Abstract
This study investigated features of the acid tolerance response (ATR) in Lactobacillus casei ATCC 334. To optimize ATR induction, cells were acid adapted for 10 or 20 min at different pH values (range, 3.0 to 5.0) and then acid challenged at pH 2.0. Adaptation over a broad range of pHs improved acid tolerance, but the highest survival was noted in cells acid adapted for 10 or 20 min at pH 4.5. Analysis of cytoplasmic membrane fatty acids (CMFAs) in acid-adapted cells showed that they had significantly (P < 0.05) higher total percentages of saturated and cyclopropane fatty acids than did control cells. Specifically, large increases in the percentages of C14:0, C16:1n(9), C16:0, and C19:0(11c) were noted in the CMFAs of acid-adapted and acid-adapted, acid-challenged cells, while C18:1n(9) and C18:1n(11) showed the greatest decrease. Comparison of the transcriptome from control cells (grown at pH 6.0) against that from cells acid adapted for 20 min at pH 4.5 indicated that acid adaption invoked a stringent-type response that was accompanied by other functions which likely helped these cells resist acid damage, including malolactic fermentation and intracellular accumulation of His. Validation of microarray data was provided by experiments that showed that L. casei survival at pH 2.5 was improved at least 100-fold by chemical induction of the stringent response or by the addition of 30 mM malate or 30 mM histidine to the acid challenge medium. To our knowledge, this is the first report that intracellular histidine accumulation may be involved in bacterial acid resistance.
Lactobacillus casei is an aciduric, rod-shaped, facultatively heterofermentative lactic acid bacterium (LAB) that can be isolated from a variety of environments including raw and fermented milk and meat or plant products, as well as the oral, intestinal, and reproductive tracts of humans and animals (24). Like other LAB species, L. casei produces lactic acid as a major end product of carbohydrate fermentation during growth, and strains of L. casei are used as acid-producing starter cultures in the preparation of fermented foods, as health-enhancing probiotic cultures, and for the production of l(+)-lactic acid (18, 47, 52). In each of these applications, the industrial performance of L. casei strains is dependent, in one way or another, on their acidurance.
During fermentation, L. casei transports lactic acid outside the cell as lactate ion via an electrogenic proton-lactate symporter. As the pH of the medium (pHo) decreases or the concentration of lactate increases, the concentration of protonated (undissociated) lactic acid in the medium also increases. The undissociated form of lactic acid is membrane soluble and thus can enter the cytoplasm by simple diffusion (27). Metabolically active bacteria maintain a pH gradient (ΔpH) where the intracellular pH (pHi) is more alkaline than the pHo (4, 25), so diffusion of acid into the cytoplasm results in rapid dissociation and release of protons and anions inside the cell. If the rate of proton accumulation exceeds the cytoplasmic buffering capacity and capabilities of efflux systems, the pHi begins to fall and eventually reaches a critical point where the ΔpH can no longer be maintained and cellular functions are impaired (4, 22). Furthermore, intracellular accumulation of acid anions may be of greater importance than proton release in the inhibition of cell growth (8, 40, 41). Thus, low pH and organic acid resistance are vitally important physiological attributes of L. casei and other LAB species, in relation to both lifestyle and commercial utility, and there is considerable interest in the mechanisms used by these cells to combat acid-related stress conditions (4, 11, 22, 51).
Interestingly, differences in acidurance among LAB species have been linked to the activity and pH optima of the proton-translocating (H+)-ATPase, and this enzyme complex plays a major role in pHi regulation by these cells (11, 22, 48). However, many LAB species do not utilize the H+-ATPase to maintain a near-neutral pHi in response to acid stress. Instead, these species (including L. casei) maintain a relatively constant ΔpH as the pHo falls by allowing the pHi to decrease (25, 22, 40, 43, 45). This capability is thought to reduce the energy demand for proton translocation through the H+-ATPase (45) and has been shown to help prevent intracellular accumulation of organic acid anions (40).
In addition to H+-ATPase activity, acidurance in a LAB is also known to involve a variety of stress protection mechanisms (11, 30, 51). In particular, many cells display an inducible acid tolerance response (ATR) that protects them from acid killing. First reported by Goodson and Rowbury (19), the ATR is usually triggered by brief exposure to a sublethal pHo (“acid adaptation”), which, like other stress responses, results in transient induction of specific proteins and physiological changes that enhance cell survival during subsequent exposure to a lethal pHo. The ATR has been detected in numerous bacteria, including L. casei and other species of LAB (20, 21, 30, 31, 50, 51).
Characterization of the ATR has facilitated the identification of enzymes, proteins, and macromolecular changes (e.g., alteration of cytoplasmic membrane lipid content) that allow bacteria to combat the negative consequences of cytoplasmic acidification (11, 30, 51). Work to dissect the L. casei ATR has detected 9 to 11 acid-regulated proteins (21) and shown that cytoplasmic membrane fatty acid (CMFA) content is altered to increase the proportion of C18:1 and cyclopropane fatty acid (FA) C19:0 in response to acidification (15). However, information on the mechanisms used by L. casei to modulate ATR and CMFA content, as well as the identities and functions of acid-regulated proteins, is still lacking.
In this study, we optimized conditions for ATR induction in L. casei ATCC 334 and then analyzed its effects on membrane lipid composition and global gene expression. As expected, the membrane lipid composition of acid-adapted cells showed a dramatic increase in the ratio of saturated to unsaturated membrane FAs and cyclopropane FA content. Comparisons between the transcriptome of cells grown at pH 6.0 (control) to that of acid-adapted (5 or 20 min at pH 4.5) or acid-adapted and then acid-challenged (20 min at pH 4.5 and then 10 min at pH 2.0) cells showed differential expression of numerous genes in acid-treated versus control cells. Overall, functional predictions for these genes indicated that acid adaptation induced a stringent-type response that was accompanied by other functions, particularly malolactic fermentation and intracellular accumulation of histidine, which were important for enhanced acidurance. Validation of microarray data was provided by follow-up experiments that showed that L. casei survival at pH 2.5 was improved at least 100-fold by chemical induction of the stringent response with α-methylglucoside or by the addition of 30 mM malate or 30 mM His to the acid challenge medium.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
L. casei ATCC 334 (American Type Culture Collection, Manassas, VA) was maintained in a laboratory collection as a glycerol stock at −80°C and propagated at 37°C in MRS broth (Difco Laboratories, Detroit, MI). Working cultures were prepared from stock cultures through two successive transfers (1% inocula) in MRS broth at 37°C for 17 h.
Characterization of the acid tolerance response.
Batch cultures were prepared by 1% (vol/vol) inoculation into 1 or 2 liters of MRS in a Bioflow III fermentor (New Brunswick Scientific, Edison, NJ) or a VirTis (Gardiner, NY) Omni culture fermenter, respectively, which was equipped with pH control. The cells were incubated at 37°C with an agitation rate of 75 to 100 rpm, and a constant pH of 6.0 was maintained by automatic addition of 15% (vol/vol) NH4OH. When cells reached an A600 of 5.0 to 6.0, 200-ml samples of the suspension were aseptically harvested by centrifugation (9,000 × g, 10 min, 25°C), washed twice with 0.85% (wt/vol) NaCl (pH 7.0), and then suspended in 200 ml of fresh MRS adjusted to pH 6.0 (control), 5.0, 4.8, 4.5, 4.3, 4.0, 3.8, 3.5, or 3.0 using 10 N HCl. Cells were then acid adapted by incubation at the test pH for 10 or 20 min at 37°C and then harvested and washed as noted and suspended in 100 ml MRS adjusted to pH 2.0 with 10 N HCl (acid challenge). Acid-adapted cells were incubated under acid challenge conditions for up to 140 min with periodic sampling to evaluate viability. The bacterial suspensions were serially diluted in 0.1% Bacto peptone, plated on MRS agar, and enumerated after 48 to 72 h of anaerobic incubation at 37°C.
In some experiments, L. casei was grown in batch culture with pH control as described above and then acid adapted (20 min at pH 4.5) in the presence or absence of 30 mM sodium malate or 30 mM His. These cells were then acid challenged for up to 120 min at pH 2.0 or 2.5 in MRS that also did or did not contain an equivalent concentration of malate or His, respectively. Finally, the contribution of stringent-response induction to acid resistance was investigated by adding 1% (wt/vol) α-methylglucoside (36) to cells grown in batch culture with pH control 5 min before acid challenge at pH 2.5 in MRS that also did or did not contain 1% α-methylglucoside.
Data presented for the effects of different acid adaptation treatments on the viability of L. casei ATCC 334 during acid challenge represent mean values from two independent experiments.
Cell membrane lipid composition.
L. casei ATCC 334 was grown in batch culture with pH control and then acid adapted and acid challenged as described above. FA methyl esters (FAMEs) of CMFAs were prepared by the method of Ingham et al. (23). All solvents were procured from Sigma-Aldrich (St. Louis, MO) and were high-performance liquid chromatography grade, and gases for the gas chromatograph (GC) were over 99.9% pure (AGA Gas, Madison, WI) and the glassware used in FA determinations was washed with 10% Micro cleaner (Cole Palmer Instrument Co., Vernon Hills, IL) and rinsed repeatedly with distilled water before use.
An internal standard of 0.1 mg/ml undecanoic acid and methyl eicosanoate (Sigma-Aldrich) was diluted in diethyl ether and stored at 4°C in a bottle with a gas-tight Mininert valve (Supelco, Bellefonte, PA). One milliliter of each standard was injected into each sample with a gas-tight syringe (Hamilton Co., Reno, NV) prior to methyl transesterification. A Hewlett-Packard (Avondale, PA) model 5890 GC equipped with a flame ionizing detector and a 30-m by 0.25-mm by 0.25-μm film DB23 50% cyanopropyl-50% dimethylpolysiloxane column (Agilent Technologies, Inc., Santa Clara, CA) was used for FAME separation. A sample (1.0 μl) was injected in the split mode with a ratio of 100:1. The flow rate of hydrogen, the carrier gas, was adjusted to 1.00 ml/min. The injector and detector temperatures were maintained at 250°C and 300°C, respectively. The column temperature was held at 100°C for 2 min, raised to 220°C at a rate of 10°C/min, and then held at 220°C for 5 min. FAMEs were identified by comparison of sample retention times to a qualitative standard bacterial FAME mixture (CP mix; Matreya, Inc., Pleasant Gap, PA) analyzed under the same conditions, and results represent mean values from two independent experiments.
Cell treatments for transcriptome studies.
