Abstract
The phylogeny of the large bacterial class Gammaproteobacteria has been difficult to resolve. Here we apply a telescoping multiprotein approach to the problem for 104 diverse gammaproteobacterial genomes, based on a set of 356 protein families for the whole class and even larger sets for each of four cohesive subregions of the tree. Although the deepest divergences were resistant to full resolution, some surprising patterns were strongly supported. A representative of the Acidithiobacillales routinely appeared among the outgroup members, suggesting that in conflict with rRNA-based phylogenies this order does not belong to Gammaproteobacteria; instead, it (and, independently, “Mariprofundus”) diverged after the establishment of the Alphaproteobacteria yet before the betaproteobacteria/gammaproteobacteria split. None of the orders Alteromonadales, Pseudomonadales, or Oceanospirillales were monophyletic; we obtained strong support for clades that contain some but exclude other members of all three orders. Extreme amino acid bias in the highly A+T-rich genome of Candidatus Carsonella prevented its reliable placement within Gammaproteobacteria, and high bias caused artifacts that limited the resolution of the relationships of other insect endosymbionts, which appear to have had multiple origins, although the unbiased genome of the endosymbiont Sodalis acted as an attractor for them. Instability was observed for the root of the Enterobacteriales, with nearly equal subsets of the protein families favoring one or the other of two alternative root positions; the nematode symbiont Photorhabdus was identified as a disruptor whose omission helped stabilize the Enterobacteriales root.
Although Gammaproteobacteria has only the taxonomic rank of class within the phylum Proteobacteria, it is richer in genera (∼250) than all bacterial phyla except Firmicutes (11). It includes the paradigmatic bacterium Escherichia coli, well-known pathogens Salmonella, Yersinia, Vibrio, and Pseudomonas, additional pathogens occurring at the more basal and less well-resolved positions in the phylogeny like Coxiella and Francisella, and endosymbionts that can be required for survival of their insect hosts. Members exhibit broad ranges of aerobicity, of trophism, including chemoautotrophism and photoautotrophism, and of temperature adaptation (31). Morphologies include rods, curved rods, cocci, spirilla, and filaments, and the class contains the largest known bacterial cells (30). Interesting combinations of phenotypes occur, such as terrestrial bioluminescence with pathogenicity (toward insects and humans) and symbiosis (with nematodes) in the genus Photorhabdus (37). One feature alone, 16S rRNA sequence relationship, has been used to define the class (11).
Previous phylogenetic studies of this group indicate deep branches that make it difficult to obtain a large well-resolved phylogeny (10, 40). Taking advantage of the large number of genomes available for the class, we have followed a multiprotein approach to gammaproteobacterial phylogeny (39). A potential challenge to the validity of phylogenetic reconstruction in bacteria is the view that horizontal transfer is so pervasive that the tree interpretation is artifactual (2). However, a central trend in the phylogenetic signals from multiple bacterial protein families can be detected that probably represents their shared vertical inheritance (25). Our methodology sought to include single-copy families most likely to reflect this central trend, applying a filter to remove the families most discordant with the majority. The resulting tree breaks with current taxonomy.
In contrast to previous studies with a small number of genomes, indicating high phylogenetic concordance among gammaproteobacterial protein families (20, 24), we found that nearly equal numbers of protein families favored one or the other of two roots for Enterobacteriales. However, much of this effect could be ascribed to a single taxon that was absent from the earlier studies.
MATERIALS AND METHODS
Genome selection by collapse of a preliminary tree.
On 27 July 2007, there were 215 complete or incomplete genomes available for nominal Gammaproteobacteria at NCBI. Because the phylogenetic distribution was biased, we selected a smaller number with balanced distribution. Protein sets were taken for the 215 Gammaproteobacteria and for two outgroup species each from the Alphaproteobacteria and Betaproteobacteria.
An initial set of protein families that approximated one occurrence per genome among 97 Gammaproteobacteria were selected from the GeneTrees database (36), and the alignments were used for HMMsearch using HMMer (3) to find instances in the full set of 215 genomes. The 20 gene families with the lowest standard deviation of members per genome, the closest to single copy, were processed further. Phylogenetic trees were built for each family to resolve 54 cases of duplicate or split genes. A multiprotein species tree was prepared, aligning sequences with MUSCLE version 3.6 (7) in default mode and removing ambiguous portions of alignments with Gblocks version 0.91b (35) in default mode except for using the intermediate setting for gap tolerance; the concatenation of masked alignments containing 5,538 characters was used to prepare the tree applying MrBayes version 3.2 (28) in model-jumping mode as described previously (39). A process of sequential tip pair collapse was applied to this tree, in which at each stage the terminal node with the shortest total length of its two leaves was collapsed into a single leaf whose length was the average of the two. The distance between the pair of collapsed tips, a measure of diversity reduction, was plotted against the stage of collapse (Fig. 1). The stage with 104 clusters was chosen because it preceded a phase of steep loss of genome diversity and yet yields a reasonable number for phylogenetic analysis. One genome was selected from within each of the 104 clusters, favoring complete genomes and better-known taxa. The total was raised to 110 with additional genomes: three from unrepresented genera that had become available at NCBI more recently, two that allowed comparison to a classical set of 13 taxa (20), and a nonpublic genome that became available to us. Later, two incomplete genomes were rejected because as described below their rRNA sequences suggested that they were not bacteriologically pure. The remaining 108 taxa contained four outgroup taxa, at least one member from each of the 14 gammaproteobacterial orders, and 11 taxa classified at NCBI as Gammaproteobacteria but not assigned to an order (see Table S1 in the supplemental material).
FIG. 1.
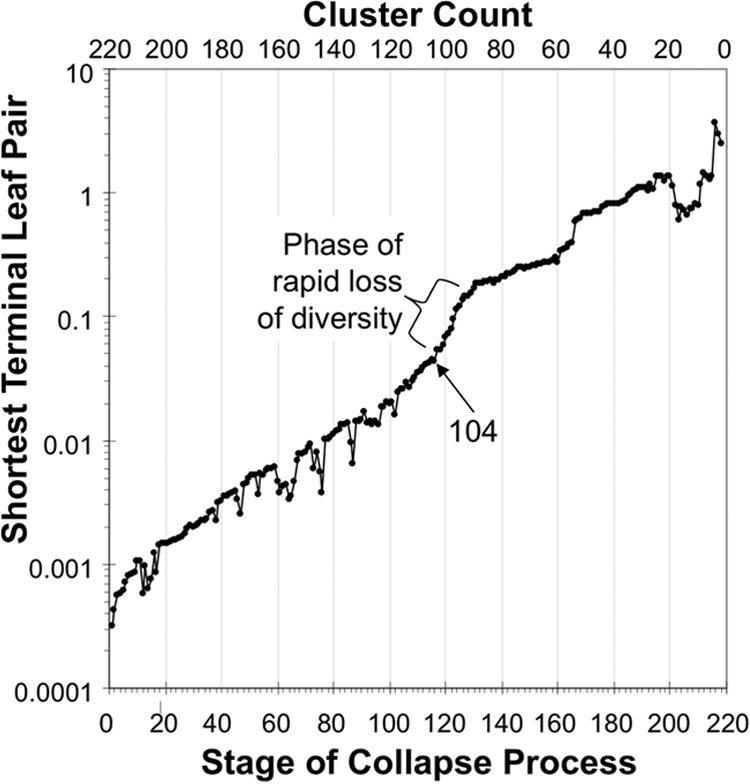
Taxon selection by collapse of a preliminary tree. Collapse was a sequential process of identifying the shortest leaf pair and replacing it with a single representative leaf. This maximized diversity between clusters of the available genomes. The number of genomes selected this way was 104, since this preceded a phase in the collapse process with an especially high loss of diversity.
rRNA sequence assay of genome purity.
