MH, Rhodoferax sp. P230 |
| sdpA8, sdpA5 | III | 98 | 62 (61/98) | S. herbicidovorans MH, Rhodoferax sp. P230 |
| sdpA1 | III | 97 | 63 (62/97) | S. herbicidovorans MH, Rhodoferax sp. P230 |
| sdpA4, sdpA20 | III | 98 | 58 (58/99) | D. acidovorans MC1 |
| sdpA38, sdpA25, sdpA35, sdpA28, sdpA27, sdpA23 | IV | 89 | 55 (53/95) | D. acidovorans MC1 |
Numbers indicate clustering of the sdpA clones as based on the phylogenetic tree constructed from 190-bp sdpA gene sequences (Fig. 1).
Partial amino acid sequences were translated from 269- to 296-bp nucleic acid sequences obtained by PCR-based cloning and sequencing using the sdpA primer pair sdpA_f and sdpA_r1/r2.
Percent sequence identity between sequenced clone and sequence of closest relative in GenBank (number of identical amino acid hits/total number of amino acids).
Strains represent the closest matches obtained by BLASTp analysis. Strain accession numbers for GenBank are the same as those indicated in Materials and Methods. More than one strain being listed indicates identical results.
Expression of rdpA and sdpA is regulated by (R,S)-dichlorprop exposure in soil.
Transcript levels of rdpA and sdpA mRNA in (R,S)-dichlorprop-amended soils that were not inoculated with a degrader strain were too low for quantitative detection within the time frame of this study. Therefore, D. acidovorans MC1 was used as a model strain to study regulation of rdpA and sdpA genes in agricultural soil. Figures 3 and 4 show the progress curves of rdpA and sdpA mRNA and DNA during in situ (R,S)-[14C]dichlorprop mineralization activity at first and second amendments of the soil with 2 mg kg−1 and 50 mg kg−1 of (R,S)-dichlorprop, as well as progress curves for soils receiving no herbicide. Onset of mineralization was detected immediately and without lag for soils amended with 2 mg kg−1 and 50 mg kg−1 of (R,S)-dichlorprop. Hence, the progress curves of accumulated [14C]CO2 from mineralization of 2 mg kg−1 and 50 mg kg−1 (R,S)-dichlorprop could be divided into two segments: a steep segment and a plateau.
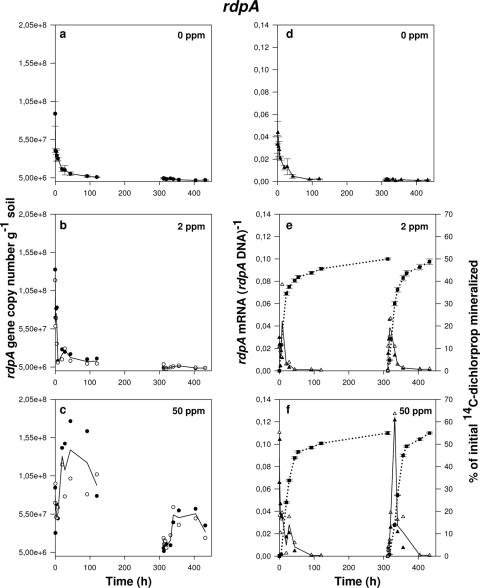
FIG. 3.
Progress curves of rdpA gene copy numbers (a to c), rdpA transcription activity represented as ratios of rdpA mRNA to rdpA DNA (d to f), and (R,S)-[14C]dichlorprop mineralization (e and f) during exposure of Delftia acidovorans MC1 to different concentrations of (R,S)-dichlorprop in soil: 0 mg kg−1 (a and d), 2 mg kg−1 (b and e), and 50 mg kg−1 (c and f). Amendment with (R,S)-dichlorprop was done at 0 h followed by a second amendment at 312 h. Data from microcosms not amended with (R,S)-dichlorprop (a and d) are shown from triplicate soil microcosms with error bars representing standard errors of the means. In panels b, c, e, and f, open and filled symbols each represent the data set from one individual soil microcosm while the lines running through the data points represent the means of the two replicates. Symbols: ○ and •, rdpA DNA; ▵ and ▴, ratio of rdpA mRNA to rdpA DNA; ▪, mineralization of (R,S)-dichlorprop.