Cells were grown in batch culture with pH control as described above, and then five 50-ml samples were aseptically harvested into sterile 200-ml centrifuge bottles and administered one of five treatments designated as control, acid adaptation for 5 or 20 min (AA5 or AA20), acid challenged (AC), and acid adaptation and then acid challenge (AA20-AC). Control cells were immediately suspended in 100 ml of RNAprotect reagent (Qiagen, Inc., Valencia, CA) plus 900 μl rifampin (25 mg/ml in methanol; Sigma-Aldrich), mixed by vortex for 15 s, and then incubated for 10 min at room temperature. After this treatment, the cells were collected by centrifugation and then the supernatant was discarded and the pellet was stored at −20°C until needed for RNA isolation.
Cells for each of the other four treatments were collected by centrifugation for 10 min in a rotor that had been stabilized at 37°C to avoid confounding from temperature stress (46). The supernatant was discarded, and the pellets were suspended in 50 ml MRS warmed to 37°C and adjusted as necessary to deliver different treatments. Cells for treatments AA5 and AA20 were suspended in MRS adjusted to a pH of 4.5 and then incubated for 5 min or 20 min, respectively, at 37°C. For AC treatment, cells were suspended in MRS adjusted to pH 2.0 and then incubated at 37°C for 10 min. Finally, cells for AA20-AC were suspended in MRS adjusted to pH 4.5, incubated for 20 min at 37°C, and then collected and suspended in MRS at pH 2.0 and incubated for another 10 min at 37°C. At the conclusion of each treatment, 100 ml of RNAprotect and 900 μl rifampin were added and the suspensions were mixed by vortex for 15 s and incubated at room temperature for 10 min. The cells were collected by centrifugation for 10 min, and the pellet was stored at −20°C until needed for RNA isolation. Cell pellets from each of the five treatments were collected from four independent experiments for microarray studies.
RNA isolation.
Cell pellets were thawed at room temperature and suspended in 1 ml of lysozyme solution (20 mg/ml in TE buffer) that also contained 20 U of mutanolysin (Sigma-Aldrich) and 75 μl of rifampin (25 mg/ml in methanol). This mixture was incubated at 37°C for 15 min in a shaker incubator at 240 rpm, and then total RNA was isolated from each of the cell samples using the Aurum Total RNA Mini Kit (Bio-Rad Laboratories, Hercules, CA) following procedures recommended by the vendor, except for the following modifications. First, cells were mixed with 3.5 ml of Aurum lysis solution supplemented with 1% beta-mercaptoethanol by pipetting up and down several times to effect lysis. Next, 2.5 ml of 70% isopropanol was added and mixed until no bilayer was visible and the viscosity was substantially reduced. Cell lysates were then transferred (∼700 μl) to a total of 10 Aurum RNA binding columns and centrifuged at 14,000 × g for 30 s (or until all of the solution had passed through). The filtrate was discarded, and the column was placed in the same wash tube. From here, the protocol was followed precisely as described by manufacturer. Once the process was complete, the RNA concentration was measured with a NanoDrop 8000 spectrophotometer (ThermoFisher Scientific, Waltham, MA), RNA quality was evaluated with an Agilent 2100 bioanalyzer, and then the RNA was stored at −80°C.
cDNA synthesis and labeling.
cDNA was synthesized and labeled as recommended by the Affymetrix (Santa Clara, CA) protocol for prokaryotic target preparation in the GeneChip Expression Analysis Technical Manual. Briefly, cDNA was synthesized from 10 μg of total RNA using random primers (Invitrogen, Carlsbad, CA) and SuperScript II reverse transcriptase (Invitrogen). After synthesis, template RNA was digested with 1 N NaOH and neutralized with 1 N HCl, and cDNA was purified using the MinElute PCR purification kit (Qiagen) using a final elution volume of 12 μl rather than the 10 μl recommended by the manufacturer. cDNA was fragmented into approximately 50 to 100 bp using DNase I (GE Healthcare, Waukesha, WI) with fragmentation efficiency determined with an Agilent 2100 bioanalyzer. Fragmented cDNA was biotin labeled with GeneChip DNA labeling reagent (Affymetrix) and terminal deoxynucleotidyl transferase (Promega, Madison, WI), and labeling efficiency was measured by gel shift assay.
Microarray design, hybridization, and data extraction.
Fragmented, labeled cDNA was taken to the Affymetrix core facility in the Center for Integrated Biosystems on the Utah State University campus for hybridization and data extraction. Hybridizations were performed against an Affymetrix custom microarray designed to include 2,674 (96%) chromosomal and 17 (85%) plasmid genes predicted to occur in L. casei ATCC 334 (7, 32). The only predicted coding sequences not included in the microarray design were redundant transposase and rRNA genes.
Data normalization and analysis.
Statistical analysis of microarray data was performed using R software (www.r-project.org). Array images were reduced to intensity values for each probe and screened for acceptable quality control criteria before further analysis. Array preprocessing (background correction, normalization, and summarization) was performed with Bioconductor software (www.bioconductor.org) using the robust multiarray average method (3). The data from all four biological replications were pooled and then filtered to remove the genes with low expression values and/or low coefficients of variability in order to restrict our focus to genes that are highly expressed and genes that show a difference in expression between treatments (42). The significance of differential gene expression between controls and treatments was calculated with the limma (linear models for microarray data) statistical package in Bioconductor. Genes were considered significantly differentially expressed if the adjusted P values were less than 0.05.
Array validation.
Validation of microarray data for 10 different genes (Table 1) was performed by real-time quantitative PCR (RT-PCR) as described previously (46), by using the same cDNA samples employed for AA5 (LSEI_0740), AA20-AC (LSEI_1586), or AA20 (all other targets) array hybridizations. Primers for RT-PCR were designed with GeneWorks software (IntelliGenetics, Inc., Mountain View, CA) and compared against the L. casei ATCC 334 genome using ERGO bioinformatic software (Integrated Genomics, Inc., Chicago, IL) to verify that each annealed to a single locus in the genome. Primer pairs were predicted to have annealing temperatures that ranged from 55 to 62°C and produce amplicons that ranged from 85 to 141 bp in length (Table 1). Template DNA from L. casei ATCC 334 was used to determine optimal RT-PCR conditions for each primer pair and to ensure the absence of nonspecific amplification, and then the fidelity of PCR products was confirmed by nucleotide sequence determination. Reactions were performed in an Opticon II thermal cycler (MJ Research, Reno, NV). Each reaction mixture consisted of 5 μl of either cDNA or water (negative controls), 5 μl of primer mix (1.2 μM each primer), and 10 μl of SYBR green mix (Bio-Rad Laboratories, Hercules, CA). Blanks contained 10 μl of SYBR green mix and 10 μl of water. RT-PCR was performed using two concentrations of cDNA (10 and 1 ng/μl) obtained from control, acid-challenged, acid-adapted, or acid-adapted and acid-challenged L. casei ATCC 334 cells as described above. Triplicate reactions were run in a 96-well plate. Amplicon quantification in RT-PCRs was performed by comparison with gene-specific standard curves constructed from known concentrations of individually purified amplicons. The obtained amplicon copy numbers were log transformed and used in the calculation of the expression change (n-fold) for a particular gene.
TABLE 1.
Target genes and oligonucleotides used for RT-PCR
| Protein function (gene ID) | Primer sequence |
Amplicon size (bp) | Annealing temp (°C) | |
|---|---|---|---|---|
| Forward | Reverse | |||
| Malolactic enzyme (LSEI_0740) | AGCAGAACGGAATCAGTATGG | CGGCTTGGTCTTTAACTGAGC | 103 | 58 |
| Two-component RR (LSEI_0460) | CTGATGAGGATAAAGTTCGTGG | GGCTTTCCCGTCTAAAGTGG | 114 | 55 |
| Sensor kinase DpiB (LSEI_2868) | CCATTGACGACATTGATTGC | TCACCATAATAGGTGCGGC | 102 | 62 |
| Transcription repressor HrcA (LSEI_1567) | CGTCTTAAAAGAGATCATCCGG | GCGAGAACTGACGTGACC | 97 | 55 |
| XRE family transcriptional regulator (LSEI_2759) | GTGGGCATGCAGTTTATTGG | GGAATCAGCAATGTAACGGC | 140 | 61 |
| Arsenate reductase (glutaredoxin) (LSEI_2761) | ACGTTCAGCACCTACACTGG | CTTCAATCTTATCGCGGACC | 141 | 55 |
| Peptide methionine sulfoxide reductase MsrA (LSEI_1393) | CCACCTATGAACAGGTTTCG | GGATAGCTAATGGTGTCGGC | 85 | 57 |
| Histidinol dehydrogenase (LSEI_1433) | GGGTCTATGACTTCGTTAAGCG | AGGGACACGTTTGATGATGG | 115 | 61 |
| Imidazole glycerol phosphate synthase, cyclase subunit (LSEI_1429) | GGATATGCACGCTTTGTTGC | AGCCAACGTGTATCAATCGC | 117 | 62 |
| 30S ribosomal protein S2 (LSEI_1586) | GGTATCGAAGACATGCCTCG | CGGTGTTAGTATCAACCATCGC | 127 | 58 |
Microarray data accession number.
Microarray hybridization data have been deposited in Gene Expression Omnibus under accession number GPL9271.
RESULTS AND DISCUSSION
The ability to survive in acidic environments is central to the lifestyle of L. casei, and it is also of paramount importance to the utility of this microorganism in bioprocessing. Although acidurance is considered an innate feature of L. casei, the molecular basis of this capability is only partly defined. L. casei has, for example, been shown to display an ATR, but few reports have addressed its effect on cell physiology (15, 21, 31, 50). Because ATR induction produces physiological changes that enhance acid resistance, more-detailed investigation of this response will reveal the molecular mechanisms used by L. casei to persist in highly acidic environments and may provide new strategies to enhance the industrial utility of this species.
Acid tolerance response of L. casei ATCC 334.
