Abstract
This work reports the characterization of the first known peptidoglycan hydrolase (Acp) produced mainly during vegetative growth of Clostridium perfringens. Acp has a modular structure with three domains: a signal peptide domain, an N-terminal domain with repeated sequences, and a C-terminal catalytic domain. The purified recombinant catalytic domain of Acp displayed lytic activity on the cell walls of several Gram-positive bacterial species. Its hydrolytic specificity was established by analyzing the Bacillus subtilis peptidoglycan digestion products by coupling reverse phase-high-pressure liquid chromatography (RP-HPLC) and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis, which displayed an N-acetylglucosaminidase activity. The study of acp expression showed a constant expression during growth, which suggested an important role of Acp in growth of C. perfringens. Furthermore, cell fractionation and indirect immunofluorescence staining using anti-Acp antibodies revealed that Acp is located at the septal peptidoglycan of vegetative cells during exponential growth phase, indicating a role in cell separation or division of C. perfringens. A knockout acp mutant strain was obtained by using the insertion of mobile group II intron strategy (ClosTron). The microscopic examination indicated a lack of vegetative cell separation in the acp mutant strain, as well as the wild-type strain incubated with anti-Acp antibodies, demonstrating the critical role of Acp in cell separation. The comparative responses of wild-type and acp mutant strains to stresses induced by Triton X-100, bile salts, and vancomycin revealed an implication of Acp in autolysis induced by these stresses. Overall, Acp appears as a major cell wall N-acetylglucosaminidase implicated in both vegetative growth and stress-induced autolysis.
Autolysins are endogenous peptidoglycan hydrolases (PGHs) that can break covalent bonds in the bacterial cell wall peptidoglycan (16, 58). Various PGHs are distinguished on the basis of their specific cleavage site in the peptidoglycan: N-acetylmuramidases, N-acetylglucosaminidases, N-acetylmuramoyl-l-alanine amidases, and endopeptidases. PGHs are involved in different physiological functions that require cell wall remodeling such as cell wall expansion, peptidoglycan turnover, daughter cell separation, or sporulation (53, 54, 60). These enzymes may also be implicated in antibiotic-induced lysis (39) and may contribute to bacterial pathogenesis by generating inflammatory cell wall degradation products (32, 40), by releasing virulence factors (4), or by mediating bacterial adherence (1, 20, 21). The roles of PGHs in bacterial physiology, and probably in bacterial pathogenicity, further reinforce the importance of understanding bacterial autolysis.
Autolytic systems of several Gram-positive low-G+C bacteria have been studied (5, 13, 34, 43, 54, 55). Belonging to this phylum is Clostridium perfringens, a common agent of food poisoning and implicated in infectious diseases initiating from the digestive tract (peritonitis, bacteremia, etc.), and it is the most common cause of clostridial gas gangrene in humans. Two PGHs have been described as implicated in the sporulation and germination of C. perfringens, an amidase (SleC) (37, 51) and a muramidase (SleM) (9), which are both produced at the early stage of sporulation, located outside the cortex in the dormant spore (9, 37, 38, 51) and involved in peptidoglycan cortex hydrolysis during germination (45). However, PGHs implicated in the vegetative growth of C. perfringens have never been characterized. In addition, the implication of PGHs in antibiotic-induced lysis of C. perfringens has never been studied.
In the present study, we identified and characterized Acp, the first known autolysin of C. perfringens produced by vegetative cells and displaying N-acetylglucosaminidase activity. Furthermore, we localized Acp at the cell septum during vegetative cell growth and constructed a knockout mutant of the acp gene to demonstrate that Acp is involved in daughter cell separation during vegetative growth. Finally, we studied the implication of Acp in autolysis induced by stresses such as bile salts and cell wall-targeting antibiotics.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
C. perfringens strain 13 (52) was used in all experiments of cloning, Acp characterization, and construction of the acp mutant and was cultivated in brain heart infusion (BHI) broth under anaerobic conditions at 37°C.
Escherichia coli strain BL21 harboring DE3-RIL (Promega), which constitutively expresses the Lac repressor protein encoded by the lacI gene, was used as a recipient for expression of the catalytic domain of Acp. E. coli TOP10 (chemocompetent cells; Invitrogen) was used to construct the pMTL007-derivated plasmid containing the retargeted intron of the acp gene. E. coli strains were, respectively, cultivated in 2× yeast extract-tryptone (YT) broth (Difco) and LB broth (Difco). When required, chloramphenicol (25 μg/ml), kanamycin (25 μg/ml; Sigma), and isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM; Sigma) were added.
Bacillus subtilis 168 HR (14) was used as a substrate to establish Acp hydrolytic activity and was cultivated in LB broth (Difco) at 37°C with shaking.
Spore counting.
Spore counting from cultures of C. perfringens was performed as follows: culture samples were incubated in ethanol 95° (vol/vol) for 30 min in order to kill vegetative cells, then aliquots of various dilutions were plated onto blood agar plates and the plates were incubated at 37°C anaerobically for 24 h.
General DNA techniques.
Chromosomal DNA from C. perfringens culture was extracted by using phenol-chloroform. DNA fragments used in the cloning procedures and PCR products were isolated from agarose gels with the Geneclean II kit (Promega), according to the manufacturer's instructions. Plasmid DNA from E. coli was isolated and purified with the QIAprep spin miniprep kit (Qiagen). PCR analyses were performed using a PTC-100 programmable thermal controller (MJ Research, Inc.) in a final volume of 50 μl containing 0.5 μM each primer, 200 μM each deoxynucleoside triphosphate (dNTP), and 1 U LA Taq DNA polymerase (Takara) in a 1× cloned LA Taq DNA polymerase reaction buffer [20 mM Tris/HCl, pH 8.8, 10 μM KCl, 2 μM MgSO4, 10 μM (NH4)2SO4]. The PCR mixtures were denatured (2 min at 94°C) and the amplification procedure followed, consisting of 30 s at 94°C, annealing for 30 s at 55°C, and ending with an extension step at 72°C for 1 min for a total of 35 cycles. DNA sequences were determined with a 3100 genetic analyzer (Applied Biosystems) sequencer using an ABI PRISM Big Dye Terminator sequencing kit (Perkin Elmer).
Cloning, expression, and purification of Acp-His-tagged fusion protein in E. coli.
Acp-His-tagged protein was expressed in E. coli BL21 codon plus (DE3)-RIL as an Acp-His-tagged fusion protein using the expression vector pET28b (Stratagene). Primers (MWG-Biotech; Invitrogen) 790 F and 790 R (Table 1; see supplemental material) were used to amplify DNA fragment encoding the catalytic domain of Acp (780 bp) from C. perfringens strain 13 total DNA. After amplification, PCR products were digested with BamHI and EcoRI and cloned in the pET28 vector, digested by the same restriction enzymes. This construction created a translational fusion adding 10 N-terminal histidine codons to the acp coding sequence and placed it under the control of the T7 promoter.
TABLE 1.
Calculated and observed m/z values for sodiated molecular ions of muropeptides obtained after hydrolysis of B. subtilis peptidoglycan by Acp or by Acp followed by mutanolysin and purification by RP-HPLCa
| Peak |
m/z |
Δm (Da) | Muropeptide identification | |
|---|---|---|---|---|
| Observed | Calculated | |||
| 1 | 892.38 | 892.37 | −0.01 | Disaccharide tripeptide (NH2) |
| 2 | 1,815.64 | 1,815.77 | −0.13 | Disaccharide tripeptide-disaccharide tetrapeptide (NH2) |
| 3 | 1,814.72 | 1,814.78 | −0.06 | Disaccharide tripeptide-disaccharide tetrapeptide (2NH2) |
| a | 689.31 | 892.37 | −203.06 | Disaccharide tripeptide (NH2) missing 1 GlcNAc |
| b | 1,409.57 | 1,815.77 | −406.2 | Disaccharide tripeptide-disaccharide tetrapeptide (NH2) missing 2 GlcNAc |
| c | 1,612.7 | 1,815.77 | −203.07 | Disaccharide tripeptide-disaccharide tetrapeptide (NH2) missing 1 GlcNAc |
| d | 1,408.59 | 1,814.18 | −405.59 | Disaccharide tripeptide-disaccharide tetrapeptide (2NH2) missing 2 GlcNAc |
| e | 1,611.66 | 1,814.78 | −203.12 | Disaccharide tripeptide-disaccharide tetrapeptide (2NH2) missing 1 GlcNAc |
NH2 indicates the presence of an amidation on the peptidic chain, most probably on mDAP according to reference 2. Δm, difference between calculated and observed m/z values; MurNAc, N-acetyl-muramic acid; GlcNAc, N-acetyl-glucosamine; disaccharide, MurNAc-GlcNAc.
