Abstract
Previous studies have demonstrated that various tricarboxylic acid (TCA) cycle genes, particularly the succinate dehydrogenase genes (sdhCAB), are upregulated in Staphylococcus aureus biofilms. To better study the role of this enzyme complex, an sdhCAB deletion mutant (Δsdh) was constructed. Compared to the wild type (wt) the mutant was impaired in planktonic growth under aerobic conditions, excreted acetic acid could not be reused and accumulated continuously, succinate was excreted and found in the culture supernatant, and metabolome analysis with cells grown in chemically defined medium revealed reduced uptake/metabolism of some amino acids from the growth medium. Moreover, the mutant was able to counteract the steadily decreasing extracellular pH by increased urease activity. The addition of fumarate to the growth medium restored the wt phenotype. The mutant showed a small-colony variant (SCV)-like phenotype, a slight increase in resistance to various aminoglycoside antibiotics, and decreased pigmentation. The decreased growth under aerobic conditions is due to the interruption of the TCA cycle (indicated by the accumulation of succinate and acetic acid) with the consequence that many fewer reduction equivalents (NADH and FADH2) can fuel the respiratory chain. The results indicate that the TCA cycle is required for acetate and amino acid catabolism; its upregulation under biofilm conditions is advantageous under such nutrient- and oxygen-limited conditions.
Staphylococcus aureus is not only a major cause of acute nosocomial and community-acquired infections (1, 25, 43), it also causes chronic infections associated with medical devices due to its biofilm-forming ability (11, 14, 29). In persisting and recurrent S. aureus infections slow-growing subpopulations, called small-colony variants (SCVs), can be frequently isolated (19, 31, 47). SCVs are distinguished by reduced respiratory activity and toxin production, a higher tolerance to aminoglycoside antibiotics, and a longer intracellular persistence. These and very likely other properties are responsible for their frequent occurrence in latent and recurrent infections as well as in device-related infections (3, 36, 37, 50).
When grown in a biofilm population (14), staphylococci must adapt to rapid changes in the environment that require equally rapid changes in the demand for energy and biosynthetic intermediates necessary for bacterial growth and survival. Many environmental and nutritional signals alter the bacterial metabolic status and also regulate most virulence determinants in staphylococci (40). The tricarboxylic acid (TCA) cycle plays a central role in metabolism. Many nutrients, like sugars, amino acids, and fatty acids, can be metabolized into TCA intermediates and enter this cycle at several points; also, intermediates can be removed from the cycle for use in biosynthetic pathways. Additionally, the TCA cycle produces reducing equivalents that generate energy through the respiratory chain when a terminal electron acceptor is present. TCA cycle activity has been associated with different effects like survival, virulence, and the production of the main biofilm slime substance, polysaccharide intercellular adhesin (PIA) (39, 51). Furthermore, different TCA cycle enzymes have been found to be upregulated in biofilm-grown cells compared to planktonically cultivated cells (33), but only the deletion of TCA cycle enzyme genes (acn, citZ, and citC) of the oxidative branch of the TCA cycle in S. aureus have been described in the literature (39; G. Somerville, personal communication).
Interruption of the respiratory chain leads to an electron transfer chain-deficient small-colony variant phenotype. Clinically occurring SCV strains require a specific substance, like hemin, menadione, and thymidine, to restore wild-type (wt) growth rates (26, 30, 31). Genetically stable mutants in the hemin biosynthetic gene, hemB, or menadione biosynthetic gene, menD, have been characterized as having lower capacities for oxidative phosphorylation. They revealed many phenotypic changes (e.g., decreased pigmentation) and unusual biochemical characteristics as consequences of altered metabolic and energy-dependent pathways (4, 5, 21, 38, 46, 48, 49).
In a previous study, it was found that some TCA cycle genes were upregulated in biofilms, particularly the succinate dehydrogenase genes (sdhCAB) but also the 2-oxoglutarate dehydrogenase (odhA) and the succinyl-coenzyme A (CoA) synthetase (sucC) genes (32, 33). The succinate dehydrogenase (EC 1.3.99.1) is part of the nonoxidative branch of the TCA cycle and is directly linked to the respiratory chain. This enzyme complex catalyzes the oxidation of succinate to fumarate, donating FADH2 for oxidative phosphorylation. It consists of three subunits: membrane-bound cytochrome b558 (SdhC), a flavoprotein containing an FAD binding site (SdhA), and an iron-sulfur protein showing a binding region signature of the 4Fe-4S type (SdhB) (15, 16).
In this study we characterized an sdh deletion mutant of S. aureus. The results suggest that upregulation of Sdh and other TCA cycle enzymes is advantageous under biofilm conditions, where nutrients and oxygen are limited. Increased TCA cycle activity provides optimal growth, excreted acetic and lactic acids as well as medium amino acids can be better consumed, oxygen consumption rate (respiration) with succinate is optimal, and a too-high acidification of the medium is circumvented by a better reconsumption of the fermentation products (increase of the urea cycle as a bypass is dispensable).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The following strains were used: Escherichia coli strain XL1-Blue (Stratagene), Staphylococcus aureus SA113, a derivative of NCTC8325 (18), HG001, also a derivative of NCTC8325 (RN1) but in which the rsbU mutation has been repaired (S. Herbert et al., submitted for publication), and S. aureus strain RN4220 (23). For DNA preparation and transformation, the strains were grown in basic medium (BM; 1% soy peptone A3 SC [Organotechnie], 0.5% yeast extract, 0.5% NaCl, 0.1% K2HPO4·3H2O, 0.1% glucose [pH 7.2 to 7.4]) or on BM plates containing 1.5% agar. Antibiotics were used, when appropriate, at the following concentrations: ampicillin at 100 μg/ml for E. coli, and erythromycin (2.5 μg/ml), spectinomycin (150 μg/ml), and chloramphenicol (10 μg/ml) for S. aureus.
Construction of the S. aureus sdhCAB deletion mutant in SA113 and construction of complementation plasmid pRBsdh.
Primers sdhCAB_B_for (5′-TATTAGGTACCGGTTCAGACCATGAAGTAG-3′; the introduced restriction site is underlined) and sdhCAB_B_rev (5′-ATTATTCTAGACCCTTGATTATTCGATGC-3′) were used to amplify the downstream flanking region (∼1.1 kb) of sdhCAB from the chromosomal DNA of S. aureus SA113. The PCR product was ligated into the Acc65I/XbaI sites of pKO2 (a derivative of pBT2 [8], constructed by B. Krismer), creating plasmid pKO22. Primers sdhCAB_A_for (5′-TATAATGAGCTCGAGTCCAGAACGATTAAAGC-3′) and sdhCAB_A_rev (5′-AATAAAAGCTTCGCTTAACCCCCTAAAGTG-3′) were used to PCR amplify the upstream flanking region (∼0.96 kb) of sdhCAB. The restricted PCR product and the spectinomycin cassette from pIC156 (HindIII/Acc65I) (42) were cloned into the SacI/Acc65I sites of pKO22, yielding pKO21spec2. All recombinant plasmids were introduced into E. coli XL1-Blue. Plasmids were introduced first into S. aureus RN4220 and then into S. aureus SA113 via electroporation (2). Allelic replacement of wild-type sdhCAB genes with spc was carried out as described previously by Brückner (8). The replacement of the genes in the correct manner to the resulting strain, S. aureus sdhCAB::spc(Δsdh), was confirmed based on the antibiotic resistance phenotype, PCR, DNA sequence analyses, and enzymatic assays.