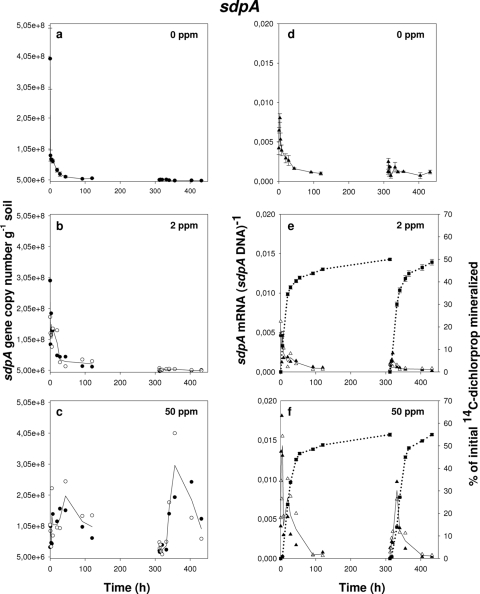
FIG. 4.
Progress curves of sdpA gene copy numbers (a to c), sdpA transcription activity represented as ratios of sdpA mRNA to sdpA DNA (d to f), and (R,S)-[14C]dichlorprop mineralization (e and f) during exposure of Delftia acidovorans MC1 to different concentrations of (R,S)-dichlorprop in soil: 0 mg kg−1 (a and d), 2 mg kg−1 (b and e), and 50 mg kg−1 (c and f). Amendment with (R,S)-dichlorprop was done at 0 h followed by a second amendment at 312 h. Data from microcosms not amended with (R,S)-dichlorprop (a and d) are shown from triplicate soil microcosms with error bars representing standard errors of the means. In panels b, c, e, and f, open and filled symbols each represent the data set from one individual soil microcosm while the lines running through the data points represent the means of the two replicates. Symbols: ○ and •, sdpA DNA; ▵ and ▴, ratio of sdpA mRNA to sdpA DNA; ▪, mineralization of (R,S)-dichlorprop.
No increase of rdpA or sdpA DNA was detected in nonamended soils or soils amended with only 2 mg kg−1 of (R,S)-dichlorprop. In these cases, both rdpA and sdpA copy numbers decreased substantially following initial cell inoculation, possibly as a result of cell death or predation by protozoa, but were still maintained throughout the experiments at around 106 copies g of soil−1 (Fig. 3a and b and 4a and b). It is also noteworthy that levels of sdpA DNA may be slightly overestimated as a result of the low background level of indigenous sdpA genes in this soil. In contrast, qPCR quantification of rdpA and sdpA DNA clearly gave evidence of D. acidovorans MC1 cell growth during exposure to 50 mg kg−1 of (R,S)-dichlorprop. Hence, rdpA gene copy number, originally ∼7 × 107 copies g of soil−1 (as verified by qPCR), reached above 108 copies g of soil−1 following 20 h of incubation (Fig. 3c). An increase in abundance of sdpA DNA was also observed starting at ∼108 copies g of soil−1 and reaching ∼2 × 108 copies g−1 (Fig. 4c). Approximately 1 week after initial mineralization had ceased, gene copy numbers remained stable at around 2 × 107 copies g and 4 × 107 copies g of soil−1 for rdpA and sdpA, respectively. A second amendment with 50 mg kg−1 (R,S)-dichlorprop at 312 h caused rdpA to increase to ∼5 × 107 copies g−1 while sdpA underwent a higher increase compared to the first amendment, this time reaching ∼3 × 108 copies g of soil−1 after 356 h of incubation.
Gene expression was calculated as mRNA/DNA ratios as a way of describing transcription dynamics independently of potential cell number increase (34). Detection of rdpA and sdpA transcripts from D. acidovorans MC1 in soil indicated similar patterns of transcription activity for the two genes. In control microcosms where D. acidovorans MC1 was inoculated but no (R,S)-dichlorprop was added, rdpA and sdpA genes were expressed at a low basal level with no apparent upregulation over the 18 days of incubation (Fig. 3d and 4d). Furthermore, no mRNA was detectable over this period for either gene in soils not amended with (R,S)-dichlorprop and without the degrader strain (data not shown), indicating that indigenous background expression was absent or below the detection limit. No clear increase in transcription activity could be detected in response to the first amendments with 2 mg kg−1 and 50 mg kg−1 of (R,S)-dichlorprop (Fig. 3e and f and 4e and f). More precisely, expression of rdpA and sdpA started off high during the first amendment with (R,S)-dichlorprop and slowly decreased over time to reach a minimal level of expression at around 100 h. This progressive decrease coincided well with the beginning of the plateau phase of (R,S)-[14C]dichlorprop mineralization when most of the (R,S)-dichlorprop had been degraded in the soil. At that point, low transcript levels were comparable to those in control microcosms.