E. coli BL21 codon plus (DE3)-RIL electrocompetent cells were transformed with the resultant plasmid (pCD470) by electroporation (200 Ω; 2.5 kV; 25 μF). Nucleotide sequencing of plasmids from recombinant clones confirmed the insertion of a 780-bp fragment encoding the catalytic domain of Acp. An E. coli recombinant strain was grown at 22°C overnight in 2× YT medium containing selective agents. Protein expression was achieved by induction of cells with 1 mM IPTG followed by subsequent incubation for 5 h at 22°C to avoid formation of inclusion bodies. Acp-His-tagged protein was purified by affinity chromatography on Ni-nitrilotriacetic acid (NTA) columns (Qiagen) under native conditions. Purity of the His-tagged protein was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then dialyzed against sodium phosphate buffer (pH 8.0).
Detection of cell wall lytic enzymes in SDS-PAGE renaturing gel.
Proteins were extracted from bacteria with an SDS treatment as described by Leclerc and Asselin (31). Briefly, the bacterial pellet of 100 ml of the C. perfringens strain 13 cell culture was resuspended in 25 ml of 4% (wt/vol) SDS solution. The suspension was shaken for 120 min and sonicated twice on ice for 1 min. The extract was heated at 90°C for 15 min and centrifuged at 9,500 × g for 20 min, and the supernatant was stored at −20°C. Lytic activity was detected by using SDS-polyacrylamide gels (31) containing 0.2% (wt/vol) Micrococcus lysodeikticus ATCC 4698 (Sigma), Bacillus subtilis 168 HR (14), Clostridium difficile 630, and C. perfringens strain 13 lyophilized or autoclaved cells (121°C for 20 min). SDS-PAGE was performed as described by Laemmli (30) with 15% polyacrylamide. After electrophoresis, the gel was gently shaken at 37°C for 16 h in 50 ml of 25 mM Tris-HCl (pH 8.0) solution containing 1% (vol/vol) Triton X-100 to allow protein renaturation. Clear bands resulting from lytic activity were visualized after staining with 1% (wt/vol) methylene blue (Sigma) in 0.01% (wt/vol) KOH and subsequent destaining with distilled water.
Determination of the hydrolytic bond specificity of Acp on peptidoglycan.
Peptidoglycan from B. subtilis 168 HR vegetative cells was prepared with the protocol described previously for Lactococcus lactis (36) with some modifications. Briefly, pelleted cells were resuspended in 10% (wt/vol) SDS and boiled for 25 min. Insoluble material was recovered by centrifugation (20,000 × g for 10 min at 20°C) and boiled again in 4% (wt/vol) SDS for 15 min after resuspension. The resulting insoluble wall preparation was then washed with hot distilled water (60°C) six times to remove SDS. The covalently attached proteins were removed by treatment with pronase (2 mg/ml) for 90 min at 60°C, then by trypsin (200 mg/ml) for 16 h at 37°C. The walls were then recovered by centrifugation (20,000 × g for 10 min at 20°C), washed once in distilled water, and resuspended in hydrofluoric acid (HF) (48% [vol/vol] solution); the mixture was incubated at 4°C for 24 h. The insoluble material was collected by centrifugation (20,000 × g for 10 min at 20°C) and washed repeatedly by centrifugation and resuspension twice with Tris-HCl buffer (250 mM, pH 8.0) and four times with distilled water until the pH reached 5.0. The material was lyophilized and then stored at −20°C. Peptidoglycan extract (2 mg) was incubated overnight at 37°C with purified Acp-His recombinant protein (160 μg) in a final volume of 250 μl of sodium phosphate buffer (100 mM, pH 8.0). Samples were boiled for 3 min to stop the reaction, and the insoluble material was removed by centrifugation at 14,000 × g for 15 min. Half of the soluble muropeptide fraction was further digested with mutanolysin (2,500 U/ml) (Sigma). The soluble muropeptides obtained after digestion were reduced with sodium borohydride, and the reduced muropeptides were then separated by reverse phase-high-pressure liquid chromatography (RP-HPLC) with an LC module I system (Waters) and a Hypersyl ODS C18 column (250 by 4.6 mm; particle size, 5 μm) (ThermoHypersil-Keystone) at 50°C using ammonium phosphate buffer and methanol linear gradient (11). Muropeptides were analyzed without desalting by MALDI-TOF MS using a Voyager-DE STR mass spectrometer (Applied Biosystems) as reported previously (11).
Preparation of anti-Acp polyclonal antibodies.
Polyclonal antibodies were obtained by BALB/c mouse immunization (agreement number B 41-245-4; AgroBio) consisting of three injections with 75 μg of the Acp-purified catalytic domain.
Western blot analysis.
Proteins separated by SDS-PAGE were electroblotted onto Hybond-enhanced chemiluminescence (ECL) nitrocellulose membranes (4°C for 1 h, 100 V) (Amersham Biosciences). Filters were probed first with autolysin mouse antiserum (or control serum) used at a 1/5,000 dilution and then with goat anti-mouse immunoglobulin G (IgG) conjugated to horseradish peroxidase (GE Healthcare) diluted at 1/5,000. Immunodetection of protein was performed with the SuperSignal West Femto kit (Thermo Scientific) according to the manufacturer's recommendations.
Cell microscopy analysis.
For electron microscopy analysis, bacterial colonies were suspended in 0.1 M sodium cacodylate buffer. The cells were fixed in 2.5% glutaraldehyde-0.1 M sodium cacodylate buffer overnight at 4°C. The resulting pellets were washed twice with 0.1 M sodium cacodylate buffer, and the cells were let to adhere on poly-lysine-precoated coverslips. The specimens were postfixed in 1% osmium teroxyde-0.1 M sodium cacodylate buffer for 1 h at room temperature and dehydrated in graded series of ethanol, followed by critical point drying with CO2 in a Bal-Tec critical point dryer apparatus. The dried specimens were mounted on stubs with carbon tape, and ions were splattered with 15 nm of platin/carbone using a high-resolution ion beam coater (Gatan model 681). Analysis of secondary electron images (SEI) was performed with a Jeol JSM-6700F scanning microscope with a field emission gun operating at 5 kV.
For immunofluorescence assays, C. perfringens strain 13 and C. perfringens strain 13 acp::erm were grown upon end exponential phase (3 h at 37°C in an anaerobic atmosphere) in BHI broth. Samples were fixed aerobically for 1 h at 4°C in 2% paraformaldehyde (PFA). The fixative was removed, the pellets were resuspended in 400 μl of PFA, and 30 μl of the samples was adsorbed on a poly-lysine-precoated slide for 30 min at room temperature. Free aldehyde groups were blocked with 30 μl of NH4Cl2 (50 mM) for 15 min at room temperature and washed twice with 0.5% gelatin-phosphate-buffered saline (PBS). Preimmune serum and immunoserum were depleted for 1 h at 37°C with a mid-exponential acp mutant culture (1:5 dilution). The samples were then incubated with depleted anti-Acp mouse polyclonal antibodies (final dilution of 1:10 in BHI) for 30 min at room temperature, washed twice with BHI, and incubated with donkey anti-mouse IgG (1:200 dilution in BHI) conjugated to Alexa Fluor 488 (Molecular Probes) for 30 min at room temperature. After two washes with BHI to remove unbound antibodies, nuclear staining was performed with To-Pro-3 (1:500) for 10 min and rinsed twice in MilliQ water, and finally a drop of Vectashield mounting medium was added to cover the sample. Samples were visualized on a Zeiss Axiovert 200 M inverted microscope piloted by Zeiss Axiovision 4.4 software (Carl Zeiss, Inc.), operating a black-and-white CoolSNAP HQ charge-coupled device camera (Photometrics).
Cell fractionation.