All complete or incomplete 16S and 23S rRNA sequences were collected from 110 genomes using a curated set of seed sequences and a BLASTN-based script, resulting in 1,135 sequences (529 of 16S rRNA and 606 of 23S rRNA), of which 791 were unique. These sets were aligned using MUSCLE. For 54 rRNA fragments, sequences from the ends of a contig that aligned poorly to whole rRNAs were trimmed, leaving 337 16S rRNA and 452 23S rRNA unique sequences that were realigned using MUSCLE, manually correcting its error of splitting off small terminal portions of incomplete sequences. For all intragenomic pairs whose alignment overlapped >40 nt, the subalignment for the overlap region was taken for all available rRNA sequences (78 subalignments for 16S rRNA, 124 for 23S rRNA). Intragenomic sequence pairs for 16S and 23S rRNA were 409 and 658 for the full alignments and 228 and 558 for the subalignments, respectively. For each full and subalignment, a distance matrix was taken using distmat (26). All cases where an intragenomic pair had a distance score above the tenth percentile (39) for a partner were investigated further.
In the 23S rRNA subalignment of positions 808 to 1075, a fragment from Beggiatoa sp. strain SS had a higher distance score to its intragenomic partner than to 99% of all its distance scores and also ranked highly in other 23S subalignments. BLAST found one of the pair to have 83% identity to Coxiella, but the other had 96% identity to Haemophilus influenzae PittGG. Because this incomplete genome, derived from a single bacterial filament isolated from sea sediment, contained highly divergent 23S fragments, we considered that its protein sequences might be contaminated with those from additional microbes, and it was rejected from our analysis.
A 23S rRNA fragment from the incomplete Azotobacter vinelandii genome had high-ranking intragenomic distance scores and in one subalignment ranked as high as 98% among all distance scores. This fragment showed 96% identity to Desulfovibrio 23S rRNA, while the other eight sequences from this genome matched Azotobacter 23S rRNA. We rejected this genome from our analysis due to the potential for contamination with protein sequences from other organisms. The complete A. vinelandii genome now available no longer has this rRNA contamination.
A short high-scoring 23S rRNA fragment from the complete Chromohalobacter genome proved to be a likely regulatory region of an operon encoding proteins known to bind that region of 23S rRNA (38).
One high-scoring intragenomic rRNA distance was ascribed to misalignment by MUSCLE.
The remaining high-scoring intragenomic rRNA distances were from incomplete genomes, and the top BLAST hit of each contender was to the nominal genus though low scoring. These cases could be ascribed to low-quality sequence data.
rRNA trees.
Analysis began with the rRNA sequences from 108 genomes remaining after the filters of the genomic purity assay. In cases of multiple rRNA sequences, a single full-length sequence was chosen as having the shortest branch length in a preliminary tree or, if only fragments, were available, they were merged. With single representative 16S and 23S rRNA sequences for each genome, the MUSCLE alignments were further evaluated using the secondary structure models available at the comparative RNA web site (5). Sequences were manually adjusted to adhere to the predicted helical and unpaired regions of the rRNAs, using compensatory base change evidence to support homology assignment (17). Regions of the alignment that included length heterogeneity were minimized by flanking structural support (i.e., compensatory base change evidence) with remaining regions of ambiguous alignment masked (12). In several cases of mismatch to the structural model, the flanking ends of partial sequences were also masked. Resulting alignments were processed using tools from the jRNA web site (13) to create alignments, base pair analyses, and input files for MrBayes annotated for use of the doublet model, which assigns an RNA substitution matrix to base pairs.
Bayesian phylogenetic trees were generated for the 16S and 23S rRNA masked alignments and for a concatenation of the two. For each, MrBayes was run three times as a single MCMC chain for 2 × 106 generations, with a GTR + Γ + I model (Γ distribution approximated by 4 rate categories) for among-site rate variation, differing at each site, and the doublet substitution model for base pairs. In all cases, burn-in was attained well before 1.5 × 106 generations, and the trees from the final 0.5 × 106 generations were pooled from triplicate runs (7,500 trees total) to build a consensus tree.
Multiprotein data sets.
An automated workflow for gene family selection, discordance filtering, and tree building was implemented through a series of Perl scripts. All proteins for a group of genomes were sorted into families using all-versus-all BLASTP followed by OrthoMCL version 1.4 (21). Approximately single-copy protein families were selected by choosing (i) a core set of complete and well-behaved ingroup genomes from among the taxa and tolerances for (ii) the number of core genomes missing from the family and (iii) the number of genomes with multiple members in the family (see Table 2, footnote a).
TABLE 2.
Selection of protein families
| Step | No. of families remaining (by genome group) |
||||
|---|---|---|---|---|---|
| All-taxon | Basal | PO | VAAP | Entero | |
| OrthoMCL | 25,292 | 8,227 | 12,727 | 12,482 | 7,365 |
| Near-single-copy filtera | 405 | 684 | 679 | 712 | 1,403 |
| Masking filterb | 404 | 684 | 679 | 712 | 1,403 |
| High-support bipartition filterc | 396 | 680 | 678 | 712 | 1,402 |
| Discordance filter | 356 | 616 | 611 | 640 | 1,262 |
This filter removed families exceeding the tolerance for either absent or multiple representation of core taxa in the family membership. The following tolerances were used (absent/multiple): all-taxon, 2/6; Basal, 5/1; PO, 1/1; VAAP, 2/1; Entero, 0/0.
Families with no sequence remaining after masking by Gblocks was removed.
Taxon bipartitions for a protein family were counted as highly supported when they appeared among ≥75% of the bootstrap trees for the family (except for the all-taxon families, where a 95% cutoff was applied); the filter removed families with no high-support bipartitions.
For the full taxon set of 108 study taxa, this process yielded 404 families, with two taxa especially poorly represented: Candidatus Carsonella in 69 families and Candidatus Endoriftia in 58. Since only 182 proteins are annotated for the extremely small Ca. Carsonella genome, its low family representation was not pursued further. The incomplete Ca. Endoriftia genome had many genes split among different contigs, frameshifted by apparent sequencing errors or simply uncalled. This genome was queried by all members of its unrepresented families using TBLASTN, and 317 of the unrepresented families received good hits, of which the 148 most reliable models for missing proteins were added to the corresponding families and to the Ca. Endoriftia protein file.