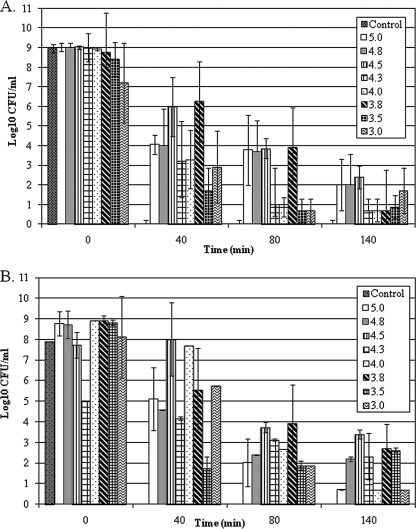
Acid adaptation experiments with L. casei ATCC 334 demonstrated that ATR induction could be triggered by transient exposure to a wide range of sublethal pH values (Fig. 1). Although all of the acid-adapted L. casei ATCC 334 cultures suffered a greater-than-4-log decrease in viability after 140 min of acid challenge, numbers of viable control cells (acid adaptation treatment for 10 or 20 min at pH 6.0) fell below the limit of detection after only 40 min. In contrast, survival after 140 min of acid challenge by cells that were acid adapted for 10 min at lower pH values ranged from 0.7 to 2.4 log CFU/ml and from 0.7 to 3.4 log CFU/ml for ATCC 334 cells that had been acid adapted at lower pH values for 20 min (Fig. 1). Data presented in Fig. 1 also illustrate the importance of optimizing conditions for ATR induction in a particular strain, as some adaptation treatments were less effective at protecting cells against a lethal acid challenge or simply showed greater variability in their results. Data presented in Fig. 1 revealed that cells that had been acid adapted at pH 4.5 for 10 or 20 min displayed superior survival (with good experimental reproducibility) during a 140-min acid challenge at pH 2.0.
FIG. 1.
Influence of different acid adaptation treatments on the viability of L. casei ATCC 334 during acid challenge at pH 2.0. L. casei ATCC 334 was propagated at pH 6.0 and then incubated for 10 min (panel A) or 20 min (panel B) at pH 6.0 (control), 5.0, 4.8, 4.5, 4.3, 4.0, 3.8, 3.5, or 3.0 and then acid challenged at pH 2.0. Values represent the mean number of recovered CFU calculated from duplicate analysis of duplicate experiments. Error bars indicate the standard error of the mean. If error bars are not visible, the standard error was less than 2%.
Membrane lipid composition of acid-adapted L. casei ATCC 334.
When confronted with acid stress, bacteria may act to counter proton influx by increasing the rigidity and compactness of the cytoplasmic membrane (11, 14, 51, 54). Such structural changes are accomplished, in part, by changing the FA species within the cytoplasmic membrane. The major CMFAs of Lactobacillus spp. are myristic acid (C14:0), palmitic acid (C16:0), palmitoleic acid (C16:1n(9)), stearic acid (C18:0), oleic acid (C18:1n(9)), cis-vaccinic acid (C18:1n(11)), and the cyclopropanes dihydrosterculic acid [cis-9,10-methyleneoctadecanoic acid; C19:0(9c)] and lactobacillic acid [cis-11,12-methyleneoctadecanoic acid; C19:0(11c)] (38). As shown in Table 2, these acids made up over 87% of the total CMFA content of early-stationary-phase L. casei ATCC 334 propagated at pH 6.0 in MRS.
TABLE 2.
Effect of acid adaptation and acid challenge on the cell membrane FA composition of early-stationary-phase L. casei ATCC 334a
| FA or parameter | Mean % of each FA species in total cytoplasmic membrane lipid pool ± SEM after treatment |
||||
|---|---|---|---|---|---|
| Control | AA10 | AA20 | AA10-AC | AA20-AC | |
| C14:0 | 3.7 ± 0.5 | 40.0 ± 0.1 | 39.0 ± 1.1 | 36.0 ± 1.1 | 20.0 ± 0.3 |
| C16:1n(9) | 0.6 ± 0.3 | 3.3 ± 0.1 | 2.0 ± 0.0 | 3.0 ± 0.0 | 5.8 ± 1.0 |
| C16:0 | 15.0 ± 0.1 | 22.0 ± 0.1 | 27.0 ± 0.1 | 30.0 ± 0.1 | 25.0 ± 0.3 |
| C17:0(9c) | 0.6 ± 0.5 | BQLb | BQL | 1.9 ± 0.0 | 4.2 ± 1.8 |
| C18:1n(9) | 41.0 ± 0.3 | 7.6 ± 0.0 | 7.0 ± 0.1 | 6.8 ± 0.0 | 11.0 ± 0.1 |
| C18:1n(11) | 23.0 ± 1.3 | 8.1 ± 0.0 | 7.0 ± 0.0 | 7.7 ± 0.0 | 8.3 ± 2.6 |
| C18:0 | 1.3 ± 1.1 | 1.7 ± 0.0 | BQL | BQL | 15.0 ± 0.6 |
| C19:0(11c) | 5.0 ± 3.1 | 17.0 ± 0.0 | 18.0 ± 0.1 | 15.0 ± 0.1 | 15.0 ± 2.2 |
| % Cyclopropanec | 5.6 | 17 | 18 | 16.9 | 19.2 |
| % Saturatedd | 26 | 80 | 84 | 83 | 79 |
| % Unsaturatede | 65 | 19 | 16 | 17 | 25 |
| Saturated/unsaturated ratio | 0.4 | 4.2 | 5.3 | 4.9 | 3.2 |
Cells were propagated at pH 6.0 and then acid adapted by incubation for 10 min (AA10) or 20 min (AA20) at pH 4.5 or acid adapted for 10 or 20 min and then acid challenged at pH 2.0 for 140 min (AA10-AC or AA20-AC, respectively).
BQL, below quantifiable limit (0.002%).
Percentage of cyclopropane FAs [C17:0(9c) and C19:0(11c)] in the membrane.
Percentage of saturated FAs [C14:0, C16:0, C17:0(9c), C18:0, and C19:0(11c)] in the membrane.
Percentage of unsaturated FAs (C16:1 and C18:1) in the membrane.
To decrease fluidity, cells may increase the concentration of saturated FAs or cyclopropane FAs (10, 15, 16), and both events were prominent in the CMFA profiles of acid-adapted versus control cells analyzed in this study (Table 2). The saturated-to-unsaturated FA ratio increased from 0.4 in nonadapted control cells to 4.2 or 5.3, respectively, after 10 or 20 min of acid adaptation at pH 4.5 because acid-adapted cells had a higher total percentage of saturated FAs and cyclopropane than did control cells (Table 2). Specifically, the FAs C14:0 and C19:0(11c) exhibited the largest increase in acid-adapted cells, and proportions of C16:1n(9) and C16:0 also increased, while C18:1n(9) and C18:1n(11) exhibited the greatest decrease in comparison to control cells. Similar trends were noted for these FAs in cells that were acid adapted and acid challenged, where the ratio of saturated to unsaturated FAs was 4.9 or 3.2, respectively, for cells that had been acid adapted for 10 or 20 min prior to acid challenge (Table 2). Levels of C17:0(9c) were also higher in acid-challenged, but not acid-adapted, ATCC 334. However, interpretation of CMFA changes in acid-challenged cells is subject to question because these cells are rapidly losing viability and the observed changes may be a result of the erratic metabolic activity of dying cells.
Fozo et al. (15) have previously reported that the CMFA composition of L. casei was altered if cells were incubated at pH 5.0 rather than pH 7.0. Specifically, those authors noted that incubation at the lower pH resulted in increased proportions of C18:1 and cyclopropane C19:0, with concomitant decreases in C16:0 and cyclopropane C17:0 (15). As shown in Table 2, acid adaptation of L. casei ATCC 334 for 10 or 20 min at pH 4.5 also produced a sizable increase in the proportion of cyclopropane C19:0 and a decrease in C17:0. However, results from this work also differed from those of Fozo et al. (15) in that we observed a small increase in C16:0 and a dramatic decrease in C18:1n(9) and C18:1n(11) in acid-adapted versus control cells. Moreover, the proportion of C14:0 was substantially increased in acid-adapted cells, and increased levels of C16:1n(9) and C16:0 were also noted (Table 2). Finally, the ratio of saturated to unsaturated FAs decreased from 4.6 to 2.4 when L. casei was incubated at pH 5.0 versus pH 7.0 in the work of Fozo et al. (15), but an opposite and more dramatic change was noted between cells incubated at pH 6.0 (control) and those that had been acid adapted for 10 or 20 min at pH 4.5 in this study (Table 2). The basis for these discrepancies is unclear, but they are most likely a reflection of the different growth media, pHs, and incubation conditions used in the two studies.
Influence of acid stress on global gene expression.
The effect of ATR induction at pH 4.5 on transcription was investigated using an Affymetrix custom microarray designed for L. casei ATCC 334. Normalization and analysis of microarray hybridization signals revealed significant (P < 0.05) changes in the expression of 15 genes after comparisons between the transcriptomes of control and AA5-treated cell preparations (grown at pH 6.0, collected, washed, and acid adapted for 5 min in fresh MRS adjusted to pH 4.5). Twelve (80%) of the 15 genes had known or predicted functions (Table 3; for a complete list of the differentially expressed genes, see Table S1 in the supplemental material). One gene, encoding malolactic enzyme, was upregulated more than 16-fold, while 14 other genes for proteins involved in metabolism, information processing, and other processes were significantly downregulated in AA5 cells (Table 3).
TABLE 3.
Numbers of genes with known or predicted functions that were significantlya up- or downregulated in acid-adapted L. casei ATCC 334b
| General functional category | AA-5 |
AA-20 |
AA20-AC |
|||
|---|---|---|---|---|---|---|
| Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | |
| Metabolism | ||||||
| Energy production, conversion | 1 | 1 | 3 | 2 | 1 | 1 |
| Carbohydrate transport, metabolism | —c | 1 | 12 | 9 | 2 | 3 |
| Amino acid transport, metabolism | — | — | 19 | 5 | 1 | 2 |
| Nucleotide transport, metabolism | — | 4 | — | 25 | — | 14 |
| Coenzyme metabolism | — | — | — | 1 | — | 1 |
| Secondary metabolites biosynthesis, transport, catabolism | — | — | 5 | — | 1 | — |
| Information storage, processing | ||||||
| Translation, ribosomal structure, biogenesis | — | 2 | — | 58 | — | 3 |
| Transcription | — | — | 3 | 13 | — | — |
| DNA replication, recombination, repair | — | — | — | 5 | — | 2 |
| Mobile DNA elements | — | — | 12 | 1 | 1 | — |
| Cellular processes | ||||||
| Protein turnover, stress response | — | 2 | 1 | 13 | — | 3 |
| Signal transduction | — | — | 2 | 1 | — | — |
| Cell secretion | — | — | — | 7 | — | 2 |
| Cell envelope biogenesis | — | 1 | — | 11 | — | 2 |
| Cell division, chromosome partitioning | — | — | 2 | 7 | — | 1 |
| Poorly characterized, general function prediction only | — | — | 13 | 25 | 2 | 7 |
P < 0.05.
