Abstract
The abg locus of the Escherichia coli chromosome includes three genes encoding proteins (AbgA, AbgB, and AbgT) that enable uptake and utilization of the folate breakdown product, p-aminobenzoyl-glutamate (PABA-GLU). We report on the purification and characterization of the p-aminobenzoyl-glutamate hydrolase (PGH) holoenzyme encoded by abgA and abgB. One-step purification was accomplished using a plasmid carrying abgAB with a hexahistidine tag on the carboxyl terminus of AbgB and subsequent metal affinity chromatography (MAC). Analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) revealed two subunits (∼53-kDa and ∼47-kDa proteins) of the expected masses of AbgB and AbgA; N-terminal sequencing confirmed the subunit identification, and amino acid analysis yielded a 1:1 ratio of the subunits. Size exclusion chromatography coupled with light-scattering analysis of purified PGH revealed a predominant molecular mass of 206 kDa and a minor component of 400 to 500 kDa. Both peaks contained PGH activity, and SDS-PAGE revealed that fractions containing activity were composed of both AbgA and AbgB. MAC-purified PGH was highly stimulated by manganese chloride. Kinetic analysis of MAC-purified PGH revealed a Km value for PABA-GLU of 60 ± 0.08 μM and a specific activity of 63,300 ± 600 nmol min−1 mg−1. Folic acid and a variety of dipeptides served as poor substrates of PGH. This locus of the E. coli chromosome may encode a portion of a folate catabolism pathway.
Reduced derivatives of folic acid are required for biosynthesis of DNA, RNA, amino acids, and other important cellular components (14). Folic acid is an essential dietary supplement for humans, while both microorganisms and plants can synthesize this vitamin de novo. The Escherichia coli folic acid biosynthetic pathway is composed of proteins encoded by genes scattered across the chromosome (9); these genes appear to be constitutively expressed at low levels (26, 27). The genes and enzymes involved in folate catabolism in E. coli remain largely unidentified.
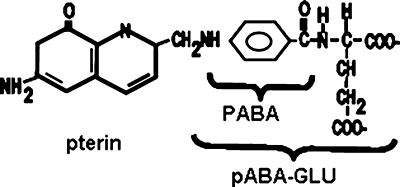
The abg region of E. coli was first identified in a search for mutants able to grow on folic acid in order to circumvent p-aminobenzoate (PABA) auxotrophy; characterization showed that the auxotrophs were utilizing the folate breakdown product, p-aminobenzoyl-glutamate (PABA-GLU), not folic acid (Fig. 1) (11). The original mutations were point mutations in the intergenic region between abgA and abgR, and it was hypothesized that this resulted in increased expression of the abgABT genes. The abg region, named for enhanced growth on p-aminobenzoyl-glutamate, was mapped to 30 min and was found to consist of a potential operon including abgA, abgB, and abgT. Divergently oriented from abgABT, abgR encodes AbgR, which has homology to LysR-type regulatory proteins (21). Sequence analysis of the putative gene products revealed that AbgA and AbgB were similar to one another and to aminoacyl aminohydrolases and that AbgT was similar to transport proteins (11).
FIG. 1.
The structures of folic acid, PABA, and PABA-GLU.
Previously, we had found that wild-type cells transformed with a high-copy-number plasmid carrying abgT demonstrated saturable uptake of PABA-GLU (KT [transport constant] = 123 μM); control cells harboring the vector alone demonstrated negligible uptake (4). Tritiated PABA-GLU taken in by cells expressing large amounts of AbgT was not rapidly metabolized to a form that was trapped in the cell, as addition of nonradioactive PABA-GLU to these cells resulted in rapid loss of intracellular label. Addition of nonradioactive PABA had no effect. However, experiments with cells harboring complementary plasmids carrying abgT and abgAB, respectively, demonstrated initial uptake of radiolabeled PABA-GLU followed by loss of intracellular label over time. These findings were consistent with the hypothesis that AbgT catalyzed the uptake of PABA-GLU into the cell and that when sufficient amounts of PABA-GLU accumulated, AbgAB catalyzed cleavage to form PABA and glutamate. Since PABA enters cells in a nonsaturable manner consistent with diffusion (27), once the radioactive PABA is generated, it can depart the cell. Taken together, these data supported a model of PABA-GLU utilization in which AbgT catalyzes the import of PABA-GLU, which is subsequently cleaved to PABA by a protein composed of subunits encoded by abgA and abgB.
The focus of this study was the purification and characterization of E. coli p-aminobenzoyl-glutamate hydrolase (PGH), encoded by abgA and abgB. This is the first description of an enzyme involved in a potential folate catabolic pathway in E. coli.
MATERIALS AND METHODS
Materials.
Ampicillin sodium salt, chloramphenicol, ammonium bicarbonate, folic acid, N-(p-aminobenzoyl)-glutamic acid, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), p-aminobenzoic acid, calcium chloride, manganese chloride (MnCl2), magnesium chloride, nickel sulfate, ferrous nitrate, lithium chloride, copper sulfate, zinc chloride, alanyl-glutamine, aspartyl-glutamate, glutamyl-glutamine, glycyl-glutamate, phenylalanyl-alanine, phenylalanyl-valine, prolyl-glycine, prolyl-leucine, serinyl-glutamate, threonyl-glutamate, tryptophanyl-glycine, tyrosinyl-alanine, tyrosinyl-leucine, and valinyl-glutamine were obtained from Sigma (St. Louis, MO). [3H]folic acid diammonium salt (3′,5′,7,9-3H) (1 mCi/ml; 64 Ci/mmol) was obtained from Moravek Biochemicals (Brea, CA). Radiolabeled PABA-GLU was synthesized from radiolabeled folate as previously described (4).