Cell fractions were prepared as described by Candela and Fouet (8) with some modifications. Mid-exponential (2 h) and late stationary (24 h) phase cultures of C. perfringens strain 13 were centrifuged, and the resulting pellet was suspended in 50 mM Tris-HCl (pH 7.4) and then sonicated (three times for 20 s each time) to disrupt cells. Cell envelope components were separated by centrifugation (8,000 × g for 20 min at 4°C), and the pellet was resuspended in 50 mM Tris-HCl (pH 7.4) containing 5 mM EDTA and 1% Triton X-100, incubated for 1 h at 4°C and centrifuged again (20,000 × g) for 1 h at 4°C in order to separate the membrane (supernatant) and the cell wall (pellet) components.
RNA isolation and real-time quantitative reverse transcription (qRT-PCR).
RNA protection solution (acetone-ethanol 1:1; 20 ml) was immediately added to 20 ml-samples of C. perfringens strain 13 taken at various times points of cell growth and stored at −80°C before its use. After centrifugation, the pellet was washed with Tris-EDTA (10 and 1 mM, pH 8.0) buffer and lysed mechanically with glass beads. The samples were further purified with an RNeasy mini kit (Qiagen) in succeeding steps with spin columns, and then samples were treated first with DNase I (Sigma) and then with a Turbo DNA-free kit (Ambion) according to the manufacturer's recommendations. cDNA was synthesized from two micrograms of total RNA using the Omniscript enzyme (Qiagen) and random 15-mer primers (MWG). A total of 6 ng of cDNA was used for subsequent PCR amplification with primers designed using Beacon Designer software (Premier Biosoft International) (Table 1; see supplemental material), targeting the 16S rRNA (rrn) and acp genes. PCR amplification was performed in a final volume of 15 μl, including 0.5 μM for each couple of primers in an IQ SYBR green Supermix (Bio-Rad). Thermal conditions of the CFX96 real-time PCR detection system (Bio-Rad) were as follows: 10 min at 95°C, followed by 50 repeats of 15 s at 95°C and 1 min at 55°C. A melting curve analysis was done at the end of each run for all primer sets. This resulted in single-product-specific melting curves, and no primer dimers were generated during the runs. A “no-template control” (distilled H2O) and an “RT-negative control” (RNA samples which had not undergone the reverse transcription step) were included in each run in order to confirm the absence of DNA contamination. SYBR green PCRs were performed in duplicate, and for each condition the experiments were done independently in triplicate.
The housekeeping 16S rRNA gene, whose expression is constant during cell growth, was used to normalize the results. The cycle threshold (CT), used to determine the fold change of acp gene expression, was calculated using the comparative critical threshold method (2−ΔΔCT) described by Livak and Schmittgen (33).
5′/3′ RACE PCRs.
Total RNA of growing cells was extracted as described above, and the mRNA 5′ end determination was performed using the 5′/3′ rapid amplification of cDNA ends (RACE) PCR kit, second generation (Roche Applied Science). Three antisense gene-specific primers (SP1, SP2, and SP3) were designed (Table 1; see supplemental material) in order to produce the cDNA and to prepare DNA for sequence analysis.
Obtention of acp gene knockout mutants.
Sigma TargeTron Design predicted 7 TargeTron insertion sites in the Acp C-terminal-encoding gene (corresponding to the catalytic domain). The insertion site in the antisense strand at position 3129 to 3130 in the acp open reading frame (ORF) was preferentially chosen to generate ClosTron-intron modifications. Retargeted region L1.LtrB intron of the ClosTron, responsible for target specificity, was obtained by PCR from primers designed by Sigma TargeTron, as seen on their website (Table 1; see supplemental material). The 350-bp product was then digested and ligated into the pMTL007 ClosTron-shuttle vector and transformed by heat shock into E. coli TOP10 in order to verify sequence of the retargeted intron specified for acp insertion.
The recombinant pMTL007 containing the modified acp intron (pCD405) was then electroporated into electrocompetent C. perfringens strain 13 as described previously (28). The transformation mixture was plated onto BHI agar supplemented with thiamphenicol (15 μg/ml) and cycloserine (Cycloserin; 250 μg/ml) and left overnight at 37°C in anaerobic conditions to select clones of C. perfringens transformed by pCD405. These selected clones were then plated on BHI agar supplemented with erythromycin (5 μg/ml) and cycloserine (250 μg/ml) and incubated overnight at 37°C in anaerobic conditions to select clones harboring the spliced erythromycin retrotransposition activated marker (ErmRAM), which indicates intron integration.
PCR analyses were performed to verify the acp intron insertion from genomic extracts (QIAamp DNA mini kit; Qiagen) of the selected clones, using the following different combinations of primers: target-R primer (790 R) and EBS universal primer to demonstrate the intron insertion in acp and ErmRAM-F and ErmRAM-R primers to demonstrate the ErmRAM splicing. Amplification was performed on a PTC-100 programmable thermal controller (Bio-Rad) in a final volume of 50 μl containing 0.5 μM each primer, 200 μM dNTPs, 2.5 μM MgCl2, and 1 U LA Taq DNA polymerase (Takara) in a 1× cloned LA Taq DNA polymerase buffer [20 mM Tris-HCl, pH 8.8, 10 μM KCl, 2 μM MgSO4, 10 μM (NH4)2SO4]. After denaturation (1 min at 94°C), DNA was amplified according to the following procedure: denaturation for 30 s at 94°C, annealing for 30 s at 50°C, and extension at 72°C for 1 min and 30 s, for a total of 35 cycles, and a final extension step at 72°C for 10 min.
MIC determination.
MICs of vancomycin, teicoplanin, penicillin G, and amoxicillin against C. perfringens strain 13 acp::erm and C. perfringens strain 13 were determined by the Etest method from bacterial suspensions at 0.5 McFarland turbidity according to the manufacturer's recommendations (bioMérieux).
Autolysis assays.
For Triton X-100-induced autolysis, overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.1 in BHI broth and grown at 37°C in an anaerobic atmosphere until the OD600 reached 1.0. Cells were harvested, washed twice, and suspended in 50 mM potassium phosphate buffer containing 0.05% of Triton X-100. Cells were incubated at 37°C, and the lysis was measured by the OD600 of the bacterial suspension with an Ultrospec 1100 pro spectrophotometer (Amersham Biosciences) every 30 min to follow cell lysis.
For bile salts autolysis assay, bovine bile (Sigma) was added to growing cells (OD600 = 1.0) to a final concentration of 0.3%, and the autolysis was checked by measuring the OD600 every 30 min.
For antibiotic-induced autolysis, overnight cultures were diluted to an OD600 of 0.1 in BHI broth and grown at 37°C in an anaerobic atmosphere until the OD600 reached 1.0. Vancomycin, teicoplanin, penicillin G, and amoxicillin were added to a final concentration corresponding to 3× MIC (2.25 μg/ml, 0.096 μg/ml, 0.141 μg/ml, and 0.060 μg/ml, respectively). Cells were incubated at 37°C, and the OD600 of the bacterial suspension was measured every 30 min following cell lysis. For inhibition assays, polyclonal anti-Acp antibodies or preimmune serum was added to the growing culture as it reached an OD600 of 0.3.
Nucleotide sequence accession number.
The GenBank accession number for the acp sequence reported in this paper is GU192369.
RESULTS
Identification of a putative PGH-encoding gene in the C. perfringens strain 13 genome sequence.
We identified a 3,390-bp ORF in the C. perfringens strain 13 genome sequence encoding a putative PGH through sequence similarity analysis with other PGHs of Gram-positive bacteria, including Acd of C. difficile 630 (13). We amplified and sequenced the corresponding gene, named acp, and using RACE PCR analysis we found that the mRNA 5′ end of acp was located 113 bp upstream of the initiation codon. The potential −35 (TTGGCT) and −10 (TATAAT) boxes were located 7 bp upstream of the deduced 5′ end of the acp transcript.
