Abstract
Salmonella pathogenicity island 1 (SPI1) and SPI4 have previously been shown to be jointly regulated. We report that SPI1 and SPI4 gene expression is linked through a transcriptional activator, SprB, encoded within SPI1 and regulated by HilA. SprB directly activates SPI4 gene expression and weakly represses SPI1 gene expression through HilD.
Salmonella enterica serovar Typhimurium is a common food-borne pathogen that invades intestinal epithelial cells using a type three secretion system (T3SS) encoded within Salmonella pathogenicity island 1 (SPI1) (8, 12, 15, 22). The SPI1 master regulator, HilA, activates the expression of the genes encoding the T3SS structural proteins, chaperones, and secreted effectors (16). Three AraC-like transcription factors—HilC, HilD, and RtsA—positively regulate HilA expression in response to various intracellular and environmental signals (5, 7, 17). In addition to these positive regulators, the genes within SPI1 are negatively regulated by HilE (2).
Also encoded within SPI1 is a transcription factor, SprB, from the LuxR/UhaP family of transcription factors. Eichelberg and coworkers (4) previously found the gene for SprB to be expressed under conditions similar to those under which other SPI1 genes are expressed. However, they found that SprB was not involved in regulating the expression of SPI1 genes and speculated that it may instead regulate novel SPI1 T3SS substrates. In this work, we demonstrate that SprB regulates the expression of the genes within SPI4.
SPI4 encodes a nonfimbrial adhesin (9) that is involved in the intestinal phase of infection (13, 14, 19). Previously, a number of studies have shown that there is a link between SPI1 and SPI4 gene expression (1, 3, 6, 9, 18). In particular, they demonstrated that SPI4 gene expression is HilA dependent. However, Main-Hester and coworkers showed that, in the absence of the SPI1 locus, HilA is unable to activate SPI4 gene expression (18). Their results suggest that some other HilA-dependent regulator within SPI1 may be involved. Here, we show that this other regulator is SprB.
SprB transcriptionally regulates SPI4 gene expression.
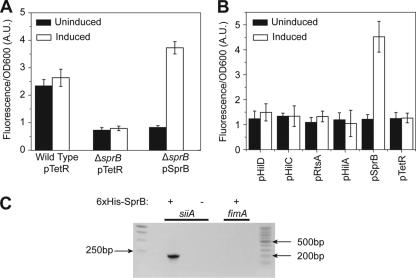
The SPI4 locus encodes six genes within a single operon under the control of the PsiiA promoter (9). To test whether SprB in fact regulates SPI4 gene expression, we measured PsiiA promoter activity using a plasmid-based Venus transcriptional reporter (20) in the wild type and a ΔsprB mutant. As shown in Fig. 1A, deleting sprB causes a threefold decrease in PsiiA promoter activity as determined by changes in fluorescence. Moreover, we can complement this mutant by expressing SprB from an anhydrotetracycline (aTc)-inducible promoter on a compatible plasmid (21).
FIG. 1.
SprB regulates SPI4 gene expression. (A) Comparison of PsiiA promoter activity in the wild type, a ΔsprB mutant, and a ΔsprB mutant expressing SprB from an aTc-inducible promoter on a plasmid (pSprB). pTetR denotes the empty plasmid control. Expression was induced with 50 ng/ml aTc. (B) SprB can activate the PsiiA promoter in the absence of other SPI1 genes (ΔSPI1 ΔrtsAB); the other SPI1 regulators—HilA, HilC, HilD, and RtsA—cannot. Activators were expressed from an aTc-inducible promoter on a plasmid. Expression was induced with 50 ng/ml aTc. Fluorescence values were normalized with the optical density at 600 nm (OD600; A.U., arbitrary units) to account for cell density. Error bars indicate standard deviations. (C) SprB binds to the PsiiA promoter region as determined by a coprecipitation assay using 6×His-tagged SprB. PCR was used to determine whether the PsiiA promoter region is in the coprecipitated DNA. The PfimA promoter region was included as a negative control. For an expanded description of the experimental procedures, see the supplemental material.