Strains and plasmids.
E. coli MG1655 and BN1103 were obtained from the laboratory of Brian Nichols, University of Illinois at Chicago, and JM109 was from New England Biolabs (Ipswich, MA). The plasmids used included pUC19, pECABT19, and pLenABHis; the cloning of the last two plasmids is described here.
Microbiological and molecular methods.
Bacteria were maintained in Luria broth (LB). Ampicillin was used at 100 μg ml−1 for maintenance of plasmids (1). E. coli chromosomal-DNA purification, calcium chloride-based transformations, and agarose gel electrophoresis were performed as described previously (1). Plasmids were purified using QIAprep kits (Qiagen, Valencia, CA). All reactions and procedures were performed in accordance with the manufacturer's recommendations.
PCR amplification and subsequent cloning of abgABT and abgAB.
Primers were designed for cloning using published nucleotide sequences for the abgABT region of E. coli (2). For cloning of the combined abgABT region into pUC19, we used the following primers: 5′-GATCAAGCTTATGGAGTCTTTGAATCAATTT-3′ (abgAforHindIII) and 5′-GATCGGATCCTTAAGACAAACGTGGGTAAATACC-3′ (abgTrevBamHI). These primers encompassed the abgABT genomic fragment from 1402589 to 1398271 (ASAP) (8). PCR amplification was performed using chromosomal DNA from wild-type MG1655 as a template and Taq DNA polymerase. The PCR conditions were as follows: 94°C for 4 min and 35 cycles of denaturing at 95°C for 30 s, annealing at 66°C for 1 min, and extension at 72°C for 6 min. The entire PCR mixture was subjected to agarose gel electrophoresis. The band containing the abgABT PCR product was excised from the gel, and the DNA was eluted using the QIAEXII gel extraction kit (Qiagen, Valencia, CA). The purified PCR product was digested with BamHI and HindIII and ligated to similarly restricted pUC19 using T4 DNA ligase. The ligation mixtures were transformed into JM109, and white colonies were selected for on LB plates containing ampicillin and X-Gal (1). Plasmid DNA was purified with Qiaquick miniprep kits (Qiagen, Valencia, CA). Candidate clones were confirmed by restriction mapping and sequence analysis (Molecular Cloning Laboratories, South San Francisco, CA). The resultant plasmid was named pECABT19.
For cloning of the combined abgAB genes, with incorporation of a hexahistidine tag on the carboxyl terminus of abgB, we used the following primers: abgAforHindIII and 5′-GATCGGATCCTTAGTGGTGGTGGTGGTGGTGTTTTAAAGGTGACGGTG-3′ (abgBBamHIhisrev). These primers encompassed the abgAB genomic fragment from 1402589 to 1399834 (ASAP) (8). PCR amplification was performed using plasmid DNA from pECABT19 as a template and AccuPrime Pfx DNA polymerase. The PCR conditions were as follows: 95°C for 2 min and 30 cycles of denaturing at 95°C for 15 s, annealing at 55°C for 30 s, and extension at 68°C for 2.0 min. The remainder of the cloning and selection was as described above for pECABT19, except that the plasmid was named pLenABHis.
Purification of PGH.
E. coli MG1655 transformed with pLenABHis was grown for 8 h in LB containing ampicillin and then inoculated 1:200 into LB-ampicillin (0.5 liter) and grown at 37°C with shaking for 16 h. The initial preparations of cells did not include MnCl2 in the growth medium; after it was discovered that the enzyme was activated by manganese chloride, it was included in the growth medium to a final concentration of 10 μM. The cells were pelleted by centrifugation, washed with saline, and frozen at −20°C. Thawed cells were resuspended in 10 ml of lysis buffer (50 mM sodium phosphate, pH 8.0, 300 mM sodium chloride, 10 mM imidazole). The cells were disrupted using glass beads (10 ml; 0.1 mm) in a BeadBeater. The resulting cell extract was centrifuged at 4°C for 15 min at 14,000 × g, and the supernatant (volume, typically 5.5 ml) was added to an equal volume of Ni-nitrilotriacetic acid (NTA) agarose slurry preequilibrated with lysis buffer (Qiagen, Valencia, CA). This mixture was stirred slowly at 4°C for 1 h. The Ni-NTA agarose was poured into a column and washed with 8 bed volumes of 50 mM sodium phosphate, pH 8.0, 300 mM sodium chloride, 20 mM imidazole. The enzyme was eluted with 50 mM sodium phosphate, pH 8.0, 300 mM sodium chloride, 250 mM imidazole. Fractions were collected, and the protein content was measured using the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA). The purified enzyme was brought to either 50% glycerol or 10% glycerol, and MnCl2 was added to a final concentration of 0.5 mM prior to storage at −20°C.
Denaturing PAGE.
SDS-PAGE was performed using 7.5% ready-made gels subjected to electrophoresis for 60 min at 100 V according to the manufacturer's instructions (Bio-Rad, Hercules, CA) and published protocols (1). The gels were stained with Gelcode Blue (Thermo Scientific, Rockford, IL). The retardation factor (Rf) was measured for the standards, and the relation to their masses was analyzed with GraphPad 5 using a second-order polynomial fit; the Rfs of the unknowns were used to determine their masses.