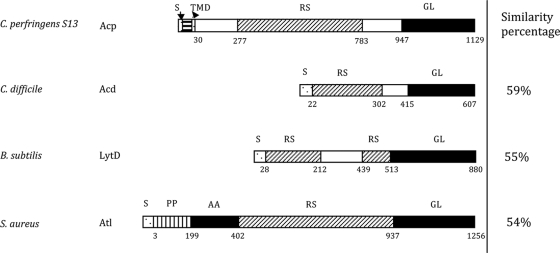
The acp gene encodes a putative protein of 1,129 amino acids with an expected molecular mass of 122,388 Da with a pI of 8.79. Acp protein has a structural organization with three main domains, a signal sequence domain, an N-terminal domain exhibiting 10 repeated sequences (of 51 or 52 amino acids in length), and a putative C-terminal catalytic domain (182 amino acids) (Fig. 1). The first 30 N-terminal residues of Acp were determined as a putative signal peptide sequence by SignalP (http://www.cbs.dtu.dk/services/SignalP/), with a possible cleavage site between amino acid residues 30 and 31. They could also constitute an N-terminal signal anchor, since a transmembrane helix is predicted in positions 8 to 24 by TMpred software (www.ch.embnet.org/software/TMPRED_form.html). Alignment of the Acp sequence with the sequences of C. difficile Acd (13), B. subtilis LytD (48), and Staphylococcus aureus Atl (43) (Fig. 1) revealed higher similarities in the C-terminal amino acid regions, suggesting that the acp gene encodes a putative PGH with N-acetylglucosaminidase activity, as described for Acd, LytD, and Atl.
FIG. 1.
Modular organization of C. perfringens Acp compared to C. difficile Acd, B. subtilis LytD, and S. aureus Atl. Percent similarity between the catalytic domain of Acp (black rectangles) and the three other autolysins are indicated to the right. S, signal sequence; RS, repeated sequence; GL, N-acetylglucosaminidase; AA, l-alanyl-amidase; TMD, transmembrane domain.
Expression, purification, and bacteriolytic activity of the Acp catalytic domain.
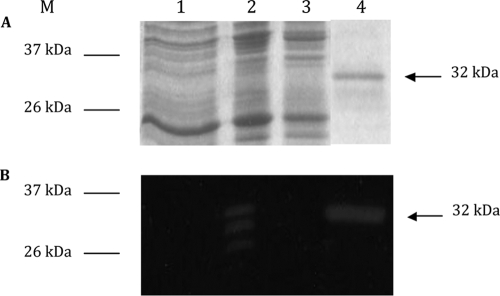
We initially intended to purify the entire Acp protein to demonstrate the activity of Acp. Because of unsuccessful experiments, we chose to clone a truncated fragment of acp (780-bp) corresponding to the C-terminal catalytic domain of Acp. The resulting recombinant pCD470 plasmid was transformed in E. coli BL21 codon plus (DE3)-RIL to overexpress the C-terminal domain of Acp by IPTG induction. His-tagged Acp protein was visualized as a single 32-kDa protein band by using SDS-PAGE after Coomassie blue staining and produced a clear hydrolysis band (deduced protein size, 32.5 kDa) in renaturing SDS-PAGE experiments with renaturation buffer at pH 8.0 containing M. lysodeikticus lyophilized cells as substrate (Fig. 2). The same activity of the 32-kDa protein was also detected with C. perfringens strain 13, C. difficile 630, and B. subtilis 168 HR lyophilized and autoclaved cells as substrates (data not shown).
FIG. 2.
Purification of His-tagged Acp catalytic domain. Analysis of protein extracts on SDS-PAGE (A) and renaturing SDS-PAGE (B) containing 0.2% Micrococcus lysodeikticus cell wall (zymogram). Lane 1, crude cell extract of E. coli BL21 carrying pET28; lane 2, crude cell extract of E. coli BL21 carrying pCD470 induced by 1 mM IPTG; lane 3, crude cell extract of E. coli BL21 carrying noninduced pCD470; and lane 4, purified His-tagged Acp catalytic domain under native conditions. M, benchmark protein ladders (Invitrogen).
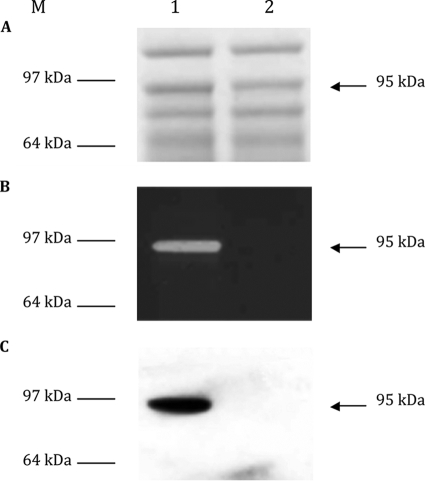
Proteins extracted from C. perfringens strain 13 (Fig. 3A, lane 1) were also assayed for bacteriolytic activity on renaturing SDS-PAGE gels containing M. lysodeikticus, C. difficile strain 630, or C. perfringens strain 13 lyophilized or autoclaved cells. A hydrolysis band was detected in SDS extracts with a molecular mass of 95 kDa (Fig. 3B, lane 1), lower than the expected protein, suggesting that this band should correspond to an active degradation product of Acp. Western blot analysis with specific anti-Acp polyclonal antibody revealed only one band at 95 kDa (Fig. 3C, lane 1).
FIG. 3.
Detection of Acp in the total crude extract of C. perfringens strain 13 (lane 1) and C. perfringens strain 13 acp::erm (lane 2). (A) Coomassie blue-stained SDS-PAGE. (B) Methylene blue-stained zymogram containing M. lysodeikticus lyophilized cells. (C) Western blot. M, molecular mass marker (SeeBlue Plus 2 pre-stained standard; Invitrogen).
Determination of Acp hydrolytic bond specificity.
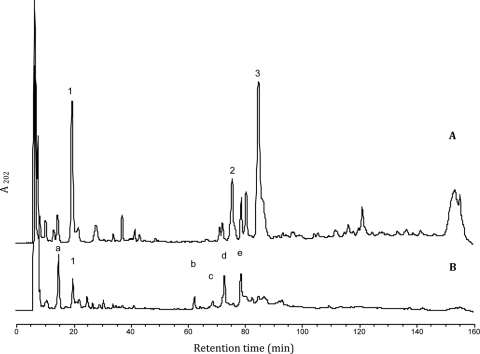
Sequence homology analysis of Acp with other PGHs of Gram-positive bacteria (Fig. 1) suggested that Acp might be an N-acetylglucosaminidase. In order to establish the hydrolytic specificity of Acp, the His-tagged Acp-purified protein was used to digest cell walls of B. subtilis 168 HR. Mutanolysin, a PGH with muramidase activity, was used as a digestion control. The RP-HPLC profile analysis showed that soluble muropeptides released by Acp digestion (Fig. 4A) were different from those released by mutanolysin (data not shown), indicating that Acp does not possess muramidase activity. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis of peaks “1,” “2,” and “3” generated molecular ions with m/z values of 892.37, 1815.77, and 1814.78, respectively (Table 1). According to previous data (2, 25), these m/z values correspond to a disaccharide tripeptide muropeptide with one amidation for peak 1 and to a disaccharide tripeptide-disaccharide tetrapeptide with one or two amidations for peaks 2 and 3, respectively (Table 1; Fig. 5). The soluble muropeptide fraction obtained by Acp digestion was further incubated with mutanolysin and then analyzed by RP-HPLC. The obtained profile revealed new peaks (Fig. 4B, a to e), which were analyzed by MALDI-TOF MS. The observed m/z values (Table 1) indicate the loss of one or two N-acetylglucosamine residues from the muropeptides detected in peaks 1, 2, and 3 identified in Fig. 4A. The deduced structures are presented in Fig. 5. These results reveal that the muropeptides generated by Acp hydrolysis could be further cleaved by a muramidase (mutanolysin), indicating that N-acetylglucosamine is present on the reducing end of the disaccharide of these muropeptides. Finally, these results demonstrate that Acp has N-acetylglucosaminidase specificity.
FIG. 4.
RP-HPLC analysis of the soluble muropeptides released from B. subtilis vegetative peptidoglycan after incubation with Acp (A) or with Acp and mutanolysin (B). The numbers and letters indicate the peaks analyzed by MALDI-TOF MS.
FIG. 5.
Structure of the muropeptides from B. subtilis peptidoglycan obtained after Acp digestion (peaks 1, 2, and 3) or Acp plus mutanolysin digestion (peaks a to e). Peak numbers or letters refer to the peaks on the chromatograms presented in Fig. 4. According to their mass (Table 1), peaks 2, b, and c bear one amidation whereas peaks 3, d, and e bear two amidations, located most probably on mDAP (2).
Analysis of the acp gene in various strains of C. perfringens.