A second amendment with 2 mg kg−1 (R,S)-dichlorprop at 312 h caused a clear and transient increase in rdpA gene expression with maximal mRNA abundance being reached within 5 h to 8 h after respiking (Fig. 3e). This was in accordance with the high activity of the mineralization curve that quickly reached the plateau phase. However, no upregulation was observed for the sdpA gene in comparison to the control microcosms (Fig. 4e). In comparison, a second amendment with 50 mg kg−1 (R,S)-dichlorprop at 312 h caused a dramatic and transient increase in rdpA gene expression that reached a maximum within 20 h after respiking and decreased thereafter to levels similar to the control at 400 h of incubation (Fig. 3f). This time shift in maximal expression of rdpA in comparison to the 2-mg kg−1 (R,S)-dichlorprop amendment corresponded well to the larger amount of (R,S)-dichlorprop available for degradation in the soil. A similar progress curve was obtained for sdpA gene expression during the second amendment with 50 mg kg−1 (R,S)-dichlorprop, although abundance was clearly lower than that with rdpA transcripts (Fig. 4f).
DISCUSSION
The microbial degradation of (R,S)-dichlorprop and (R,S)-mecoprop is not yet fully understood and has previously been proposed to be linked to the action of microbial consortia (21, 41) with reports of some degraders carrying tfdA or tfdA-like genes and degradation pathways similar to those of 2,4-D and MCPA (38, 42). However, the recent characterization of phenoxypropionate-degrading strains that carry enantiospecific dioxygenases apart from TfdA highlights the importance of developing appropriate molecular tools for detection of these genes in the environment.
As sequence data from (R)- and (S)-dichlorprop cleaving dioxygenases have only recently become available, only one primer set for each gene has been used (39). Theoretically, the published primers for rdpA can amplify all sequences described to date, as they are nearly 100% identical in composition. However, rdpA primers were tested here and found unsuitable for use in SYBR green qPCR due to unsatisfactory amplification efficiency during qPCR of the standard curve (data not shown). In the case of sdpA, Schleinitz et al. (39) designed primers based on the peptide sequences obtained from D. acidovorans MC1 (43) and first reported that although S. herbicidovorans MH and Rhodoferax sp. P230 did exhibit (S)-dichlorprop cleaving dioxygenase activity, neither strain carried sdpA-like genes. Conversely, Müller et al. (30) successfully isolated and sequenced sdpA from S. herbicidovorans MH and reported that its predicted amino acid sequence shared only 63% identity to that of D. acidovorans. A closer look at this target sequence reveals that the primers designed by Schleinitz et al. (39) were unsuitable for detection of sdpA in strain S. herbicidovorans MH and that the inability to first detect sdpA, even under low-stringency conditions, was most likely a result of high nucleotide diversity found at the primer hybridization sites. In this work, rdpA and sdpA primers were designed based on the alignment of all available sequences and were shown to successfully amplify rdpA and sdpA from both S. herbicidovorans MH and D. acidovorans MC1.
We further report the first case of sdpA gene detection in natural soils even without prior (R,S)-dichlorprop exposure; the qPCR assay developed consistently detected levels of approximately 103 copies of sdpA DNA g of soil−1. The absence of rdpA in the same soil was unexpected as both genes and corresponding enzymes have always been reported together in previously characterized strains (30, 39). However, given the very limited amount of information available on the distribution of these genes in the environment and the few strains isolated to date that harbor them, it is feasible that some organisms may carry only rdpA, sdpA, or a combination of both. Alternatively, rdpA genes could have been present but at levels below the detection limit of our method. The only other study to target rdpA and sdpA DNA in environmental samples used the primers designed by Schleinitz et al. (39) to screen mecoprop-degrading soil enrichment cultures (46). In that case, rdpA was detected only when cultures were enriched on (R)-mecoprop; although sdpA could not successfully be PCR amplified, dot blot hybridization was able to detect the presence of sdpA from the enrichment cultures grown on (R)- and (R,S)-mecoprop. The authors did not report the presence of sdpA or rdpA genes from samples not exposed to mecoprop. It can be presumed that the full extent of sdpA genes was not detected given the limited range of the primer set. Furthermore, no sdpA sequence data were provided.
