Abstract
Tannerella forsythia is a key contributor to periodontitis, but little is known of its virulence mechanisms. In this study we have investigated the role of sialic acid in biofilm growth of this periodontal pathogen. Our data show that biofilm growth of T. forsythia is stimulated by sialic acid, glycolyl sialic acid, and sialyllactose, all three of which are common sugar moieties on a range of important host glycoproteins. We have also established that growth on sialyllactose is dependent on the sialidase of T. forsythia since the sialidase inhibitor oseltamivir suppresses growth on sialyllactose. The genome of T. forsythia contains a sialic acid utilization locus, which also encodes a putative inner membrane sialic acid permease (NanT), and we have shown this is functional when it is expressed in Escherichia coli. This genomic locus also contains a putatively novel TonB-dependent outer membrane sialic acid transport system (TF0033-TF0034). In complementation studies using an Escherichia coli strain devoid of its outer membrane sialic acid transporters, the cloning and expression of the TF0033-TF0034 genes enabled an E. coli nanR nanC ompR strain to utilize sialic acid as the sole carbon and energy source. We have thus identified a novel sialic acid uptake system that couples an inner membrane permease with a TonB-dependent outer membrane transporter, and we propose to rename these novel sialic acid uptake genes nanO and nanU, respectively. Taken together, these data indicate that sialic acid is a key growth factor for this little-characterized oral pathogen and may be key to its physiology in vivo.
Tannerella forsythia is a Gram-negative obligate anaerobic pathogen that causes periodontitis in humans (26, 30, 31). This is a globally important disease that affects over 300 million people and is increasingly linked with a number of systemic sequelae (11). However, little is known regarding its physiology or virulence factors (30).
Accumulation of subgingival plaque is considered a prerequisite to periodontal disease, and there is much evidence that T. forsythia is one of the dominant organisms in the plaque biofilm in periodontitis (26, 30, 31). The link between biofilm formation and virulence has been established for a number of important pathogens, such as uropathogenic Escherichia coli and Pseudomonas aeruginosa (12), and so it is likely that the ability to adapt to life within the plaque biofilm (interact with other species present, evade host defenses, and obtain local nutrients) is closely linked to the pathogenicity of T. forsythia. Thus, an understanding of how the physiology of this periodontal pathogen is adapted to the biofilm lifestyle is key to gaining a clearer picture of disease.
T. forsythia has notoriously fastidious growth requirements, and the original growth studies indicated that the only compound tested which stimulates growth of T. forsythia in broth and plate culture was the monomeric form of the bacterial cell wall component N-acetylmuramic acid (NAM) (26, 31, 36). However, it is not known how relevant this is to the bacteria's growth in vivo where one assumes that the only sources of NAM are other autolyzed bacteria in the oral community.
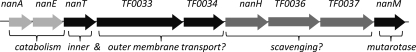
The genome of T. forsythia encodes a large putative sialic acid utilization and transport locus in the genome of T. forsythia (TF0030-TF0038) (Fig. 1) (Oral Pathogen Sequence Databases [http://www.oralgen.lanl.gov]). This locus encodes putative catabolic enzymes involved in the breakdown of sialic acid to ManNac and pyruvate (the neuraminate lyase [TF0031]) and then conversion into GlcNac-6P by neuraminate epimerase (TF0030) (33, 34). In addition, the putative sialic acid locus also codes for transport and scavenging enzymes and includes a newly characterized NanH sialidase enzyme that might be involved in the cleavage of sialic acid from host glycoproteins (33), a putative NanT inner membrane permease that has 45% amino acid sequence similarity with the E. coli nanT gene, and a predicted outer membrane TonB-dependent transport system with 43% amino acid sequence similarity to the starch utilization SusC system of intestinal Bacteroides species. Sialic acid is an important biomolecule that is present on the surface of many mammalian glycoproteins (10, 22), and the oral environment is particularly rich in sialylated glycoproteins, such as salivary mucins and fibronectin (13). Recent work has also highlighted the presence of sialic acid on the surface of several bacteria and indicated a role in immune evasion (25) and biofilm formation (20, 27).
FIG. 1.
Genetic organization of putative sialic acid utilization locus of T. forsythia. The nanA (neuraminate lyase) and nanE (N-acylglucosamine 2-epimerase) genes encode cytoplasmic enzymes required for sialic acid utilization (light gray), while nanH, TF0036, and TF0037 encode a potentially secreted sialidase, a beta-hexosaminidase, and a sialic acid-specific 9-O-acetyl esterase, respectively, which might be involved in scavenging sialic acid from the oral environment. Transport is most likely achieved by the putative inner membrane permease nanT and the potential TonB-dependent outer membrane-specific sialic acid transporters TF0033 and TF0034. The putative mutarotase nanM is responsible for the interconversions of isomers of sialic acid, making the uptake process more efficient. Genes were identified from http://www.oralgen.lanl.gov/.
The original growth studies on T. forsythia also tested sialic acid as a growth factor for plate and broth culture but did not test its effect on biofilm growth (36). Thus, given the presence of this locus and the fact that several human-pathogenic bacteria can cleave or utilize sialic acid, we considered the possibility that sialic acid might be an alternative growth factor for T. forsythia when it is grown as a biofilm. In addition, we aimed to determine the functionality of the putative transport systems identified in the genome.
MATERIALS AND METHODS
Bacterial growth and strains.
T. forsythia ATCC 43037 was maintained in an anaerobic cabinet (containing CO2, H2, and N2) at 37°C on Fastidious Anaerobe agar supplemented with 5% horse blood containing 0.17 mM (wt/vol) NAM. Where indicated, NAM was replaced with sialic acid (Neu5Ac; Carbosynth United Kingdom), sialyllactose (mixture of 2,6- and 2,3-sialyllactose [Sigma A0828]), or glycolyl sialic acid [NeuGc; Carbosynth, United Kingdom) at the concentrations indicated in the relevant figures and legends. Liquid cultures were grown in tryptic soy broth ([TSB] supplemented with 0.4% yeast extract, 1 mg/ml hemin, and 1 mg/ml vitamin K [15]) and the appropriate sugar addition.