The S. aureus sdhCAB genes (based on the sequence of S. aureus NCTC8325; gene ID SAOUHSC_01103-01105) with their putative native promoter were amplified from chromosomal DNA of S. aureus SA113 by PCR using the primers sdhCAB_C_for (5′-TTATAGAGCTCGAAGGAAGCTACACTTGAAG-3′) and sdhCAB_C_rev (5′-TTATTAAGCTTCCAAGTTATTGTGTATCTCCATTC-3′). The resulting PCR product was cloned into SacI/HindIII sites of the medium-copy-number plasmid pRB473 (9), generating plasmid pRBsdh. The plasmid was introduced into E. coli XL1-Blue and transferred into S. aureus Δsdh by electroporation to complement the mutant. The PCRs were carried out with High-Fidelity PCR enzyme mix (Fermentas); used primers were obtained from biomers.net. Computer sequence analyses were performed with MacDNASIS Pro (Hitachi Software Engineering).
Creation of an sdhCAB deletion mutant in HG001 by phage transduction.
The sdhCAB::spc region of the SA113Δsdh mutant was transduced into HG001 by phage transduction using φ11. The HG001 sdhCAB::spc mutant was verified by PCR and DNA sequencing. This mutant was also complemented with the plasmid pRBsdh.
Phage transduction.
For generating phage φ11 lysates, the staphylococcal donor strains were resuspended in 500 μl phage buffer (1 mM MgSO4, 4 mM CaCl2, 0.59% NaCl, 0.1% gelatin, 50 mM Tris-HCl, pH 7.8), infected with φ11 (50 μl), and grown for 5 to 6 h in 4 ml soft tryptic soy agar (TSA) top agar (tryptic soy broth [TSB] and 0.5% agar) on CaCl2-agar plates (TSB, 0.5 mM CaCl2, 1.5% agar). Then, the soft TSA top agar was resuspended in 4 ml TSB, warmed for 10 min at 50°C, and sterilized by passage through 0.45-μm-pore-size filters after centrifugation (5,000 × g, 10 min, 4°C). For transduction, S. aureus cells from a fresh overnight plate were resuspended in 0.2 ml phage buffer, infected with φ11 lysates (100 μl), and incubated for 10 min at 37°C. Afterwards, 3.5 ml of phage top agar (3 g/liter Casamino Acids, 3 g/liter yeast extract, 5.9 g/liter NaCl, 7.5 g/liter agar) was added and poured on TSA-agar plates supplemented with appropriate antibiotic (2.5 μg/ml erythromycin, 150 μg/ml spectinomycin).
Enzymatic assays.
TSB (Sigma) was inoculated from overnight cultures to an optical density at 578 nm (OD578) of 0.07. Six-well cell culture plates (Greiner) were filled with bacterial suspension and incubated under static conditions at 37°C for 24 h. Then the cells were scraped off the plates, harvested by centrifugation, washed, resuspended in buffer (20 mM Tris-HCl, pH 8), and disrupted by shaking with glass beads in a tissue lyser (Qiagen). After a first centrifugation step (15 min, 2,000 × g, 4°C), the supernatants were ultracentrifuged at 30,000 rpm for 1 h at 4°C. The membrane fractions were washed and resuspended, and protein concentrations were determined by the method of Bradford (7). Different protein patterns in Coomassie brilliant blue-stained SDS-PAGE gels according to the method of Schägger and von Jagow (35) indicated successful separation of cytoplasmic and membrane fractions. To reaction tubes containing 100-μl aliquots of diluted membrane fractions, 300 μl of reaction mixture (each at 1 part: 0.2 M sucrose, 0.1 M Tris-HCl [pH 7.5], 10 mM sodium azide, 8 mM iodonitrotetrazolium chloride; 2 parts H2O) was added. The reaction was started by addition of 0.5 M sodium succinate, pH 7.5 (100 μl) and incubated at 37°C, and when a change in color was visible 1.5 ml of ethanol was added and well mixed. The samples were incubated on ice for 10 min and centrifuged (10 min, 800 rpm), and the absorption at 458 nm was determined. Samples that either exchanged the enzyme fraction or the substrate against buffer served as controls. The specific activity was calculated, and the wild-type activity was set as 100%.
Growth in TSB and determination of extracellular metabolites.
For growth analysis, TSB overnight cultures were diluted to an OD578 of 0.07 (∼1:7 culture-to-flask volume ratio) and incubated at 37°C and 150 rpm; no antibiotics were added. CFU were determined by a modified drop plate method (17). For determination of metabolites the culture supernatant was heated to 80°C for 15 min and then stored at −20°C until further analysis. Glucose, acetate, d/l-lactate and succinate were determined enzymatically (R-Biopharm) in flat-bottom 96-well microtiter plates, and the A340 was determined in a microtiter plate reader (SpectraMax 340; Molecular Devices); the metabolite concentration was calculated from a standard curve.
Growth in CDM and determination of extracellular metabolites.
Nuclear magnetic resonance (NMR) analysis of extracellular metabolites is more reliable in a chemically defined medium (CDM). The used CDM consisted of 10 mM Na2HPO4, 10 mM KH2PO4, 0.5 g/liter NH4Cl, 0.5 g/liter NaCl, 0.2 g/liter MgSO4·7H2O, 0.14 mM trisodium citrate dihydrate, and 7.47 mM glucose. This was supplemented with amino acids (l-alanine, l-arginine, l-aspartic acid, l-cysteine, l-glutamic acid, l-histidine, l-isoleucine, l-leucine, l-lysine, l-phenylalanine, l-proline, l-serine, l-threonine, l-tryptophan, l-valine; all at 1 mM), the vitamins cyanocobalamine (0.05 μg/ml), 4-aminobenzoic acid (0.04 μg/ml), biotin (0.01 μg/ml), 0.1 μg/ml nicotinic acid, 0.1 μg/ml Ca-d-pantothenic acid, 0.16 μg/ml pyridoxamine dihydrochloride, 0.1 μg/ml thiamine hyrdochloride, and 0.1 μg/ml riboflavine, and with 0.75 μM FeCl3, 0.069 μM ZnCl2, 0.099 μM MnCl2, 0.006 μM boric acid, 0.35 μM CoCl2, 0.002 μM CuCl2, 0.24 μM NiCl2, and 0.036 μM Na2MoO4.
For growth analysis CDM overnight cultures were diluted to an OD578 of 0.2 (∼1:7 culture-to-flask volume ratio) and incubated at 37°C and 100 rpm; no antibiotics were added. After 4 h, 6 h, 9 h, and 24 h samples were taken and filter sterilized, the resulting supernatants were stored at −20°C. The metabolites were analyzed with 1H-NMR as described elsewhere (24).
Urease activity.
Cells were grown in either TSB or CDM supplemented with 100 mM sodium fumarate as described above. After different times the cells were pelleted, washed, resuspended in 50 mM potassium phosphate buffer (pH 8), and disrupted by shaking with glass beads in a tissue lyser (Qiagen). After centrifugation (0.5 min, 13,500 rpm) the protein concentrations of the cell lysates were determined by the method of Bradford (7). Two different protein concentrations of each sample were used for urease activity determination. Cell lysate and buffer were mixed to a final volume of 800 μl in 1.5-ml reaction tubes. Incubations (30 min to 4 h) were started by the addition of 200 μl of 250 mM urea to the reaction tubes. The reaction was stopped by heat inactivation at 95°C for 1.5 min, and time zero samples (t0) were treated the same way and used as background controls. Ammonium was determined by using Nessler reagent.