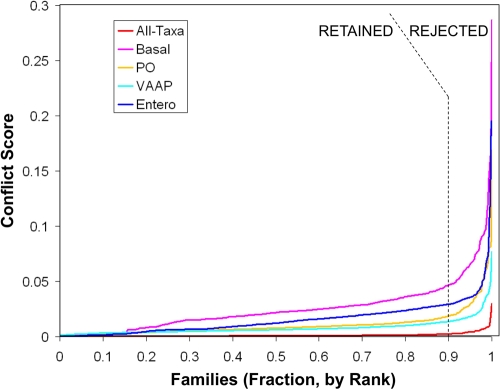
The resulting families were subjected to discordance filtering, that is, removal of the 10% with the most anomalous phylogenetic signal (e.g., potential cases of lateral gene transfer). Family members were aligned (MUSCLE) and the alignments were masked to remove unreliably aligned regions (Gblocks, with intermediate gap tolerance); families obliterated by Gblocks were rejected. For each family, the preferred amino acid substitution model among the 20 available in RAxML 7.0.4 was chosen, most frequently the WAG model, based on a script provided with RAxML (34). Fifty quick bootstraps (RAxML option -x) were taken for the family to determine high-support bipartitions (HSBs) of the taxa according to a chosen cutoff (see Table 2). Families with no HSBs were immediately rejected; otherwise, pairwise normalized HSB conflict counts were taken for all pairwise comparisons of the families, after modifying HSBs for shared taxa. Unshared taxa were removed, causing some HSBs to merge and others to become trivial. Conflicting HSBs were counted and normalized, dividing by the product of the two families' HSB counts. A conflict score was assigned to each family (its average pairwise normalized HSB conflict count), which had little correlation with numbers of characters, taxa, or HSBs (data not shown). A given fraction (10%) of families with the highest conflict scores were removed. Figure 2 shows that 10% is an effective and safe rejection rate. Note that discordance filtering does not refer to (and thereby circularly reinforce) any particular phylogenetic tree topology.
FIG. 2.
Family conflict. The conflict scores for each set of protein families identified those with egregious phylogenetic signals. Families scoring above the 90th percentile were rejected.
Phylogenetic analysis.
The masked alignments for the remaining families were concatenated. We found that each individual MrBayes run would canalize into a single tree topology, but duplicate runs would find slightly different topologies. We also found that the product of the faster (MIX) mode of RAxML depended on the starting tree provided. Instead we used the slowest RAxML mode, with gamma-distributed rate heterogeneity, which yielded a single final tree whether we started with random trees or with the products from the MIX-mode runs. Protein families were assigned their preferred substitution models found previously. To provide support values, tree building was repeated for 50 whole-protein jackknife (random removal of half the protein families) resamplings and for all single-taxon jackknife resamplings.
The above approach was applied to the full taxon set and to the four main taxon subgroups as detailed in Table 1. In addition to the use of multiple amino acid substitution matrices with the gene-partitioned concatenations, we tested uniform usage of the new LG matrix (18). Maximum likelihood trees were prepared with uniform (unpartitioned) use of LG for the concatenations of the full taxon set and all four subgroups using RAxML version 7.2.3. All trees had topologies identical to those of their counterparts that had been produced with multimodel partitioning, except that the Basal subset and the full taxon set (in the Basal region) each had a single case of a branch crossing a single node. Both cases involved the node with the poorest jackknife support in the whole tree (represented by multifurcation in Fig. 3). Comparing the two different trees in these two cases, using each of the two substitution matrix sets by the Approximately Unbiased test, the differences were not significant at the 0.05 level, and the trees from multimodel partitioning scored higher than those from uniform application of the LG matrix.
TABLE 1.
Selection of genomes
| Step | No. of genomes remaining |
||
|---|---|---|---|
| Ingroup | Corea | Outgroup | |
| Collect gammaproteobacterial genomes from NCBI | 215 | 0 | |
| Select alpha-/betaproteobacterial outgroups | 215 | 4 | |
| Collapse preliminary 20-protein tree | 100 | 4 | |
| Complete the “classical” genome set (20) | 102 | 4 | |
| Add recently available genomes from unrepresented genera | 106 | 4 | |
| Apply rRNA purity filter | 104 | 4 | |
| Select core genomesa for family selection | 104 | 31 | 4 |
| Generate main tree, split into isolated subgroups | |||
| Basal subgroup | 19 | 14 | 3 |
| PO subgroup | 27 | 17 | 2 |
| VAAP subgroup | 37 | 24 | 2 |
| Entero subgroup | 19 | 8 | 2 |
The core genomes are a subset of the ingroup that have complete sequences and were further selected based on their diversity and their low compositional bias; these were used in the near-single-copy filter for protein families.
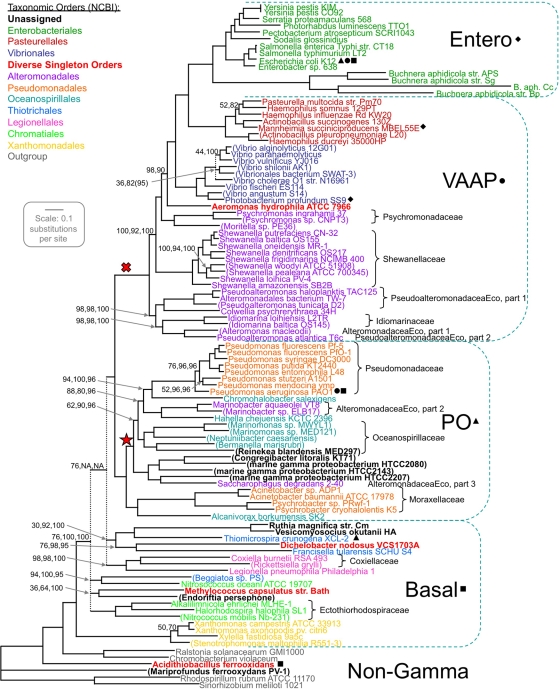
FIG. 3.
Gammaproteobacterial phylogeny. Four subgroups of the taxa (excepting Ca. Carsonella, which is omitted here) were found not to mingle, by protein-jackknife subsampling of an all-taxon concatenation of 356 protein alignments. Longer concatenations were prepared for each taxon subgroup (Table 1), and tree topologies were built for each; small black geometric symbols link each subgroup to the outgroups used in building its subtree. The four regional trees were merged back into the topology for the all-taxon data set. Branch lengths for the merged topology were computed based on the all-taxon data set, and the four of its bifurcations that had less than 25% protein-jackknife support were collapsed, leaving three multifurcating nodes (dotted lines). Support values are shown (in the following order: all-taxon protein 50% jackknifing, taxon-subgroup protein 50% jackknifing, and taxon-subgroup single-taxon jackknifing) when any of these are <100%; cases marked NA (not applicable) were for nodes not included in taxon subgroup studies. The Entero region subtree was prepared by a two-step procedure described in the text that makes the usual support values inapplicable; however, for the first-step subtree prepared for the core Enterobacteriales taxa whose topology persists in this figure, protein and taxon jackknifing gave 100% support at each node. Taxon names are given in parentheses for incomplete genomes and are color coded according to the taxonomic order designation at NCBI. Taxonomic families represented by more than one taxon, except in cases in which the lone family represents the order, are bracketed, extending brackets to include unassigned taxa as necessary; only two of these nine families are split in our tree. Two nodes of interest are marked: the X indicates a node supported by a rare indel (10), and the star indicates a fully supported node that groups members of Pseudomonadales, Alteromonadales, and Oceanospirillales while excluding other members of each order. All known members of the latter clade, and no other known bacteria, autoregulate a ribosomal protein operon through strong mimicry of a 23S rRNA domain (38).
Amino acid bias.
To examine amino acid bias, the 61 proteins from the 356 all-taxon set that included Ca. Carsonella were used to measure amino acid bias for each taxon relative to all the gammaproteobacterial taxa according to the method of Karlin (15) (Fig. 4). To examine the significance of amino acid bias, the chi-squared test implemented in TreePuzzle version 5.2 (29) was applied; 79 of 108 genomes were biased at the 5% level. As a second test of the significance of amino acid bias, we generated an artificial distribution of amino acid bias scores by making 1 million simulated 61-protein concatenations where each character state was chosen randomly from each column of the study supermatrix; bias against the gammaproteobacterial taxa was calculated for each, and significance levels were taken from the sorted list of scores. According to this test, 105 of the 108 genomes were biased at the P < 0.05 level, and 81 (which included all 79 of those identified by TreePuzzle) were biased at the P < 0.000001 level.