E. coli strains were maintained on LB agar with appropriate concentrations of antibiotic (50 μg/ml ampicillin [Amp], 10 μg/ml tetracycline [Tet], and 50 μg/ml kanamycin [Kan]). Strains are listed in Table 1. For assessment of the use of sialic acid as a sole carbon and energy source, strains were grown in M9 salts medium supplemented with glucose (0.5%) or sialic acid (6 mM). For E. coli MC1000, leucine was included at a concentration of 50 mg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Reference or source |
|---|---|
| Strains | |
| T. forsythia 43037 | W. Wade (Kings College London, UK) |
| E. coli | |
| MG1655 | 2 |
| MG1655 nanR | |
| (amber mutation) ΔnanC | I. Blomfield (University of Kent, UK) |
| MC1000 ompR::Tn10(tet) | I. Blomfield (University of Kent, UK) |
| MG1655 nanR | |
| (amber mutation) ΔnanC ompR::Tn10(tet) | This study |
| MC1000 | 6 |
| MC1000 ΔnanT::FRT::Km::FRT | This study |
| TOP10 | Invitrogen |
| Plasmids | |
| pCR2.1-TOPO | Invitrogen |
| pBAD18 | 14 |
| pKD13 | 9 |
| pKD46 | 9 |
| pBAD18-nanT-coli | This study |
| pBAD18-nanT-TF | This study |
| pCR2.1-TOPO-TF0033-34 | This study |
Biofilm growth of T. forsythia.
Briefly, T. forsythia was grown on Fastidious Anaerobe agar as above for 4 to 5 days before being harvested and washed twice in supplemented TSB. For biofilm growth bacteria were inoculated at a final optical density at 600 nm (OD600) of 0.05 into the same medium containing either 0.005% (wt/vol) NAM or sialic acid (Neu5Ac), at the concentrations indicated in the relevant figures and legends, in polystyrene 96- or 24-well plates. Cell numbers for mature biofilms were assessed after 4 days of growth by removal of medium (and suspended bacteria); samples were washed two to three times in sterile phosphate-buffered saline (PBS), followed by rigorous resuspension of biofilms in PBS before cells were counted microscopically using a Helber counting chamber (Hawksley) under a phase-contrast lens (magnification, ×1,000). For growth curve experiments cells were harvested by using an identical method at 24-h intervals and then enumerated. The harvesting method was verified by crystal violet staining of the harvested wells to assess residual cells. T. forsythia biofilms were also grown in the presence of the indicated amounts of N-glycolylneuraminic acid, sialic acid, or 2,3- and 2,6-sialyllactose as a growth inducer in place of NAM.
The effect of the influenza virus neuraminidase inhibitor oseltamivir on T. forsythia biofilm growth was assessed after inclusion of various concentrations (1 to 10 mM) in the medium and enumeration of cells after 4 to 5 days.
Live/dead staining of biofilm cells.
To assess the viability of the cells, T. forsythia biofilms were grown on sterile 1-mm borosilicate glass coverslips (Fisher, United Kingdom) in 24-well tissue culture plates in TSB to an A600 of 0.05 and incubated for 5 days. Planktonic cultures were grown in glass universal tubes without shaking. Before staining, the medium from the 24-well tissue culture plates was removed and replaced with a similar volume of PBS (pH 7.2), whereas for the planktonic culture, 1 ml of sample was diluted with PBS (pH 7.2) in a fresh tube. The samples were stained with 3 μl of a 1:1 mixture of 3 μg/ml propidium iodide (PI) (Invitrogen) and 8.33 μg/ml bis-benzimide Hoechst trihydrochloride (Hoechst; Invitrogen, OR) and incubated anaerobically at 37°C in the dark for 10 min. Three microliters of planktonic cells stained with the above mixture was placed on a microscope slide and examined. For the biofilm, the coverslip with the biofilm cells was inverted onto a microscope slide. Slides were observed at a magnification of ×1,000 with a Zeiss Axiovert 200 M inverted fluorescence microscope with an integrated high-resolution digital camera (AxioCam MRm; Zeiss) equipped with filter sets 00 (PI) and 02 (Hoechst). Pictures were processed with AxioVision, version 4.6, software (Imaging Associates Limited, Bicester, United Kingdom). PI staining was visualized in the red channel, and Hoechst staining was visualized in the blue channel.
Sialidase assay.
Sialidase activity of T. forsythia cells (1 × 107 per well) was detected using the fluorogenic substrate 4-methylumbelliferyl-α-d-neuraminic acid (1.5 mM; Sigma) in 50 mM Tris buffer (pH 7.5). The reaction mixtures were incubated for 2 h at 37°C before the fluorescence intensity was measured at 355-nm excitation and 430 nm (± 10 nm) emission wavelengths on a BMG Labtech microplate reader. Sialidase assays with different concentrations (1 mM, 2 mM, 5 mM, and 10 mM) of oseltamivir (Sigma) were performed to quantify the inhibition of neuraminidase activity. All assays were performed in triplicate on three separate occasions.
Molecular biology techniques.
Standard molecular cloning techniques were performed according to Sambrook and Russell (24). All cloning experiments were performed using the electrocompetent recA mutant cloning strain E. coli TOP10. Transformation of plasmids into MG1655 and MC1000 was achieved by electroporation using a Bio-Rad Micropulser according to the manufacturer's instructions. Phage P1vir transductions were carried out according to Miller (19).
Mutagenesis and heterologous complementation of E. coli.
A nanT deletion strain of E. coli MC1000 was constructed using primers delnanTfrontcoli and delnanTrevcoli (Table 1) with plasmid pKD13 as the template for PCR using Phusion polymerase (NEB) and selection on LB Kan plates (9). Mutations were confirmed by sequencing flanking regions and by the loss of the ability to utilize sialic acid as a sole carbon and energy source on M9 salts medium. Heterologous expression was achieved by cloning the nanT gene of T. forsythia into the expression vector pBAD18 (14) with an optimized E. coli ribosome binding site under the control of Para using primers nanT-xba-for-tf and nanT-hin-rev-tf (Table 2). Proofreading PCR amplification was achieved using Phusion polymerase with T. forsythia genomic DNA as the template. After restriction with XbaI and HinDIII and ligation with calf intestinal alkaline phosphatase (CIAP)-treated plasmids, the constructs were confirmed by sequencing. Similarly the nanT gene of E. coli MG1655 was amplified as a positive control and cloned using primers nanT-xba-for-coli and nanT-hin-rev-coli.