Construction of hemB and menD mutants in S. aureus SA113.
The hemB::ermB mutation region in S. aureus 8325-4 (48) and the menD::ermC allele of S. aureus DB24 (4) were transduced with the help of phage φ11 into SA113. The successful integration of the erythromycin resistance cassettes in the two SA113 mutants was verified by PCR.
Colony size on Mueller-Hinton agar plates.
Flasks filled with fresh Mueller-Hinton broth (MH; Applichem) were inoculated with overnight cultures to an OD578 of 0.07, grown to mid-exponential phase, and plated onto MH agar plates. Growth was assayed with and without supplementation of either glucose (5 mM), sodium fumarate (5 mM), or sodium succinate (20 mM). The plates were incubated at 37°C and checked with respect to colony size and pigmentation.
Pigment extraction.
S. aureus produces staphyloxanthin and diaponeurosporen as the main carotenoids (10, 28). Strains were cultivated for 24 h in TSB, and the pelleted cells were treated with ethanol (2.4 μl per mg [wet weight] of cells) and incubated for 20 min at 40°C in the dark. Cells were pelleted, and the majority of the pigments was in the ethanol phase, which was used to record an absorption spectrum (range, 350 to 550 nm). The absorption maxima of staphyloxanthin and intermediary products are between 450 and 470 nm, depending on the oxidation state of the carotenoids.
Determination of oxygen uptake.
For determination of oxygen uptake, S. aureus clones were grown in TSB for 20 to 24 h. Cells were centrifuged, washed three times, and resuspended in 33 mM potassium phosphate buffer (pH 7.0) to an OD578 of 20. This suspension (1.8 ml) was analyzed for oxygen consumption using a Clark-type oxygen electrode. Respiration was initiated by the addition of either 100 mM succinate or 1 mM glucose as electron donor. The basal oxygen consumption rate (in the absence of electron donor) was subtracted. The oxygen consumption rate of the wt was set as 100%.
Susceptibility to antibiotics.
Antibiotic susceptibility was determined in two ways: (i) agar diffusion assay and (ii) microdilution method. In the agar diffusion assay the following antibiotics (and concentrations) were tested: gentamicin, tobramycin, and kanamycin (10 μg, 5 μg, 2.5 μg, 1.25 μg, and 0.625 μg), chloramphenicol (10 μg), erythromycin (5 μg), fusidic acid (10 μg), mupirocin (5 μg), novobiocin (5 μg), ofloxacin (5 μg), oxacillin (1 μg), penicillin (1 μg), rifampin (5 μg), tetracyclin (5 μg), trimethoprim (2.5 μg), and the combination of trimethoprim (1.25 μg) and sulfamethoxazole (23.75 μg). For the aminoglycosides gentamicin, tobramycin, and kanamycin, the MICs were additionally determined by the microdilution method.
Statistical analyses.
For determination of P values, paired Students t tests were performed.
RESULTS
Construction and characterization of the Δsdh mutant.
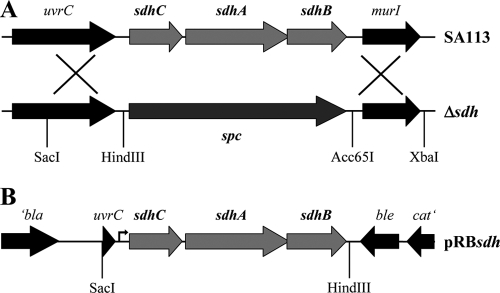
In S. aureus succinate dehydrogenase (Sdh) of the TCA cycle shows the typical subunit organization of SdhA, -B, and -C. As Sdh is involved in the electron transfer of the respiratory chain, it is postulated that the enzyme is membrane associated. The corresponding genes are organized in an operon that in S. aureus SA113 was completely deleted and replaced by a spectinomycin resistance cassette (Fig. 1A). The ΔsdhCAB mutant (designated herein as the Δsdh mutant) showed no Sdh activity in either the cytoplasm or membrane fraction, indicating that no other Sdh-like enzyme is present, while both SA113 and the complemented mutant showed activity in the membrane fraction (data not shown). For complementation, plasmid pRBsdh was constructed (Fig. 1B). However, the plasmid was not able to fully restore the Δsdh mutant, although it encodes the structural genes sdhCAB together with a 408-bp upstream region with the putative native promoter region. To rule out a potential influence of second-site mutations in Δsdh, the sdhCAB::spc allele was transduced into HG001 as an alternative strain; this strain has never undergone chemical mutagenesis and, in contrast to SA113, the global regulators Agr and RsbU (SigB) are functional. However, most of the experiments were carried out with SA113.
FIG. 1.
Construction of the succinate dehydrogenase deletion mutant and complementation. (A) Location of the sdh operon in the chromosome of SA113 (upper panel) and the sdhCAB deletion mutant (Δsdh; lower panel). (B) Gene arrangement in the complementation plasmid pRBsdh, including the putative native promoter region upstream (arrow). Plasmid-encoded genes: bla, beta-lactamase gene; ble, bleomycin resistance gene; cat, chloramphenicol resistance gene; uvrC, 3′ end of the sdh upstream gene.
Growth and pH profile.
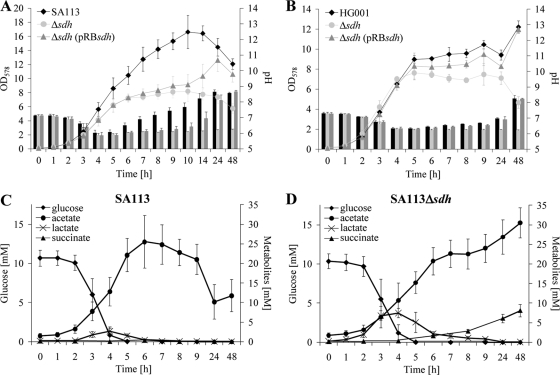
Following growth in TSB medium under aerobic conditions (vigorous shaking) for over 48 h, one can see that SA113Δsdh reached only half the OD values as SA113 (Fig. 2A). Also interesting was the course of the extracellular pH values. With SA113, the pH decreased steadily from 7.2 to 6.0 to reach a minimum at the end of the exponential growth phase, and then the pH values climbed to 8.4 in the late stationary growth phase. The pH profile of the mutant was significantly different. Like SA113, it decreased steadily from 7.2 to 6.0, but the pH remained at this level. After some delay, the complemented mutant reached the wt phenotype; however, the tendency can be clearly seen in later growth stages, where both pH and growth increased as in SA113 (Fig. 2A). A comparison of growth curves and pH courses revealed some differences in SA113 and HG001 (Fig. 2A and B). SA113 reached a higher OD value than HG001, while on the other hand HG001 grew faster (the generation time was only 46 ± 3 min, compared to 59 ± 7 min in SA113). The onset of alkalization of the culture supernatant was earlier in SA113 than in HG001. These differences very likely reflect the influence of the global regulators. Despite these general differences in parent strains, the corresponding Δsdh mutants were impaired in growth and realkalization of the culture medium. In both Δsdh mutants the complementing plasmid pRBsdh was unable to fully restore the wt phenotype, ruling out a side effect by point mutations in SA113Δsdh.
FIG. 2.