FIG. 4.
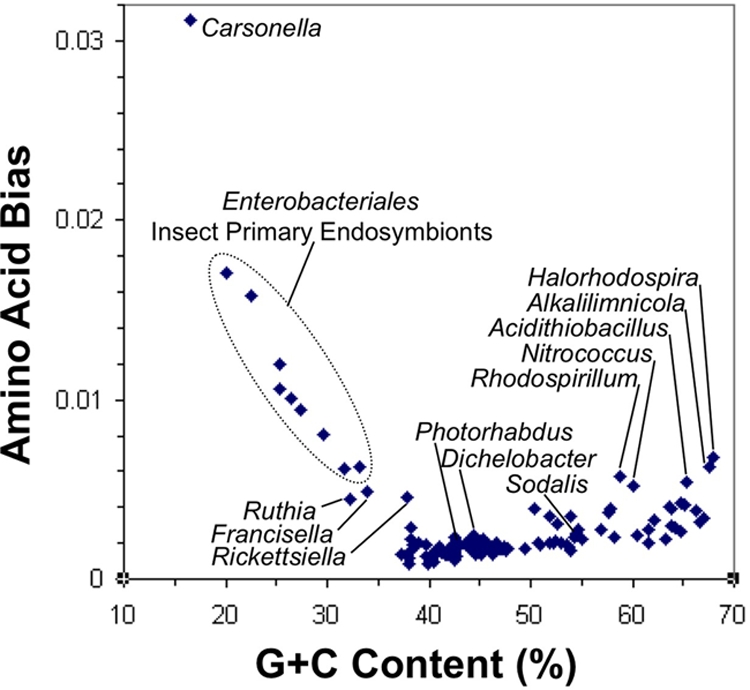
Compositional bias. Amino acid bias was measured according to the method of Karlin (15) for the set of 61 all-taxon protein families that contain Ca. Carsonella and plotted against nucleotide composition, producing a horn of A+T-rich endosymbionts with Ca. Carsonella at its tip.
Statistical tree tests.
The Approximately Unbiased test as implemented in CONSEL v0.1j (32) was applied, and site likelihood scores were prepared with RAxML.
RESULTS
Taxon selection.
Over 200 complete and incomplete gammaproteobacterial genomes were available when our project began, but they had a biased taxonomic distribution; the six genera Escherichia, Haemophilus, Pseudomonas, Shewanella, Vibrio, and Yersinia accounted for over half the genomes. These near-duplicate genomes contain little additional phylogenetic information; we selected a subset of the genomes with maximized diversity by (i) building a preliminary phylogenetic tree, (ii) sequentially clustering close relatives, and (iii) choosing a stage in the clustering process that preceded a phase of rapid loss of diversity (Fig. 1). Two incomplete genomes were rejected because analysis of rRNA sequences suggested that these two projects might be contaminated with foreign DNA. The final set of 108 genomes included four outgroup taxa (two each from the sister class Betaproteobacteria and the subtending class Alphaproteobacteria), at least one representative of each of the 14 orders listed in Bergey's Manual (11), and 11 taxa that had been assigned at NCBI to the class Gammaproteobacteria but not to a particular order (see Table S1 in the supplemental material). This genome selection process is summarized in Table 1.
rRNA trees.
As important a tool as 16S rRNA phylogeny has been in bacteriology, we and others have found that this single gene contains insufficient phylogenetic information to robustly recover deep relationships, such as those of interest in this work. For our 108 selected genomes, we made trees for 16S and 23S rRNAs (see Fig. S1 and S2 in the supplemental material). Both trees were questionable because numerous ingroup genomes (21 and 17, respectively) were placed between the two Alphaproteobacteria and Betaproteobacteria outgroups; moreover, the two trees differed substantially at the deeper nodes. A tree for the concatenated alignment of the two rRNAs had the same problems (see Fig. S3 in the supplemental material). In a multiprotein study of alphaproteobacterial phylogeny, it was found that the combined rRNAs had the phylogenetic inaccuracy (measured as the level of disagreement between two different phylogenetic reconstruction methods) of ∼2.5 single-copy proteins combined; moreover, accuracy increased as more proteins were employed, at least up to 100 proteins (39). Therefore, we took a multiprotein approach to gammaproteobacterial phylogeny.
Telescoping multiprotein approach.
The first step in our multiprotein approach (Table 2) was to select a large number of protein families with (nearly) single-copy distribution patterns, because these are likely to have similar histories that reflect the organismal phylogeny. A strict single-copy criterion would only have allowed one gene family, so the criterion was slightly relaxed. A set of 31 core taxa was selected that had complete genomes of ≥1 Mbp and were phylogenetically representative (see Table S1 in the supplemental material), and the relaxed single-copy criterion allowed families with up to two core taxa unrepresented and up to six core taxa with duplicate membership. Cases of multiple membership for a taxon were expediently resolved by removing both (or all) members from that taxon in that family. This yielded 405 gene families.
The alignments for these families were masked by removing ambiguously aligned regions. Families with no sequence remaining after masking were rejected, as were families whose bootstrap trees produced no highly supported taxon bipartitions. These two filters eliminated families that were grossly deficient in phylogenetic signal.
To further refine the gene families and minimize the potential influence of horizontal transfer, a phylogenetic discordance filter was imposed to eliminate the 10% of protein families least concordant with the others. The set of highly supported taxon bipartitions for each family were compared to generate a normalized conflict matrix (Fig. 2). Families with high average discordance were eliminated without referring to (and thereby circularly reinforcing) an a priori tree topology, leaving 356 families (see Table S2 in the supplemental material). Fifteen of 19 COG (clusters of orthologous groups) categories were significantly overrepresented (translation proteins especially) or underrepresented (uncategorized proteins especially) among these families (see Fig. S4 in the supplemental material).
The masked alignments for these families were concatenated into a supermatrix, and a maximum likelihood tree was produced. Its robustness was evaluated by whole-protein 50% jackknifing, that is, generating subsets of the data from which a random half of the protein families had been removed and computing a tree for each. Support for each node in the main tree was then evaluated as the fraction of protein jackknife trees that produced the node. A rationale for the use of this support measure is presented in the Discussion. Several nodes received very low support. However, the protein jackknife data revealed five regions of the tree whose boundaries were never crossed (except by the anomalous taxon Ca. Carsonella, which we discarded, as further described below), indicating isolated groups of taxa that could be studied separately, reuniting the resulting telescoped trees into a better-resolved composite tree. One group was the Enterobacterales clade (“Entero” in Fig. 3). The second group, termed “VAAP,” consists mainly of Vibrionales, Alteromonadales, Aeromonadales, and Pasteurellales; the Entero clade emerges from the VAAP group. A sister clade to the VAAP and Entero groups is the “PO” group, consisting mainly of Pseudomonadales and Oceanospirillales. The “Basal” region is a paraphyletic group of the most anciently diverging lineages in the class. Finally, the “Non-Gamma” region contains the alpha- and betaproteobacterial outgroups and additionally contains “Mariprofundus,” which had been assigned to the Gammaproteobacteria by the NCBI taxonomy but to a novel lineage outside the Gammaproteobacteria by rRNA analysis (8), and, surprisingly, Acidithiobacillus, which has been considered a member of the Gammaproteobacteria (16). The Acidithiobacillus result is not due to an anomaly of the particular genome project studied here; a second Acidithiobacillus project has since become available, and it shows near-identity to the earlier project in all the protein families employed here.