TABLE 2.
Primers used in this study
| Primer | Sequencea |
|---|---|
| delnanTfrontcoli | CTGTACCCTACAATTTCATACCAAAGCGTGTGGGCATCGCCCACCGCGGGAGGCTCACATGTGTAGGCTGGAGCT |
| delnanTrevcoli | TTGTGCAAGTAACGACATACATCTTCCCTTAGCGAAAGGCCCGGTACATAGACCGGGCAACAGGATTCCGGGGATCCGTCGACC |
| nanT-xba-for-coli | AAATCTAGACTGTACCCTACAATTTTC |
| nanT-hin-rev-coli | AAAAAGCTTCGGTACATAGACCGGGCA |
| nanT-xba-for-tf | AAATCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGAAAAACTCAAAGTATTAC |
| nanT-hin-rev-tf | AAAAGCTTAGTTATTGATTAATGAAG |
| tf0033-xba-for | AATCTAGATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGAAAGGAATTTTAAAAAAAT |
| tf0034-xba-rev | AAATCTAGATTATTCATACCCCGGAGT |
Restriction sites are underlined.
For experiments on the heterologous expression of TF0033 and TF0034, we created a nanR nanC ompR triple mutant of MG1655. The nanC nanR mutant was kindly supplied by Ian Blomfield of the University of Kent and acted as the recipient strain for a P1vir transduction of the ompR::Tn10(tet) allele from MC4100 ompR::Tn10(tet). A similar triple mutant strain was previously shown to be unable to utilize sialic acid as a sole carbon and energy source in M9 liquid medium (8). We confirmed the phenotype of the transductants before proceeding with complementation experiments. The TF0033 and TF0034 genes were cloned in tandem into the cloning vector pCRTOPO2.1 with an optimized ribosome binding site after amplification using Phusion polymerase (NEB) and the addition of poly(A) tails using Taq polymerase.
RESULTS
Sialic acid stimulates growth of T. forsythia biofilms.
To test the influence of sialic acid on biofilm growth, our normal biofilm growth medium was supplemented with various concentrations of sialic acid in place of NAM under the same conditions, i.e., in polystyrene microtiter well plates incubated anaerobically at 37°C. The range of sialic acid concentrations used was chosen based upon the concentration required to support growth of E. coli in minimal medium with sialic acid as a sole carbon source (3 mM) (28, 35).
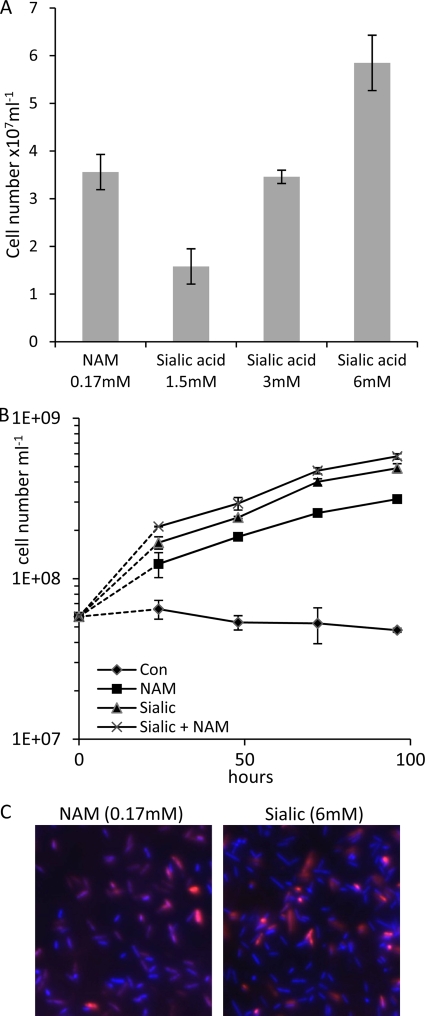
After the biofilms were washed and bacteria were harvested from the plates, the growth of bacteria was enumerated by repeated pipetting and counting microscopically. To ensure that all bacteria were removed from the microtiter plates, they were stained with crystal violet and visualized under a light microscope. Figure 2A illustrates that the cell numbers after 4 days increased with increasing sialic acid concentrations and were comparable to the numbers obtained when NAM was used as a substrate although at a higher total carbon concentration. In order to further confirm this observation, we also assessed growth using the same method over a 5-day period (Fig. 2B). These data illustrate similar biofilm growth curves for T. forsythia on NAM and sialic acid although it is interesting that growth on 6 mM sialic acid produced a higher final cell number (4.9 × 107 ml−1) than 0.17 mM NAM alone (3.1 × 107 ml−1). However, growth on a combination of sialic acid and NAM did not produce a substantially higher final cell number than growth on sialic acid alone (5.8 × 107 ml−1), indicating that the bacteria may prefer to use sialic acid for growth in biofilm culture. The result, however, is hard to interpret since the concentration of NAM (170 μM) required in the growth medium is much lower than the concentration of sialic acid (6 mM). In contrast, while T. forsythia growth was stimulated in biofilm culture, we did not see a corresponding stimulation of growth either in planktonic culture or on agar plates. This observation of a lack of growth stimulation in either planktonic or plate culture was expected since the original work determining NAM as a growth factor for T. forsythia tested growth on sialic acid in broth and plate culture with similar results (36).
FIG. 2.