The sequence diversity in sdpA found here corroborates well with previous, albeit limited, knowledge of sdpA gene sequences. As mentioned above, the DNA sequence of sdpA from S. herbicidovorans MH and Rhodoferax sp. P230 compared to D. acidovorans MC1 exhibits considerable diversity. Interestingly, the partial sdpA sequences that we recovered from natural soils demonstrated a high diversity not only in composition but also in length. Furthermore, phylogenetic analysis of a fixed 190-bp region for all sequences consistently divided the partial sdpA genes sequenced into four separate clusters. Although analysis was done on a fixed-length region, two of these clusters (cluster I and cluster IV) could easily be distinguished based on amplicon length, each being exclusively composed of 281-bp and 269-bp sequences, respectively. The diversity obtained in the nucleic acid sequences of sdpA was maintained when translated into partial putative amino acid sequences, indicating that it was not merely a consequence of synonymous mutations. Müller et al. (30) noted that the predicted SdpA amino acid sequence of S. herbicidovorans MH was 63% identical to that of D. acidovorans MC1 and 37% identical to the canonical TfdA sequence from C. necator JMP134(pJP4). In our analysis, all predicted amino acid sequences shared the closest identity to one of the previously characterized SdpA sequences and showed a fully conserved predicted α-ketoglutarate binding site as well as a Fe(II)-binding residue (31, 39). The sequences with the highest similarity to SdpA of S. herbicidovorans MH and Rhodoferax sp. P230 shared between 78 and 85% identity while the ones with the lowest similarity were the shortest sequences and shared only 55% identity to SdpA of D. acidovorans MC1. Comparison of the amino acid sequences to the sdpA gene phylogenetic tree revealed that similarly to nucleic acid sequences, sequence length could not help in distinguishing between all four clusters. However, all four clusters could be resolved by percent identities obtained through BLASTp analysis of the amino acid sequences. Hence, each of the four clusters was composed of sequences within a well-defined range of identities to their closest relative in GenBank. Nevertheless, the database of sdpA sequences is still too small to make any conclusions regarding the full gene diversity and the possibility of dividing sequences into different classes, as has been the case for the tfdA genes (20, 24).
Apart from the already described SdpA, the sequences obtained here shared some resemblance to genes belonging to the tfdA-α group. Beta- and Gammaproteobacteria involved in 2,4-D degradation typically carry the tfdA gene, which can be divided into three classes (classes I to III) (24), while tfdA-α is a tfdA homologue usually found in Alphaproteobacteria (18, 19). Interestingly, tfdA genes are widespread in the environment, even in pristine soils with no previous exposure to phenoxy acid herbicides (17, 20) and in soil organisms unable to degrade 2,4-D (15). Recently, Bælum et al. (2) detected low background levels of tfdA in soil prior to addition of 2,4-D and MCPA. It has been postulated that the TfdA enzyme may have undiscovered natural substrates which could help explain its widespread distribution in the environment (8, 15). Although previously characterized SdpA enzymes have a wider substrate range than RdpA and the closest relatives of SdpA are members of the TfdA enzyme family, it is unknown whether the sdpA gene pool detected in this soil is a result of selective pressure caused by the presence of natural substrates for SdpA. Our report on the natural abundance and diversity of sdpA genes in addition to the detection of rdpA and sdpA genes from soil enrichment cultures reported by Zakaria et al. (46) underscores the importance of further investigating the role of these genes during in situ phenoxypropionate herbicide degradation.