The variability of acp was studied in 20 strains of C. perfringens (clinical isolates from feces, suppurations, or blood, or strains from the Collection Institut Pasteur Paris). The acp gene was detected in the 20 strains, and the catalytic domain was found conserved by nucleotidic sequencing. Conversely, the N-terminal part of Acp displayed 7 to 10 repeated sequences as revealed by PCR experiments.
Transcriptional and translational analysis of acp during growth of C. perfringens strain 13.
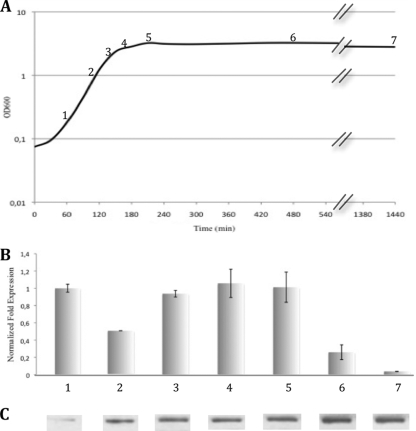
Transcriptional analysis of acp at different stages of growth (Fig. 6A) revealed that the acp gene is expressed constitutively during vegetative growth, with a 4-fold decrease at the end of the stationary phase (Fig. 6B). The expression of acp at time “24 h,” which corresponds to the beginning of sporulation (as revealed by spore detection) decreased dramatically. Western blot analysis revealed that Acp, accumulated during growth of C. perfringens strain 13, including the stationary phase (Fig. 6C), is highly stable.
FIG. 6.
Analysis of acp during growth of C. perfringens strain 13 (BHI medium at 37°C under anaerobic conditions). (A) Growth of C. perfringens strain 13 followed by OD600; numbers 1 to 7 represent the different times of total RNA and protein preparation used for qRT-PCR and Western blot analysis. (B) qRT-PCR analysis results showing relative expression of acp normalized by the housekeeping gene 16S rRNA during the different growth phases of C. perfringens strain 13 and compared to the early exponential phase. Error bars indicate standard deviation. (C) Western blot of Acp with polyclonal anti-Acp antibodies.
Chromosomal acp mutagenesis using group II intron strategy.
Mobile group II introns are site-specific retroelements that use a retrohoming mechanism to directly insert the excised intron lariat RNA into a specific DNA target site and reverse transcribe into DNA that inactivates the gene of interest (19). Among the 7 potential sites of insertion in the catalytic domain of acp, we chose the antisense site at the beginning of the catalytic domain. Positive mutants were selected by PCR screening, allowing determination of the directional intron insertion (data not shown). Proteins from C. perfringens strain 13 and C. perfringens strain 13 acp::erm were extracted by SDS treatment and analyzed by zymography and Western blot analysis with anti-Acp immunoserum. No lysis band or immunoreactive protein was detected from C. perfringens strain 13 acp::erm extract (Fig. 3), confirming the acp knockout mutant of C. perfringens strain 13 and indicating that Acp is the major active autolysin in growing C. perfringens strain 13.
Implication of Acp in cell separation.
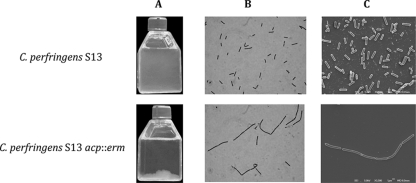
C. perfringens strain 13 and C. perfringens strain 13 acp::erm showed the same growth curve (see Fig. 11A). However, the culture sedimentation of the acp mutant is different compared to that of the wild type as shown in Fig. 7A. Light microscopy examination and scanning electron microscopy revealed long chains for C. perfringens strain 13 acp::erm compared to those for the parental C. perfringens strain 13 (Fig. 7B), suggesting that septum formation and/or cell separation was defective (Fig. 7C).
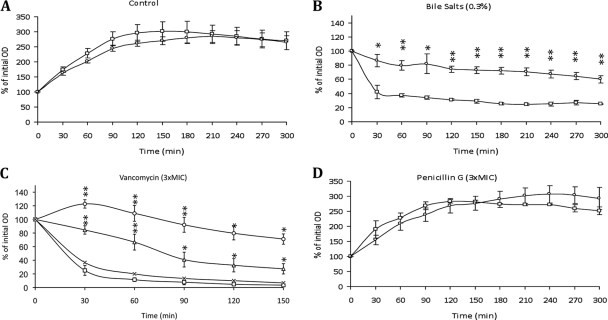
FIG. 11.
Impact of acp inactivation on stress-induced autolysis of C. perfringens strain 13. Growth of mid-exponential phase cultures was determined at 37°C in the absence (A) or presence (B) of 0.3% of bile salts, 3× MIC of vancomycin (C), or 3× MIC of penicillin G (D) by measuring the optical densities (expressed as percent initial absorbance at an optical density at 600 nm). Shown are results for C. perfringens strain 13 (□), C. perfringens strain 13 acp::erm (○), C. perfringens strain 13 plus preimmune serum (×), and C. perfringens strain 13 plus polyclonal anti-Acp antibodies (▵). Error bars indicate standard deviations, and asterisks indicate statistically significant differences compared to C. perfringens strain 13 (*, P < 0.05; **, P < 0.01).
FIG. 7.
Macroscopic and microscopic observations of C. perfringens strain 13 and C. perfringens strain 13 acp::erm. (A) Overnight BHI broth culture. (B) Gram staining microscopy view. (C) Scanning electron microscopy.
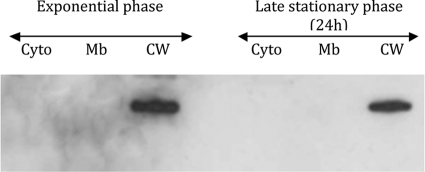
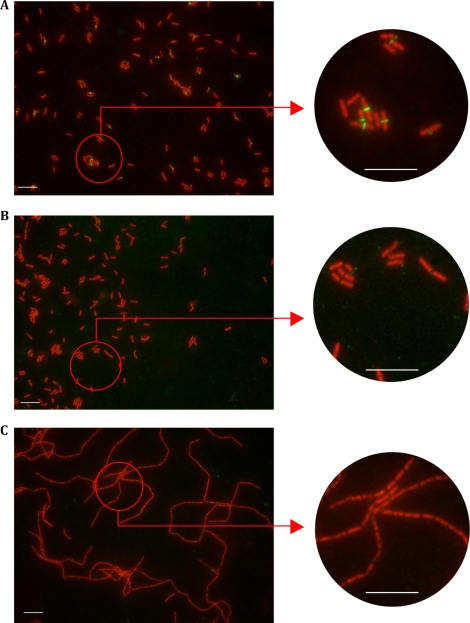
Cell fractionation of C. perfringens strain 13 showed that Acp is located in the cell wall fraction of C. perfringens (Fig. 8). Using indirect immunofluorescence staining with anti-Acp antibodies, we localized Acp at the division septum. Of note, many cells show fluorescence at a single pole, suggesting that these represent recently divided septa. No staining was observed with the preimmune serum or with C. perfringens strain 13 acp::erm incubated with anti-Acp polyclonal antibodies (Fig. 9).
FIG. 8.
Cell fractionation localization of Acp during exponential and late stationary growth phases of C. perfringens as follows: cytoplasm (Cyto), membrane (Mb), and cell wall (CW). Protein extracts were analyzed by Western blotting with specific anti-Acp antibodies.
FIG. 9.
Immunolocalization of Acp on C. perfringens strain 13 during exponential growth phase by indirect immunofluorescence. Bar, 10 μM. Red staining fluorescence is due to the monomeric cyanin nucleic acid stain To-Pro-3, and green fluorescence is due to the secondary anti-mouse IgG coupled with Alexa Fluor 488 fluorophore. (A) C. perfringens strain 13 stained with depleted anti-Acp immune serum. (B) C. perfringens strain 13 stained with depleted anti-Acp preimmune serum as a control. (C) C. perfringens strain 13 acp::erm stained with depleted anti-Acp immune serum.
We also examined the possible implication of Acp in the sporulation of C. perfringens. No significant difference in spore counting at time 24 h was found between the wild type (6.2 ± 0.1 per 1,000 cells) and acp mutant strains (10.0 ± 5.0 per 1,000 cells), indicating that Acp is not involved in sporulation of C. perfringens. Overall, Acp appears as the main autolysin implicated in the cell separation during the vegetative growth of C. perfringens.
Triton X-100-induced autolysis assay.