Versus control cells grown at pH 6.0. AA5 or AA20, cells grown at pH 6.0 and then acid adapted for 5 or 20 min, respectively, in fresh MRS adjusted to pH 4.5; AA20-AC, cells grown at pH 6.0, incubated for 20 min in MRS adjusted to pH 4.5, and then suspended in MRS at pH 2.0 for 10 min. See Table S1 in the supplemental material for full details.
—, no gene found.
After 20 min of acid adaptation at pH 4.5 (AA20), the number of genes whose expression was significantly (P < 0.05) altered had increased to 320, with 104 genes upregulated and 216 significantly downregulated (see Table S1 in the supplemental material). The 15 genes whose expression was significantly altered in the control-to-AA5 comparison were also differentially expressed in the same manner (up- or downregulated) by this treatment. Of the total of 320 genes affected by this treatment, 254 (79%) encode proteins with known or predicted functions (Table 3).
No significant changes in gene expression were detected in acid-challenged cells (pH 2.0 for 10 min) compared to control cells, but the transcriptome of cells that were acid adapted for 20 min at pH 4.5 before acid challenge (AA20-AC) showed differential expression of 66 genes. Forty-nine (74%) of these genes encode proteins with known or predicted functions (Table 3), and all but two (LSEI_0561 and LSEI_1487) had been detected and showed a similar response in the control-to-AA20 comparison (see Table S1 in the supplemental material).
RT-PCR validation.
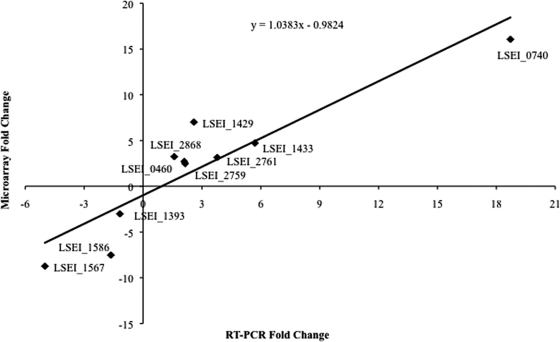
Of the 10 genes selected for analysis by RT-PCR, 7 were predicted by microarray data to be upregulated (LSEI_0740, LSEI_0460, LSEI_2868, LSEI_2759, LSEI_2761, LSEI_1433, and LSEI_1429) and 3 were downregulated (LSEI_1586, LSEI_1393, and LSEI_1567). As shown in Fig. 2, no contradictions between the two platforms were detected, and there was a strong positive correlation (r = 0.84) between the fold change in gene induction or repression predicted from the microarray and the respective values determined by RT-PCR.
FIG. 2.
Correlation of fold change values from microarray results and RT-PCR. Total RNA was extracted from both controls and all four acid-treated L. casei ATCC 334 cell groups and used as the template for cDNA synthesis to be used for microarray hybridizations and RT-PCR experiments. The fold change values were obtained for the 10 genes used in the RT-PCR experiments (Table 1). The best-fit curve is shown along with the calculated equation. The r value is 0.84.
FA biosynthesis.
Alteration of CMFA composition in response to acid stress has been observed in gram-positive and gram-negative bacteria (10, 11, 14, 15, 51) and likely occurs through a combination of de novo FA biosynthesis and modification of existing lipid membrane phospholipid acyl chains (54, 55). Because FA biosynthesis in bacteria is an energy-intensive process (55), the capability to modify existing CMFAs offers cells a less taxing means to adjust membrane structure, and at least three distinct enzymes have been found to catalyze these reactions in bacteria: phospholipid acyl desaturase, cyclopropane synthase (Cfa), and FA cis-trans isomerase (54). The L. casei ATCC 334 genome appears to encode only one of these enzymes, Cfa, which catalyzes the addition of a methylene residue across the cis double bond of C16:1n(9), C18:1n(9), or C18:1n(11) unsaturated FAs to form an unsaturated cyclopropane derivative (49). Thus, the concomitant decrease in C18:1n(11) and increase in C19:0(11c) detected in the cytoplasmic membrane content of acid-adapted cells (Table 2) can be directly attributed to Cfa activity on existing C18:1n(11). Other changes that occurred in the membrane lipid content of acid-adapted cells, however, are more likely explained by adjustments in the de novo synthesis of long fatty acyl chains and phospholipid turnover. The large reduction in C18:1n(9) noted in acid-adapted cells, for example, occurred without a corresponding increase in C18:0 and supports our conclusion that L. casei ATCC 334 does not possess phospholipid acyl desaturase activity. Because most of the changes that were detected in the CMFA composition of acid-adapted L. casei appear to be dependent on de novo lipid synthesis, acid adaptation likely places a significant demand on the energy resources of the cell.
Inspection of the L. casei ATCC 334 genome for genes involved in FA (fab) and phospholipid (pls) biosynthesis revealed that fab genes are located in a large, 15-gene cluster (LSEI_2121 to LSEI_2107) that is structurally unique among all of the sequenced LAB species by virtue of its inclusion of the gene for Cfa. In contrast, genes that encode the three enzymes needed to transfer long-chain FAs to glycerol-3-phosphate and form phosphatidic acid (plsX, plsY, and plsC) are distributed across three different loci (LSEI_1614, LSEI_1407, and LSEI_1589, respectively). Although CMFA data indicate that enzymes for FA and phospholipid biosynthesis are involved in the L. casei ATR, microarray data indicate that the genes that encode those enzymes are not differentially expressed during ATR induction. This finding indicates that the constitutive expression levels of these genes are sufficient to meet cellular needs during ATR induction or that the activity of enzymes involved in these processes is not regulated at the transcriptional level. Even though the fab and pls genes were not affected by acid adaptation, significant upregulation of several genes that might be involved in phospholipid turnover was detected in AA20 cells. These included LSEI_0140, which encodes a putative membrane-associated phospholipid phosphatase, and LSEI_2773 and LSEI_1868, which are predicted to code for an esterase and an acetyltransferase, respectively ( Table 4).
TABLE 4.
Examples of differentially regulateda genes in acid-adapted L. casei ATCC 334
| General functional category and gene IDc | Predicted function | Fold change vs controlb |
||
|---|---|---|---|---|
| AA5 | AA20 | AA20-AC | ||
| Metabolism | ||||
| Energy production and conversion | ||||
| LSEI_0740 | Malolactic enzyme | 16.06 | 7.35 | NS |
| LSEI_0741 | Malate/lactate antiporter | NS | 5.99 | NS |
| LSEI_1161 | ATP synthase C chain (EC 3.6.3.14) | NS | −5.00 | NS |
| Carbohydrate transport, metabolism: LSEI_2764 | sn-Glycerol-3-phosphate transport system permease protein UgpA | NS | 2.71 | NS |
| Amino acid transport, metabolism | ||||
| LSEI_2061 | Oligopeptide transport ATP-binding protein OppF | NS | 3.27 | NS |
| LSEI_2062 | Oligopeptide transport ATP-binding protein OppD | NS | 5.49 | NS |
| LSEI_2063 | Oligopeptide transport system permease protein OppC | NS | 3.06 | NS |
| LSEI_2064 | Oligopeptide transport system permease protein OppB | NS | 3.39 | NS |
| LSEI_0175 | Oligopeptide-binding protein OppA | NS | 3.64 | NS |
| LSEI_1890 | Oligopeptide transport system permease protein OppB | NS | 2.34 | NS |
| LSEI_0046 | Branched-chain amino acid transport protein AzlD | NS | 3.01 | NS |
| LSEI_1260 | Histidine transport system permease protein HisM | NS | 2.17 | NS |
| LSEI_1426 | Histidinol-phosphate aminotransferase (2.6.1.9) | NS | 7.52 | NS |
| LSEI_1427 | Phosphoribosyl-ATP pyrophosphatase (EC 3.6.1.31) | NS | 4.76 | NS |
| LSEI_1429 | Imidazole glycerol phosphate synthase, cyclase subunit (EC 4.1.3.-) | NS | 7.01 | NS |
| LSEI_1430 | 1-(5-Phosphoribosyl)-5-[(5-phosphoribosylamino) methylideneamino]imidazole-4-carboxamide isomerase (EC 5.3.1.16) | NS | 3.61 | NS |
| LSEI_1431 | Imidazole glycerol phosphate synthase, glutamine amidotransferase subunit (EC 2.4.2.-) | NS | 5.43 | NS |
| LSEI_1432 | Imidazoleglycerol-phosphate dehydratase (EC 4.2.1.19) | NS | 3.98 | NS |
| LSEI_1433 | Histidinol dehydrogenase (EC 1.1.1.23) | NS | 4.72 | NS |
| LSEI_1434 | ATP phosphoribosyltransferase (EC 2.4.2.17) | NS | 5.00 | NS |
| LSEI_0078 | Anthranilate phosphoribosyltransferase (EC 2.4.2.18) | NS | 2.68 | NS |
| LSEI_0098 | Diaminopimelate decarboxylase (EC 4.1.1.20) | NS | 2.80 | 3.01 |
| LSEI_2796 | Acetylornithine deacetylase (EC 3.5.1.16)/succinyl-diaminopimelate desuccinylase (EC 3.5.1.18), related deacylases | NS | 2.36 | NS |
| LSEI_1345 | Arginine transport ATP-binding protein ArtP | NS | −3.00 | −3.83 |
| LSEI_1486 | Aspartate aminotransferase (EC 2.6.1.1) | NS | −3.21 | NS |
| LSEI_1643 | Xaa-Pro dipeptidase (EC 3.4.13.9) | NS | −3.23 | NS |
| LSEI_1652 | Glutamine synthetase (EC 6.3.1.2) | NS | −4.47 | NS |
| LSEI_1653 | Transcriptional regulator, MerR family | NS | −5.52 | NS |
| LSEI_1288 | 5′-Methylthioadenosine nucleosidase (EC 3.2.2.16)/ S-adenosylhomocysteine nucleosidase (EC 3.2.2.9) | NS | −4.54 | −4.40 |
| Nucleotide transport and metabolism | ||||