Patterns of rdpA and sdpA gene expression during the degradation of (R,S)-dichlorprop were determined from D. acidovorans MC1, which is able to fully degrade the herbicide following the modified ortho-cleavage pathway (29). This strain was chosen because of its well-characterized behavior in pure culture, while data on its activity under in situ soil conditions remain limited. Besides, sdpA and rdpA mRNA transcript levels were too low for detection from natural uninoculated soils during the 18 days of incubation, thus explaining the need to inoculate a degrader strain for expression studies. A time course analysis of rdpA and sdpA transcription dynamics during microbial degradation of (R,S)-dichlorprop was undertaken to achieve understanding of how these genes are regulated in the environment. Previously, pure culture studies with S. herbicidovorans MH grown on pyruvate in mineral medium showed that expression of both rdpA and sdpA genes was constitutive regardless of exposure to (R)-dichlorprop, (S)-dichlorprop, 2,4-D, or DMSO (used as a noninducing control) (30). Although the presence of mRNA was constitutive, the study did not provide information on the actual regulation of these genes, as no quantitative expression data were presented and conclusions were based on data collected from a single time point. Constitutive gene expression was also reported for pure culture induction studies of D. acidovorans MC1 based on the etherolytic degradation of both enantiomers of (R,S)-dichlorprop but was not supported by accompanying data on mRNA transcripts (29). Nevertheless, our results also support the idea that expression of both rdpA and sdpA genes in D. acidovorans MC1 was constitutive and maintained at a low basal level when the strain was inoculated into soil without exposure to (R,S)-dichlorprop.
As observed in similar studies involving MCPA, 2,4-D, and expression of the tfdA genes in soil (2, 34), regulation of rdpA and sdpA gene expression may be largely dependent upon exposure to (R,S)-dichlorprop. Soil microcosms receiving the first amendment with 2 mg kg−1 or 50 mg kg−1 of (R,S)-dichlorprop did not immediately result in clear upregulation of gene expression. Hence, rdpA and sdpA mRNA (expressed as ratios of mRNA normalized to DNA) started off quite high followed by a progressive decrease until levels fell to those similar to those for unamended control soils. The preinoculation step which consisted of growth of D. acidovorans MC1 in (R,S)-dichlorprop-amended medium followed by overnight growth in nonselective PYE medium prior to soil inoculation might have been responsible for the high abundance of rdpA and sdpA transcripts detected at time zero before the first amendment. Although such pregrown cells were washed, they may have retained significant levels of mRNA as a consequence of being grown in a selective medium prior to microcosm experiments. Nonetheless, we chose this setup as a consequence of reports indicating the spontaneous loss of genes involved in etherolytic cleavage of (R,S)-dichlorprop when D. acidovorans MC1 is grown under nonselective conditions for extended periods (29, 39).
After the second amendment, however, an obvious increase in gene expression was observed in response to repeated amendments with (R,S)-dichlorprop, particularly in soils receiving 50 mg kg−1 of the herbicide. Progress curves of mRNA formation showed a sharp and transient maximum corresponding to the active phase of (R,S)-dichlorprop mineralization. As noted by Nicolaisen et al. (34), the subsequent decreasing levels of mRNA most likely indicate a progressive degradation of transcripts as a result of an adequate pool of degradative enzymes. Both genes were clearly upregulated in response to reexposure to high concentrations of (R,S)-dichlorprop, although the abundance of mRNA transcripts was higher for rdpA than for sdpA. Repeated treatment with the low concentration of herbicide (2 mg kg−1) produced similar results, although expression of sdpA was not markedly different from that in control soils.
This study has demonstrated that rdpA and sdpA gene expression is constitutive at a basal level but is induced in response to (R,S)-dichlorprop degradation under in situ conditions. Although a basal constitutive level is not detected for the tfdA gene (2, 34), other degradative enzyme-encoding genes such as atz, involved in the initial steps of atrazine degradation, have been reported to be continuously and basally expressed with transient increases in gene expression following herbicide exposure (5). Further studies on the abundance of the rdpA and sdpA genes in various soils as well as in-depth studies of their expression in indigenous soil microbial populations exposed to (R,S)-dichlorprop over extended periods of time should help to gain a better understanding of how these genes are regulated in the environment.
Acknowledgments
We thank R. H. Müller (Helmholtz Centre for Environmental Research, Leipzig, Germany) for kindly donating strain Delftia acidovorans MC1.
This study was supported by the Center for Environmental and Agricultural Microbiology (CREAM) funded by the Villum Kann Rasmussen Foundation.