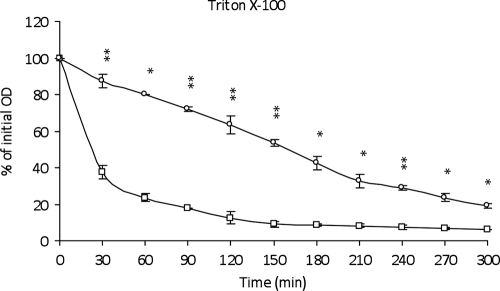
Triton X-100 is a nonionic detergent that forms micelles with lipoteichoic acids (LTA) which are known to inhibit the autolytic activity in the peptidoglycan (41). Then Triton X-100 can, by its interaction with LTA, reveal the general bacterial autolytic system. The effect of 0.05% of Triton X-100 upon autolysis was checked in the mid-exponential phase of both parental and acp mutant strains of C. perfringens strain 13. The parental strain lysed significantly more rapidly than the acp mutant, indicating that Acp is strongly involved in the Triton X-100-induced autolysis (Fig. 10) during the growth of C. perfringens.
FIG. 10.
Autolytic activities of C. perfringens strain 13 during Triton X-100-induced autolysis. The autolysis is expressed in percent initial absorbance at an optical density of 600 nm. Shown are results for C. perfringens strain 13 (□) and C. perfringens strain 13 acp::erm (○). Error bars indicate standard deviations, and asterisks indicate statistically significant differences (*, P < 0.05; **, P < 0.01).
Bile salts autolysis assay.
Bile salts are responsible for phospholipid solubilization, allowing the disappearance of the membrane and then the higher turgor pressure that can make the cell wall more fragile (23). We examined if Acp was implicated in the autolysis induced by a physiological concentration of bile salts (0.3%) added in the growth medium (Fig. 11B). The acp mutant appeared to be significantly more resistant to this autolysis stress than the parental strain, suggesting that Acp has a role in the bile salt-induced autolysis in C. perfringens strain 13.
Antibiotic-induced lysis.
Autolysins have been reported to be implicated in antibiotic-induced lysis (7, 15). We studied the possible role of Acp in antibiotic-induced lysis using two glycopeptides (vancomycin and teicoplanin) and two β-lactams (penicillin G and amoxicillin) which interfere with the peptidoglycan biosynthesis and are known to be bactericidal antibiotics. The acp mutant was more resistant to vancomycin-induced lysis than the parent strain (Fig. 11C). However, no difference was found between parental and acp mutant strains for the teicoplanin-induced autolysis (data not shown). Assays with penicillin G (Fig. 11D) or amoxicillin did not show any lysis for both parental and acp mutant strains.
Inhibition assays.
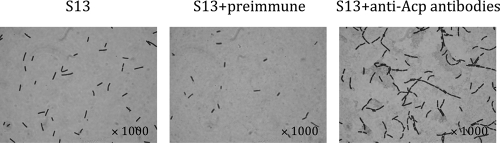
We initially intended to complement the acp mutant but were unable, despite repeat experiments, to clone the entire acp gene for this purpose. Consequently we decided to perform antibodies inhibition assays as previously described for the characterization of AtlA in Streptococcus mutans (50). C. perfringens strain 13 grown with the anti-Acp antibodies formed longer chains than when grown with the corresponding preimmune serum (Fig. 12), as with the acp mutant. Furthermore, when C. perfringens strain 13 was grown with anti-Acp antibodies and then incubated with vancomycin, a reduced cell lysis was observed (Fig. 11C), as with the acp mutant. Based on these results, we conclude that the phenotype of the acp mutant was due to acp inactivation.
FIG. 12.
Effect of anti-Acp polyclonal antibodies on chain length. C. perfringens strain 13 was grown in BHI broth including polyclonal anti-Acp antibody or preimmune serum (1/50).
DISCUSSION
The aim of this study was to characterize the first known autolysin involved in peptidoglycan hydrolysis during vegetative growth of C. perfringens. Like most previously described bacterial PGHs (13, 18, 35), Acp has a modular organization with three main domains consisting of a signal peptide, an N-terminal domain characterized by repeated sequences, and a C-terminal catalytic domain conferring the hydrolytic activity. The first 30 amino acid residues of the N-terminal domain of Acp might constitute a putative signal sequence and probably a retention site with a transmembrane domain, as described for cell wall hydrolases in B. subtilis (56).
The C-terminal domain (residues 947 to 1113) of Acp, which is highly conserved, exhibited significant homology with the catalytic domain of several N-acetylglucosaminidases of low-G+C Gram-positive bacteria, such as Acd of C. difficile (13), LytD of B. subtilis (48), and Atl of S. aureus (43). However, Acp could have a different activity than that predicted by sequence homology, as it was reported for the B. subtilis autolysin LytG (24). Since the purified recombinant catalytic domain of Acp was found to hydrolyze the B. subtilis vegetative cell wall in renaturing SDS-PAGE experiments, we investigated the hydrolytic bond specificity of Acp on the B. subtilis vegetative peptidoglycan, whose molecular structure has been previously studied in detail (2). RP-HPLC and MALDI-TOF analyses of muropeptides generated by Acp hydrolysis concluded that Acp has N-acetylglucosaminidase activity. According to the protein organization (Fig. 1), Acp is a monofunctional type of PGH, whereas others were described as bifunctional autolysins, as was Atl of S. aureus (43), AtlL of Staphylococcus lugdunensis (5), and Aas of Staphylococcus saprophyticus (21), which exhibit two catalytic domains.
Repeated sequences are known to be involved in cell wall targeting, such as peptidoglycan binding (3), although some of them seem to be implicated in the virulence of bacteria (22, 26, 47, 59). The N-terminal domain of Acp contains 10 putative SH3 modules of 51 or 52 amino acids. Although the function of SH3 modules is not exactly known, it is tempting to speculate that they are involved in attaching cell wall-degrading enzymes to their substrate by binding directly either to murein or to other cell wall components, such as carbohydrates and/or polyproline stretches (27). We are now interested to know if putative SH3 modules of Acp display such an anchoring function.
The acp gene is constitutively transcribed during the exponential growth phase with a decrease in the late stationary phase. In addition, the expression of acp decreases drastically at the beginning of sporulation, and the quantification of spores is not affected by acp mutation. Overall, these data demonstrate that Acp is specifically implicated in vegetative growth and not in sporulation.
The zymographic analysis showed a unique hydrolytic band for C. perfringens strain 13 but not with the acp mutant, indicating that Acp is the major active PGH expressed during vegetative growth. The localization of Acp into the cell wall fraction, and further at the separation septum, suggests that Acp might be implicated in septum formation and/or separation of cells during vegetative growth. The acp mutant grows in long chains (as the wild-type strain incubated with anti-Acp antibodies), but the presence of normal septa on dividing cells strongly supports the hypothesis that Acp is most probably implicated in the separation of the daughter cells, unlike LytR which is essential for normal septum formation during exponential growth of Streptococcus pneumoniae (29). Thin-section transmission electron micrographs did not reveal any significant difference in the wall thickness of both parental and mutant strains (data not shown), indicating that Acp does not play a critical function in cell wall remodeling. Thus, Acp is involved mainly in daughter cell separation, as already described for LytB of S. pneumoniae (12). It is the first PGH characterized as implicated in cell separation during vegetative growth of C. perfringens, whereas autolysins implicated in sporulation and germination (SleC and SleM) had been previously described (9, 37, 38, 45).
Autolysins, belonging to the PGH family, are characterized by their ability to induce bacterial autolysis. Triton X-100 autolysis is one of the most recognized tests showing an overview of bacterial autolytic systems. This nonionic detergent induces the release of acylated lipoteichoic acids and excretion of membrane lipids (49), which are known to regulate the autolytic system in Gram-positive bacteria (10, 17) by inhibiting the cell wall autolysin activity. The resistance of the acp mutant to Triton X-100-induced lysis demonstrates that Acp supports a critical function in the C. perfringens autolytic system during vegetative growth.
This led us to test the implication of Acp in the following different types of stresses potentially inducing lysis: oxidative stress (H2O2, oxygen), ethanol (7.5% and 15%), osmotic stress (NaCl), acid pH (from 3 to 6), bile salts, and cell wall-targeting antibiotics (two β-lactams and two glycopeptides). Oxidative, ethanol, osmotic, and acid pH stresses did not reveal a significant difference in the responses of wild-type and acp mutant strains (data not shown). Conversely, bile salts and vancomycin exposure displayed a significant difference in lysis of parental and acp mutant strains.