N-terminal sequence analysis.
Purified PGH was subjected to SDS-PAGE; the proteins were transferred to a polyvinylidene difluoride (PVDF) membrane and then stained with Coomassie blue according to established methods (13, 25). Bands were excised and outsourced for N-terminal sequence analysis (Protein Facility of Iowa State University, Ames, IA).
Radioactive assay for PGH.
The radioactive assay for PGH was an adaptation of that for the plant PGH (3). Reaction mixtures consisted of 50 mM Tris, pH 8.5, 10 mM β-mercaptoethanol, 5 mM MnCl2, and various amounts of tritiated PABA-GLU synthesized as described previously (9,400 dpm/nmol) (4); the final volume was 100 μl. The reaction was initiated by the addition of enzyme, and the reaction mixtures were incubated for varying periods of time and quenched by the addition of 100 mM sodium citrate, pH 5.5 (1.0 ml). The reaction mixtures were then extracted with ethyl acetate (2 ml) and vortexed briefly, and the phases were allowed to separate. Samples of the upper ethyl acetate layer (1 ml) were placed in 5 ml scintillation fluid, and the radioactivity was measured.
Spectrophotometric assay for PGH.
The biochemical basis for the spectrophotometric assay was similar to that of the radioactive assay, except that the absorbance of the product, PABA, was measured rather than the radioactivity. The reaction mixtures (5 ml) included the buffer described above and various amounts of PABA-GLU. The reaction was initiated by the addition of enzyme. Aliquots (0.5 ml) were taken over time, typically at 1, 2, 3, 4, and 5 min, and quenched with 0.5 ml 1.0 M sodium citrate, pH 5.5. The reaction mixtures were extracted into 2 ml of ethyl acetate as described above. Absorbance of the ethyl acetate layer, which contained the product p-aminobenzoic acid, was measured at 284 nm (ɛ = 13,400 M−1 cm−1, determined in this study). The velocity was calculated as the slope of the line (nanomoles of product versus time in minutes). The molar absorptivity coefficient for PABA in ethyl acetate was measured using stock mixtures of PABA in water, in which PABA has published absorbance properties (ɛ266 = 15,000 M−1 cm−1 [6]). We then determined the molar absorptivity coefficient for PABA in acidified ethyl acetate to be 13,400 M−1 cm−1 at the peak of 284 nM.
Determination of the amino acid yield from cleavage of PABA-GLU, dipeptides, and folic acid by HPLC.
Enzyme reaction mixtures contained 50 mM phosphate, pH 8.5, 2 mM MnCl2, 1 mM substrate, and purified PGH (1 to 2 μg) in a final volume of 600 μl. The reaction mixtures were incubated at 37°C for various times, and the reaction was terminated by placement in a boiling water bath for 1 min. Protein was removed from the reaction mixtures with a Micron Ym-10 filter device (Millipore, Billerica, MA). The reaction mixtures were subjected to modification by dabsylation and then analyzed by high-pressure liquid chromatography (HPLC). Dabsylation was performed as an adaptation of the method described by Knecht and Chang (12). Briefly, the reaction mixtures (400 μl) were mixed with 200 μl of 0.2 M sodium bicarbonate, pH 9.0, and 20 μl of dimethyl-aminoazobenzenesulfonyl chloride (DABS-CL) (0.5 mg/ml in acetone). Samples were heated for 30 min at 70°C, dried under nitrogen, and resuspended in 200 μl of 70% ethanol. Samples (20 μl) were analyzed by reverse-phase HPLC using a 5 μm Supelcosil LC-18 column (25 cm by 4.6 mm; Supelco, Bellefonte, PA) and a Pelliguard LC-18 guard column (2 cm; Supelco, Bellefonte, PA). The elution was performed as described previously (10). Amino acids were identified and quantified by comparison to known samples using standard curves.
Determination of molecular mass by SEC-LS analysis.
The experiments to determine molecular mass were performed at the Keck Biotechnology Resource Laboratory at Yale University. The light-scattering (LS) data were collected using a Superdex 200 HR 10/300 size exclusion chromatography (SEC) column (GE Healthcare, Piscataway, NJ) connected to an Agilent 1200 (Agilent Technologies, Wilmington, DE) HPLC system equipped with an autosampler. The elution from SEC was monitored by a photodiode array (PDA) UV/visible-light (VIS) detector (Agilent Technologies, Wilmington, DE); a differential refractometer (Opti-Lab rEx; Wyatt Corp., Santa Barbara, CA); and a static and dynamic, multiangle laser LS detector (Heleos II with QELS capability; Wyatt Corp., Santa Barbara, CA). The SEC-UV/LS/refractive index (RI) system was equilibrated in 20 mM HEPES, pH 7.3, 150 mM NaCl, 1 mM MnCl2, 10% glycerol at a flow rate of 0.5 ml/min. Standard gel filtration molecular weight markers were used, and all proteins were analyzed under the same experimental conditions. Two software packages were used for data collection and analysis: Chemstation software (Agilent Technologies, Wilmington, DE) controlled the HPLC operation and data collection from the multiwavelength UV/VIS detector, while ASTRA software (Wyatt Corp., Santa Barbara, CA) collected data from the refractive index detector and the light-scattering detectors and recorded the UV trace at 280 nm sent from the PDA detector. The weight-averaged molecular weights (Mw) were determined across the entire elution profile in the intervals of 1 s from static LS measurement using ASTRA software as previously described (7). Fractions (20 s; ∼170 μl) were collected, frozen, and sent to Midwestern University for PGH spectrophotometric enzyme assays and SDS-PAGE analysis.