Footnotes
Published ahead of print on 19 March 2010.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bælum, J., M. H. Nicolaisen, W. E. Holben, B. W. Strobel, J. Sørensen, and C. S. Jacobsen. 2008. Direct analysis of tfdA gene expression by indigenous bacteria in phenoxy acid amended agricultural soil. ISME J. 2:677-687. [DOI] [PubMed] [Google Scholar]
- 3.Buerge, I. J., T. Poiger, M. D. Müller, and H. R. Buser. 2003. Enantioselective degradation of metalaxyl in soils: chiral preference changes with soil pH. Environ. Sci. Technol. 37:2668-2674. [DOI] [PubMed] [Google Scholar]
- 4.Buser, H. R., M. D. Müller, and N. Theobald. 1998. Occurrence of the pharmaceutical drug clofibric acid and the herbicide mecoprop in various Swiss lakes and in the North Sea. Environ. Sci. Technol. 32:188-192. [Google Scholar]
- 5.Devers, M., G. Soulas, and F. Martin-Laurent. 2004. Real-time reverse transcription PCR analysis of expression of atrazine catabolism genes in two bacterial strains isolated from soil. J. Microbiol. Methods 56:3-15. [DOI] [PubMed] [Google Scholar]
- 6.Diao, J. L., C. G. Lv, X. Q. Wang, Z. H. Dang, W. T. Zhu, and Z. Q. Zhou. 2009. Influence of soil properties on the enantioselective dissipation of the herbicide lactofen in soils. J. Agric. Food Chem. 57:5865-5871. [DOI] [PubMed] [Google Scholar]
- 7.Don, R. H., A. J. Weightman, H. J. Knackmuss, and K. N. Timmis. 1985. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4). J. Bacteriol. 161:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunning Hotopp, J. C., and R. P. Hausinger. 2001. Alternative substrates of 2.4-dichlorophenoxyacetate/alpha-ketoglutarate dioxygenase. J. Mol. Catal. B 15:155-162. [Google Scholar]
- 9.Ehrig, A., R. H. Müller, and W. Babel. 1997. Isolation of phenoxy herbicide-degrading Rhodoferax species from contaminated building material. Acta Biotechnol. 17:351-356. [Google Scholar]
- 10.European Commission. 2002. Commission regulation (EC) no. 2076/2002 of 20 November 2002 extending the time period referred to in article 8(2) of council directive 91/414/EEC and concerning the non-inclusion of certain active substances in annex I to that directive and the withdrawal of authorisations for plant protection products containing these substances. Off. J. Eur. Union 319:3-11. [Google Scholar]
- 11.Fukumori, F., and R. P. Hausinger. 1993. Purification and characterization of 2,4-dichlorophenoxyacetate/alpha-ketoglutarate dioxygenase. J. Biol. Chem. 268:24311-24317. [PubMed] [Google Scholar]
- 12.Garrison, A. W. 2006. Probing the enantioselectivity of chiral pesticides. Environ. Sci. Technol. 40:16-23. [DOI] [PubMed] [Google Scholar]
- 13.Garrison, A. W., P. Schmitt, D. Martens, and A. Kettrup. 1996. Enantiomeric selectivity in the environmental degradation of dichlorprop as determined by high-performance capillary electrophoresis. Environ. Sci. Technol. 30:2449-2455. [Google Scholar]
- 14.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 15.Hogan, D. A., D. H. Buckley, C. H. Nakatsu, T. M. Schmidt, and R. P. Hausinger. 1997. Distribution of the tfdA gene in soil bacteria that do not degrade 2,4-dichlorophenoxyacetic acid (2,4-D). Microb. Ecol. 34:90-96. [DOI] [PubMed] [Google Scholar]
- 16.Horvath, M., G. Ditzelmüller, M. Loidl, and F. Streichsbier. 1990. Isolation and characterization of a 2-(2,4-dichlorophenoxy) propionic acid-degrading soil bacterium. Appl. Microbiol. Biotechnol. 33:213-216. [DOI] [PubMed] [Google Scholar]
- 17.Itoh, K., R. Kanda, Y. Momoda, Y. Sumita, Y. Kamagata, K. Suyama, and H. Yamamoto. 2000. Presence of 2,4-D-catabolizing bacteria in a Japanese arable soil that belong to BANA (Bradyrhizobium-Agromonas-Nitrobacter-Afipia) cluster in α-Proteobacteria. Microb. Environ. 15:113-117. [Google Scholar]