Stimulation of T. forsythia biofilm growth by sialic acid. (A) The ability of sialic acid to stimulate growth of T. forsythia biofilms was tested by replacing NAM (0.17 mM) with various concentrations of sialic acid (1.5 to 6 mM). Biofilms were incubated for 4 days before cell counting. Means and standard deviations of three independent wells are shown. The experiment was conducted on three separate occasions with essentially identical results. (B) Growth rates were assessed by inoculating identical wells of a 96-well plate and harvesting the biofilm at the time points shown. These experiments used sialic acid and NAM at concentrations of 6 mM and 0.17 mM, respectively. Control samples contained only the TSB growth medium with no growth factors added (Con). Growth was assessed by cell counting of washed and harvested biofilms that had been resuspended in PBS; complete harvesting was established by crystal violet staining of harvested wells. Means of three wells are shown with standard deviations illustrated. The first data point illustrates the initial cell number prior to inoculation while the second and subsequent points relate to cell numbers of harvested biofilms. (C) Live/dead staining of NAM or sialic acid biofilms grown on glass coverslips. Hoechst (blue) was used to stain all bacteria and propidium iodide (red) was used to highlight dead bacteria. Individual channels were enumerated using ImageJ software. Merged images are shown.
To assess the viability of the sialic acid-grown biofilm cells, we performed live/dead staining on biofilms grown on glass coverslips under the same growth conditions (Fig. 2C). After 4 days the cell viability was assessed by live/dead staining using Hoechst as a stain for all for nucleic acids (live and dead cells; blue) and propidium iodide (red) to highlight dead cells (as this cannot cross intact cell membranes). As illustrated in Fig. 2C there were identical ratios of live/dead cells in the biofilms grown on either NAM or sialic acid (i.e., approximately 80:20, live to dead). Taken together these data indicate that T. forsythia is able to utilize this biologically relevant potential carbon and nitrogen source to stimulate biofilm growth.
Ability of T. forsythia to utilize alternative forms of sialic acid.
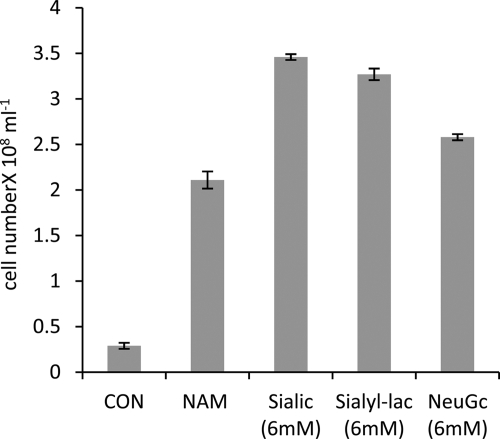
Recent work by others highlighted the ability of a putative exported sialidase to release free sialic acid from sialyllactose (33). Sialyllactose is a common moiety on glycoproteins and notably is the sugar head group on the GM3 ganglioside that is ubiquitous on mammalian plasma cell membranes (18). We therefore assessed the ability of T. forsythia to grow in a biofilm using sialyllactose (a mixture of 2,3- and 2,6-sialyllactose) as a growth stimulant. Figure 3 illustrates that the growth yield was similar to that obtained with equimolar amounts of free sialic acid while lactose had no effect (this is a product of the sialidase cleavage of sialyllactose) (data not shown). Again live/dead staining revealed comparable viability of the sialic acid-grown cells to the wild-type strain (data not shown).
FIG. 3.
Biofilm growth yield of T. forsythia on alternative sialic acid sources. The ability to use sialic acid and the alternative forms sialyllactose (sialyl-lac) and glycolyl sialic acid (NeuGc) to stimulate growth of T. forsythia was tested by replacing NAM (0.17 mM) with 6 mM concentrations of the other substrates. Control wells (CON) were incubated under identical conditions with TSB lacking any additional growth factors. Biofilms were incubated for 4 days before cells were washed, harvested, and counted. Means and standard deviations of three independent wells are shown. The experiment was conducted on three separate occasions with essentially identical results.
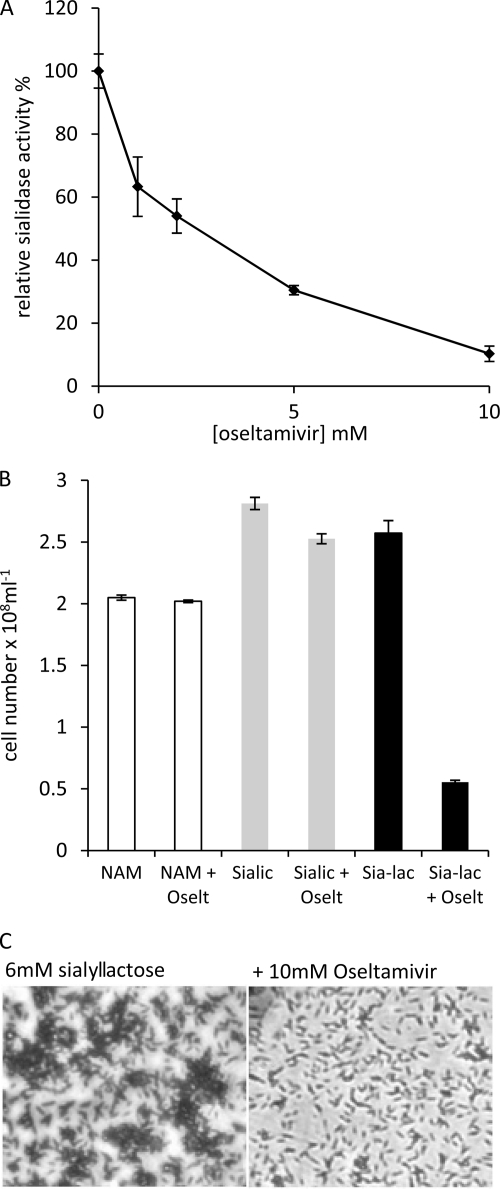
Since the release of sialic acid from sialyllactose probably requires the action of the secreted sialidase enzyme, we first searched for an inhibitor of whole-cell sialidase activity. Figure 4A illustrates the effect on whole-cell sialidase activity using the fluorescent substrate 4-methylumbelliferyl-α-d-neuraminic acid and various concentrations (1 to 10 mM) of the neuraminidase inhibitor oseltamivir (Tamiflu). The experiment reveals that the whole-cell activity is inhibited in a dose-dependent manner by oseltamivir. We then assessed the effects of inhibiting the sialidase activity on the ability of T. forsythia to utilize sialyllactose (6 mM) in biofilm growth experiments. Figure 4B illustrates that while the inclusion of 10 mM oseltamivir in the medium containing NAM or sialic acid had no effect on biofilm growth, it did inhibit biofilm growth with sialyllactose as the growth substrate, resulting in 4-fold lower cell numbers (Fig. 4B and C). This shows that the sialidase enzyme activity is required for the use of sialyllactose for growth and suggests that it may be important for utilization of sialic acid contained on the surface of host glycoproteins, on which sialyllactose is a common terminal sugar group.