Amino acid analysis.
A sample of metal affinity chromatography (MAC)-purified PGH (0.38 mg/ml, dialyzed into 25 mM phosphate buffer, pH 7.0) was subjected to amino acid hydrolysis/analysis by the Molecular Biology Core Facilities of the Dana-Farber Cancer Institute (Boston, MA).
Data analysis.
Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA).
RESULTS
Cloning and purification of histidine-tagged PGH.
Based on previous growth experiments on strains transformed with various plasmids carrying abgA, abgB, and abgAB (4), we hypothesized that abgA and abgB encoded subunits of a single protein. Initial efforts to measure the cleavage of PABA-GLU using mixed crude extracts of strains transformed with plasmids carrying abgA and abgB separately were unsuccessful (data not shown). Using PCR, we cloned the entire abgABT region into pUC19; the resulting plasmid was named pECABT19. We then transformed wild-type MG1655 with pECABT19 and obtained ∼2 g of cells from overnight growth in 500 ml of LB-ampicillin. The cells were disrupted and centrifuged, and the supernatant was used as the source of enzyme in the radioactive assay for PGH. Because abgA and abgB have homology to carboxypeptidase G, which requires zinc (20, 22), and because the plant PGH requires manganese (3), we tested the enzyme activity of the supernatant with a variety of divalent cations and observed that manganese chloride stimulated activity ∼3-fold over reaction mixtures containing Ca2+, Cu2+, Fe2+, Li2+, Mg2+, Ni2+, or Zn2+ or with no added cation (data not shown). We concluded that our pECABT19 clone encoded a functional PGH and that the enzyme required a divalent cation, most likely manganese, for optimal activity.
Our next goal was to generate a histidine-tagged protein to facilitate purification using metal affinity chromatography. Using DNA from pECABT19 as a template, we used PCR to amplify the abgAB fragment and to incorporate a hexahistidine tag on the carboxyl terminus of AbgB, as described above. We performed spectrophotometric PGH assays of supernatants from cells (MG1655) transformed with the resulting plasmid, pLenABHis, and confirmed that activity was present in the histidine-tagged enzyme supernatant. Kinetic experiments performed in triplicate with the supernatants revealed that the Km values for the supernatant containing non-histidine-tagged PGH and the histidine-tagged enzyme were 71.0 ± 14.0 μM (standard deviation [SD]) and 90.8 + 13.0 μM (SD), respectively (data not shown). We concluded that the hexahistidine tag did not adversely affect PGH function and proceeded with purification of PGH from E. coli MG1655 transformed with pLenABHis using Ni-NTA agarose.
Manganese requirement of PGH.
Using the Ni-NTA-purified PGH, we measured changes in activity with the addition of a variety of divalent cations (Table 1). MnCl2 was essential for activity and was included in all further experiments and as a supplement to the growth medium of our cells (10 μM). Although manganese has been shown to inhibit the growth of E. coli, the concentration at which this occurs is much higher (0.1 to 1 mM) than that used in our growth medium (23).
TABLE 1.
Effects of divalent cations on NTA agarose-purified PGH activity
| Contents of assay mixturea | No. of dpm ± SE | Relative % |
|---|---|---|
| Enzyme, no added metal | 105 ± 3.5 | 1.5 |
| No enzyme, no added metal | 91.0 ± 4.7 | 1.3 |
| Enzyme, Mn2+ | 6,960 ± 244 | 100 |
| No enzyme, Mn2+ | 92.0 ± 2.3 | 1.3 |
| Enzyme, Ca2+ | 140 ± 4.4 | 2.0 |
| No enzyme, Ca2+ | 100 ± 5.9 | 1.4 |
| Enzyme, Cu2+ | 98 ± 4.5 | 1.4 |
| No Enzyme, Cu2+ | 103 ± 3.0 | 1.5 |
| Enzyme, Fe2+ | 111 ± 2.5 | 1.6 |
| No enzyme, Fe2+ | 100 ± 5.0 | 1.4 |
| Enzyme, Li2+ | 96.0 ± 6.9 | 1.4 |
| No enzyme, Li2+ | 102 ± 8.7 | 1.5 |
| Enzyme, Mg2+ | 465 ± 88 | 6.7 |
| No enzyme, Mg2+ | 91.0 ± 3.1 | 1.3 |
| Enzyme, Ni2+ | 213 ± 35 | 3.1 |
| No enzyme, Ni2+ | 103 ± 8.9 | 1.5 |
| Enzyme, Zn2+ | 100 ± 7.1 | 1.4 |
| No enzyme, Zn2+ | 104 ± 5.8 | 1.5 |
Reaction mixtures (100 μl) consisted of 50 mM Tris, pH 8.5, 10 mM β-mercaptoethanol, 100 μM PABA-GLU, 1 mM divalent cation [MnCl2, CaCl2, Cu(SO4)2, Fe(NO3)2, LiCl2, MgCl2, Ni(SO4)2, or ZnCl2], and NTA agarose-purified PGH (150 ng) or buffer; triplicate tubes were incubated at 37°C for 15 min and then analyzed as described in Materials and Methods.
Identification of reaction products and kinetic constants using PABA-GLU.