- 18.Itoh, K., R. Kanda, Y. Sumita, H. Kim, Y. Kamagata, K. Suyama, H. Yamamoto, R. P. Hausinger, and J. M. Tiedje. 2002. tfdA-like genes in 2,4-dichlorophenoxyacetic acid-degrading bacteria belonging to the Bradyrhizobium-Agromonas-Nitrobacter-Afipia cluster in α-Proteobacteria. Appl. Environ. Microbiol. 68:3449-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh, K., Y. Tashiro, K. Uobe, Y. Kamagata, K. Suyama, and H. Yamamoto. 2004. Root nodule Bradyrhizobium spp. harbor tfdAα and cadA, homologous with genes encoding 2,4-dichlorophenoxyacetic acid-degrading proteins. Appl. Environ. Microbiol. 70:2110-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamagata, Y., R. R. Fulthorpe, K. Tamura, H. Takami, L. J. Forney, and J. M. Tiedje. 1997. Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl. Environ. Microbiol. 63:2266-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, M. P., M. R. Heniken, and O. H. Tuovinen. 1991. Dechlorination and spectral changes associated with bacterial degradation of 2-(2-methyl-4-chlorophenoxy) propionic acid. J. Ind. Microbiol. 7:137-145. [Google Scholar]
- 22.Lewis, D. L., A. W. Garrison, K. E. Wommack, A. Whittemore, P. Steudler, and J. Melillo. 1999. Influence of environmental changes on degradation of chiral pollutants in soils. Nature 401:898-901. [DOI] [PubMed] [Google Scholar]
- 23.Ma, Y., C. Xu, Y. Z. Wen, and W. P. Liu. 2009. Enantioselective separation and degradation of the herbicide dichlorprop methyl in sediment. Chirality 21:480-483. [DOI] [PubMed] [Google Scholar]
- 24.McGowan, C., R. Fulthorpe, A. Wright, and J. M. Tiedje. 1998. Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl. Environ. Microbiol. 64:4089-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgenstern, B. 2004. DIALIGN: multiple DNA and protein sequence alignment at BiBiServ. Nucleic Acids Res. 32:W33-W36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller, M. D., and H. R. Buser. 1997. Conversion reactions of various phenoxyalkanoic acid herbicides in soil. 1. Enantiomerization and enantioselective degradation of the chiral 2-phenoxypropionic acid herbicides. Environ. Sci. Technol. 31:1953-1959. [Google Scholar]
- 27.Müller, R. H. 2007. Activity and reaction mechanism of the initial enzymatic step specifying the microbial degradation of 2,4-dichlorophenoxyacetate. Eng. Life Sci. 7:311-321. [Google Scholar]
- 28.Müller, R. H., S. Jorks, S. Kleinsteuber, and W. Babel. 1999. Comamonas acidovorans strain MC1: a new isolate capable of degrading the chiral herbicides dichlorprop and mecoprop and the herbicides 2,4-D and MCPA. Microbiol. Res. 154:241-246. [DOI] [PubMed] [Google Scholar]
- 29.Müller, R. H., S. Kleinsteuber, and W. Babel. 2001. Physiological and genetic characteristics of two bacterial strains utilizing phenoxypropionate and phenoxyacetate herbicides. Microbiol. Res. 156:121-131. [DOI] [PubMed] [Google Scholar]
- 30.Müller, T. A., S. A. Byrde, C. Werlen, J. R. van der Meer, and H. P. E. Kohler. 2004. Genetic analysis of phenoxyalkanoic acid degradation in Sphingomonas herbicidovorans MH. Appl. Environ. Microbiol. 70:6066-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller, T. A., T. Fleischmann, J. R. van der Meer, and H. P. E. Kohler. 2006. Purification and characterization of two enantioselective α-ketoglutarate-dependent dioxygenases, RdpA and SdpA, from Sphingomonas herbicidovorans MH. Appl. Environ. Microbiol. 72:4853-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller, T. A., and H.-P. E. Kohler. 2004. Chirality of pollutants—effects on metabolism and fate. Appl. Microbiol. Biotechnol. 64:300-316. [DOI] [PubMed] [Google Scholar]