| LSEI_1557 | Adenine phosphoribosyltransferase (EC 2.4.2.7) | NS | −3.19 | −3.26 |
| LSEI_1118 | Xanthine permease | NS | −3.16 | −4.12 |
| LSEI_1119 | Phosphoribosylaminoimidazole carboxylase NCAIR mutase subunit (EC 4.1.1.21) | NS | −3.33 | NS |
| LSEI_1120 | Adenylosuccinate lyase (EC 4.3.2.2) | NS | −3.27 | −3.99 |
| LSEI_1286 | ADP-ribose pyrophosphatase (EC 3.6.1.13) | −4.27 | −3.72 | −5.39 |
| LSEI_1746 | Phosphoribosylamine-glycine ligase (EC 6.3.4.13) | NS | −3.34 | NS |
| LSEI_1747 | Phosphoribosylaminoimidazolecarboxamide formyltransferase (EC 2.1.2.3)/IMP cyclohydrolase (EC 3.5.4.10) | NS | −9.35 | −8.18 |
| LSEI_1748 | Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) | NS | −5.18 | NS |
| LSEI_1749 | Phosphoribosylformylglycinamidine cycloligase (EC 6.3.3.1) | NS | −4.32 | −4.65 |
| LSEI_1750 | Amidophosphoribosyltransferase (EC 2.4.2.14) | NS | −4.48 | NS |
| LSEI_1751 | Phosphoribosylformylglycinamidine synthase (EC 6.3.5.3) | NS | −4.47 | NS |
| LSEI_1752 | Phosphoribosylformylglycinamidine synthase (EC 6.3.5.3) | NS | −4.75 | −4.77 |
| LSEI_1753 | Phosphoribosylformylglycinamidine synthase, PurS component (EC 6.3.5.3) | −5.71 | −7.49 | −8.75 |
| LSEI_1754 | Phosphoribosylamidoimidazole-succinocarboxamide synthase (EC 6.3.2.6) | NS | −7.16 | −6.86 |
| LSEI_1755 | Phosphoribosylaminoimidazole carboxylase NCAIR mutase subunit (EC 4.1.1.21) | NS | −6.98 | −7.55 |
| LSEI_1756 | Phosphoribosylaminoimidazole carboxyltransferase subunit (EC 4.1.1.21) | −5.09 | −7.49 | −8.35 |
| LSEI_1979 | GMP synthase (glutamine hydrolyzing) (EC 6.3.5.2) | NS | −3.82 | NS |
| LSEI_1468 | Ribonucleoside-diphosphate reductase alpha chain (EC 1.17.4.1) | NS | −3.44 | NS |
| LSEI_2482 | Adenylate kinase (EC 2.7.4.3)/nucleoside-diphosphate kinase (EC 2.7.4.6) | NS | −10.06 | NS |
| LSEI_1450 | Orotidine 5′-phosphate decarboxylase (EC 4.1.1.23) | NS | −2.61 | −3.08 |
| LSEI_1806 | Nicotinate phosphoribosyltransferase (EC 2.4.2.11) | NS | −2.80 | −3.46 |
| LSEI_1378 | Cytidylate kinase (EC 2.7.4.14) | NS | −3.13 | NS |
| LSEI_1669 | Uridine kinase (EC 2.7.1.48) | −4.10 | −3.16 | −4.59 |
| LSEI_1668 | Transcription elongation factor | NS | −5.27 | NS |
| LSEI_1159 | Uracil phosphoribosyltransferase (EC 2.4.2.9) | NS | −3.29 | NS |
| LSEI_1584 | Uridylate kinase | NS | −4.94 | NS |
| Information storage and processing | ||||
| Translation, ribosomal structure and biogenesis | ||||
| LSEI_0977 | tmRNA-binding protein | NS | −2.62 | NS |
| LSEI_1057 | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C (EC 6.3.5.-) | NS | −3.32 | NS |
| LSEI_1058 | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit A (EC 6.3.5.-) | NS | -3.24 | NS |
| LSEI_1292 | tRNA (5-methylaminomethyl-2-thiouridylate)methyltransferase (EC 2.1.1.61) | NS | −4.24 | −4.78 |
| LSEI_1579 | Prolyl-tRNA synthetase (EC 6.1.1.15) | NS | −2.96 | NS |
| LSEI_1280 | Isoleucyl-tRNA synthetase (EC 6.1.1.5) | NS | −2.97 | NS |
| LSEI_1485 | Asparaginyl-tRNA synthetase (EC 6.1.1.22) | NS | −3.37 | NS |
| LSEI_1703 | Threonyl-tRNA synthetase (EC 6.1.1.3) | NS | −3.80 | NS |
| LSEI_0883 | Leucyl-tRNA synthetase (EC 6.1.1.4) | NS | −4.05 | NS |
| LSEI_2569 | LSUd ribosomal protein L31P | NS | −2.78 | NS |
| LSEI_1697 | LSU ribosomal protein L20P | NS | −4.60 | NS |
| LSEI_1642 | Protein translation elongation factor P (EF-P) | NS | −5.55 | NS |
| LSEI_1641 | General stress protein, Gls24 family | NS | −5.18 | NS |
| LSEI_1640 | Transcription antitermination factor | NS | −4.38 | NS |
| LSEI_1644 | LSU ribosomal protein L27P | NS | −5.09 | NS |
| LSEI_1645 | Hypothetical ribosome-associated protein | NS | −8.30 | NS |
| LSEI_1847 | LSU ribosomal protein L33P | NS | −6.38 | NS |
| LSEI_1572 | Ribosome-binding factor | NS | −6.49 | NS |
| LSEI_1573 | Protein translation initiation factor 2 (IF-2) | NS | −4.76 | NS |
| LSEI_1574 | LSU ribosomal protein L7AE | −7.33 | −7.55 | −5.64 |
| LSEI_1575 | Hypothetical cytosolic protein | NS | −4.91 | NS |
| LSEI_1576 | Transcription termination factor | NS | −3.88 | NS |
| LSEI_1577 | Hypothetical cytosolic protein | NS | −3.84 | NS |
| LSEI_2272 | LSU ribosomal protein L12P (L7/L12) | NS | −13.08 | NS |
| LSEI_2273 | LSU ribosomal protein L10P | NS | −10.70 | NS |
| LSEI_2476 | LSU ribosomal protein L17P | NS | −7.52 | NS |
| LSEI_2478 | SSUe ribosomal protein S11P | NS | −6.88 | NS |
| LSEI_2479 | SSU ribosomal protein S13P | NS | −10.19 | NS |
| LSEI_2480 | LSU ribosomal protein L36P | NS | −19.62 | NS |
| LSEI_2481 | Protein translation initiation factor 1 (IF-1) | NS | −9.48 | NS |
| LSEI_2484 | LSU ribosomal protein L15P | NS | −11.94 | NS |
| LSEI_2485 | LSU ribosomal protein L30P | NS | −9.30 | NS |
| LSEI_2486 | SSU ribosomal protein S5P | NS | −8.51 | NS |
| LSEI_2487 | LSU ribosomal protein L18P | NS | −12.10 | NS |
| LSEI_2488 | LSU ribosomal protein L6P | NS | −8.62 | NS |
| LSEI_2489 | SSU ribosomal protein S8P | NS | −12.20 | NS |
| LSEI_2490 | SSU ribosomal protein S14P | NS | −12.31 | NS |
| LSEI_2491 | LSU ribosomal protein L5P | NS | −7.17 | NS |
| LSEI_2492 | LSU ribosomal protein L24P | −7.42 | −17.81 | NS |
| LSEI_2493 | LSU ribosomal protein L14P | NS | −10.32 | NS |
| LSEI_2494 | SSU ribosomal protein S17P | NS | −14.50 | NS |
| LSEI_2495 | LSU ribosomal protein L29P | NS | −9.62 | NS |
| LSEI_2496 | LSU ribosomal protein L16P | NS | −8.59 | NS |
| LSEI_2497 | SSU ribosomal protein S3P | NS | −8.83 | NS |
| LSEI_2499 | SSU ribosomal protein S19P | NS | −7.34 | NS |
| LSEI_2500 | 50S ribosomal protein L2 | NS | −7.79 | NS |
| LSEI_2501 | LSU ribosomal protein L23P | NS | −9.67 | NS |
| LSEI_2502 | LSU ribosomal protein L1E (= L4P) | NS | −6.11 | NS |
| LSEI_2503 | Ribosomal protein L3 | NS | −5.57 | NS |
| LSEI_2508 | Protein translation elongation factor G (EF-G) | NS | −3.38 | NS |
| LSEI_2509 | SSU ribosomal protein S7P | NS | −7.19 | NS |
| LSEI_1379 | SSU ribosomal protein S1P | NS | −6.04 | NS |
| LSEI_1244 | SSU ribosomal protein S4P | NS | −7.14 | NS |
| LSEI_1601 | SSU ribosomal protein S16P | NS | −14.28 | NS |
| LSEI_1327 | SSU ribosomal protein S20P | NS | −15.68 | NS |
| LSEI_1328 | SSU ribosomal protein S15P | NS | −4.43 | NS |
| LSEI_1332 | Protein translation elongation factor Tu (EF-TU) | NS | −8.07 | NS |
| LSEI_0009 | SSU ribosomal protein S6P | NS | −8.45 | NS |
| LSEI_1583 | Ribosome recycling factor (RRF) | NS | −5.90 | NS |
| LSEI_1585 | Protein translation elongation factor Ts | NS | −8.36 | NS |
| LSEI_1586 | 30S ribosomal protein S2 | NS | −13.17 | −7.52 |
| LSEI_1303 | Peptide deformylase (EC 3.5.1.88) | NS | −2.65 | NS |
| LSEI_1393 | Peptide methionine sulfoxide reductase MsrA (EC 1.8.4.11) | NS | −3.02 | NS |
| Mobile DNA elements | ||||
| LSEI_1333 | Transposase (transposase_12 superfamily) | NS | 3.79 | NS |
| LSEI_1101 | Transposase (transposase_12 superfamily) | NS | 3.20 | NS |
| LSEI_1103 | Transposase (transposase_12 superfamily) | NS | 3.62 | NS |
| LSEI_2689 | Transposase, IS30 family | NS | 2.43 | NS |
| LSEI_2691 | Transposase, IS30 family | NS | 3.58 | NS |
| LSEI_2008 | Transposase, IS30 family | NS | 2.21 | NS |
| LSEI_1907 | Transposase, IS3 family | NS | 3.42 | 3.57 |
| LSEI_0597 | Transposase, IS3 family | NS | 3.03 | NS |
| LSEI_0590 | Transposase, IS3 family | NS | 2.65 | NS |
| LSEI_0580 | Transposase, IS5 family | NS | 3.37 | NS |
| LSEI_0230 | Transposase, IS5 family | NS | 2.28 | NS |
| LSEI_2166 | Transposase, IS66 family | NS | 2.56 | NS |
| LSEI_0787 | Resolvase | NS | −3.32 | NS |
| Cellular processes | ||||
| Protein turnover, stress response | ||||
| LSEI_1617 | General stress protein, Gls24 family | NS | −2.45 | NS |
| LSEI_1281 | Cold shock protein | −3.71 | −2.80 | −3.52 |
| LSEI_1848 | Superoxide dismutase (EC 1.15.1.1) | NS | −3.64 | NS |
| LSEI_0963 | ATP-dependent endopeptidase Clp proteolytic subunit ClpP (EC 3.4.21.92) | NS | −3.67 | NS |
| LSEI_1467 | Glutaredoxin | NS | −5.19 | NS |
| LSEI_1762 | ATP-dependent endopeptidase Clp ATP-binding subunit ClpE | −5.54 | −6.19 | −5.08 |
| LSEI_1338 | FKBP-type peptidyl-prolyl cis-trans isomerase (trigger factor) (EC 5.2.1.8) | NS | −9.26 | NS |
| LSEI_2239 | Chaperonin GroES | NS | −29.35 | NS |
| LSEI_2238 | 60-kDa chaperonin GroEL | NS | −8.56 | NS |
| LSEI_1567 | Heat-inducible transcription repressor HrcA | NS | −8.73 | −6.61 |
| LSEI_1566 | Molecular chaperone GrpE | NS | −16.72 | −11.27 |
| LSEI_1565 | Molecular chaperone DnaK | NS | −13.71 | NS |
| LSEI_1564 | Small hypothetical protein | −14.36 | −12.59 | −9.71 |
| LSEI_1563 | Chaperone protein DnaJ | NS | −5.22 | NS |
| Signal transduction | ||||
| LSEI_2868 | Sensor kinase DpiB (EC 2.7.3.-) | NS | 3.24 | NS |
| LSEI_0460 | Two-component RR | NS | 2.71 | NS |