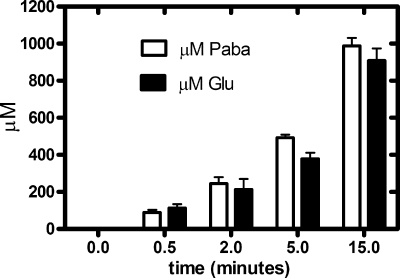
We developed the spectrophotometric assay based on the absorbance properties of PABA. This assay offered several advantages over the radioactive assay, in that it was less expensive and safer, and higher substrate concentrations could be achieved. To identify the products of the cleavage of PABA-GLU, we performed parallel analyses for PABA and glutamate detection from a single reaction mixture using spectrophotometric and HPLC techniques, respectively (Fig. 2). The reaction went to completion, and PABA and glutamate were generated in equal amounts.
FIG. 2.
Analysis of the formation of products from the reaction of PGH with PABA-GLU. Reactions containing 50 mM phosphate buffer, pH 8.5, 2 mM MnCl2, and 1 mM PABA-GLU were initiated by the addition of purified PGH. At various time points, samples (500 μl) were quenched and analyzed for PABA as described in the text. A parallel sample (600 μl) was inactivated by placement in a boiling water bath for 1 min; enzyme was removed using a Microcon YM-10 filter device with a 10,000-molecular-weight cutoff (Millipore, Billerica, MA), and 400 μl of this sample was subjected to dabsylation and analysis by HPLC as described in the text. Samples were done in triplicate, repeated three times, and analyzed using GraphPad Prism 5. The data shown are averages plus standard errors (SE).
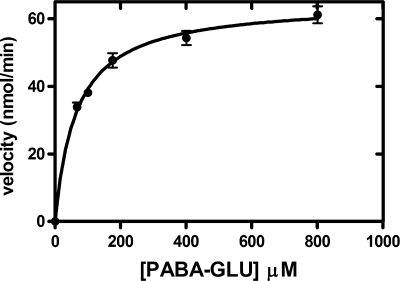
Using the purified PGH in the spectrophotometric assay, kinetic experiments yielded a Km value for PABA-GLU of 60.0 ± 0.08 μM (SD) and a specific activity of 63,300 ± 600 nmol min−1 mg−1 (SD) (Fig. 3).
FIG. 3.
Kinetics of PGH activity from E. coli. Shown is the effect of the PABA-GLU concentration on the initial velocity of PGH activity of purified enzyme isolated from E. coli MG1655 transformed with pLenABHis. A spectrophotometric assay was utilized, as described in Materials and Methods. The reaction mixtures (1.6 ml) consisted of buffer and various concentrations of PABA-GLU (67, 100, 175, 400, and 800 μM). The reaction was initiated by the addition of Ni-NTA agarose-purified enzyme (3.1 μg), and the mixture was incubated at 37°C. The concentration of PABA-GLU is plotted versus the initial velocity. The data were analyzed using GraphPad Prism 5. The error bars indicate standard deviations.
Assessing dipeptides and folate as potential substrates of PGH.
Because the bond between PABA and glutamate in PABA-GLU resembles a peptide bond, it was plausible that the proteins encoded by the abg region were involved in salvage of peptides (15). Using an HPLC system capable of simultaneous separation and detection of multiple amino acids, we measured the ability of PGH to cleave a variety of dipeptides chosen because of their resemblance to PABA-GLU; except for serinyl-glutamate and glycyl-glutamate, which served as poor substrates (exhibiting about 10% cleavage), all amino acids tested failed to act as substrates of PGH (Table 2). Finally, we tested the ability of PGH to cleave folic acid and found that only 2.5% folate was cleaved under conditions in which 100% of PABA-GLU was cleaved. This activity can be explained by slight (<5%) contamination of the folic acid with PABA-GLU; we observed a small peak in the HPLC chromatographs of the folate samples that had the same elution time as commercial PABA-GLU. Given these data, it seems unlikely that the physiological role of PGH is to cleave peptides or folic acid.
TABLE 2.
Activities of PGH with selected substrates
| Substrate | Activity (μM)a |
||
|---|---|---|---|
| NH2-terminal amino acid | COOH-terminal amino acid | GLU | |
| Dipeptide | |||
| Ala-Gln | ND | ND | |
| Asp-Glu | ND | ND | |
| Glu-Gln | ND | ND | |
| Gly-Glu | 36 ± 7.5 | 42 ± 3.7 | |
| Phe-Ala | ND | ND | |
| Phe-Val | ND | ND | |
| Pro-Gly | ND | ND | |
| Pro-Leu | ND | ND | |
| Ser-Glu | 114 ± 8 | 117 ± 18 | |
| Thr-Glu | ND | ND | |
| Trp-Gly | ND | ND | |
| Tyr-Ala | ND | ND | |
| Tyr-Leu | ND | ND | |
| Val-Gln | ND | ND | |
| Folate derivatives | |||
| PABA-GLU | 1,072 ± 15.8 | ||
| Folate | 25 ± 11.3 | ||
Substrate (1 mM) was incubated with purified PGH (3.2 μg), heat inactivated after incubation for 15 min at 37°C, and then analyzed as described in Materials and Methods. ND, not detectable. Average values (±SE) are given.
Analysis of PGH by SDS-PAGE, N-terminal sequence analysis, and amino acid analysis.