FIG. 4.
Effect of oseltamivir on whole-cell sialidase activity and biofilm growth on sialyllactose. (A) Four-day-old T. forsythia biofilm cells were harvested, washed with PBS, and adjusted to a cell density at A600 of 0.05. The sialidase substrate 4-methylumbelliferyl-α-d-neuraminic acid was then added to a final concentration of 1.5 mM. Oseltamivir was then added at the concentrations shown, and the reaction was allowed to proceed for 2 h at 37°C anaerobically before the release of free umbelliferyl was measured using a fluorimeter. Assays were performed in triplicate, and the readings are presented as a percentage of the cell activity without any oseltamivir added; standard error is shown. (B) T. forsythia biofilms were set up as before in triplicate wells supplemented with either 0.17 mM NAM, 6 mM sialic acid, or 6 mM sialyllactose (Sia-lac) with the addition of 10 mM oseltamivir (Oselt) to the medium as indicated at the time of inoculation. After 4 days of growth, biofilm cells were washed, harvested, and enumerated. Data are presented with standard deviations. (C) Micrographs of crystal violet-stained biofilms of T. forsythia grown on sialyllactose (6 mM) with and without the addition of 10 mM oseltamivir and visualized at a magnification of ×400 on a Nikon TS100 inverted light microscope attached to a Coolpix P1500 digital camera.
Recent work has implicated the product of the yhcH gene of Haemophilus influenzae as an epimerase involved in the metabolism of an alternate but naturally abundant form of sialic acid known as glycolyl sialic acid (glycolylneuraminic acid [NeuGc]) (32). We therefore screened the T. forsythia genome for a homologue of this gene, and even though the TF1464 gene is not in the same region as the other putative sialic acid utilization genes, it has 46% similarity at the amino acid level to the yhcH gene. In light of this we tested the ability of T. forsythia to grow using equimolar amounts of NeuGc as a growth stimulant in place of sialic acid (Neu5Ac). The data presented in Fig. 3 illustrate that the growth yields were similar on NeuGc and Neu5Ac, indicating that T. forsythia can efficiently utilize and most likely transport NeuGc into the cell and thus expanding the possible range of in vivo substrates for T. forsythia growth.
T. forsythia contains a functional inner membrane sialic acid permease.
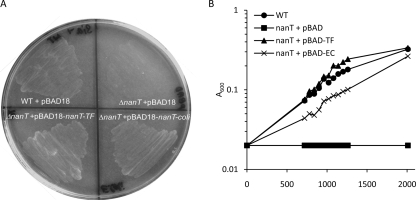
Our data so far indicate that T. forsythia has the ability to utilize sialic acid while evidence from other investigators has indicated that this bacterium contains a catalytically active sialic acid-scavenging sialidase enzyme originating from the same area of the chromosome as nanA and nanE (33). We now aimed to test the functionality of the putative nanT sialic acid permease (TF0031) as a sialic acid transporter. Despite reports of the production of deletion mutants of T. forsythia by other laboratories (15, 23), it is still very difficult to construct targeted chromosomal mutants in T. forsythia. Therefore, in order to examine the function of the putative nanT permease, we decided to follow a heterologous expression route and cloned the T. forsythia nanT gene into the arabinose-inducible expression vector pBAD18 (14) to create plasmid pBAD18-nanT-TF. We then tested its ability to complement the null phenotype of an E. coli MC1000 (araD mutant) ΔnanT strain compared to the corresponding nanT gene from E. coli (pBAD18-nanT-coli) (Fig. 5). Figure 5 illustrates that in contrast to the empty pBAD18 plasmid, the pBAD18-nanT-TF plasmid was able to restore the ability of MC1000 to grow on sialic acid as the sole carbon source without induction with arabinose, thus indicating that in addition to being able to utilize and scavenge sialic acid, this organism also contains a functional inner membrane sialic acid transporter. In fact induction of pBAD18-nanT-TF and pBAD18-nanT-coli with arabinose (which MC1000 is unable to use due to a mutation in araD) at concentrations as low as 0.01% (wt/vol) resulted in cell death with both the E. coli and T. forsythia constructs (data not shown).
FIG. 5.
Complementation of E. coli MC1000 ΔnanT by T. forsythia nanT. (A) E. coli MC1000 ΔnanT was transformed with either pBAD18 or derivatives containing the nanT gene from T. forsythia (pBAD18-nanT-TF:pBAD-TF) or E. coli (pBAD18-nanT-coli:pBAD-EC), and growth was assessed on M9 agar plates containing (0.5%; 15 mM) sialic acid. All strains showed identical growth with glucose (0.5%) as the carbon source (data not shown). (B) The same strains were grown in M9 liquid medium, and growth was monitored by measuring the A600 at the time points indicated. Experiments were repeated on four separate occasions with essentially identical results. WT, wild type.
Our data above indicated that T. forsythia is able to utilize NeuGc as a growth stimulant. The ability to utilize NeuGc requires it to be transported into the cell by the cell's sialic acid permease (35). In order to examine the specificity of the T. forsythia NanT for NeuGc, we tested the ability of the nanT permease of T. forsythia to restore growth on sialic acid to our nanT mutant on NeuGc. While the nanT mutant strain is unable to utilize NeuGc as a carbon source, the strain complemented with the nanT gene of T. forsythia is restored in its ability to use NeuGc for growth (data not shown). These data are consistent with our demonstration that NeuGc can stimulate growth of T. forsythia biofilms and suggest that, as in E. coli, the KD (equilibrium dissociation constant) values of the nanT permease are similar for NeuGc and Neu5Ac.
TF0033 and TF0034 form a novel outer membrane TonB-dependent sialic acid uptake system.