Comparison of growth parameters and extracellular metabolites in S. aureus and the Δsdh mutant. (A and B) Growth curves and pH profile (bars) of SA113 (A) and HG001 (B). (C and D) Metabolite concentrations in the culture supernatant of SA113 (C) and its Δsdh mutant (D). Strains were aerobically cultivated in TSB at 37°C. Error bars indicate standard deviations from the means of three experiments.
Extracellular metabolites.
To obtain more information regarding the metabolic state of the cells, we also determined the concentrations of extracellular metabolites during growth of SA113 and its isogenic Δsdh mutant. In TSB, all strains consumed glucose within the first 5 h. Glucose consumption was correlated with an increase of acetate and lactate in both strains (Fig. 2C and D). The major difference was that in SA113 acetate is catabolized after prolonged growth, while the mutant was unable to utilize acetate that accumulated further to finally reach a concentration of 30 mM. The complemented mutant showed a similar acetate course as the wt. In contrast to SA113 and SA113Δsdh(pRBsdh), SA113Δsdh accumulated succinate in the culture supernatant, which was expected, as Sdh is missing in the mutant. In both SA113 and SA113Δsdh, maximal lactate concentrations in the supernatants were reached after 4 h and declined gradually with time. When cultivated in TSB under aerobic conditions, the wt reached maximal lactic acid content after 4 h of incubation with 1.4 ± 0.3 mM d-lactic acid and 1.3 ± 0.5 mM l-lactic acid; obviously, lactate production is counteracted by lactate reoxidation. The SA113Δsdh mutant reached a much higher lactic acid peak (2.9 ± 0.5 mM d-lactate and 5.0 ± 0.6 mM l-lactate). After 4 h the complemented mutant [SA113Δsdh(pRBsdh)] was not yet able to reach the wt pattern; it still revealed a similar high lactic acid content as the mutant (data not shown).
By 1H-NMR measurements of extracellular medium, the number of small-molecule metabolites could be extended. As such studies were not possible in detail with a complex medium like TSB, we used a CDM without interfering peptides or other macromolecules, as are included in TSB. Growth in this medium was poor for both SA113 and the mutant; a maximal OD578 of only 2.2 could be reached. In contrast to TSB, all strains grown in CDM maintained low extracellular pH values after the exponential growth phase (data not shown). The extracellular concentrations after 24 h of some selected metabolites are shown in Table 1. After 9 h of cultivation under aerobic growth, glucose (8 mM) was entirely consumed, while citrate, the second carbon source in the chemically defined medium, was hardly degraded after 24 h (data not shown). In CDM there was no marked difference in acetate and lactate accumulation, which is different from the results obtained in TSB (Table 1); even SA113 was unable to reutilize acetate. Only the accumulation of succinate in the culture supernatant of the mutant was comparable with the results in TSB. Other typical fermentation products, such as acetoine, 2,3-butanediol, ethanol, and formate, were not altered in SA113Δsdh. The highest accumulated metabolite beside acetate was acetoine, with a concentration of circa 2 mM; there was no significant difference between SA113 and its mutant. In all strains traces of 2-oxoglutarate, fumarate, and ornithine as well as some amino acid degradation products (isovaleric acid, 2-methyl propionic acid, and 2-methyl butyric acid) were detected in similar amounts in the culture supernatant.
TABLE 1.
Accumulated metabolites in culture supernatant of CDM after 24 h
| Metabolite | Concn (mM) in culture supernatanta |
||
|---|---|---|---|
| SA113 | Δsdh | Δsdh (pRBsdh) | |
| Acetate | 10.7 ± 0.7 | 9.4 ± 0.23 | 10.7 ± 0.6 |
| Succinate | 0.0 ± 0.0** | 1.2 ± 0.2** | 0.2 ± 0.2 |
| Lactate | 0.4 ± 0.1 | 0.6 ± 0.1 | 0.4 ± 0.1 |
| Acetoine | 1.8 ± 0.1 | 2.1 ± 0.2 | 2.0 ± 0.1 |
Metabolite concentrations were determined in the culture supernatant of cultures grown aerobically in CDM for 24 h. Values are the averages of three independent experiments ± standard deviations. **, P < 0.05 for the Δsdh mutant versus SA113.
The CDM contained 15 amino acids, each at 1 mM. Cysteine, phenylalanine, and tryptophan concentrations stayed more or less constant in the culture supernatants, indicating that they were not metabolized. Histidine, lysine, valine, leucine, isoleucine, proline, alanine, arginine, aspartate, glutamate, serine, and threonine concentrations were decreased in the culture supernatants after 24 h. The SA113Δsdh mutant showed a decreased consumption of some amino acids, particularly of proline (Table 2).
TABLE 2.
Remaining amino acids in culture supernatant of CDM after 24 h
| Amino acid | Concn (%)a in culture supernatant |
||
|---|---|---|---|
| SA113 | Δsdh | Δsdh (pRBsdh) | |
| Alanine | 22.0 ± 4.7** | 47.4 ± 2.0** | 33.8 ± 4.0 |
| Glutamate | 19.1 ± 6.0** | 43.8 ± 3.3** | 30.0 ± 5.3 |
| Histidine | 76.5 ± 0.4* | 90.7 ± 7.1* | 83.9 ± 6.5 |
| Isoleucine | 30.4 ± 2.7* | 48.0 ± 5.4* | 40.5 ± 0.7 |
| Proline | 29.0 ± 14.6** | 91.4 ± 13.9** | 52.9 ± 12.9 |
Amino acid concentrations in the culture supernatant of cultures grown aerobically in CDM for 24 h. Of the 15 amino acids contained in CDM, the differentially consumed amino acids are listed. An initial amino acid concentration of 1 mM was set as 100%. Values are the averages of three independent experiments ± standard deviations. *, P < 0.1; **, P < 0.05 (Δsdh mutant versus SA113).
Stationary-phase survival assay.
Inactivation of the TCA cycle enzyme aconitase resulted in enhanced stationary-phase survival (39). To address the question of whether this also occurs in the Δsdh mutant we performed a long-term survival assay. SA113, its Δsdh mutant, and the complemented mutant were cultivated in TSB medium aerobically for 7 days, and the CFU were determined daily. In the course of this study, SA113Δsdh survived better than wt or the complemented mutant (Fig. 3). A similar effect was also seen with HG001 and its mutant (data not shown).
FIG. 3.
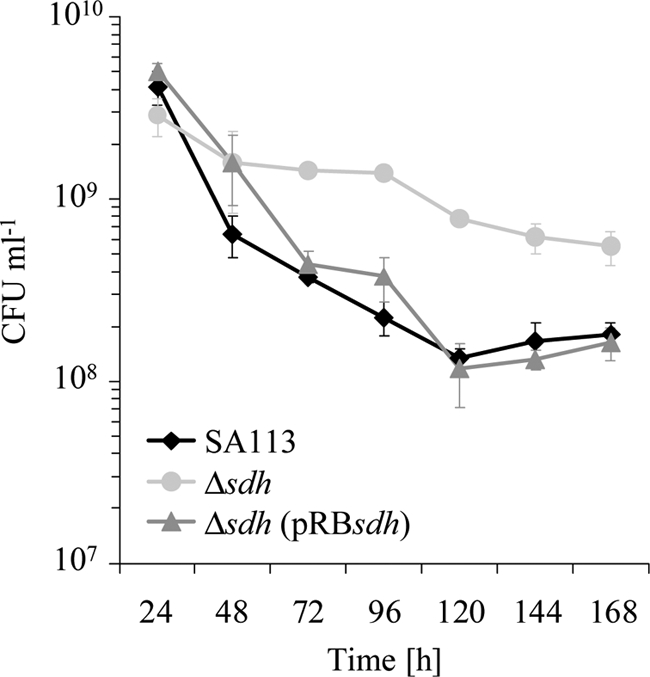
Long-term survival assay results. Strains were aerobically cultivated in TSB at 37°C for 7 days. Cultural aliquots were removed daily, spotted on TSB agar plates, and incubated at 37°C for 24 h. The values (CFU per milliliter) were determined by the drop plate method (17) in six replicates. Error bars indicate standard deviations from the means of five experiments.