It has been reported that S. pneumoniae strains are resistant to bile salts when the major autolysin LytA exhibits a 6-bp deletion in the binding domain (42). In our study, the acp mutant appears more resistant to bile salts than the parental strain, suggesting a role of Acp in bile-induced lysis of C. perfringens. It will be interesting to test strains with a variable number of repeated sequences in Acp to determine if the binding domain of Acp can be implicated in bile sensitivity.
Autolytic enzymes are also involved in cell wall turnover and cell lysis induced by cell wall-targeting antibiotics; mutants defective in autolysin show reduced rates of cell wall turnover and/or absence of lysis in the presence of such antibiotics in S. aureus or in S. pneumoniae (44, 57). Concerning β-lactams, penicillin G or amoxicillin exposure did not show any significant difference between both parental and acp mutant strains, suggesting that Acp does not interfere with β-lactam activity in C. perfringens. Of note, AtlA, another PGH with N-acetylglucosaminidase activity, has been reported to contribute to bactericidal activity of amoxicillin (7) and penicillin G (46) in Enterococcus faecalis, suggesting that PGH exhibiting similar hydrolytic activity may have different implications in the response to antibiotic exposure.
Concerning glycopeptides, we observed that Acp was implicated in vancomycin-induced lysis but not, or at a lower rate, in teicoplanin-induced lysis. We previously reported such a difference between vancomycin and teicoplanin bactericidal activities in Staphylococcus lugdunensis (6). Transcriptional (qRT-PCR) and translational (Western blot) analyses did not reveal any difference in Acp expression when cells were exposed to vancomycin at a subinhibitory concentration (data not shown). This indicates that acp is not induced by vancomycin and suggests that the activity of Acp causes the lysis of C. perfringens strain 13 when exposed to vancomycin.
In conclusion, Acp appears as the major PGH expressed in vegetative growth of C. perfringens and is the first known N-acetylglucosaminidase autolysin implicated in daughter cell separation in this species. Moreover, Acp appears implicated in lysis induced by bile salts and vancomycin. Further studies should also explore the possible contribution of Acp to the virulence of C. perfringens; for example, through adhesive properties, as reported for staphylococci (20), or by facilitating the release of intracellular toxins, as suggested for S. pneumoniae (4).
Supplementary Material
Acknowledgments
This work was supported by resources provided by University of Rouen, Rouen University Hospital, Institut Pasteur, Paris, France, and the research grant (A1057637) from the U.S. Public Health Service.
We thank Nigel P. Minton and John T. Heap for providing the ClosTron gene knockout system and Agnès Fouet and Eliette Touati for helpful discussions.
Footnotes
Published ahead of print on 26 February 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allignet, J., P. England, I. Old, and N. El Solh. 2002. Several regions of the repeat domain of the Staphylococcus caprae autolysin, AtlC, are involved in fibronectin binding. FEMS Microbiol. Lett. 213:193-197. [DOI] [PubMed] [Google Scholar]
- 2.Atrih, A., G. Bacher, G. Allmaier, M. P. Williamson, and S. J. Foster. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., and M. Bycroft. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113-1119. [DOI] [PubMed] [Google Scholar]
- 4.Berry, A. M., R. A. Lock, D. Hansman, and J. C. Paton. 1989. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect. Immun. 57:2324-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgeois, I., E. Camiade, R. Biswas, P. Courtin, L. Gibert, F. Gotz, M. P. Chapot-Chartier, J. L. Pons, and M. Pestel-Caron. 2009. Characterization of AtlL, a bifunctional autolysin of Staphylococcus lugdunensis with N-acetylglucosaminidase and N-acetylmuramoyl-l-alanine amidase activities. FEMS Microbiol. Lett. 290:105-113. [DOI] [PubMed] [Google Scholar]
- 6.Bourgeois, I., M. Pestel-Caron, J. F. Lemeland, J. L. Pons, and F. Caron. 2007. Tolerance to the glycopeptides vancomycin and teicoplanin in coagulase-negative staphylococci. Antimicrob. Agents Chemother. 51:740-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bravetti, A. L., S. Mesnage, A. Lefort, F. Chau, C. Eckert, L. Garry, M. Arthur, and B. Fantin. 2009. Contribution of the autolysin AtlA to the bactericidal activity of amoxicillin against Enterococcus faecalis JH2-2. Antimicrob. Agents Chemother. 53:1667-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Candela, T., and A. Fouet. 2005. Bacillus anthracis CapD, belonging to the gamma-glutamyltranspeptidase family, is required for the covalent anchoring of capsule to peptidoglycan. Mol. Microbiol. 57:717-726. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Y., S. Miyata, S. Makino, and R. Moriyama. 1997. Molecular characterization of a germination-specific muramidase from Clostridium perfringens S40 spores and nucleotide sequence of the corresponding gene. J. Bacteriol. 179:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleveland, R. F., A. J. Wicken, L. Daneo-Moore, and G. D. Shockman. 1976. Inhibition of wall autolysis in Streptococcus faecalis by lipoteichoic acid and lipids. J. Bacteriol. 126:192-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courtin, P., G. Miranda, A. Guillot, F. Wessner, C. Mezange, E. Domakova, S. Kulakauskas, and M. P. Chapot-Chartier. 2006. Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an L, D-carboxypeptidase involved in peptidoglycan maturation. J. Bacteriol. 188:5293-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Las Rivas, B., J. L. Garcia, R. Lopez, and P. Garcia. 2002. Purification and polar localization of pneumococcal LytB, a putative endo-beta-N-acetylglucosaminidase: the chain-dispersing murein hydrolase. J. Bacteriol. 184:4988-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhalluin, A., I. Bourgeois, M. Pestel-Caron, E. Camiade, G. Raux, P. Courtin, M. P. Chapot-Chartier, and J. L. Pons. 2005. Acd, a peptidoglycan hydrolase of Clostridium difficile with N-acetylglucosaminidase activity. Microbiology 151:2343-2351. [DOI] [PubMed] [Google Scholar]
- 14.Foster, S. J. 1991. Cloning, expression, sequence analysis and biochemical characterization of an autolytic amidase of Bacillus subtilis 168 trpC2. J. Gen. Microbiol. 137:1987-1998. [DOI] [PubMed] [Google Scholar]
- 15.Gazzola, S., and P. S. Cocconcelli. 2008. Vancomycin heteroresistance and biofilm formation in Staphylococcus epidermidis from food. Microbiology 154:3224-3231. [DOI] [PubMed] [Google Scholar]
- 16.Ghuysen, J. M., D. J. Tipper, and J. L. Strominger. 1966. Enzymes that degrade bacterial cell walls. Methods Enzymol. 8:685-699. [Google Scholar]
- 17.Ginsburg, I. 2002. Role of lipoteichoic acid in infection and inflammation. Lancet Infect. Dis. 2:171-179. [DOI] [PubMed] [Google Scholar]
- 18.Goda, H. M., K. Ushigusa, H. Ito, N. Okino, H. Narimatsu, and M. Ito. 2008. Molecular cloning, expression, and characterization of a novel endo-alpha-N-acetylgalactosaminidase from Enterococcus faecalis. Biochem. Biophys. Res. Commun. 375:441-446. [DOI] [PubMed] [Google Scholar]
- 19.Gupta, P., and Y. Chen. 2008. Chromosomal engineering of Clostridium perfringens using group II introns. Methods Mol. Biol. 435:217-228. [DOI] [PubMed] [Google Scholar]
- 20.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 21.Hell, W., H. G. Meyer, and S. G. Gatermann. 1998. Cloning of aas, a gene encoding a Staphylococcus saprophyticus surface protein with adhesive and autolytic properties. Mol. Microbiol. 29:871-881. [DOI] [PubMed] [Google Scholar]
- 22.Hirst, R. A., B. Gosai, A. Rutman, C. J. Guerin, P. Nicotera, P. W. Andrew, and C. O'Callaghan. 2008. Streptococcus pneumoniae deficient in pneumolysin or autolysin has reduced virulence in meningitis. J. Infect. Dis. 197:744-751. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann, A. F. 1994. Bile acids, p. 677-718. In I. M. Arias, J. L. Boyer, N. Fausto, W. B. Jackoby, D. A. Schachter, and D. A. Shafritz (ed.), The liver: biology and pathobiology. Raven Press, New York, NY.