As illustrated in Fig. 1, the genome of T. forsythia contains a large sialic acid metabolism locus that also contains a putative outer membrane transport system encoded by the genes TF0033 and TF0034 (see the URL of the Oral Pathogen Sequence Databases above). Sequence analysis of TF0033 indicates that it is a putative TonB-dependent receptor protein (see URL of the Oral Pathogen Sequence Databases) while the TF0034 gene possesses 40% homology with the SusD protein of several Bacteroides spp. (21). These proteins are usually involved in the transport of iron-containing compounds across the outer membrane into the periplasm of Gram-negative bacteria (3, 16).
To enable us to test the function of the TF0033-TF0034 gene products, we constructed an nanC ompR nanR triple mutant that lacks the outer membrane sialic acid-transporting NanC, OmpC, and OmpF proteins and contains an nanR mutation to allow constitutive expression of the sialic acid catabolism genes in E. coli MG1655, and we confirmed that this strain is unable to grow using sialic acid as a sole carbon and energy source (Fig. 6). To test the functionality of the TF0033 and TF0034 genes, we cloned them in tandem into the E. coli cloning vector pCRTOPO-2.1. Figure 6 illustrates that in the presence of the pCRTOPO2.1-TF0033-34 construct, the nanC ompR nanR strain regained the ability to grow using sialic acid as a sole carbon and energy source in liquid medium and that the growth rate was similar to that of the wild type. This indicates that this is a putative high-affinity sialic acid uptake complex that seems to be functional in tandem with the E. coli TonB-ExbB-ExbD complex which it requires to provide energy for the transport process.
FIG. 6.
Ability of T. forsythia TF0033-TF0034 to support growth on sialic acid. E. coli MG1655 (wild type [WT]) containing the empty pCR2.1-TOPO vector and the MG1655 nanR nanC ompR mutant containing either the empty pCR2.1-TOPO vector or pCR2.1-TOPO containing the TF0033-TF0034 genes of T. forsythia were grown in M9 salts medium containing 15 mM sialic acid, and growth was monitored by measuring the A600 at the time points indicated. Experiments were repeated on four separate occasions with essentially identical results. A representative experiment is shown.
Our data also highlight the ability of NeuGc to stimulate biofilm growth of T. forsythia, so we wished to examine whether the putative outer membrane sialic acid transporter encoded by the TF0033-TF0034 genes also had specificity for NeuGc. We therefore examined the ability of TF0033-TF0034 to support growth of the nanC nanR ompR mutant strain of MG1655 and found that while the nanC nanR ompR strain is unable to grow with NeuGc as the sole carbon and energy source, when TF0033-TF0034 is provided in trans in plasmid pCR2.1-TF0033-34, this ability is gained (data not shown).
DISCUSSION
The findings presented here show for the first time that the fastidious anaerobe T. forsythia is able to utilize sialic acid as a growth factor to specifically stimulate biofilm growth. However, we found no sialic acid-dependent growth stimulation on agar plates or in liquid broth. At this time we cannot explain why sialic acid can substitute for NAM to stimulate growth of T. forsythia only in a biofilm, but it is worth noting that in vivo this organism would most commonly inhabit a biofilm. It is possible that the higher cell density in a biofilm triggers a response similar to a quorum-sensing mechanism or that some other regulatory mechanism responding to levels of metabolic by-products stimulates expression of the nan genes under biofilm conditions. We have preliminary proteomic (G. Stafford and P. Wright, personal communication) and real-time PCR data that seem to confirm this idea with upregulation at both the protein and transcript levels of the nan genes in mature biofilms compared to planktonic cultures (data not shown). This is currently under investigation alongside the role of sialic acid itself in inducing expression of these genes (as occurs in E. coli) (34, 35). Nevertheless, our data indicate that sialic acid is a growth stimulant of T. forsythia biofilms. While several other organisms, such as E. coli and H. influenzae, are capable of utilizing sialic acid as a growth substrate (34), this is the only indication of such a metabolic capability in a Gram-negative oral anaerobe.
As illustrated in Fig. 1, the genome of T. forsythia contains homologues of the nanA (neuraminate lyase) and nanE (N-acetylmannosamine epimerase) genes but lacks a nanK (N-acetylmannosamine kinase) homologue. The latter is required for phosphorylation of N-acetylmannosamine (ManNac) to ManNac-6P before conversion into N-acetylglucosamine-6-phosphate (NAG-6P) by NanE in E. coli and H. influenzae (34). However, sequence analysis of the nanE gene of T. forsythia reveals that it has 83% amino acid homology with the nanE gene from Bacteroides fragilis. Recent work by other investigators has shown that even though B. fragilis also lacks an nanK homologue, it is capable of utilizing sialic acid (4). The NanE from B. fragilis is capable of converting ManNac to NAG, which is then phosphorylated by a hexokinase called RokA before being processed by the rest of the pathway (4). A genome search reveals that the open reading frame (ORF) TF1997 has 82% amino acid homology with RokA from B. fragilis, further strengthening the idea that T. forsythia might use a similar sialic acid metabolic pathway as that of B. fragilis. Based on this information, we have outlined a proposed sialic acid metabolism pathway in T. forsythia, which is illustrated in Fig. 7A. This putative pathway also proposes the nagA and nagB genes for the processing of N-acetylglucosamine.
FIG. 7.
Schematic of putative sialic acid utilization and uptake pathways. (A) Predicted sialic acid utilization pathway of T. forsythia. The enzymes shown are predicted from the genome searches and by comparison with the pathway proposed for B. fragilis (diagram adapted from reference 4). The gene identification numbers from the T. forsythia genome annotation are indicated in parentheses. NanA, neuraminate lyase; NanE, N-acetylmannosamine epimerase; RokA, hexokinase; NagA, N-acetylglucosamine-6-phosphate deacetylase; NagB, glucosamine-6-phosphate isomerase. (B) Proposed sialic acid uptake pathway. The NanOU (TF0033-TF0034) outer membrane (OM) transport system and the NanT(TF0032) inner membrane (IM) permease that function when transplanted into E. coli in trans are shown in light gray while the TonB (TF1354/TF1960/TF0783)-ExbB (TF0785)-ExbD (TF0785) complex that probably energizes this transport is shown in dark gray (1).