Urease activity.
As mentioned above, in SA113Δsdh the decrease in medium pH corresponded with glucose consumption. While in the wt the pH increased with time, it stayed constant in the mutant, at approximately 6.2, after depletion of glucose (Fig. 2A, C, and D). This was astonishing, as in the mutant acetic acid constantly increased with time. During exponential growth, lactic acid was excreted and reused, very likely to form acetic acid via oxidation to pyruvate. In the course of this process, the medium pH should become more alkaline as the comparatively strong lactic acid (pKa 3.9) is replaced by the weaker acetic acid (pKa 4.8). Furthermore, the glucose (10 mM) in the medium allows theoretically only the formation of 20 mM acetate. As we found, however, of the 30 mM acetate produced, part of the acetate must be derived from pathways such as amino acid or fatty acid catabolism. The ammonia concentration in TSB culture supernatants of SA113 and its Δsdh mutant were increased only after 48 h (wt, 1.24 ± 0.29 mM, versus Δsdh, 2.99 ± 0.66 mM per OD578 unit of 1.0). In the complemented Δsdh mutant the ammonia concentration was between that of the wt and the mutant (1.85 mM ± 0.32 mM per OD578 unit of 1.0). One explanation for the continuous low pH levels in SA113Δsdh could be the upregulation of a pathway influencing medium pH. One promising candidate is urease, which produces ammonia and CO2 from urea as a substrate. Indeed, we found that in cell lysates of the mutant the urease activity was 2- to 5-fold increased compared to the wt. In the presence of fumarate, this effect was largely relieved. The enhanced urease activity in the mutant was more pronounced in TSB than in CDM (Table 3). The complemented mutant showed a wt phenotype when grown in CDM, but not in TSB.
TABLE 3.
Specific urease activity in cell lysates
| Medium | Culture time (h) | Urease sp act (mU/mg)a |
|||||
|---|---|---|---|---|---|---|---|
| Without fumarate |
With 100 mM fumarate |
||||||
| SA113 | Δsdh | Fold difference, Δsdh vs SA113 | SA113 | Δsdh | Fold difference, Δsdh vs SA113 | ||
| TSB | 3 | 1.8 ± 0.8 | 3.7 ± 1.9 | 2.2 ± 0.8 | 1.6 ± 0.7 | 2.9 ± 1.8 | 1.8 ± 0.4 |
| 5 | 2.4 ± 0.9 | 8.7 ± 1.6 | 4.0 ± 1.0 | 1.9 ± 0.4 | 5.5 ± 2.1 | 2.8 ± 0.6 | |
| 7 | 13.6 ± 7.3 | 50.4 ± 11.6 | 4.3 ± 1.7 | 9.9 ± 1.4 | 19.0 ± 6.8 | 1.9 ± 0.4 | |
| 9 | 14.5 ± 7.9 | 69.2 ± 16.1 | 5.3 ± 1.4* | 7.6 ± 1.3 | 20.5 ± 3.3 | 2.7 ± 0.1* | |
| 24 | 16.7 ± 8.8 | 59.6 ± 2.5 | 4.2 ± 1.7* | 7.4 ± 1.6 | 15.3 ± 2.2 | 2.1 ± 0.2* | |
| CDM | 7 | 5.4 ± 1.2 | 7.7 ± 1.5 | 1.5 ± 0.6 | 2.5 ± 0.9 | 3.5 ± 0.4 | 1.5 ± 0.4 |
| 9 | 6.2 ± 1.6 | 9.1 ± 0.7 | 1.5 ± 0.3 | 2.9 ± 1.0 | 3.7 ± 0.9 | 1.3 ± 0.1 | |
| 24 | 3.3 ± 0.6 | 8.6 ± 0.7 | 2.7 ± 0.3** | 6.8 ± 1.9 | 4.9 ± 0.8 | 0.7 ± 0.1** | |
Specific urease activities (with 1 U defined as 1 μmol NH4+ min−1 mg of protein−1) were determined in cell lysates from cultures grown in TSB and CDM either with or without addition of 100 mM fumarate. Values are the averages of three independent experiments ± standard deviations. *, P < 0.1; **, P < 0.05 (urease activity ratio of cells cultivated in medium without fumarate versus activity ratio of cells cultivated in medium with 100 mM fumarate added).
We assume that in Δsdh the urea cycle (all genes annotated in S. aureus NCTC8325) is more active. This makes sense, as the urea cycle produces fumarate and urea, thus closing the metabolic gap in the TCA cycle, and delivers at the same time the substrate for urease.
In the medium supplemented with fumarate (100 mM), urease activity was decreased in the Δsdh mutant (Table 3). Furthermore, the addition of fumarate led to markedly better growth of SA113Δsdh in TSB and to an increase in external pH values comparable to the wt without fumarate (data not shown). Taken together, we showed that the interruption of the TCA cycle in the Δsdh mutant led to an enhanced stationary-phase survival, an impaired catabolism of acetate and succinate as well as some amino acids, and a temporary shift to lactate fermentation. Moreover, the mutant revealed an increased urease activity, which could be indicative for an increased urea cycle.
SA113Δsdh has an SCV-like phenotype.
Due to the linkage of Sdh to the respiratory chain, we asked if the Δsdh mutant shows a SCV phenotype. As controls, we constructed hemB and menD insertion mutants in SA113 to generate stable SCVs. In these mutants the biosynthesis of hemin and menadione, respectively, is interrupted, which causes an inactivation of respiration. The two mutants showed the typical pinpoint colonies on agar plates, with colonies of the hemB mutant slightly smaller than colonies of the menD mutant. Both mutants also showed impaired growth in liquid medium under aerobic conditions. Supplementation of the culture medium for the hemB mutant with 1 μg/ml hemin restored wt growth, as previously shown (48).
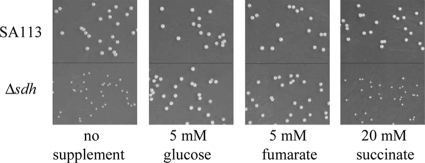
The SA113Δsdh mutant also displayed markedly smaller colonies than the wt on Mueller-Hinton agar plates with no glucose. While wt colonies showed yellowish pigmentation, the Δsdh mutant colonies appeared white. However, the small colonies and the decreased pigmentation of SA113Δsdh were restored to the wt phenotype by the addition of either glucose or fumarate to the MH medium. The supplementation of succinate had no influence, confirming the finding that the Δsdh mutant is unable to use succinate (Fig. 4). The colonies of the complemented mutant did not quite reach wild-type size (data not shown).
FIG. 4.
Size of colonies on Mueller-Hinton agar plates. The SA113Δsdh mutant displayed a SCV phenotype. Supplementation of the agar with 5 mM glucose or 5 mM sodium fumarate, but not with 20 mM sodium succinate, restored wild-type colony size.