| LSEI_1679 | CsrR-like RR | NS | −2.77 | NS |
| Cell envelope biogenesis | ||||
| LSEI_1033 | Glycosyltransferase family 8 | NS | −2.54 | NS |
| LSEI_1808 | N-Acetylglucosamine-6-phosphate deacetylase (EC 3.5.1.25) | NS | −2.85 | NS |
| LSEI_2012 | dTDP-4-dehydrorhamnose reductase (EC 1.1.1.133) | NS | −3.00 | NS |
| LSEI_2014 | dTDP-4-dehydrorhamnose 3,5-epimerase (EC 5.1.3.13) | NS | −2.90 | NS |
| LSEI_2015 | Glucose-1-phosphate thymidylyltransferase (EC 2.7.7.24) | NS | −2.74 | NS |
| LSEI_1117 | Xanthine phosphoribosyltransferase (EC 2.4.2.-) | NS | −3.04 | NS |
| LSEI_1395 | Carboxy-terminal processing protease precursor (EC 3.4.21.102) | NS | −3.80 | NS |
| LSEI_0796 | d-Alanyl carrier protein | NS | −4.53 | −5.63 |
| LSEI_0797 | d-Alanyl transfer protein DltD precursor | NS | −4.07 | NS |
| LSEI_0020 | Cell wall hydrolase (amidase family) | NS | −5.57 | NS |
| LSEI_2029 | Cell wall hydrolase (amidase family) | −5.54 | −11.28 | −12.11 |
| Cell division and chromosome partitioning | ||||
| LSEI_2441 | CrcB family protein | NS | 2.47 | NS |
| LSEI_2442 | CrcB family protein | NS | 2.41 | NS |
| LSEI_0932 | Cell division protein FtsX | NS | −2.58 | NS |
| LSEI_1268 | Cell division protein FtsL | NS | −2.66 | NS |
| LSEI_1274 | Cell division protein FtsA | NS | −2.46 | NS |
| LSEI_1275 | Cell division protein FtsZ | NS | −4.89 | NS |
| LSEI_1279 | Cell division initiation protein DivIVA | NS | −4.27 | NS |
| LSEI_1478 | Cell division initiation protein DivIVA | NS | −7.37 | −9.28 |
| LSEI_0931 | Cell division ATP-binding protein FtsE | NS | −4.89 | NS |
| Poorly characterized, general function prediction only | ||||
| LSEI_1868 | Acetyltransferase, GNAT family | NS | 2.85 | NS |
| LSEI_2773 | Esterase | NS | 2.64 | NS |
| LSEI_0140 | Putative membrane-associated phospholipid phosphatase | NS | 2.57 | NS |
P < 0.05.
Fold change in normalized microarray signal intensity represents the average value calculated from four independent replicates. Control, cells grown at pH 6.0; AA5 or AA20, cells grown at pH 6.0 and then acid adapted for 5 or 20 min, respectively, in fresh MRS adjusted to pH 4.5; AA20-AC, cells grown at pH 6.0, incubated for 20 min in MRS adjusted to pH 4.5, and then suspended in MRS at pH 2.0 for 10 min.
Gene ID represents the locus tag used in GenBank for chromosomal or plasmid-carried genes in L. casei ATCC 334 (NC 008526 and NC 008502, respectively).
LSU, large subunit.
SSU, small subunit.
Acid adaptation invokes a stringent response.
In addition to the cytoplasmic membrane, acid stress has a deleterious effect on a variety of other cellular functions (20), so it was not unexpected to find significant changes between the transcriptomes of control and AA20-treated L. casei cells (see Table S1 in the supplemental material). The largest fraction (>35%) of downregulated genes was involved in cellular information processing, and those that contribute to the translational machinery of L. casei were especially targeted for repression (Table 4; Fig. 2). The latter observation is of particular interest because transcriptional repression of genes involved in the translational apparatus of cells is a hallmark of the bacterial stringent response to nutrient deprivation, which couples that action to the upregulation of genes associated with metabolic activities, especially amino acid biosynthesis, as well as some transport functions (9, 13). Other characteristics of the stringent response include the repression of genes needed for growth and reproduction (e.g., cell wall or DNA synthesis) and weak or no induction of chaperone/chaperonin proteins (9, 13). Very similar phenomena were observed in the transcriptional profile of AA20-treated versus control cells, except that the transcription of genes for chaperones, chaperonins, and other stress proteins was actually repressed (Table 4; Fig. 2).
Transcriptional changes that accompany the stringent response are triggered by Rel-dependent synthesis of (p)ppGpp, which interacts with RNA polymerase to effect gene repression or induction (9, 13). Rallu et al. (36) demonstrated that (p)ppGpp is also a signal for the induction of acid resistance in the LAB Lactococcus lactis and showed that insertion mutations in genes for purine biosynthesis such as guaA, which encodes GMP synthase (EC 6.3.5.2), dramatically enhanced acid resistance in this species. The L. casei ATCC 334 genome includes genes for two (p)ppGpp synthetases, relA (LSEI_1539) and relQ (LSEI_0901). Microarray data showed no significant change in the expression of relA or relQ in AA20 versus control cells, but transcriptional repression of 24 genes involved in nucleotide metabolism, including guaA (LSEI_1979), was detected (Table 4).
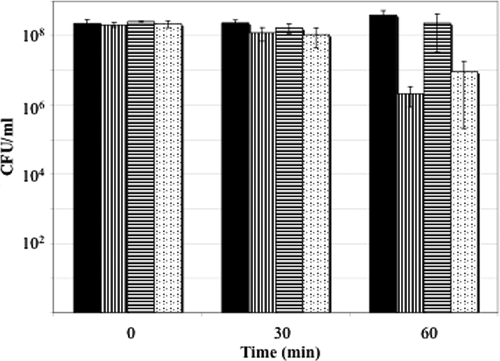
As a whole, these observations strongly suggested that (p)ppGpp also serves as the signal for ATR induction in L. casei and that the ATR in this species involves mechanisms that overlap the stringent response. To test this hypothesis, we performed acid challenge studies with cells that were treated with 1% (wt/vol) α-methylglucoside to induce the stringent response (36). As demonstrated in Fig. 3, exposure to α-methylglucoside before and during acid challenge resulted in a 100-fold increase in cell survival. This result confirmed that the stringent response confers acid resistance on L. casei and strongly suggested that (p)ppGpp accumulation is the signal for ATR induction in this species.
FIG. 3.
Influence of a chemically induced stringent response on the viability of L. casei ATCC 334 during acid challenge at pH 2.5. Cells were grown in batch culture with pH control (pH of 6.0) and incubated for 5 min in the presence (▤, ░⃞) or absence (▥) of 1% (wt/vol) α-methylglucoside to induce the stringent response and then acid challenged in MRS adjusted to pH 2.5 that also did (▤) or did not (░⃞, ▥) contain 1% α-methylglucoside. Control cells (▪) were incubated in MRS adjusted to pH 6.0 for 60 min. Data represent the mean values from two independent experiments.
While the ATR and stringent responses of L. casei appear to employ the same effector molecule, logic dictates that differences must exist in the mechanisms used to trigger Rel-dependent (p)ppGpp synthesis and in the downstream physiological consequences of each stress response. In the classic example of nitrogen-starved cells, induction of the stringent response is precipitated by entry of deacylated tRNA into an empty ribosomal A site, which triggers RelA-dependent (p)ppGpp synthesis (9). Not surprisingly, one of the most important physiological consequences of the stringent response involves the upregulation of genes that encode functions (e.g., amino acid biosynthesis and transport) that can help the cell escape amino acid starvation.
The trigger for ATR induction in L. casei is unknown, but a role for deacylated tRNA seems unlikely since cells were subject to acid adaptation treatment in a nutritionally rich laboratory medium (MRS). Because the L. casei ATR is accompanied by rapid and wholesale changes in CMFA composition (Table 2), and most of those changes appear to be dependent on de novo CMFA synthesis, one plausible hypothesis is that (p)ppGpp synthesis is triggered by a transient limitation for FAs (35). Although fab or pls gene expression was not affected by acid adaptation treatment, significant upregulation of the gene that encodes UgpA (LSEI_2764), the permease for an sn-glycerol-3-phosphate transport system, was detected in AA20 versus control cells (Table 4). The latter observation may be significant because glycerol-3-phosphate is a key precursor for phospholipid biosynthesis, but the relationship of this molecule or CMFA synthesis in general to ATR induction in L. casei requires further investigation.
Malate and His contribute to acid adaptation in L. casei.
If the pool of genes that were upregulated as a consequence of ATR induction in L. casei are viewed in parallel to those induced in other bacteria by nitrogen starvation, then malolactic fermentation (MLF) and intracellular pools of His may also play key roles in acid adaptation (Table 4). In MLF, l-malate is decarboxylated in the cytoplasm by the malolactic enzyme to produce l-lactate and CO2 (37). Decarboxylation contributes to alkalinization of the cytoplasm and allows ATP generation through H+-ATPase (34). The electrogenic potential created by lactate efflux through a malate/lactate antiporter, whose gene is commonly organized in an operon structure with that which encodes malolactic enzyme, may also facilitate energy production (34). MLF has not been associated with the stringent response, but it has been linked to LAB survival under acidic conditions (17, 34, 37, 44).