The ability to utilize sialic acid as a growth factor is particularly pertinent, given the range of sialylated glycoproteins present both in saliva (e.g., mucins) and on the surface of epithelial cells (e.g., fibronectin and integrins). Other human-pathogenic bacteria such as E. coli, H. influenzae, and B. fragilis can utilize sialic acid as a growth substrate (34). In addition E. coli is able to utilize glycolyl sialic acid (NeuGc) (35), which humans are unable to synthesize de novo but which is acquired from dietary sources (29) and processed into human glycoproteins on the surface of gut (and presumably oral) epithelial cells in vivo (5, 17). It is therefore of no surprise that NeuGc can act in a manner similar to Neu5Ac to stimulate biofilm growth of T. forsythia.
As part of this sialic acid utilization locus (Fig. 1), T. forsythia contains an nanH sialidase gene which was recently expressed in E. coli and shown to cleave 2,3- and 2,6-sialyl-linked sialyllactose and has the potential to recover sialic acid from host-glycoproteins on which sialyllactose is a common sugar motif (33). The importance of this sialidase in sialic acid utilization was illustrated in our work when we inhibited this enzyme with oseltamivir and thus abrogated the ability of T. forsythia biofilms to utilize sialic acid linked to lactose. However, in contrast to findings in Streptococcus pneumoniae and P. aeruginosa, inhibition of the sialidase activity did not completely inhibit biofilm growth per se, i.e., with NAM or sialic acid as growth factors (Figure 4B and C) (20, 27). However, the sialidase activity is likely to play a role in sialic acid scavenging in vivo, and we are currently investigating this with a range of physiologically relevant glycoproteins, including mucins.
In order to be able to utilize sialic acid, T. forsythia must transport sialic acid into the cell across both the outer and inner membranes. Our findings have established the functionality of the major facilitator family protein homologue, nanT, in E. coli. We have also established that the novel outer membrane transport genes TF0033 and TF0034 encode a functional sialic acid transport system that is also able to complement sialic acid growth defects in an E. coli strain unable to transport sialic acid across its outer membrane. Given that our data indicate that these two genes encode an outer membrane sialic acid transporter pair, we propose to follow the current naming convention and name TF0033 nanO (neuraminate outer membrane permease) and TF0034 nanU (extracellular neuraminate uptake protein). The NanO amino acid sequence indicates that it is a TonB-dependent receptor (TBDR) family protein possessing 43% amino acid sequence homology with the susC gene of Bacteroides thetaiotaomicron, which is involved in the uptake of starch and oligomeric maltose sugars in a mechanism that is energized by the TonB-ExbB-ExbD (TBDR) protein complex (1, 3, 37). In common with other members of the Bacteroidetes, T. forsythia contains over 60 TBDRs in its genome of 3.4 Mbp, indicating that this class of proteins is overrepresented (1). This overrepresentation may reflect the fact that T. forsythia, in the oral environment, has evolved to utilize a wide range of dietary sugars like other Bacteroides species from the gut (38).
In contrast to NanO, NanU has homology to members of the SusD protein family in the closely related Bacteroides spp. (e.g., B. thetaiotaomicron and B. fragilis) that are all predicted to be involved in nutrient utilization of a range of carbohydrates. It possesses a type II signal recognition particle signal and has approximately 40% amino acid similarity and predicted structural similarity to the SusD protein from B. thetaiotaomicron (21) (G. P. Stafford, unpublished data). There are a number of homologues of SusD in the T. forsythia genome, again indicating that like other Bacteroides species this may be an important mechanism for nutrient uptake.
Based on our data, we propose the tentative model illustrated in Fig. 7B for sialic acid transport mechanisms in T. forsythia. Free sialic acid or that released from glycoproteins is bound by TF0034 (NanU) and, by a similar mechanism, to the SusCD complex (7); this interacts with TF0033 (NanO, which passes the sialic acid on for transport into the periplasm. The latter mechanism requires energization by the TonB-ExbB-ExbD complex. The sialic acid is then transported from the periplasm into the cytoplasm by the inner membrane permease, NanT, before utilization by the sialic acid metabolism pathway. There are clearly a number of parts of this model that require investigation, such as the putative protein interactions between NanO and NanU, the binding of sialic acid to NanU, and the role of TonB in energizing the process (there are three TonB homologues in the T. forsythia genome) (see the URL of the Oral Pathogen Sequence Databases above).
Our overall findings indicate that sialic acid may represent a more relevant in vivo growth factor than NAM, the only previously established growth factor for T. forsythia. However, to what extent sialic acid is used as a sole carbon and energy source is not known. Pertinently, the requirement for NAM is in itself not fully understood since the small amount of NAM (0.17 mM) required to stimulate growth argues against its use as a sole carbon source but rather that T. forsythia lacks some biosynthetic capability to produce NAM or other cell wall building blocks. It is intriguing, therefore, that sialic acid stimulates growth of T. forsythia biofilms since it is metabolized into N-acetylglucosamine, which in many bacteria is converted into NAM and which is also a building block in cell wall biosynthesis. The importance of sialic acid to this pathogen is reinforced since we have also identified and established the functionality of a novel outer membrane sialic acid transport system that might be important for pathogenicity. Our work highlights several important avenues for further investigation of the mechanistic and physiological aspects of this system, particularly its role in biofilm growth. These findings expand our knowledge of this poorly understood oral pathogen and highlight the possible importance of sialic acid as a growth factor or substrate in the oral microbial ecosystem.
Acknowledgments
We thank William Wade (Kings College London) for the initial gift of T. forsythia and advice on its growth. We also thank Ian Blomfield for the gift of E. coli nanR nanC and ompR strains.
This work was funded by grants from the Royal Society and British Oral and Dental Research Trust to G.S. and by a University of Sheffield Oral Disease cluster studentship to S.R.
Footnotes
Published ahead of print on 26 February 2010.