Decreased pigmentation in the Δsdh mutants of SA113 and HG001.
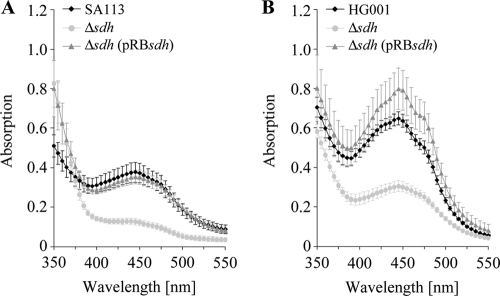
The Δsdh mutants of both strains produced much less carotenoids than the corresponding wt, as shown in the absorption spectra of crude ethanol extracts. Due to the rsbU defect in SA113, pigmentation in this strain was generally weaker than in HG001 (6). In both complemented mutants the wt carotenoid content was fully reached (Fig. 5).
FIG. 5.
Absorption spectrum of extracted pigments. SA113 (A) and HG001 (B) along with corresponding Δsdh mutants were aerobically cultivated for 24 h in TSB. Absorption spectra were determined in ethanol extracts (2.4 μl ethanol per mg [wet weight] of cells). Error bars indicate standard deviations from the means of four experiments.
Impairment of succinate-dependent oxygen consumption rate in SA113Δsdh.
We also compared the oxygen consumption rate of resting cell suspensions with a Clark-type oxygen electrode with either glucose or succinate as electron donor. With succinate, the oxygen consumption rate was strong and rapid in SA113, while in the mutant it was less than 10% (Fig. 6A). With glucose the oxygen consumption rate was even slightly higher with the Δsdh mutant, indicating that the general electron transfer was not affected in SA113Δsdh (Fig. 6B).
FIG. 6.
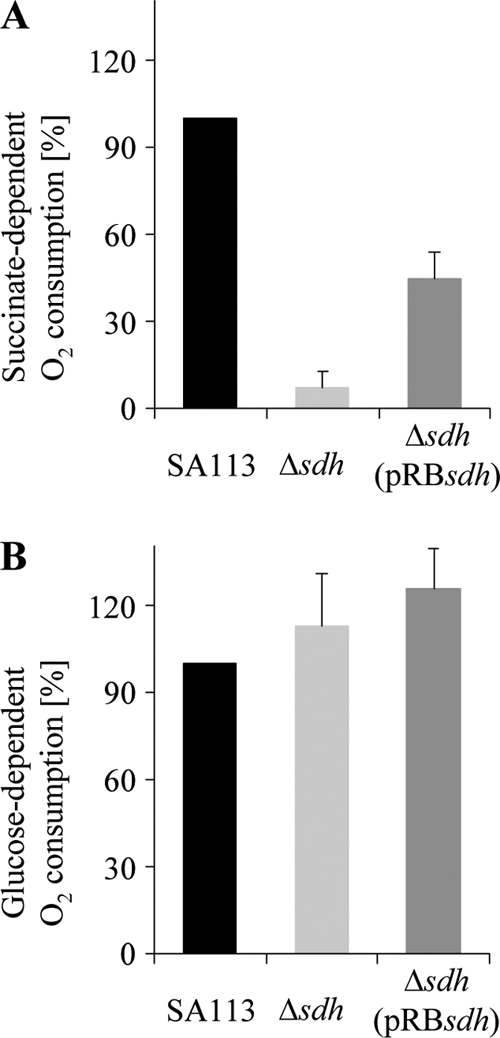
Polarographic measurement of the oxygen consumption rate, determined with a Clark-type oxygen electrode. SA113 and isogenic Δsdh mutants were grown aerobically for 20 to 24 h. Respiration of washed cell suspensions was initiated either with 100 mM succinate (A) or 1 mM glucose (B) as a substrate. The oxygen consumption rate of SA113 was set as 100%. Error bars indicate standard deviations from the means of three experiments.
Increased aminoglycoside antibiotic resistance.
The SA113Δsdh mutant was slightly (two to four times) more resistant to various aminoglycoside antibiotics, such as gentamicin, tobramycin, or kanamycin, than the wt. This is far below the high resistance that was observed with a hemB or menD mutants, for which the MIC values were increased 32- to 64-fold (Table 4). In summary, the deletion of sdh genes in S. aureus leads to some properties similar to electron-deficient SCVs, even though the effects are not so pronounced as in classic SCVs.
TABLE 4.
MICs of different aminoglycoside antibiotics
| Antibiotic | MIC (μg/ml) for strain |
||||||
|---|---|---|---|---|---|---|---|
| SA113 | Δsdh (sdhCAB::spc)a | Δsdh (pRBsdh)a | Δlip (lip::spc)a | Δfdh (fdh::spc)a | hemB | menD | |
| Gentamicin | 1 | 2-4 | 2-4 | 1 | 1 | 32 | 32 |
| Tobramycin | 0.5-1 | 2 | 2 | 0.5-1 | 0.5-1 | 32 | 32 |
| Kanamycin | 2-4 | 8 | 8 | 2-4 | 2-4 | 64-128 | 64-128 |
These strains contain the same spectinomycin resistance cassette on the chromosome. The deletion mutants for the lipase gene (SA113Δlip) and formate dehydrogenase gene (SA113Δfdh) were constructed by G. Thumm and M. Leibig in our laboratory.
DISCUSSION
In a staphylococcal biofilm, TCA cycle enzymes (32) and corresponding genes (33) are upregulated compared to levels under planktonic growth, which suggests that this pathway is important in biofilm formation. Among the genes which were upregulated 3- to 4-fold in biofilms were the three subunit genes of the succinate dehydrogenase complex (sdhCAB). Sdh is the only enzyme of the TCA cycle that can directly feed electrons to the respiratory chain. It was therefore of general interest what impact this enzyme of the nonoxidative part of the TCA cycle has on the physiology in staphylococci. This could best be seen when an sdhCAB deletion mutant of S. aureus was compared with the wt. The most eye-catching differences of the sdhCAB mutant under aerobic growth conditions in TSB were decreased growth, inability to reuse exported acetate, leading to its continuous accumulation, and accumulation of succinate. There was also transiently a higher amount of lactate exported than from the wt, but as with the wt, lactate could be reused and disappeared with time from the culture supernatant.