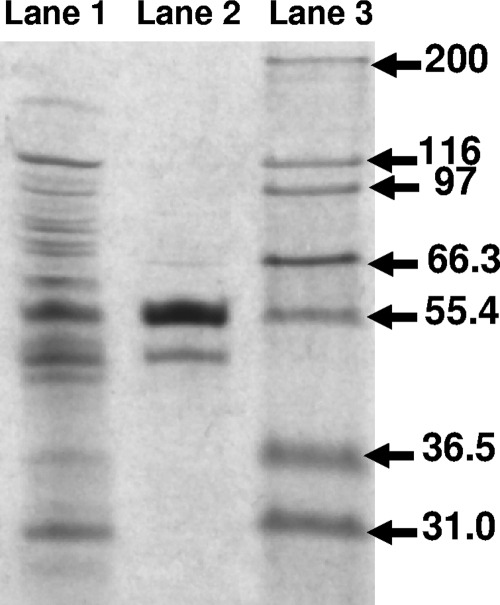
To measure the purity of PGH, as well as to estimate the subunit stoichiometry, we performed SDS-PAGE (Fig. 4). The two bands were correlated with the predicted sizes of AbgA and AbgB, 47 kDa and 53 kDa, respectively. N-terminal sequencing revealed that the 53-kDa and 47-kDa bands had sequences of M-Q-E-I-Y-R-F-I-D-D- and M-E-(S + S′)-L-N-Q-F-V-N-(S + S′), thereby confirming the identities of subunits AbgB and AbgA, respectively. While the bands appeared to be in a 2:1 ratio (AbgB/AbgA), protein binding to dye is not sufficiently quantitative to be used as a measure of relative abundance. In order to better determine the subunit ratio, a sample of MAC-purified PGH was subjected to amino acid analysis; the known amino acid compositions of the two proteins enabled calculation of the ratio of subunits within the sample, and the data were consistent with a 1:1 ratio of AbgB to AbgA (data not shown).
FIG. 4.
SDS-PAGE of cell supernatant and Ni-NTA agarose-purified PGH. Histidine-tagged PGH was purified with Ni-NTA agarose, and samples of the cell supernatant and purified enzyme were subjected to electrophoresis on a 7.5% ready-made gel and stained as described in Materials and Methods. Lane 1, supernatant (5 μg); lane 2, purified PGH (1 μg); lane 3, Mark12 Unstained Protein Standards (Invitrogen, Carlsbad, CA). Molecular masses in kilodaltons are indicated on the right.
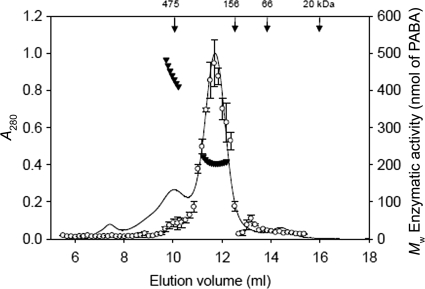
Assessment of the molecular mass of PGH by gel filtration/light scattering.
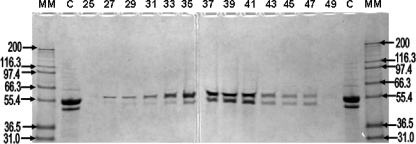
SEC-LS analysis was performed to identify the molecular mass of the active holoenzyme (Fig. 5). When PGH was subjected to SEC-LS, its activity was primarily associated with a molecular mass of 206 kDa, with a secondary peak associated with a mass between 400 and 500 kDa. SDS-PAGE of the fractions revealed that both AbgA and AbgB were present in all fractions containing PGH activity (Fig. 6). When SEC-LS was performed with dilutions of PGH, the distribution between the different oligomeric forms did not change with the concentration (data not shown); this indicated that the 400- to 500-kDa and 206-kDa forms are not in dynamic equilibrium.
FIG. 5.
Determination of the oligomeric state of PGH from SEC-UV/LS/RI analyses and its correlation with enzymatic activity. Ni-NTA agarose-purified PGH (0.8 mg) was analyzed on a Superdex 200 HR 10/300 SEC column as described in Materials and Methods. Aliquots from fractions (5 μl) were incubated for 15 min using 200 mM PABA-GLU in the spectrophotometric assay, as described in Materials and Methods. The vertical arrows indicate the elution positions of molecular mass markers. Weight-averaged molecular weights (MW) ▾ and PGH activity ○ are plotted; the error bars represent standard deviations. The line corresponds to the absorbance (280 nM) trace of the protein eluting from the SEC column. Molecular weights were recorded every 5 μl across the elution profile (for clarity, only every 10th measurement is plotted). The average for the major eluting peak, from three independent SEC-UV/LS/RI analyses, was 206,000 ± 2,000 g/mol.
FIG. 6.
SDS-PAGE of SEC-LS fractionation of PGH. Samples (20 μl) of odd-numbered SEC fractions (25 to 49) spanning the peaks containing PGH activity in Fig. 5 (elution volumes, 9.5 to 13.7) were applied to a 7.5% polyacrylamide gel and stained as described in Materials and Methods. Molecular masses (MM) in kilodaltons are indicated.
In order to compare the kinetic characteristics of the PGH corresponding to the major peak (206 kDa) and the minor peak (400 to 500 kDa), we pooled the fractions comprising these peaks and performed protein and kinetic assays in duplicate. We found that the pooled PGH from the 400- to 500-kDa peak fractions was characterized by a Km value of 157 ± 2 (SD) μM with a specific activity of 42,000 ± 7,000 (SD) nmol min−1 mg−1; in comparison, the pooled PGH from the 200-kDa peak fractions had a Km value of 40 ± 1.4 μM with a specific activity of 72,000 ± 14,000 nmol min−1 mg−1.