We next tested how individually expressing the SPI1 regulators—HilA, HilC, HilD, RtsA, and SprB—from an aTc-inducible promoter would affect PsiiA promoter activity in a ΔSPI1 ΔrtsAB mutant. Of the five, only SprB was capable of activating the PsiiA promoter (Fig. 1B). We performed similar experiments with Escherichia coli and obtained identical results (data not shown).
To test whether SprB directly binds to the PsiiA promoter, we used a coprecipitation assay. In this experiment, we first expressed 6×His-tagged SprB from the constitutive PLtetO-1 promoter on a plasmid in a ΔsprB mutant and then passed the cross-linked cell lysate over a Ni2+ column (see the supplemental material for more details). Upon elution from the column and reverse cross-linking, we found by PCR that SprB directly binds the PsiiA promoter region (Fig. 1C). As a negative control, we also tested whether SprB binds to the PfimA promoter region, a promoter whose activity is unaffected by SprB, and found that it did not. Collectively, these results demonstrate that SprB binds the PsiiA promoter and activates SPI4 gene expression.
HilA transcriptionally regulates SprB expression.
Multiple studies have previously shown that SPI4 gene expression is HilA dependent (1, 3, 6, 9, 18). We therefore hypothesized that HilA regulates SprB expression, as this would explain the decrease in PsiiA promoter activity previously observed when HilA is deleted. It would also explain why other SPI1 regulators—namely HilC, HilD, and RtsA—affect SPI4 gene expression, as they in turn regulate HilA expression.
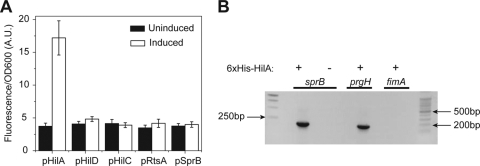
To test this hypothesis, we measured the effect of individually expressing the SPI1 regulators—HilA, HilC, HilD, RtsA, and SprB—from an aTc-inducible promoter in a ΔSPI1 ΔrtsAB mutant on PsprB promoter activity. Similar to the experiments described above, we used a plasmid-based Venus transcriptional reporter to measure PsprB promoter activity. Of the five, only HilA was capable of activating the PsprB promoter (Fig. 2A). Identical results were obtained when we performed similar experiments with E. coli (data not shown). To test whether this effect is direct, we used a coprecipitation assay with 6×His-tagged SprA and found that HilA does indeed bind the PsprB promoter (Fig. 2B). As respective positive and negative controls, we used the PprgH and PfimA promoter regions. These results demonstrate that HilA transcriptionally regulates SprB expression.
FIG. 2.
HilA regulates SprB expression. (A) HilA can activate the PsprB promoter in the absence of other SPI1 genes (ΔSPI1 ΔrtsAB); the other SPI1 regulators—HilC, HilD, and RtsA—cannot. SprB was also found not to regulate its own expression. Activators were expressed from an aTc-inducible promoter on a plasmid. Expression was induced with 50 ng/ml aTc. Fluorescence values were normalized with the optical density at 600 nm (OD600; A.U., arbitrary units) to account for cell density. Error bars indicate standard deviations. (B) HilA binds to the PsprB promoter region as determined by a coprecipitation assay with 6×His-tagged HilA. PCR was used to determine whether the PsprB promoter region is in the coprecipitated DNA. The PprgH and PfimA promoter regions were included as positive and negative controls, respectively. For an expanded description of the experimental procedures, see the supplemental material.
SprB transcriptionally represses HilD expression.