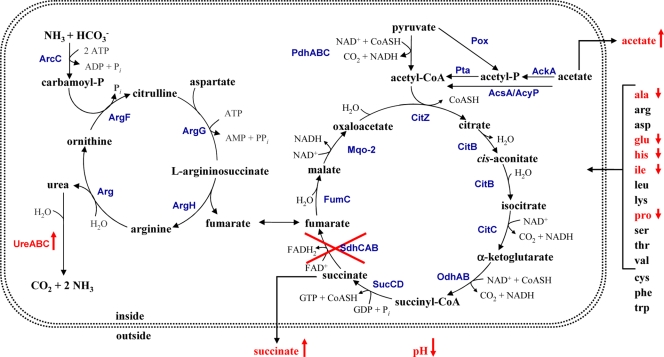
It is well known that in staphylococci under aerobic conditions glucose is oxidized in two steps. In the first step glucose is only partially oxidized to acetate, which accumulates in the supernatant; in the second step, when glucose is almost completely consumed, the excreted acetate disappears from the medium and is further oxidized by the TCA cycle (13, 39, 41, 44). As shown in Fig. 2C the consumption of 12.6 mM glucose was correlated with an accumulation of 25 mM acetate in the supernatant, indicating that almost all glucose is converted to acetate. The TCA cycle comes into play after glucose consumption. The decreased growth of the Δsdh mutant under aerobic conditions is due to the interruption of the TCA cycle (indicated by the further accumulation of acetate and succinate) with the consequence that much less reduction equivalents (NADH and FADH2) can fuel the respiratory chain. The glyoxylate pathway as a short-circuit of the TCA cycle also produces succinate, but there are no genomic or physiological hints for this pathway in S. aureus (51). As compensation for the limited reducing equivalents in the mutant, lactic acid production increased transiently, with a maximum after 4 h growth, when d- and l-lactic acids amounted to 3 mM and 5 mM, respectively (Fig. 2D). However, the excreted lactic acid can be reused by the mutant, and the question arises as to how this is done. It has been shown that l-lactate oxidation occurred via two terminal menaquinol oxidases, the ba3 type, which is sensitive to low KCN, and the bo type, which is insensitive to cyanide, and that ATP synthesis with l-lactate was coupled only to the bo-type oxidase (45). Whether l- and d-lactic acids are actually directly oxidized by the bo-type oxidase or via lactate dehydrogenase (LDH) oxidation to yield NADH is unknown. However, in both cases the respiratory chain is fueled with reducing equivalents and is accompanied by pyruvate production. What is the fate of pyruvate? In the wt, pyruvate can be fed into the TCA cycle via the pyruvate dehydrogenase (PDH) complex. In the mutant, the TCA cycle is a dead end, and part of the pyruvate is metabolized and the larger portion is converted to acetic acid, which increased steadily in the culture supernatant (Fig. 2D). Pyruvate oxidation occurs probably via the phosphate acetyltransferase (Pta) and acetate kinase (AckA) pathways, generating reducing potential and ATP. Another candidate for such a pyruvate oxidase is CidC, which catalyzes the oxidative decarboxylation of pyruvate, yielding acetate and CO2 (27). In a nonfunctional TCA cycle S. aureus is unable to further metabolize or to oxidize acetic acid, and it has been mentioned that accumulation of acetic acid contributes to cell death and lysis in stationary-phase cultures (27). We found, however, in the long-term stationary-phase survival assay that the viable counts of the Δsdh mutant were increased compared to wt. A similar observation was also made with an aconitase mutant (39). Although acetic acid continuously accumulated in the Δsdh mutant during growth, the medium pH stayed constant around 6.2. We think that the steady pH value was achieved by upregulation of urease, which counteracts further acidification via excretion of ammonia. Very likely not only urease but also probably the entire urea cycle is upregulated, although we have not yet examined this (Fig. 7). This is another example of how efficiently S. aureus can adapt survival strategies in cases of emergency.
FIG. 7.
Schematic representation of observed metabolic differences in the Δsdh mutant. In the Δsdh mutant urease activity was increased, succinate and acetate were excreted in higher amounts, the pH of the culture supernatant stayed low, and consumption of the amino acids alanine (ala), glutamate (glu), histidine (his), isoleucine (ile), and proline (pro) was affected. The corresponding genes of the indicated enzymatic reactions are annotated in S. aureus NCTC8325: ArcC, carbamate kinase (SAOUHSC_01129 and SAOUHSC_02965); ArgF, ornithine carbamoyltransferase (SAOUHSC_01128); ArgG, argininosuccinate synthase (SAOUHSC_00899); ArgH, argininiosuccinate lyase (SAOUHSC_00898); Arg, arginase (SAOUHSC_02409); UreA, urease subunit gamma (SAOUHSC_02558); UreB, urease subunit beta (SAOUHSC_02559); UreC, urease subunit alpha (SAOUHSC_02561); PdhA, pyruvate dehydrogenase complex E1 component, alpha-subunit (SAOUHSC_01040); PdhB, pyruvate dehydrogenase complex E1 component, beta-subunit (SAOUHSC_01041); PdhC, pyruvate dehydrogenase component E2 (SAOUHSC_01042); CitZ, citrate synthase (SAOUHSC_01802); CitB, aconitate hydratase (SAOUHSC_01347); CitC, isocitrate dehyrogenase (SAOUHSC_01801); OdhA/Kgd, 2-oxoglutarate dehydrogenase E1 component/alpha-ketoglutarate decarboxylase (SAOUHSC_01418); OdhB, 2-oxoglutarate dehydrogenase E2 component/dihydrolipoamide acetyltransferase (SAOUHSC_01416); SucC, succinyl-CoA synthetase subunit beta (SAOUHSC_01216); SucD, succinyl-CoA synthetase subunit alpha (SAOUHSC_01218); SdhC, succinate dehydrogenase cytochrome b558 subunit (SAOUHSC_01103); SdhA, succinate dehydrogenase flavoprotein subunit (SAOUHSC_01104); SdhB, succinate dehydrogenase iron-sulfur subunit (SAOUHSC_01105); FumC, fumarate hydratase class II (SAOUHSC_01983); Mqo-2, malate:quinone oxidoreductase (SAOUHSC_02647 and SAOUHSC_02927); Pox, pyruvate oxidase (SAOUHSC_02849); AckA, acetate kinase (SAOUHSC_01820); AcyP, acylphosphatase (SAOUHSC_01406); Pta, phosphate acetyltransferase (SAOUHSC_00574); AcsA, acetyl-CoA synthetase (SAOUHSC_01846).
In CDM, growth was severely affected, with a maximal OD of only 2.2. This was expected, as glucose was present at a lower concentration (7.5 mM versus 12.6 mM) to enhance amino acid consumption. In CDM, the growth and accumulation of fermentation products (e.g., acetic acid, lactic acid, and acetoin) were very similar between the wt and mutant strains (Table 1). Surprisingly, in both strains acetic acid increased continuously throughout growth over 24 h, indicating that in CDM the wt was unable to reuse acetic acid. To drive the TCA cycle, anaplerotic reactions are needed to offset the extraction of intermediates for anabolic pathways. If there is a limited supply of C4 or C5 compounds, the TCA cycle shuts down, which could be an explanation for the inability to reuse acetate. Consistent with our observations in TSB, succinate accumulated only in the mutant. Another difference in the mutant was the retarded amino acid consumption of alanine, glutamate, histidine, isoleucine, and proline (Table 2).
In Δsdh, the respiratory chain is still intact. NADH formed during glycolysis could fuel respiration. Even lactic acid, which is typically transiently excreted during aerobic growth in staphylococci, can directly feed the bo-type oxidase as mentioned above. Even glutamate oxidation via the bo-type oxidase and subsequent ATP synthesis have been described (45). There are alternative pathways and substrates by which the respiratory chain can be fueled even in the absence of a functional TCA cycle. Nevertheless, the loss of Sdh is sufficient to cause an SCV-like phenotype. We name it SCV-like because the effect is not that pronounced as that described for the hemB (48) or menD (20) mutants, in which the respiratory chain is completely disrupted. These mutants have a typical anaerobic phenotype even under aerobic conditions, consistent with high accumulation of lactic acid and low excretion of acetic acid (21). Typically, the SCV phenotype displays an increased tolerance for aminoglycoside antibiotics, which require a certain membrane potential (Δψ) for uptake (5, 31, 48). The Δsdh mutant shows only a 2- to 4-fold-higher resistance to aminoglycoside antibiotics, but this is much lower than the 32- to 64-fold increase in resistance observed for hemB and menD mutants. This indicates that the membrane potential of Δsdh cannot be that low. Another characteristic feature of SCVs is decreased pigmentation (48). Indeed, the absorption spectra of wt (SA113 and HG001) and corresponding Δsdh mutants showed that carotenoid content in the mutants was significantly decreased; in the complemented mutants the wt level was restored.