The sequenced genomes of L. casei and several other Lactobacillus species demonstrate that genes for malolactic enzyme (mleS) and malate/lactate antiporter (mleP) are arranged in tandem and presumably cotranscribed under the control of a LysR-type regulatory protein, MleR, whose gene is present immediately upstream and in divergent orientation with respect to mleS. An identical structure for MLF genes has been described for Oenococcus oeni, where mleSP cotranscription has been demonstrated by Northern hybridization (26).
mleS (LSEI_0740) was the only significantly upregulated gene in L. casei ATCC 334 AA5 cells, while both mleS and mleP (LSEI_0741) were among the most strongly upregulated genes in AA20 cells (Table 4; Fig. 2). No change was detected in the expression level of mleR (LSEI_0739) after any treatment, and differential expression of mleS or mleP was not detected in AA20-AC cells. Transient, high-level induction of mleSP in response to acid adaptation suggests that MLF is a critical component of the L. casei ATR, and its known physiological consequences are consistent with our hypothesis that acid adaptation requires a significant energy investment from cells.
The importance of MLF to L. casei biology is also illustrated by recent comparative genome hybridization data which demonstrated that mleS, mleP, and mleR were present in all of the 21 L. casei strains examined, which were isolated from cheese, plant material, or human sources (6, 7). Furthermore, Sheng and Marquis (44) found that L. casei had the highest specific MLF activity and the lowest pH optimum among five different species of oral LAB. More importantly, malate addition has been shown to enhance the survival of Lactobacillus plantarum and Streptococcus mutans during acid challenge at low pH values (17, 44).
Unlike MLF, induction of genes for His biosynthesis has been reported as part of the stringent response in gram-negative and gram-positive bacteria (5, 39). In this study, the transcriptome of AA20-treated cells exhibited a stringent-response-like response that included significant upregulation of an eight-gene cluster for His biosynthesis (LSEI_1426 to LSEI_1434) (Table 4; Fig. 2). Significant upregulation of a gene predicted to encode HisM (LSEI_1260), a permease that forms part of a His transport system, was also detected in these cells. In view of the fact that acid adaptation was performed in a nutritionally rich medium, these observations suggest that intracellular pools of His may also contribute to the ATR in L. casei.
One possibility is that His contributes to intracellular buffering capacity. With a pKa value near 6.0, the imidazole groups of His and His-containing peptides have been shown to contribute to intracellular buffering in higher cells (1). Len et al. (29) have suggested that acid-induced genes for branched-chain amino acid synthesis in S. mutans might contribute to alkalinization of the cytoplasm as a consequence of NH3 production. In L. casei, His biosynthesis may have a comparable function by virtue of its ability to function as a base.
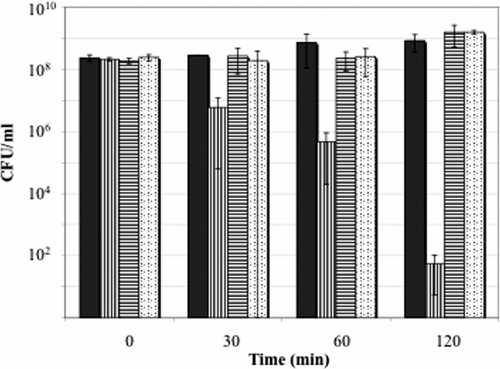
To determine whether the presence of malate or His would influence acid tolerance in L. casei, we performed acid challenge studies at pHs 2.0 and 2.5 using cells that were either acid adapted (20 min at pH 4.5) or acid challenged in MRS that contained 30 mM sodium malate or 30 mM His. The viability of L. casei ATCC 334 was not improved by the addition of malate or His at either phase of treatment when acid challenge was performed at pH 2.0, and results were highly variable during acid challenge at pH 2.5 when the compounds were present during acid adaptation (data not shown). However, the presence of either malate or His during acid challenge at pH 2.5 resulted in a more-than-100-fold increase in cell survival after 60 min of incubation and a greater-than-107-fold improvement after 2 h (Fig. 4). To our knowledge, this is the first reported evidence that intracellular His accumulation may contribute to acid resistance in bacteria.
FIG. 4.
Influence of malate or histidine addition on the viability of L. casei ATCC 334 during acid challenge at pH 2.5. Cells were acid adapted for 20 min in MRS broth adjusted to pH 4.5 and then acid challenged at pH 2.5 in regular MRS (▥) or MRS that contained 30 mM sodium malate (▤) or 30 mM histidine (░⃞). Control cells (▪) were incubated in MRS adjusted to pH 6.0. Data represent the mean values from two independent experiments.
atpC is downregulated during acid adaptation.
Acidurance in several LAB species has been directly linked to the activity and pH optima of H+-ATPase (11, 22, 48), and increased activity has sometimes been correlated with elevated transcription of atp genes (33). However, ATR induction in some species is not accompanied by a significant change in H+-ATPase activity or transcription of atp genes that encode its cognate proteins (33). In L. casei ATCC 4646, H+-ATPase activity was significantly reduced when cells were grown at pH 5.0 versus pH 7.0 (R. A. Burne, personal communication), and microarray data from this study demonstrated that atpC (LSEI_1161) was actually downregulated in AA20 versus control cells (Table 4). Though perhaps unexpected, these observations are, in fact, consistent with the view that H+-ATPase activity is more important for energy production (via MLF) than proton extrusion in acid-adapted L. casei, assuming that the former need can be readily met by existing enzymes.
Differential expression of two-component systems.
The molecular sensor(s) and regulators that modulate the L. casei ATR remain unknown, but AA20 treatment did affect the expression levels of genes associated with two-component regulatory systems (2CRS) and ABC-type oligopeptide transport proteins (Opp), which are known to function as sensors for environmental change (12, 28), as well as several transcriptional regulators (Table 4; Fig. 2). The L. casei ATCC 334 genome is predicted to encode 16 complete 2CRS, more than are found in any other sequenced Lactobacillus sp., as well as at least 11 ABC-type transporters for the uptake of peptides or amino acids and 124 transcriptional regulators (7, 32). While some of the 2CRS, Opp, and transcriptional regulators whose cognate genes were induced or repressed by acid adaptation could be involved in ATR modulation, their specific functions have not yet been determined. One possible exception is LSEI_1679, which encodes the response regulator (RR) for a 2CRS implicated in the acid resistance of Lactobacillus acidophilus (2). In that study, inactivation of the associated histidine kinase (HK) dramatically increased acid sensitivity in logarithmic-phase L. acidophilus and produced greater-than-2-fold changes in the expression of 80 genes. In particular, the authors noted upregulation in HK mutants incubated at pH 5.5 of numerous genes associated with the LAB proteolytic enzyme system, including two Opp operons (2).
In this work, significant downregulation of the RR (LSEI_1679) was detected in AA20-treated L. casei (Table 4). Because inactivation of the cognate HK gene in L. acidophilus increased the acid sensitivity of log-phase cells, and loss of HK activity would be predicted to cause RR downregulation at the protein level, transcriptional repression of LSEI_1679 might not be an expected component of ATR induction. However, repression of LSEI_1679 during acid adaptation of L. casei was only transient (Table 4), and Azcarate-Peril et al. (2) found that an L. acidophilus HK mutant could still mount an ATR that was just as effective as that produced by wild-type cells for protection against lethal acid challenge at pH 3.5 over a 2-h period. Although viability of the mutant began to decline faster than that of wild-type cells between 2 to 2.5 h of acid challenge, the results clearly demonstrated that ATR induction in L. acidophilus involves mechanisms that are not exclusively controlled by this particular 2CRS (2). Thus, our discovery that LSEI_1679 was transiently repressed during acid adaptation of L. casei is not inconsistent with the findings of Azcarate-Peril et al. (2) and supports a role for this 2CRS in the acid resistance of L. casei.
Differential expression of transposase genes.
An unexpected finding from this work involved the transient upregulation of 12 genes that encode transposase proteins from four different insertion sequence families in AA20 cells (Table 4). None of these genes was differentially expressed in AA5-treated cells, and only one (LSEI_1907) was still upregulated in AA20-AC cells. We are unaware of any similar observation in LAB species, but the histone-like nucleoid structuring protein of gram-negative bacteria, which is an important regulator of stress responses, has been shown to promote the transposition of Tn10, IS903, Tn552, and IS1 (53). Cellular mechanisms to stimulate transposition in response to environmental change could provide an evolutionary advantage to the host and the mobile DNA element. From that perspective, it is very interesting that even though transposon-related genes are quite abundant in L. casei ATCC 334 (over 3% of the total open reading frames) (32), 9 of the 12 transposase genes that were significantly upregulated in AA20 cells are either located in (LSEI_1101, LSEI_1103, LSEI_2008, and LSEI_0580) or proximate to (LSEI_2689, LSEI_2691, LSEI_1907, LSEI_0597, and LSEI_0590) discrete genomic regions that were likely acquired through horizontal gene transfer (7).
In summary, survival in acidic environments is critical to both the lifestyle and industrial performance of L. casei, so detailed understanding of its ATR holds fundamental and applied value. Physiological and transcriptional data presented in this report describe important features of the L. casei ATR and provide compelling evidence that acid adaption invokes a stringent-type response which is accompanied by other changes, including MLF and intracellular His accumulation, which promote cell survival at lower pH values. In combination, results from cell incubations with α-methylglucoside, malate, and His provide strong validation of our interpretation of microarray data and underscore the value of this technology for research on complex physiological processes such as acid resistance.
The overall nature of acid-induced changes in L. casei suggests that the ATR is an energy-intensive process whose cost may be driven by wholesale changes in CMFA composition that largely appear to rely on de novo FA biosynthesis. Efforts are currently under way in our laboratories to develop effective tools for gene inactivation in L. casei ATCC 334 so that we can further validate and characterize the contributions of enzymes and metabolic pathways described in this report to acid resistance in this species.
Supplementary Material
Acknowledgments
We thank Theresa Walunas and Integrated Genomics for assistance in designing the L. casei microarray, the Utah State University Center for Integrated Biosystems for microarray hybridizations and scanning, and John R. Stevens of the USU Mathematics and Statistics Department for help with R bioinformatic software.
This research was supported by the United States Department of Agriculture and by the Utah Agricultural Experiment Station, Utah State University, Logan.
Footnotes
Published ahead of print on 5 March 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
Approved as Utah Agricultural Experiment Station journal paper 8129.