DISCUSSION
In this report, we describe the first purification of an enzyme in E. coli involved in catabolism of a folate breakdown product. The activity seems to be specific for PABA-GLU, as folic acid and a group of dipeptides were poor substrates. The major end products of folate catabolism in humans are PABA-GLU and its derivatives; these catabolic end products are present in both urine and fecal matter and presumably are available to our resident bacteria, including E. coli (24).
It is interesting to compare E. coli PGH to a similar plant enzyme that has been partially characterized. Plants have high rates of folate catabolism, degrading approximately 10% of their total folates per day, and there is evidence that plants may salvage these breakdown products (3, 16-19). Bozzo et al. identified a PGH activity in pea (Pisum sativum L.) leaves and in Arabidopsis roots that has some similarities to the E. coli enzyme: it hydrolytically cleaves PABA-GLU and is stimulated by manganese (3). Size exclusion chromatography for the plant enzyme was consistent with a size of ∼ 200,000 kDa, which is similar to the predominant species observed in SEC-LS experiments with the E. coli enzyme; the subunit structure was not determined. While they were not able to purify the protein to homogeneity or clone the gene, they measured a Km value for PABA-GLU of 370 μM, somewhat higher than the Km value measured in this study (∼60 μM).
We have now characterized in part the functions of three of the proteins encoded by the abg region in E. coli: AbgA, AbgB, and AbgT. AbgT imports PABA-GLU with a KT of 123 μM (4). AbgA and AbgB comprise subunits of a manganese-dependent enzyme that hydrolyzes PABA-GLU. E. coli PGH, when assayed in cell supernatants, showed stimulation (to ∼300%) when MnCl2 was included in the assay mixture (data not shown). This suggests that some enzyme lacks the manganese cofactor. After purification by metal affinity chromatography, the enzyme displayed no activity unless the reaction mixture was supplemented with manganese chloride (Table 1); loss of manganese is common among enzymes that utilize divalent manganese owing to the fact that it forms relatively weak bonds to ligands (5). While purified E. coli PGH was activated only by addition of manganese chloride, the cell supernatant showed some minor stimulation by other divalent cations, including zinc and calcium. It is possible that other enzymes present in the extract have some ability to cleave PABA-GLU and may utilize different cations. This is consistent with prior studies, which showed that strains in which abgA or abgB were interrupted still maintained some PABA-GLU cleavage ability, as measured in crude extracts (11).
Our data indicate that PGH is a multisubunit enzyme composed of AbgB and AbgA in a 1:1 ratio. SEC-LS revealed that most activity was associated with a molecular mass of ∼206 kDa, with a secondary peak corresponding to 400 to 500 kDa. Given that the known amino acid sequence and SDS-PAGE experiments both yielded consistent molecular masses of 53.1 kDa and 47.1 kDa for AbgB (hexahistidine tagged) and AbgA, respectively, one can calculate the theoretical molecular masses for various tetramers as follows: AbgA4, 188.5 kDa; AbgA2-AbgB2, 200.5 kDa; AbgB4, 212.5 kDa. Our current hypothesis is that the predominant tetramer may be composed of AbgB2-AbgA2, since AbgA copurifies with histidine-tagged AbgB on a Ni-NTA agarose column. One cannot, however, exclude the possibility of a higher-order structure composed of mixed tetramers, such as AbgA4 in association with AbgB4, that may dissociate to form homotetramers. This prediction would be consistent with the higher-molecular-mass species (400 to 500 kDa) observed in the SEC-LS experiments, as well as the averaged molecular mass of 206 kDa that constituted the major peak in the same experiments.
Kinetic experiments with pooled fractions corresponding to the 400- to 500-kDa peak and the 206-kDa peak revealed that these samples possessed different kinetic properties. Compared to the 206-kDa peak, the material corresponding to the 400- to 500-kDa peak possessed a higher Km value (∼160 μM versus 40 μM) and a lower specific activity (only ∼60% of the value for the tetramer). It is possible that the various species represent a regulatory mechanism.
Here, we have described the purification and characterization of PGH from E. coli. This manganese-dependent enzyme seems to be specific for cleavage of PABA-GLU, a product of folate catabolism. We are continuing our studies of the abg region to better understand the roles of these genes and proteins in E. coli physiology.
Acknowledgments
We gratefully acknowledge the expert technical assistance of Al Campione, who assisted with the HPLC experiments at Midwestern University, and Ewa Folta-Stogniew of the William M. Keck Foundation Biotechnology Resource Laboratory at Yale University, who collected and analyzed the SEC-LS data.
This work was supported by funds from Midwestern University and by award number R15 GM085760 from the National Institute of General Medical Sciences; the SEC-LS/UV/RI instrumentation was supported by NIH award number 1S10RR023748-01.
The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print on 26 February 2010.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1999. Short protocols in molecular biology, 4th ed. John Wiley & Sons, Inc., New York, NY.