- 24.Horsburgh, G. J., A. Atrih, M. P. Williamson, and S. J. Foster. 2003. LytG of Bacillus subtilis is a novel peptidoglycan hydrolase: the major active glucosaminidase. Biochemistry 42:257-264. [DOI] [PubMed] [Google Scholar]
- 25.Huard, C., G. Miranda, F. Wessner, A. Bolotin, J. Hansen, S. J. Foster, and M. P. Chapot-Chartier. 2003. Characterization of AcmB, an N-acetylglucosaminidase autolysin from Lactococcus lactis. Microbiology 149:695-705. [DOI] [PubMed] [Google Scholar]
- 26.Humann, J., R. Bjordahl, K. Andreasen, and L. L. Lenz. 2007. Expression of the p60 autolysin enhances NK cell activation and is required for Listeria monocytogenes expansion in IFN-gamma-responsive mice. J. Immunol. 178:2407-2414. [DOI] [PubMed] [Google Scholar]
- 27.Humann, J., and L. L. Lenz. 2009. Bacterial peptidoglycan degrading enzymes and their impact on host muropeptide detection. J. Innate Immun. 1:88-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jirásková, A., L. Vitek, J. Fevery, T. Ruml, and P. Branny. 2005. Rapid protocol for electroporation of Clostridium perfringens. J. Microbiol. Methods 62:125-127. [DOI] [PubMed] [Google Scholar]
- 29.Johnsborg, O., and L. S. Havarstein. 2009. Pneumococcal LytR, a protein from the LytR-CpsA-Psr family, is essential for normal septum formation in Streptococcus pneumoniae. J. Bacteriol. 191:5859-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Leclerc, D., and A. Asselin. 1989. Detection of bacterial cell wall hydrolases after denaturing polyacrylamide gel electrophoresis. Can. J. Microbiol. 35:749-753. [DOI] [PubMed] [Google Scholar]
- 32.Lenz, L. L., S. Mohammadi, A. Geissler, and D. A. Portnoy. 2003. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 100:12432-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 34.Margot, P., M. Pagni, and D. Karamata. 1999. Bacillus subtilis 168 gene lytF encodes a gamma-D-glutamate-meso-diaminopimelate muropeptidase expressed by the alternative vegetative sigma factor, sigmaD. Microbiology 145:57-65. [DOI] [PubMed] [Google Scholar]
- 35.Mesnage, S., F. Chau, L. Dubost, and M. Arthur. 2008. Role of N-acetylglucosaminidase and N-acetylmuramidase activities in Enterococcus faecalis peptidoglycan metabolism. J. Biol. Chem. 283:19845-19853. [DOI] [PubMed] [Google Scholar]
- 36.Meyrand, M., A. Boughammoura, P. Courtin, C. Mezange, A. Guillot, and M. P. Chapot-Chartier. 2007. Peptidoglycan N-acetylglucosamine deacetylation decreases autolysis in Lactococcus lactis. Microbiology 153:3275-3285. [DOI] [PubMed] [Google Scholar]
- 37.Miyata, S., S. Kozuka, Y. Yasuda, Y. Chen, R. Moriyama, K. Tochikubo, and S. Makino. 1997. Localization of germination-specific spore-lytic enzymes in Clostridium perfringens S40 spores detected by immunoelectron microscopy. FEMS Microbiol. Lett. 152:243-247. [DOI] [PubMed] [Google Scholar]
- 38.Miyata, S., R. Moriyama, N. Miyahara, and S. Makino. 1995. A gene (sleC) encoding a spore-cortex-lytic enzyme from Clostridium perfringens S40 spores; cloning, sequence analysis and molecular characterization. Microbiology 141:2643-2650. [DOI] [PubMed] [Google Scholar]
- 39.Moreillon, P., Z. Markiewicz, S. Nachman, and A. Tomasz. 1990. Two bactericidal targets for penicillin in pneumococci: autolysis-dependent and autolysis-independent killing mechanisms. Antimicrob. Agents Chemother. 34:33-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myhre, A. E., J. F. Stuestol, M. K. Dahle, G. Overland, C. Thiemermann, S. J. Foster, P. Lilleaasen, A. O. Aasen, and J. E. Wang. 2004. Organ injury and cytokine release caused by peptidoglycan are dependent on the structural integrity of the glycan chain. Infect. Immun. 72:1311-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neuhaus, F. C., and J. Baddiley. 2003. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:686-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obregón, V., P. Garcia, E. Garcia, A. Fenoll, R. Lopez, and J. L. Garcia. 2002. Molecular peculiarities of the lytA gene isolated from clinical pneumococcal strains that are bile insoluble. J. Clin. Microbiol. 40:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oshida, T., M. Sugai, H. Komatsuzawa, Y. M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. U. S. A. 92:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oshida, T., and A. Tomasz. 1992. Isolation and characterization of a Tn551-autolysis mutant of Staphylococcus aureus. J. Bacteriol. 174:4952-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paredes-Sabja, D., P. Setlow, and M. R. Sarker. 2009. SleC is essential for cortex peptidoglycan hydrolysis during germination of spores of the pathogenic bacterium Clostridium perfringens. J. Bacteriol. 191:2711-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin, X., K. V. Singh, Y. Xu, G. M. Weinstock, and B. E. Murray. 1998. Effect of disruption of a gene encoding an autolysin of Enterococcus faecalis OG1RF. Antimicrob. Agents Chemother. 42:2883-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin, Z., Y. Ou, L. Yang, Y. Zhu, T. Tolker-Nielsen, S. Molin, and D. Qu. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153:2083-2092. [DOI] [PubMed] [Google Scholar]
- 48.Rashid, M. H., M. Mori, and J. Sekiguchi. 1995. Glucosaminidase of Bacillus subtilis: cloning, regulation, primary structure and biochemical characterization. Microbiology 141:2391-2404. [DOI] [PubMed] [Google Scholar]
- 49.Raychaudhuri, D., and A. N. Chatterjee. 1985. Use of resistant mutants to study the interaction of triton X-100 with Staphylococcus aureus. J. Bacteriol. 164:1337-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shibata, Y., M. Kawada, Y. Nakano, K. Toyoshima, and Y. Yamashita. 2005. Identification and characterization of an autolysin-encoding gene of Streptococcus mutans. Infect. Immun. 73:3512-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimamoto, S., R. Moriyama, K. Sugimoto, S. Miyata, and S. Makino. 2001. Partial characterization of an enzyme fraction with protease activity which converts the spore peptidoglycan hydrolase (SleC) precursor to an active enzyme during germination of Clostridium perfringens S40 spores and analysis of a gene cluster involved in the activity. J. Bacteriol. 183:3742-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimizu, T., S. Ohshima, K. Ohtani, T. Shimizu, and H. Hayashi. 2001. Genomic map of Clostridium perfringens strain 13. Microbiol. Immunol. 45:179-189. [DOI] [PubMed] [Google Scholar]
- 53.Shockman, G. D., and J.-V. Höltje. 1994. Microbial peptidoglycan (murein) hydrolases, p. 131-166. Elsevier Science B. V., Amsterdam, Netherlands.
- 54.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249-262. [DOI] [PubMed] [Google Scholar]
- 55.Thomasz, A. 2000. The staphylococcal cell wall, p. 351-360. In V. A. Fischetti (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, DC.
- 56.Tjalsma, H., H. Antelmann, J. D. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J. Y. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. van Dijl. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomasz, A., P. Moreillon, and G. Pozzi. 1988. Insertional inactivation of the major autolysin gene of Streptococcus pneumoniae. J. Bacteriol. 170:5931-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vollmer, W., B. Joris, P. Charlier, and S. Foster. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32:259-286. [DOI] [PubMed] [Google Scholar]
- 59.Wang, L., and M. Lin. 2008. A novel cell wall-anchored peptidoglycan hydrolase (autolysin), IspC, essential for Listeria monocytogenes virulence: genetic and proteomic analysis. Microbiology 154:1900-1913. [DOI] [PubMed] [Google Scholar]
- 60.Ward, J. B., and R. Williamson. 1984. Bacterial autolysins: specificity and function, p. 159-166. In C. Nombela (ed.), Microbial cell wall synthesis and autolysis. Elsevier Science Publishers, Amsterdam, Netherlands.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.