REFERENCES
- 1.Blanvillain, S., D. Meyer, A. Boulanger, M. Lautier, C. Guynet, N. Denance, J. Vasse, E. Lauber, and M. Arlat. 2007. Plant carbohydrate scavenging through tonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS ONE. 2:e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Braun, V. 2003. Iron uptake by Escherichia coli. Front Biosci. 8:s1409-s1421. [DOI] [PubMed] [Google Scholar]
- 4.Brigham, C., R. Caughlan, R. Gallegos, M. B. Dallas, V. G. Godoy, and M. H. Malamy. 2009. Sialic acid (N-acetyl neuraminic acid) utilization by Bacteroides fragilis requires a novel N-acetyl mannosamine epimerase. J. Bacteriol. 191:3629-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byres, E., A. W. Paton, J. C. Paton, J. C. Lofling, D. F. Smith, M. C. Wilce, U. M. Talbot, D. C. Chong, H. Yu, S. Huang, X. Chen, N. M. Varki, A. Varki, J. Rossjohn, and T. Beddoe. 2008. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature 456:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 7.Cho, K. H., and A. A. Salyers. 2001. Biochemical analysis of interactions between outer membrane proteins that contribute to starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 183:7224-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Condemine, G., C. Berrier, J. Plumbridge, and A. Ghazi. 2005. Function and expression of an N-acetylneuraminic acid-inducible outer membrane channel in Escherichia coli. J. Bacteriol. 187:1959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dwek, R. A. 1996. Glycobiology: toward understanding the function of sugars. Chem. Rev. 96:683-720. [DOI] [PubMed] [Google Scholar]
- 11.El-Kheshen, M. 2008. Plaque plague. Dentist 2008:48-50. [Google Scholar]
- 12.Fux, C. A., J. W. Costerton, P. S. Stewart, and P. Stoodley. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 13:34-40. [DOI] [PubMed] [Google Scholar]
- 13.Gabriel, M. O., T. Grunheid, and A. Zentner. 2005. Glycosylation pattern and cell attachment-inhibiting property of human salivary mucins. J. Periodontol. 76:1175-1181. [DOI] [PubMed] [Google Scholar]
- 14.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honma, K., S. Inagaki, K. Okuda, H. K. Kuramitsu, and A. Sharma. 2007. Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb. Pathog. 42:156-166. [DOI] [PubMed] [Google Scholar]
- 16.Koebnik, R. 2005. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13:343-347. [DOI] [PubMed] [Google Scholar]
- 17.Martin, M. J., J. C. Rayner, P. Gagneux, J. W. Barnwell, and A. Varki. 2005. Evolution of human-chimpanzee differences in malaria susceptibility: relationship to human genetic loss of N-glycolylneuraminic acid. Proc. Natl. Acad. Sci. U. S. A. 102:12819-12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masud, M. M., M. Kuwahara, H. Ozaki, and H. Sawai. 2004. Sialyllactose-binding modified DNA aptamer bearing additional functionality by SELEX. Bioorg. Med. Chem. 12:1111-1120. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Parker, D., G. Soong, P. Planet, J. Brower, A. J. Ratner, and A. Prince. 2009. The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect. Immun. 77:3722-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reeves, A. R., G. R. Wang, and A. A. Salyers. 1997. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J. Bacteriol. 179:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudd, P. M., M. R. Wormald, R. L. Stanfield, M. Huang, N. Mattsson, J. A. Speir, J. A. DiGennaro, J. S. Fetrow, R. A. Dwek, and I. A. Wilson. 1999. Roles for glycosylation of cell surface receptors involved in cellular immune recognition. J. Mol. Biol. 293:351-366. [DOI] [PubMed] [Google Scholar]
- 23.Sakakibara, J., K. Nagano, Y. Murakami, N. Higuchi, H. Nakamura, K. Shimozato, and F. Yoshimura. 2007. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology 153:866-876. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Severi, E., G. Randle, P. Kivlin, K. Whitfield, R. Young, R. Moxon, D. Kelly, D. Hood, and G. H. Thomas. 2005. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol. Microbiol. 58:1173-1185. [DOI] [PubMed] [Google Scholar]
- 26.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 27.Soong, G., A. Muir, M. I. Gomez, J. Waks, B. Reddy, P. Planet, P. K. Singh, Y. Kaneko, M. C. Wolfgang, Y. S. Hsiao, L. Tong, and A. Prince. 2006. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J. Clin. Invest. 116:2297-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steenbergen, S. M., J. L. Jirik, and E. R. Vimr. 2009. YjhS (NanS) is required for Escherichia coli to grow on 9-O-acetylated N-acetylneuraminic acid. J. Bacteriol. 191:7134-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tangvoranuntakul, P., P. Gagneux, S. Diaz, M. Bardor, N. Varki, A. Varki, and E. Muchmore. 2003. Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc. Natl. Acad. Sci. U. S. A. 100:12045-12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanner, A. C., and J. Izard. 2006. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol. 2000. 42:88-113. [DOI] [PubMed] [Google Scholar]
- 31.Tanner, A. C., P. M. Milgrom, R. Kent, Jr., S. A. Mokeem, R. C. Page, C. A. Riedy, P. Weinstein, and J. Bruss. 2002. The microbiota of young children from tooth and tongue samples. J. Dent. Res. 81:53-57. [DOI] [PubMed] [Google Scholar]
- 32.Teplyakov, A., G. Obmolova, J. Toedt, M. Y. Galperin, and G. L. Gilliland. 2005. Crystal structure of the bacterial YhcH protein indicates a role in sialic acid catabolism. J. Bacteriol. 187:5520-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson, H., K. A. Homer, S. Rao, V. Booth, and A. H. Hosie. 2009. An orthologue of Bacteroides fragilis NanH is the principal sialidase in Tannerella forsythia. J. Bacteriol. 191:3623-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vimr, E. R., K. A. Kalivoda, E. L. Deszo, and S. M. Steenbergen. 2004. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68:132-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vimr, E. R., and F. A. Troy. 1985. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J. Bacteriol. 164:845-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyss, C. 1989. Dependence of proliferation of Bacteroides forsythus on exogenous N-acetylmuramic acid. Infect. Immun. 57:1757-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 38.Xu, J., M. A. Mahowald, R. E. Ley, C. A. Lozupone, M. Hamady, E. C. Martens, B. Henrissat, P. M. Coutinho, P. Minx, P. Latreille, H. Cordum, B. A. Van, K. Kim, R. S. Fulton, L. A. Fulton, S. W. Clifton, R. K. Wilson, R. D. Knight, and J. I. Gordon. 2007. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 5:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]