Aiming to improve resolution, each of the four gammaproteobacterial regions (Basal, PO, VAAP, and Entero) of the tree were separately reanalyzed with larger numbers (611 to 1,262) of protein families, by the same selection/discordance-filtering/tree-building approach described above, and as detailed in Table 2. Even the very large Entero collection (1,262 protein families) shows functional bias compared to all 4,149 encoded proteins of E. coli; overrepresented COG categories were translation, coenzyme metabolism, cell division, DNA metabolism, and protein processing (chi-squared P values of <10−10, <10−10, <10−7, <10−7, <10−4, and <10−3, respectively), while the uncategorized, carbohydrate metabolism, and transcription genes were underrepresented (<10−10, <10−7, and <0.03, respectively) (see Fig. S4 in the supplemental material). Also, none of the 185 E. coli genes annotated in genomic islands were among the Entero families. Support values for the nodes in each regional tree were then evaluated by whole-family 50% jackknifing as described above and also by single-taxon jackknifing, in which, for each ingroup taxon, a tree was built after that taxon was removed. Before reuniting these telescoped regional trees, the Enterobacteriales region was examined further, initially because it contained the anomalous Ca. Carsonella genome.
Compositional attraction.
The Ca. Carsonella genome (22) is unusual for many reasons: it has an extremely small and A+T-rich genome and is represented in only 61 of our 356 families. It was located on an extremely long branch in our original tree within a group of other insect endosymbionts, themselves (except for Sodalis) on long branches and with small, A+T-rich genomes. It was the only taxon that crossed a tree regional boundary, found among the insect endosymbiont Enterobacteriales in 48 of the whole-protein jackknife replicates but with the A+T-rich Francisella or Rickettsiella genomes (Basal region) in the other two. Either or both of these tree positions may have been artifacts resulting from amino acid bias, in turn arising from nucleotide bias in these genomes. Using the protein families that contain Ca. Carsonella, we measured the amino acid bias of each taxon relative to all Gammaproteobacteria combined (15). In a plot of amino acid bias against nucleotide bias (Fig. 4), some taxa fell into a prominent A+T-rich horn, with Ca. Carsonella at its far tip, eight insect endosymbionts that are its usual neighbors in the trees, and three members of the Basal region: Candidatus Ruthia, Francisella, and Rickettsiella. We tested more directly the possibility that the placement of Ca. Carsonella was artifactual by building a tree after removing its eight neighboring A+T-rich taxa from the analysis; in this tree, Ca. Carsonella jumped to the group with the next-most-similar composition, appearing with Francisella. In a similar tree built after further removing Ca. Ruthia, Francisella, and Rickettsiella, Ca. Carsonella appeared in yet a new region of the tree, among Pseudomonas species. We conclude that either (i) the placement of Ca. Carsonella among the insect endosymbionts is artifactual, (ii) its secondary placement with Francisella is artifactual, or (iii) both these placements are artifactual such that Ca. Carsonella may not even belong to the Gammaproteobacteria. Ca. Carsonella was excluded from further analysis.
The group of insect endosymbionts with small, A+T-rich genomes, consisting of Buchnera, Candidatus Baumannia, Candidatus Blochmannia, and Wigglesworthia, appeared as a single clade in the Entero region of initial trees. These genomes all have high amino acid bias relative to the remaining Gammaproteobacteria (Fig. 4), raising the possibility that the clade may have been an artifact of compositional attraction. We tested these genomes as we had Ca. Carsonella, building a tree by adding each one singly after omitting all their A+T-rich neighbors. Each member of the group was confirmed to fall among the Enterobacteriales in single-member tests using the all-taxon data set. Although each of the Ca. Baumannia-Ca. Blochmannia-Wigglesworthia strains were placed as before next to Sodalis, each Buchnera strain was placed at the base of the Enterobacteriales. These placements were reproduced when the single-member tests were repeated with the Enterobacteriales-focused data set (Fig. 5A and B, part i). This split placement supports the idea that the original grouping of all these endosymbionts was due to compositional attraction.
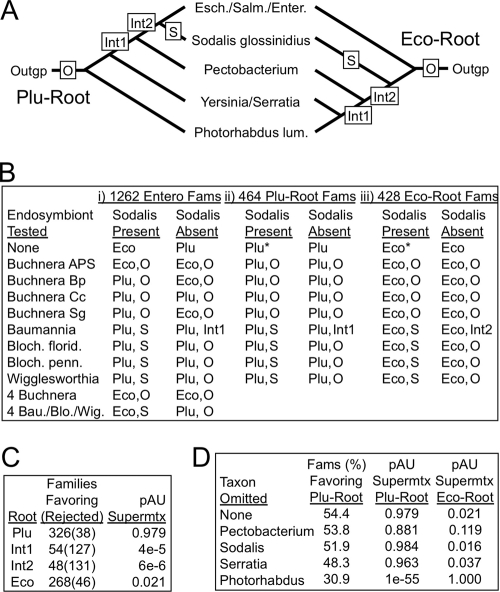
FIG. 5.
Positions of the root and endosymbionts among the Enterobacteriales. (A) Two root positions (Plu and Eco) were obtained in our study; positions taken by A+T-rich endosymbionts (O and S) and two tested intermediate root positions (Int1 and Int2) are marked. Esch., Escherichia; Salm., Salmonella; Enter., Enterobacter; lum., luminescens; outgp, outgroup. (B) Positions taken by endosymbionts and their effect on rooting. The full Enterobacteriales taxon set was reduced to a core by removing all eight A+T-rich genomes (retaining the two outgroups), then each alone, or in groups of four, was added back to the data set. The compositionally unbiased endosymbiont Sodalis was excluded or not, and either the full set of Entero protein families (Fams) (part i) or subsets with at least one outgroup and favoring either the Plu-root (part ii) or the Eco-root (part iii) were used. The resulting root position and the position taken by the tested endosymbiont is reported. *, bootstrap analysis was performed for these trees, with support measuring 100% for every node. Bau., Ca. Baumannia; Blo. or Bloch., Ca. Blochmannia; Wig., Wigglesworthia; florid., floridanus; penn., pennsylvanicus. (C) Intermediate root positions disfavored. Each of the 696 protein families containing all core taxa and both outgroups were tested for preference of four hypothetical root positions using the P value of the Approximately Unbiased test (pAU), and pAU is reported for the supermatrix (Supermtx) that combined these 696 protein families. A pAU of <0.05 is typically taken as a rejection. (D) Identifying Photorhabdus as a disruptive taxon. For each of the 696 Entero families used in panel C, the indicated core taxa were singly omitted and pAU was taken for either the Plu-Root or Eco-Root topology, to determine the topology favored by the family; pAU was also taken for the two topologies based on the corresponding supermatrix.