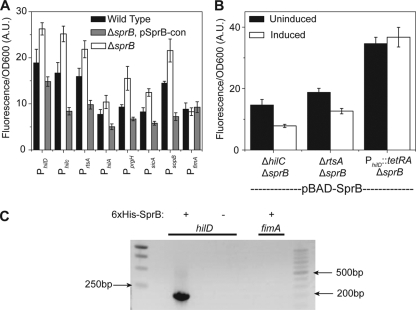
Last, we tested whether SprB regulates SPI1 gene expression. Here, we measured the activity of a number of SPI1- and SPI1-regulated promoters using plasmid-based Venus transcriptional reporters in the wild type, a ΔsprB mutant, and a ΔsprB mutant constitutively expressing sprB from the PLtetO-1 promoter on a plasmid. Our data show that deleting sprB resulted in a mild increase in the activity of all seven promoters tested, except the PfimA promoter used as a control (Fig. 3A). However, plasmid expression of SprB caused a twofold reduction in promoter activity in a ΔsprB mutant background. These results suggest that SprB is a weak negative regulator of SPI1 gene expression. The weak level of repression may explain why Eichelberg and coworkers (4) previously concluded that SprB does not regulate SPI1 gene expression. Since SPI1 gene expression is known to be growth dependent, we also checked if expressing SprB from a plasmid in a ΔsprB mutant caused any growth defect. Our data show that it has no effect (see Fig. S1 in the supplemental material).
FIG. 3.
SprB represses SPI1 gene expression through HilD. (A) Comparison of SPI1 promoter activities in the wild type, a ΔsprB mutant, and a ΔsprB mutant constitutively expressing SprB from a plasmid (pSprB-con). (B) SprB repression of SPI1 gene expression is through the PhilD promoter. Comparison of PhilA promoter activity in ΔhilC ΔsprB, ΔhilC ΔsprB, and PhilD::tetRA ΔsprB mutants when SprB is expressed from an arabinose-inducible promoter on a plasmid (pBAD-SprB). SprB expression was induced with 2 mg/ml arabinose. In the experiments involving the PhilD::tetRA ΔsprB mutant, 2 μg/ml tetracycline was added to the growth medium in order to induce HilD expression. Induction in panel B is used to denote the presence or absence of arabinose. Fluorescence values were normalized with the optical density at 600 nm (OD600; A.U., arbitrary units) to account for cell density. Error bars indicate standard deviations. (C) SprB binds to the PhilD promoter region as determined by a coprecipitation assay using 6×His-tagged SprB. PCR was used to determine whether the PhilD promoter region is in the coprecipitated DNA. The PfimA promoter region was included as a negative control. For an expanded description of the experimental procedures, see the supplemental material.
As SprB regulates multiple SPI1 promoters, we hypothesized that it likely represses the transcription of HilC, HilD, or RtsA. These three regulators are all activators of PhilA promoter activity and each other's expression as well (5, 7, 17). To identify the target for repression, we first expressed SprB from an arabinose-inducible promoter on a plasmid (10) in ΔhilC ΔsprB, and ΔrtsA ΔsprB null mutants and found that PhilA promoter activity was still repressed by SprB, as determined by using a plasmid-based Venus transcriptional reporter (Fig. 3B). These results demonstrate that repression is not HilC or RtsA dependent. Next, we investigated if SprB represses the PhilD promoter by measuring PhilA promoter activity in a strain where the PhilD promoter was replaced with the tetRA element from transposon Tn10 (11). This arrangement decouples hilD expression from its native regulation and causes it to be constitutively expressed from its native chromosomal locus in the presence of tetracycline. In the PhilD::tetRA ΔsprB mutant strain, we found that expressing SprB from an arabinose-inducible promoter plasmid (10) did not affect PhilA promoter activity (Fig. 3B), suggesting that SprB represses HilD expression. To demonstrate that the SprB-dependent repression of SPI1 gene expression is due to the binding of the SprB protein to the PhilD promoter, we used a coprecipitation assay with 6×His-tagged SprB (Fig. 3C). These results demonstrate that SprB binds to the PhilD promoter region and likely represses SPI1 gene expression by inhibiting hilD transcription.
Conclusion.