- 33.Nickel, K., M. J. F. Suter, and H. P. E. Kohler. 1997. Involvement of two α-ketoglutarate-dependent dioxygenases in enantioselective degradation of (R)- and (S)-mecoprop by Sphingomonas herbicidovorans MH. J. Bacteriol. 179:6674-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicolaisen, M. H., J. Bælum, C. S. Jacobsen, and J. Sørensen. 2008. Transcription dynamics of the functional tfdA gene during MCPA herbicide degradation by Cupriavidus necator AEO106 (pRO101) in agricultural soil. Environ. Microbiol. 10:571-579. [DOI] [PubMed] [Google Scholar]
- 35.Qin, S. J., and J. Gan. 2006. Enantiomeric differences in permethrin degradation pathways in soil and sediment. J. Agric. Food Chem. 54:9145-9151. [DOI] [PubMed] [Google Scholar]
- 36.Romero, E., M. B. Matallo, A. Peña, F. Sánchez-Rasero, P. Schmitt-Kopplin, and G. Dios. 2001. Dissipation of racemic mecoprop and dichlorprop and their pure R-enantiomers in three calcareous soils with and without peat addition. Environ. Pollut. 111:209-215. [DOI] [PubMed] [Google Scholar]
- 37.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 38.Saari, R. E., D. A. Hogan, and R. P. Hausinger. 1999. Stereospecific degradation of the phenoxypropionate herbicide dichlorprop. J. Mol. Catal. B 6:421-428. [Google Scholar]
- 39.Schleinitz, K. M., S. Kleinsteuber, T. Vallaeys, and W. Babel. 2004. Localization and characterization of two novel genes encoding stereospecific dioxygenases catalyzing 2(2,4-dichlorophenoxy) propionate cleavage in Delftia acidovorans MC1. Appl. Environ. Microbiol. 70:5357-5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), v. 4. Sinauer Associates, Sunderland, MA.
- 41.Tett, V. A., A. J. Willetts, and H. M. Lappinscott. 1994. Enantioselective degradation of the herbicide mecoprop 2-(2-methyl-4-chlorophenoxy) propionic acid by mixed and pure bacterial cultures. FEMS Microbiol. Ecol. 14:191-199. [Google Scholar]
- 42.Tett, V. A., A. J. Willetts, and H. M. Lappinscott. 1997. Biodegradation of the chlorophenoxy herbicide (R)-(+)-mecoprop by Alcaligenes denitrificans. Biodegradation 8:43-52. [Google Scholar]
- 43.Westendorf, A., D. Benndorf, R. H. Müller, and W. Babel. 2002. The two enantiospecific dichlorprop/α-ketoglutarate-dioxygenases from Delftia acidovorans MC1—protein and sequence data of RdpA and SdpA. Microbiol. Res. 157:317-322. [DOI] [PubMed] [Google Scholar]
- 44.Westendorf, A., D. Benndorf, T. Pribyl, H. Harms, and R. H. Müller. 2006. Kinetic traits and enzyme form patterns of (R)-2-(2,4-dichlorophenoxy) propionate/α-ketoglutarate dioxygenase (RdpA) after expression in different bacterial strains. Eng. Life Sci. 6:552-559. [Google Scholar]
- 45.Westendorf, A., R. H. Müller, and W. Babel. 2003. Purification and characterisation of the enantiospecific dioxygenases from Delftia acidovorans MCI initiating the degradation of phenoxypropionate and phenoxyacetate herbicides. Acta Biotechnol. 23:3-17. [Google Scholar]
- 46.Zakaria, D., H. Lappin-Scott, S. Burton, and C. Whitby. 2007. Bacterial diversity in soil enrichment cultures amended with 2 (2-methyl-4-chlorophenoxy) propionic acid (mecoprop). Environ. Microbiol. 9:2575-2587. [DOI] [PubMed] [Google Scholar]
- 47.Zipper, C., C. Bolliger, T. Fleischmann, M. J. F. Suter, W. Angst, M. D. Müller, and H. P. E. Kohler. 1999. Fate of the herbicides mecoprop, dichlorprop, and 2,4-D in aerobic and anaerobic sewage sludge as determined by laboratory batch studies and enantiomer-specific analysis. Biodegradation 10:271-278. [DOI] [PubMed] [Google Scholar]
- 48.Zipper, C., K. Nickel, W. Angst, and H. P. E. Kohler. 1996. Complete microbial degradation of both enantiomers of the chiral herbicide mecoprop [(RS)-2-(4-chloro-2-methylphenoxy)propionic acid] in an enantioselective manner by Sphingomonas herbicidovorans sp. Appl. Environ. Microbiol. 62:4318-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]