The presence of Sodalis was suspected to have influenced these results, since it is itself an insect endosymbiont (of the same insect host as Wigglesworthia) with a somewhat reduced, though compositionally unbiased (54% G+C), genome. Omitting Sodalis affected the placement of those endosymbionts that had clustered with it in the single-member tests; Ca. Blochmannia and Wigglesworthia moved to the base of the Enterobacteriales, while Ca. Baumannia moved to an intermediate position (Fig. 5A and B, part i). Thus, the basal position of the Buchnera strains was independent of Sodalis, while the positions of the other endosymbionts were Sodalis dependent and therefore uncertain.
Instability of the Enterobacteriales root.
The above experiments revealed instability in the root position of the Enterobacteriales. Our all-taxon and Enterobacteriales-focused trees, most of their taxon- or protein-jackknife support trees, and most of the endosymbiont study trees show Photorhabdus at the root (“Plu-root” in Fig. 5A), but a substantial number of members of the last group show the Escherichia-Salmonella-Enterobacter clade at the root (“Eco-root” in Fig. 5A). These two frequent rooting positions are separated by two internodes that themselves are strongly disfavored as rooting positions by the Approximately Unbiased (AU) test (32) (Fig. 5C).
The Enterobacteriales present an interesting situation where each of two different trees are supported by nearly equal subsets of the protein families. When the A+T-rich endosymbionts were excluded and the Enterobacteriales protein families for the remaining core taxa (and containing at least one outgroup) were sorted into those favoring either the Plu- or the Eco-root, the family counts were similar (464 and 428, respectively) (see Table S3 in the supplemental material). The Sodalis effect on endosymbiont placement was reexamined with the Enterobacteriales Plu- or Eco-root-favoring family subsets and found to occur with each subset (Fig. 5B, parts ii and iii). These two family subsets were not distinct in terms of functional classification (COG types) or in terms of fast- or slow-evolving genes as judged by total tree branch length for each family, nor were they substantially segregated when mapped to the E. coli genome. For each of the two family subsets, the supermatrix produced a tree with the respective root, with 100% bootstrap support (Fig. 5B, parts ii and iii). One explanation for such a situation could be a massive horizontal transfer event followed by gene sorting; however, there is no single within-Enterobacteriales genetic exchange path that would convert one tree to the other, because the alternate topologies are separated by two disfavored internodes. We identified the families among our 356-family all-gammaproteobacterial data set corresponding to the Enterobacteriales Plu- or Eco-root-favoring families, and we built all-gammaproteobacterial trees with the supermatrix for each subset. This might have identified a gammaproteobacterial source for a hypothetical massive horizontal gene transfer event; instead, the two all-taxon family subsets produced trees like that for the whole set, except again with the respective Enterobacteriales rooting. This test also ruled out the concern that the alternate-root phenomenon was an artifact of the particular two outgroup genomes chosen for the Enterobacteriales-focused data set, since the test effectively increased the outgroup from 2 to 89 members.
The observation of two large gene family subsets that favor different tree topologies appears to stand in contrast to earlier studies that found high concordance among gammaproteobacterial protein families (20, 24). These studies included only three genera (Escherichia, Salmonella, and Yersinia) from our core Enterobacteriales set; thus, the extra genomes in our study (Sodalis, Serratia, Photorhabdus, and Pectobacterium) may be responsible for the difference. Omitting each of these genomes one by one from either the Plu- or Eco-root-favoring supermatrix had no effect on remaining tree topology, except that when Photorhabdus was omitted, both supermatrices produced the Eco-root; this identified Photorhabdus as a disruptive taxon. These four taxa were also removed one by one from each of the 696 Enterobacteriales protein families that contain all core taxa and both outgroups, assessing preference for the Plu- or Eco-root (Fig. 5D). Omission of Photorhabdus made the families more concordant, shifting the fraction of Plu-favoring families from 0.54 to 0.31, confirming Photorhabdus as a disruptor.
Final tree.
Based on the difficulties with the Enterobacteriales, its regional tree was reconstructed as follows. The A+T-rich genomes and Photorhabdus were omitted, and a tree was built only for the remaining taxa; support for each node by whole-family and single-taxon jackknifing was 100%. Since the four Buchnera genomes all separately appeared in the same position of this subtree, independently of Sodalis, they were included in a second round of tree building, together with Photorhabdus; these taxa were added back to the data set to build a larger tree, constraining to the topology of the initial subtree (since without constraint the Buchnera and Photorhabdus genomes altered that topology [Fig. 5B, part i]). Since its membership in the Enterobacteriales is unconfirmed, Ca. Carsonella was excluded from the regional tree, as were the endosymbionts Ca. Baumannia, Ca. Blochmannia, and Wigglesworthia, due to uncertainty arising from their sensitivity to the presence of Sodalis.
The four regional trees were then merged back into their positions in the original all-taxon tree, yielding the final tree depicted in Fig. 3. Support values reported are the 50% whole-protein jackknife values for the all-taxon data set, the corresponding values from each regional subtree, and single-taxon jackknife values from each regional subtree. A few support values were so low as to warrant the introduction of two multifurcations in the Basal region and one in the VAAP region.
DISCUSSION
Multiprotein phylogeny produces more robust trees than single-gene approaches, which better serve bacteriologists in interpreting biological data. A concrete example is the fully supported clade marked by a star in Fig. 3 whose members (Pseudomonas, Marinomonas, Saccharophagus, etc.) share a molecular biological trait that is unique among bacteria: their S19 ribosomal protein operon contains a strong mimic of a 23S rRNA fragment that likely confers autoregulation on the operon (38). This trait would have required a more complex explanation than simple vertical inheritance according to 16S rRNA trees, since to our knowledge no such trees, including our own, recover this clade. (It was recovered with weak support in our 23S rRNA tree but was again lost in the combined 16S/23S rRNA tree.)
Although performance is improved, multiprotein phylogeny is still subject to some of the same artifacts and difficulties as single-gene approaches, such as long-branch attraction and poor resolution of branches that are deep and short. Telescoping allowed data from more protein families to be applied to isolated subgroups of taxa, but this was still insufficient to resolve all nodes or solve difficulties with highly biased genomes. We identified one genome as a phylogenetic attractor (Sodalis) and one as a disruptor (Photorhabdus); full explanation of the mechanisms of these effects will require further study and targeted genomic sequencing of more members of these clades.
This phylogenetic analysis based on hundreds of proteins for over a hundred taxa strongly supports the splitting of two families (Alteromonadaceae and Pseudoalteromonadaceae) and three orders (Alteromonadales, Pseudomonadales, and Oceanospirillales). A fourth order, Thiotrichales, also appears split but with lower support. Finally, the unity of the order Chromatiales could not be established. Thus, the classification based on 16S rRNA breaks down occasionally at the family level but more frequently at the order level.
The tree assigns taxonomy to previously unclassified genomes: Reinekea to Oceanospirillaceae and Ca. Ruthia and Candidatus Vesicomyosocius to Piscirickettsiaceae; the affiliation of Ca. Endoriftia with Methylococcales requires confirmation as more genomes become available. For four genomes (Congregibacter and the marine strains HTCC2080, HTCC2143, and HTCC2207) the closest assigned genome was Saccharophagus of the split Alteromonadaceae. Two genomes fell among the outgroup and are therefore not part of the Gammaproteobacteria. For one of these, Mariprofundus, this placement is consistent with previous analysis of its 16S rRNA sequence that placed it as a deep branch within the phylum Proteobacteria (8). The other newly identified non-gammaproteobacterium is Acidithiobacillus, surprisingly, since this genus has long been considered a member of Gammaproteobacteria (16). A recent tree (K. P. Williams, unpublished data) prepared using 173 protein families from 124 bacterium-wide genomes, chosen to represent each available bacterial order, showed the same relationships of Acidithiobacillus and Mariprofundus to other proteobacteria as depicted in Fig. 3, confirming that these two genera represent independent sister groups to the beta-/gammaproteobacteria clade that arose after divergence from the Alphaproteobacteria. All these observations suggest revisions of the taxonomy at multiple ranks in the class and phylum.