Our results demonstrate that SprB functions as a molecular link between SPI1 and SPI4 gene expression. We propose the following model (see Fig. S2 in the supplemental material). Upon induction of the SPI1 gene circuit, HilA activates SprB expression. SprB plays a dual role in regulating gene expression. First, it weakly represses SPI1 gene expression by binding to the PhilD promoter and likely inhibiting hilD transcription. Second, it activates the expression of the SPI4-encoded adhesin, presumably helping the bacterium to adhere to intestinal epithelial cells during invasion. This mode of regulation links gene expression in two distinct systems in Salmonella, enabling the coordinate regulation of adherence to and invasion of the intestinal epithelia.
Supplementary Material
Footnotes
Published ahead of print on 26 February 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahmer, B. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971-982. [DOI] [PubMed] [Google Scholar]
- 2.Boddicker, J. D., B. M. Knosp, and B. D. Jones. 2003. Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators. J. Bacteriol. 185:525-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Keersmaecker, S. C., K. Marchal, T. L. Verhoeven, K. Engelen, J. Vanderleyden, and C. S. Detweiler. 2005. Microarray analysis and motif detection reveal new targets of the Salmonella enterica serovar Typhimurium HilA regulatory protein, including hilA itself. J. Bacteriol. 187:4381-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eichelberg, K., W. D. Hardt, and J. E. Galán. 1999. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol. Microbiol. 33:139-152. [DOI] [PubMed] [Google Scholar]
- 5.Ellermeier, C. D., J. R. Ellermeier, and J. M. Slauch. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57:691-705. [DOI] [PubMed] [Google Scholar]
- 6.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellermeier, J. R., and J. M. Slauch. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10:24-29. [DOI] [PubMed] [Google Scholar]
- 8.Galán, J. E. 1999. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr. Opin. Microbiol. 2:46-50. [DOI] [PubMed] [Google Scholar]
- 9.Gerlach, R. G., D. Jackel, B. Stecher, C. Wagner, A. Lupas, W. D. Hardt, and M. Hensel. 2007. Salmonella pathogenicity island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell. Microbiol. 9:1834-1850. [DOI] [PubMed] [Google Scholar]
- 10.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlinsey, J. E. 2007. lambda-Red genetic engineering in Salmonella enterica serovar Typhimurium. Methods Enzymol. 421:199-209. [DOI] [PubMed] [Google Scholar]
- 12.Kimbrough, T. G., and S. I. Miller. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. U. S. A. 97:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiss, T., E. Morgan, and G. Nagy. 2007. Contribution of SPI-4 genes to the virulence of Salmonella enterica. FEMS Microbiol. Lett. 275:153-159. [DOI] [PubMed] [Google Scholar]
- 14.Lawley, T. D., K. Chan, L. J. Thompson, C. C. Kim, G. R. Govoni, and D. M. Monack. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, C. A., B. D. Jones, and S. Falkow. 1992. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc. Natl. Acad. Sci. U. S. A. 89:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lostroh, C. P., and C. A. Lee. 2001. The HilA box and sequences outside it determine the magnitude of HilA-dependent activation of P(prgH) from Salmonella pathogenicity island 1. J. Bacteriol. 183:4876-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lostroh, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3:1281-1291. [DOI] [PubMed] [Google Scholar]
- 18.Main-Hester, K. L., K. M. Colpitts, G. A. Thomas, F. C. Fang, and S. J. Libby. 2008. Coordinate regulation of Salmonella pathogenicity island 1 (SPI1) and SPI4 in Salmonella enterica serovar Typhimurium. Infect. Immun. 76:1024-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994-1010. [DOI] [PubMed] [Google Scholar]
- 20.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87-90. [DOI] [PubMed] [Google Scholar]
- 21.Saini, S., J. D. Brown, P. D. Aldridge, and C. V. Rao. 2008. FliZ is a posttranslational activator of FlhD4C2-dependent flagellar gene expression. J. Bacteriol. 190:4979-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sukhan, A., T. Kubori, J. Wilson, and J. E. Galán. 2001. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J. Bacteriol. 183:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.