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Bozzo, G. G., G. J. Basset, V. Naponelli, A. Noiriel, J. F. Gregory III, and A. D. Hanson. 2008. Characterization of the folate salvage enzyme p-aminobenzoylglutamate hydrolase in plants. Phytochemistry 69:29-37. [DOI] [PubMed] [Google Scholar]
- 4.Carter, E. L., L. Jager, L. Gardner, C. C. Hall, S. Willis, and J. M. Green. 2007. Escherichia coli abg genes enable uptake and cleavage of the folate catabolite p-aminobenzoyl-glutamate. J. Bacteriol. 189:3329-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crowley, J. D., D. A. Traynor, and D. C. Weatherburn. 2000. Enzymes and proteins containing manganese: an overview, p. 209-428. In A. Sigel and H. Sigel (ed.), Metal ions in biological systems: manganese and its role in biological processes, vol. 37. Marcel Dekker, Inc., New York, NY. [PubMed] [Google Scholar]
- 6.Dawson, R. M. C., D. P. Elliott, W. H. Elliott, and K. M. Jones. 1986. Data for biochemical research, 3rd ed. Oxford University Press, New York, NY.
- 7.Folta-Stogniew, E., and K. Williams. 1999. Determination of molecular masses of proteins in solution: implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J. Biomol. Tech. 10:51-63. [PMC free article] [PubMed] [Google Scholar]
- 8.Glasner, J. D., P. Liss, G. Plunkett III, A. Darling, T. Prasad, M. Rusch, A. Byrnes, M. Gilson, B. Biehl, F. R. Blattner, and N. T. Perna. 2003. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 31:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green, J. M., and R. G. Matthews. March 2007, posting date. Chapter 3.6.3.6, Folate biosynthesis, reduction, polyglutamylation and the interconversion of folate derivatives. In A. Böck, R. Curtiss III, J. B. Kaper, F. C. Neidhardt, T. Nyström, K. E. Rudd, and C. L. Squires (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. doi: 10.1128/ecosal.3.6.3.6. [DOI]
- 10.Hill, T. 1989. Techniques to enhance DABS column performance: precolumns and automated washing, ATR 89-008B. Beckman Instruments, Fullerton, CA.
- 11.Hussein, M. J., J. M. Green, and B. P. Nichols. 1998. Characterization of mutations that allow p-aminobenzoyl-glutamate utilization by Escherichia coli. J. Bacteriol. 180:6260-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knecht, R., and J. Y. Chang. 1986. Liquid chromatographic determination of amino acids after gas-phase hydrolysis and derivatization with (dimethylamino)azobenzenesulfonyl chloride. Anal. Chem. 58:2375-2379. [DOI] [PubMed] [Google Scholar]
- 13.Matsudaira, P. T. 1989. A practical guide to protein and peptide purification for microsequencing. Academic Press, Inc., San Diego, CA.
- 14.Matthews, R. G. 1996. One-carbon metabolism, p. 600-611. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. I. ASM Press, Washington, DC. [Google Scholar]
- 15.Miller, C. G. 1996. Protein degradation and proteolytic modification, p. 938-954. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. I. ASM Press, Washington, DC. [Google Scholar]
- 16.Noiriel, A., V. Naponelli, G. G. Bozzo, J. F. Gregory III, and A. D. Hanson. 2007. Folate salvage in plants: pterin aldehyde reduction is mediated by multiple non-specific aldehyde reductases. Plant J. 51:378-389. [DOI] [PubMed] [Google Scholar]
- 17.Noiriel, A., V. Naponelli, J. F. Gregory III, and A. D. Hanson. 2007. Pterin and folate salvage. Plants and Escherichia coli lack capacity to reduce oxidized pterins. Plant Physiol. 143:1101-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orsomando, G., G. G. Bozzo, R. D. de la Garza, G. J. Basset, E. P. Quinlivan, V. Naponelli, F. Rebeille, S. Ravanel, J. F. Gregory III, and A. D. Hanson. 2006. Evidence for folate-salvage reactions in plants. Plant J. 46:426-435. [DOI] [PubMed] [Google Scholar]
- 19.Orsomando, G., R. D. de la Garza, B. J. Green, M. Peng, P. A. Rea, T. J. Ryan, J. F. Gregory III, and A. D. Hanson. 2005. Plant gamma-glutamyl hydrolases and folate polyglutamates: characterization, compartmentation, and co-occurrence in vacuoles. J. Biol. Chem. 280:28877-84. [DOI] [PubMed] [Google Scholar]
- 20.Rowsell, S., R. A. Pauptit, A. D. Tucker, R. G. Melton, D. M. Blow, and P. Brick. 1997. Crystal structure of carboxypeptidase G2, a bacterial enzyme with applications in cancer therapy. Structure 5:337-347. [DOI] [PubMed] [Google Scholar]
- 21.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 22.Sherwood, R. F., R. G. Melton, S. M. Alwan, and P. Hughes. 1985. Purification and properties of carboxypeptidase G2 from Pseudomonas sp. strain RS-16. Use of a novel triazine dye affinity method. Eur. J. Biochem. 148:447-453. [DOI] [PubMed] [Google Scholar]
- 23.Silver, S., P. Johnseine, E. Whitney, and D. Clark. 1972. Manganese-resistant mutants of Escherichia coli: physiological and genetic studies. J. Bacteriol. 110:186-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suh, J. R., A. K. Herbig, and P. J. Stover. 2001. New perspectives on folate catabolism. Annu. Rev. Nutr. 21:255-282. [DOI] [PubMed] [Google Scholar]
- 25.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran, P. V., T. A. Bannor, S. Z. Doktor, and B. P. Nichols. 1990. Chromosomal organization and expression of Escherichia coli pabA. J. Bacteriol. 172:397-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran, P. V., and B. P. Nichols. 1991. Expression of Escherichia coli pabA. J. Bacteriol. 173:3680-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]