An early multiprotein study of the Gammaproteobacteria used over 200 proteins, but only 13 genomes were available at that time (20). Later studies have used far fewer protein families (10 to 35 proteins) and fewer representative genomes (28 to 55 genomes) than the present analysis (4, 6, 10, 40). All these studies and ours agree on the basal branching pattern connecting the five orders Enterobacteriales, Pasteurellales, Vibrionales, Pseudomonas, and Xanthomonadales. One of these nodes in particular (marked by an “X” in Fig. 3) has additional support from a unique 4-amino-acid deletion in RpoB; however, our results do not agree with two other suggestions based on rare indels: (i) exclusion of Francisella from the Gammaproteobacteria and (ii) a particular split of Alteromonadales (10). Some of the intermingling of bacterial orders observed here had been noted in one of these studies (40), but in the other studies the taxa employed or the extent of multifurcation precluded its detection.
Nearly half of the bacterial genomes currently available are incompletely sequenced, a fraction that may increase in the future, given short-read technologies and sequencing projects that do not include a goal of closing the genome. These incomplete genomes add greatly to the taxonomic diversity available for study and are nearly as rich in protein information as the complete ones, so they are worth including in such analyses despite the minor problems they raise. However, some incomplete genomes are contaminated with DNA from additional taxonomic sources and should be rejected; an rRNA impurity test was employed here to identify some such mixed genomes. Although highly divergent rRNA alleles can occur within a single genome (41), the caution exercised here was warranted; the highly divergent rRNA allele that we found in the incomplete Azotobacter vinelandii genome, thereby rejecting the genome, did not appear in the recently completed version of this genome project.
When our five multiprotein supermatrices (i) were partitioned according to the substitution matrix favored by each family or (ii) had the new LG substitution matrix applied uniformly, nearly identical trees resulted, with only two cases of crossing nodes, and these nodes were the least well supported in the whole tree. This agrees with a recent survey of many alignments and supermatrices that concluded that although model choice can affect tree topology, “it rarely affects evolutionary inferences drawn from the data because differences are mainly confined to poorly supported nodes” (27).
While both the jackknifing and bootstrapping approaches to determining support values remove columns from the alignment supermatrix, jackknifing does not then duplicate the remaining columns and therefore produces smaller subsamplings that can speed processing for large data sets. We prefer the random removal of whole proteins rather than single columns across all protein alignments, to better assess variation at a coarser granularity and as a double-check against possible remaining incongruity of protein families. This 50% protein jackknifing is a more stringent measure of support than the usual per-column bootstrapping, counteracting the misleadingly high support values that the latter brings to long supermatrices (23).
Compositional attraction through amino acid bias made it difficult to infer the phylogeny of the insect endosymbiont genomes, whose composition was the most biased in our taxon set. As in most previous studies, the endosymbionts joined together in a clade, with Sodalis when present, when all were included in a simple maximum likelihood analysis. When instead the endosymbionts were tested one at a time, Buchnera consistently joined at the base of the Enterobacteriales; unity of the Buchnera genomes is strongly supported by gene order studies (4). Ca. Blochmannia and Wigglesworthia may also derive from this point yet consistently branch as sister to Sodalis when that taxon is included, jumping past two tree nodes. Ca. Baumannia appeared with Sodalis when present and, unlike other endosymbionts, at nearby positions when absent, weakly supporting the idea that Ca. Baumannia has a unique origin (Fig. 5A and B, part i). Our finding of multiple origins for the endosymbionts agrees with those of other studies that have sought to avoid compositional attraction artifacts, through the use of either nonhomogeneous substitution matrices or analysis of genomic rearrangements (4, 14). It would appear that the pattern of an Enterobacteriales member adopting the endosymbiotic lifestyle and becoming highly A+T-rich has occurred in multiple independent cases.
Surprisingly, nearly equal subsets of Enterobacteriales proteins favored one or the other of two root positions. Massive horizontal transfer is a plausible explanation in principle, although we could not identify a particular single transfer path; the pattern could be a result of numerous transfers along multiple paths. It should be noted that our family selection method removed the genes best known for horizontal transfer, those on genomic islands. Earlier studies of a classical set of 13 gammaproteobacterial genomes that includes Escherichia, Salmonella, and Yersinia but not Sodalis, Pectobacterium, or Photorhabdus found very few of the single-copy genes with evidence of horizontal transfer. We found that omitting Photorhabdus shifted the support by protein families from a near balance for two root positions to a preponderance for one of these roots, identifying Photorhabdus as a disruptor. The ambiguities left by this study show that the Enterobacteriales present rich problems in phylogenetic reconstruction that are not resolved simply by accumulating larger genome-scale protein supermatrices. These problems are probably due both to horizontal transfer that is especially favored by close contact with diverse cohabitants in enteric environments and to the extensive genomic alterations in multiple isolated symbiotic lineages. Future analysis of these problems within the Enterobacteriales is favored by the detailed mechanistic information on mutation and gene transfer known from E. coli and relatives.
Much of the failure to fully resolve the deeper regions of the tree can probably be ascribed to stochastic accumulation of noise that obscures reconstruction of short and ancient internodes. It may be further compounded by residual cases of horizontal gene transfer that passed through our incongruence filter. Compositional bias may be the greatest challenge facing studies on such broad scales of bacterial phylogeny as this one. The Gammaproteobacteria make it clear that as clades develop nucleotide bias, they can concomitantly develop amino acid bias (33), which would be expected to produce local asymmetries in amino acid substitution rates. Simple statistical tests would have rejected 73% of the taxa from our study on the basis of bias. Thus, our data set (like other data sets of broad phylogenetic scope) violated the typical assumptions of phylogenetic reconstruction algorithms regarding homogeneity and reversibility of amino acid substitution. We were able to manage the problem in some cases by the approach of placing individual biased taxa in the absence of similarly biased attractors. Some possible future solutions may be to counter bias on a per-column basis in the supermatrix, to use mixture models (19), and to use maximum-likelihood programs that do not demand model homogeneity (9). Another promising area for improving multiprotein phylogeny is the use of rare indels (10), especially if the detection of these markers was automated and if tree building could properly weight indel data in combination with alignment data; most of these are removed in our current protocol.
The multiprotein approach to gammaproteobacterial phylogeny, applied here with some methodological advances, has improved resolution for this challenging group, and we anticipate that additional advances are within reach that will further improve performance of the approach. As more genomes accrue, the multiprotein approach should become a new standard, supplanting 16S rRNA as the basis for phylogenetic reconstructions (1).
Supplementary Material
Acknowledgments
This study was supported by USDA agreement 2006-55605-16655 to A.W.D., by DOD grant W911SR-04-0045 to B.W.S.S., by NIAID contract HHSN266200400035C to B.W.S.S., and by the Virginia Bioinformatics Institute. Computing support from Virginia Tech's Advanced Research Computing facility is acknowledged.
We thank David Holmes and Jorge Valdes (Universidad Nacional Andrés Bello, Santiago, Chile) for supplying the Acidithiobacillus genome prior to publication.