REFERENCES
- 1.Abe, H. 2000. Role of histidine-related compounds as intracellular proton buffering constituents in vertebrate muscle. Biochemistry 65:757-765. [PubMed] [Google Scholar]
- 2.Azcarate-Peril, M. A., O. McAuliffe, E. Altermann, S. Lick, W. M. Russell, and T. R. Klaenhammer. 2005. Microarray analysis of a two-component regulatory system involved in acid resistance and proteolytic activity in Lactobacillus acidophilus. Appl. Environ. Microbiol. 71:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 4.Booth, I. R. 1985. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 49:359-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brockmann-Gretza, O., and J. Kalinowski. 2006. Global gene expression during stringent response in Corynebacterium glutamicum in presence and absence of the rel gene encoding (p)ppGpp synthase. BMC Genomics 7:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, H., B. T. Rodríguez, W. Zhang, J. R. Broadbent, and J. L. Steele. 2007. Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiology 153:2655-2665. [DOI] [PubMed] [Google Scholar]
- 7.Cai, H., R. Thompson, M. Budinich, J. R. Broadbent, and J. L. Steele. 2009. Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol. Evol. 1:239-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter, C. E., and J. R. Broadbent. 2009. External concentration of organic acid anions and pH: key independent variables for studying how organic acids inhibit growth of bacteria in mildly acidic foods. J. Food Sci. 74:R12-R15. [DOI] [PubMed] [Google Scholar]
- 9.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 10.Chang, Y. Y., and J. E. Cronan, Jr. 1999. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol. Microbiol. 33:249-259. [DOI] [PubMed] [Google Scholar]
- 11.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Sharoud, W. M. 2005. Two-component signal transduction systems as key players in stress responses of lactic acid bacteria. Sci. Prog. 88:203-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster, J. W. 1995. Low pH adaptation and the acid tolerance response of Salmonella typhimurium. Crit. Rev. Microbiol. 21:215-237. [DOI] [PubMed] [Google Scholar]
- 15.Fozo, E. M., J. K. Kajfasz, and R. G. Quivey, Jr. 2004. Low pH-induced membrane fatty acid alterations in oral bacteria. FEMS Microbiol. Lett. 238:291-295. [DOI] [PubMed] [Google Scholar]
- 16.Fozo, E. M., and R. G. Quivey, Jr. 2004. Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl. Environ. Microbiol. 70:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia, M. J., M. Zúñiga, and H. Kobayashi. 1992. Energy production from l-malic acid degradation and protection against acidic external pH in Lactobacillus plantarum CECT 220. J. Gen. Microbiol. 138:2519-2524. [Google Scholar]
- 18.Goldin, B. R., and S. L. Gorbach. 1992. Probiotics for humans, p. 355-376. In R. Fuller (ed.), Probiotics: the scientific basis. Chapman & Hall, London, England.
- 19.Goodson, M., and R. J. Rowbury. 1989. Habituation to normal lethal acidity by prior growth of Escherichia coli at a sub-lethal acid pH. Lett. Appl. Microbiol. 8:77-79. [Google Scholar]
- 20.Hall, H. K., K. L. Karen, and J. W. Foster. 1995. Molecular responses of microbes to environmental pH stress. Adv. Microb. Physiol. 37:229-272. [DOI] [PubMed] [Google Scholar]
- 21.Hamilton, I. R., and G. Svensäter. 1998. Acid-regulated proteins induced by Streptococcus mutans and other oral bacteria during acid shock. Oral Microbiol. Immunol. 13:292-300. [DOI] [PubMed] [Google Scholar]
- 22.Hutkins, R. W., and N. L. Nannen. 1993. pH homeostasis in lactic acid bacteria. J. Dairy Sci. 76:2354-2365. [Google Scholar]
- 23.Ingham, S., J. Hassler, Y. Tsai, and B. H. Ingham. 1998. Differentiation of lactate-fermenting, gas-producing Clostridium spp. isolated from milk. Int. J. Food Microbiol. 43:173-183. [DOI] [PubMed] [Google Scholar]
- 24.Kandler, O., and N. Weiss. 1986. Genus Lactobacillus, p. 1209-1234. In P. H. A. Sheath, N. S. Maiz, M. E. Sharp, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 25.Kashket, E. R. 1987. Bioenergetics of lactic acid bacteria: cytoplasmic pH and osmotolerance. FEMS Microbiol. Rev. 46:233-244. [Google Scholar]
- 26.Labarre, C., C. Diviès, and J. Guzzo. 1996. Genetic organization of the mle locus and identification of a mleR-like gene from Leuconostoc oenos. Appl. Environ. Microbiol. 62:4493-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert, R. J., and M. Stratford. 1999. Weak-acid preservatives: modeling microbial inhibition and response. J. Appl. Microbiol. 86:157-164. [DOI] [PubMed] [Google Scholar]
- 28.Lazazzera, B. A. 2001. The intracellular function of extracellular signaling peptides. Peptides 22:1519-1527. [DOI] [PubMed] [Google Scholar]
- 29.Len, A. C., D. W. Harty, and N. A. Jacques. 2004. Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiology 150:1353-1366. [DOI] [PubMed] [Google Scholar]
- 30.Lorca, G. L., and G. Font de Valdez. 2009. Lactobacillus stress responses, p. 115-137. In A. Ljungh, and T. Wadstrőm (ed.), Lactobacillus molecular biology: from genomics to probiotics. Calister Academic Press, Norfolk, United Kingdom.
- 31.Ma, Y., T. M. Curran, and R. E. Marquis. 1997. Rapid procedure for acid adaptation of oral lactic-acid bacteria and further characterization of the response. Can. J. Microbiol. 43:143-148. [DOI] [PubMed] [Google Scholar]
- 32.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J.-H. Lee, I. Díaz-Muñiz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, L. McKay, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nascimento, M. M., J. A. Lemos, J. Abranches, R. B. Gonçalves, and R. A. Burne. 2004. Adaptive acid tolerance response of Streptococcus sobrinus. J. Bacteriol. 186:6383-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poolman, B., D. Molenaar, E. J. Smid, T. Ubbink, T. Abee, P. P. Renault, and W. N. Konings. 1991. Malolactic fermentation: electrogenic malate uptake and malate/lactate antiport generate metabolic energy. J. Bacteriol. 173:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potrykus, K., and M. Cashel. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:35-51. [DOI] [PubMed] [Google Scholar]
- 36.Rallu, F., A. Gruss, S. D. Ehrlich, and E. Maguin. 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35:517-528. [DOI] [PubMed] [Google Scholar]
- 37.Renault, P., C. Gaillardin, and H. Heslot. 1988. Role of malolactic fermentation in lactic acid bacteria. Biochimie 70:375-379. [DOI] [PubMed] [Google Scholar]
- 38.Rizzo, A. F., H. Korkeala, and I. Mononen. 1987. Gas chromatography analysis of cellular fatty acids and neutral monosaccharides in the identification of lactobacilli. Appl. Environ. Microbiol. 53:2883-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rudd, K. E., B. R. Bochner, M. Cashel, and J. R. Roth. 1985. Mutations in the spoT gene of Salmonella typhimurium: effects on his operon expression. J. Bacteriol. 163:534-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell, J. B. 1991. Resistance of Streptococcus bovis to acetic acid at low pH: relationship between intracellular pH and anion accumulation. Appl. Environ. Microbiol. 57:255-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell, J. B. 1992. Another explanation for the toxicity of fermentation acids at low pH; anion accumulation verses uncoupling. J. Appl. Microbiol. 73:363-370. [Google Scholar]
- 42.Scholtens, D., and A. von Heydebreck. 2005. Analysis of differential gene expression studies, p. 229-248. In R. Gentleman, V. Carey, W. Huber, R. Irizarry, and S. Dudoit (ed.), Bioinformatics and computational biology solutions using R and Bioconductor. Springer Science+Business Media, Inc., New York, NY.
- 43.Shabala, L., T. McMeekin, B. B. Budde, and H. Siegumfeldt. 2006. Listeria innocua and Lactobacillus delbrueckii subsp. bulgaricus employ different strategies to cope with acid stress. Int. J. Food Microbiol. 110:1-7. [DOI] [PubMed] [Google Scholar]
- 44.Sheng, J., and R. E. Marquis. 2007. Malolactic fermentation by Streptococcus mutans. FEMS Microbiol. Lett. 272:196-201. [DOI] [PubMed] [Google Scholar]
- 45.Siegumfeldt, H., K. Björn Rechinger, and M. Jakobsen. 2000. Dynamic changes of intracellular pH in individual lactic acid bacterium cells in response to a rapid drop in extracellular pH. Appl. Environ. Microbiol. 66:2330-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smeianov, V. V., P. Wechter, J. R. Broadbent, J. E. Hughes, B. Rodriguez, T. K. Christensen, Y. Ardo, and J. L. Steele. 2007. Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium. Appl. Environ. Microbiol. 73:2661-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stiles, M. E., and W. H. Holzapfel. 1997. Lactic acid bacteria of foods and their current taxonomy. Int. J. Food Microbiol. 36:1-29. [DOI] [PubMed] [Google Scholar]
- 48.Sturr, M. G., and R. E. Marquis. 1992. Comparative acid tolerances and inhibitor sensitivities of isolated F-ATPases of oral lactic acid bacteria. Appl. Environ. Microbiol. 58:2287-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suutari, M., and S. Laakso. 1992. Temperature adaptation in Lactobacillus fermentum: interconversions of oleic, vaccenic and dihydrosterulic acids. J. Gen. Microbiol. 138:445-450. [DOI] [PubMed] [Google Scholar]
- 50.Svensäter, G., U. B. Larsson, E. C. Greif, D. G. Cvitkovitch, and I. R. Hamilton. 1997. Acid tolerance response and survival by oral bacteria. Oral Microbiol. Immunol. 12:266-273. [DOI] [PubMed] [Google Scholar]
- 51.van de Guchte, M., P. Serror, C. Chervaux, T. Smokvina, S. D. Ehrlich, and E. Maguin. 2002. Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82:187-216. [PubMed] [Google Scholar]
- 52.Vijayakumar, J., R. Aravindan, and T. Viruthagiri. 2008. Recent trends in the production, purification and application of lactic acid. Chem. Biochem. Eng. Q. 22:245-264. [Google Scholar]
- 53.Wardle, S. J., M. O'Carroll, K. M. Derbyshire, and D. B. Haniford. 2005. The global regulator H-NS acts directly on the transpososome to promote Tn10 transposition. Genes Dev. 19:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, Y.-M., and C. O. Rock. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6:222-233. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, Y.-M., and C. O. Rock. 2009. Transcriptional regulation in bacterial membrane lipid synthesis. J. Lipid Res. 50(Suppl.):S115-S119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.