Footnotes
Published ahead of print on 5 March 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bapteste, E., Y. Boucher, J. Leigh, and W. F. Doolittle. 2004. Phylogenetic reconstruction and lateral gene transfer. Trends Microbiol. 12:406-411. [DOI] [PubMed] [Google Scholar]
- 2.Bapteste, E., E. Susko, J. Leigh, D. MacLeod, R. L. Charlebois, and W. F. Doolittle. 2005. Do orthologous gene phylogenies really support tree-thinking? BMC Evol. Biol. 5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, R. D. Finn, and E. L. Sonnhammer. 1999. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 27:260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belda, E., A. Moya, and F. J. Silva. 2005. Genome rearrangement distances and gene order phylogeny in gamma-Proteobacteria. Mol. Biol. Evol. 22:1456-1467. [DOI] [PubMed] [Google Scholar]
- 5.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciccarelli, F. D., T. Doerks, C. von Mering, C. J. Creevey, B. Snel, and P. Bork. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283-1287. [DOI] [PubMed] [Google Scholar]
- 7.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emerson, D., J. A. Rentz, T. G. Lilburn, R. E. Davis, H. Aldrich, C. Chan, and C. L. Moyer. 2007. A novel lineage of proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS One 2:e667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galtier, N., and M. Gouy. 1998. Inferring pattern and process: maximum-likelihood implementation of a nonhomogeneous model of DNA sequence evolution for phylogenetic analysis. Mol. Biol. Evol. 15:871-879. [DOI] [PubMed] [Google Scholar]
- 10.Gao, B., R. Mohan, and R. S. Gupta. 2009. Phylogenomics and protein signatures elucidating the evolutionary relationships among the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 59:234-247. [DOI] [PubMed] [Google Scholar]
- 11.Garrity, G. M., J. A. Bell, and T. G. Lilburn. 2005. Class III. Gammaproteobacteria class. nov., p. 1. In D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer, New York, NY. [Google Scholar]
- 12.Gillespie, J. J. 2004. Characterizing regions of ambiguous alignment caused by the expansion and contraction of hairpin-stem loops in ribosomal RNA molecules. Mol. Phylogenet. Evol. 33:936-943. [DOI] [PubMed] [Google Scholar]
- 13.Gillespie, J. J., M. J. Yoder, and R. A. Wharton. 2005. Predicted secondary structure for 28S and 18S rRNA from Ichneumonoidea (Insecta: Hymenoptera: Apocrita): impact on sequence alignment and phylogeny estimation. J. Mol. Evol. 61:114-137. [DOI] [PubMed] [Google Scholar]
- 14.Herbeck, J. T., P. H. Degnan, and J. J. Wernegreen. 2005. Nonhomogeneous model of sequence evolution indicates independent origins of primary endosymbionts within the enterobacteriales (gamma-Proteobacteria). Mol. Biol. Evol. 22:520-532. [DOI] [PubMed] [Google Scholar]
- 15.Karlin, S. 2001. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 9:335-343. [DOI] [PubMed] [Google Scholar]
- 16.Kelly, D. P., and A. P. Wood. 2000. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 50(part 2):511-516. [DOI] [PubMed] [Google Scholar]
- 17.Kjer, K. M. 1995. Use of rRNA secondary structure in phylogenetic studies to identify homologous positions: an example of alignment and data presentation from the frogs. Mol. Phylogenet. Evol. 4:314-330. [DOI] [PubMed] [Google Scholar]
- 18.Le, S. Q., and O. Gascuel. 2008. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25:1307-1320. [DOI] [PubMed] [Google Scholar]
- 19.Le, S. Q., N. Lartillot, and O. Gascuel. 2008. Phylogenetic mixture models for proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363:3965-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lerat, E., V. Daubin, and N. A. Moran. 2003. From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-Proteobacteria. PLoS Biol. 1:E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, L., C. J. Stoeckert, Jr., and D. S. Roos. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakabachi, A., A. Yamashita, H. Toh, H. Ishikawa, H. E. Dunbar, N. A. Moran, and M. Hattori. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314:267. [DOI] [PubMed] [Google Scholar]
- 23.Phillips, M. J., F. Delsuc, and D. Penny. 2004. Genome-scale phylogeny and the detection of systematic biases. Mol. Biol. Evol. 21:1455-1458. [DOI] [PubMed] [Google Scholar]
- 24.Poptsova, M. S., and J. P. Gogarten. 2007. The power of phylogenetic approaches to detect horizontally transferred genes. BMC Evol. Biol. 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puigbo, P., Y. I. Wolf, and E. V. Koonin. 2009. Search for a “Tree of Life” in the thicket of the phylogenetic forest. J. Biol. 8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 27.Ripplinger, J., and J. Sullivan. 2008. Does choice in model selection affect maximum likelihood analysis? Syst. Biol. 57:76-85. [DOI] [PubMed] [Google Scholar]
- 28.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 30.Schulz, H. N., T. Brinkhoff, T. G. Ferdelman, M. H. Marine, A. Teske, and B. B. Jorgensen. 1999. Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science 284:493-495. [DOI] [PubMed] [Google Scholar]
- 31.Scott, K. M., S. M. Sievert, F. N. Abril, L. A. Ball, C. J. Barrett, R. A. Blake, A. J. Boller, P. S. Chain, J. A. Clark, C. R. Davis, C. Detter, K. F. Do, K. P. Dobrinski, B. I. Faza, K. A. Fitzpatrick, S. K. Freyermuth, T. L. Harmer, L. J. Hauser, M. Hugler, C. A. Kerfeld, M. G. Klotz, W. W. Kong, M. Land, A. Lapidus, F. W. Larimer, D. L. Longo, S. Lucas, S. A. Malfatti, S. E. Massey, D. D. Martin, Z. McCuddin, F. Meyer, J. L. Moore, L. H. Ocampo, Jr., J. H. Paul, I. T. Paulsen, D. K. Reep, Q. Ren, R. L. Ross, P. Y. Sato, P. Thomas, L. E. Tinkham, and G. T. Zeruth. 2006. The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena XCL-2. PLoS Biol. 4:e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimodaira, H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51:492-508. [DOI] [PubMed] [Google Scholar]
- 33.Singer, G. A., and D. A. Hickey. 2000. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol. Biol. Evol. 17:1581-1588. [DOI] [PubMed] [Google Scholar]
- 34.Stamatakis, A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688-2690. [DOI] [PubMed] [Google Scholar]
- 35.Talavera, G., and J. Castresana. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56:564-577. [DOI] [PubMed] [Google Scholar]
- 36.Tian, Y., and A. W. Dickerman. 2007. GeneTrees: a phylogenomics resource for prokaryotes. Nucleic Acids Res. 35:D328-D331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterfield, N. R., T. Ciche, and D. Clarke. 2009. Photorhabdus and a host of hosts. Annu. Rev. Microbiol. 63:557-574. [DOI] [PubMed] [Google Scholar]
- 38.Williams, K. P. 2008. Strong mimicry of an rRNA binding site for two proteins by the mRNA encoding both proteins. RNA Biol. 5:145-148. [DOI] [PubMed] [Google Scholar]
- 39.Williams, K. P., B. W. Sobral, and A. W. Dickerman. 2007. A robust species tree for the alphaproteobacteria. J. Bacteriol. 189:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, M., and J. A. Eisen. 2008. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 9:R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yap, W. H., Z. Zhang, and Y. Wang. 1999. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.