The first clean TCA cycle deletion mutant was constructed in the aconitase gene (acnA::ermB) of S. aureus by Somerville and colleagues (39). This mutant shows some similarities but also differences compared to Δsdh. The Δsdh mutant appears more severely affected in growth; however, the catabolism of acetic acid was similarly affected in both mutants. Interestingly, the acnA mutant excreted no citrate, but the intracellular concentration was higher relative to the wt (G. Somerville, personal communication). In Staphlococcus epidermidis, acnA mutant citrate was also increased intracellularly (34).
The complementation of the sdh deletion mutant by pRBsdh was delayed. Such problems are often observed with complementing membrane proteins like Sdh with a medium-copy-number plasmid. Additionally, the Sdh complex consists of three subunits, which have to be organized in the right stoichiometry, which also might complicate complementation. In order to rule out that the phenotype of SA113Δsdh is not influenced by some point mutations in this strain, the Δsdh mutant was transduced into HG001. The resulting HG001Δsdh showed similar properties as SA113Δsdh: decreased growth, no alkalization of culture supernatant, decreased pigmentation, and enhanced long-term survival.
Why are various TCA cycle enzymes, particularly Sdh, upregulated in biofilms? In a biofilm nutrients are limited, oxygen is depleted, and ica genes are consequently upregulated under anoxic conditions (12, 33). Cells in a biofilm represent with respect to physiological activity a very heterogenous population due to the various gradients of oxygen, nutrients, and pH. This spectrum of different conditions affords a permanent adaptation of cells to better survive in such a complex environment. The upregulation of the TCA has the advantages (i) to make better use of traces of oxygen, (ii) to allow better metabolism of excreted fermentation products like acetic or lactic acid, and (iii) to permit better catabolism of amino acids. Staphylococcal infections where oxygen is limited are not that seldom, for example, in abscess formation or in the sputum of cystic fibrosis patients, where oxygen depletion is accelerated by the phagocyte NADPH oxidase (22).
Acknowledgments
We thank Bernhard Krismer for providing the knockout plasmid pKO2 and Rüdiger Hampp (Botany Institute, Physiological Ecology of Plants, University of Tübingen) for support with the Clark-type oxygen electrode. We thank Günther Thumm and Martina Leibig for providing the deletion mutants Δlip and Δfdh, respectively. We thank Silvia Herbert and Melanie Kull for advice and discussion as well as Detlinde Futter-Bryniok and Anne Winter for technical assistance.
This work was supported by the DFG (SFB-TR34). Manuel Liebeke was a recipient of a fellowship from the Alfried Krupp von Bohlen and Halbach-Stiftung: “A functional Genomics Approach in Infection Biology.”
Footnotes
Published ahead of print on 5 March 2010.
REFERENCES
- 1.Archibald, L. K., and R. P. Gaynes. 1997. Hospital-acquired infections in the United States. The importance of interhospital comparisons. Infect. Dis. Clin. North Am. 11:245-255. [DOI] [PubMed] [Google Scholar]
- 2.Augustin, J., and F. Götz. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 54:203-207. [DOI] [PubMed] [Google Scholar]
- 3.Baddour, L. M., and G. D. Christensen. 1987. Prosthetic valve endocarditis due to small-colony staphylococcal variants. Rev. Infect. Dis. 9:1168-1174. [DOI] [PubMed] [Google Scholar]
- 4.Bates, D. M., C. von Eiff, P. J. McNamara, G. Peters, M. R. Yeaman, A. S. Bayer, and R. A. Proctor. 2003. Staphylococcus aureus menD and hemB mutants are as infective as the parent strains, but the menadione biosynthetic mutant persists within the kidney. J. Infect. Dis. 187:1654-1661. [DOI] [PubMed] [Google Scholar]
- 5.Baumert, N., C. von Eiff, F. Schaaff, G. Peters, R. A. Proctor, and H. G. Sahl. 2002. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 8:253-260. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 9.Brückner, R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122:187-192. [DOI] [PubMed] [Google Scholar]
- 10.Clauditz, A., A. Resch, K. P. Wieland, A. Peschel, and F. Götz. 2006. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 74:4950-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Götz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cramton, S. E., M. Ulrich, F. Götz, and G. Döring. 2001. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:4079-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldschmidt, M. C., and D. M. Powelson. 1953. Effect of the culture medium on the oxidation of acetate by Micrococcus pyogenes var. aureus. Arch. Biochem. Biophys. 46:154-163. [DOI] [PubMed] [Google Scholar]
- 14.Götz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 15.Hederstedt, L., and L. Rutberg. 1980. Biosynthesis and membrane binding of succinate dehydrogenase in Bacillus subtilis. J. Bacteriol. 144:941-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hederstedt, L., and L. Rutberg. 1981. Succinate dehydrogenase: a comparative review. Microbiol. Rev. 45:542-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herigstad, B., M. Hamilton, and J. Heersink. 2001. How to optimize the drop plate method for enumerating bacteria. J. Microbiol. Methods 44:121-129. [DOI] [PubMed] [Google Scholar]
- 18.Iordanescu, S., and M. Surdeanu. 1976. Two restriction and modification systems in Staphylococcus aureus NCTC8325. J. Gen. Microbiol. 96:277-281. [DOI] [PubMed] [Google Scholar]
- 19.Kahl, B. C., A. Duebbers, G. Lubritz, J. Haeberle, H. G. Koch, B. Ritzerfeld, M. Reilly, E. Harms, R. A. Proctor, M. Herrmann, and G. Peters. 2003. Population dynamics of persistent Staphylococcus aureus isolated from the airways of cystic fibrosis patients during a 6-year prospective study. J. Clin. Microbiol. 41:4424-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohler, C., C. von Eiff, M. Liebeke, P. J. McNamara, M. Lalk, R. A. Proctor, M. Hecker, and S. Engelmann. 2008. A defect in menadione biosynthesis induces global changes in gene expression in Staphylococcus aureus. J. Bacteriol. 190:6351-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohler, C., C. von Eiff, G. Peters, R. A. Proctor, M. Hecker, and S. Engelmann. 2003. Physiological characterization of a heme-deficient mutant of Staphylococcus aureus by a proteomic approach. J. Bacteriol. 185:6928-6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolpen, M., C. R. Hansen, T. Bjarnsholt, C. Moser, L. D. Christensen, M. van Gennip, O. Ciofu, L. Mandsberg, A. Kharazmi, G. Döring, M. Givskov, N. Hoiby, and P. O. Jensen. 2010. Polymorphonuclear leukocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax 65:57-62. [DOI] [PubMed] [Google Scholar]
- 23.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 24.Liebeke, M., V. S. Brozel, M. Hecker, and M. Lalk. 2009. Chemical characterization of soil extract as growth media for the ecophysiological study of bacteria. Appl. Microbiol. Biotechnol. 83:161-173. [DOI] [PubMed] [Google Scholar]
- 25.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 26.McNamara, P. J., and R. A. Proctor. 2000. Staphylococcus aureus small colony variants, electron transport and persistent infections. Int. J. Antimicrob. Agents 14:117-122. [DOI] [PubMed] [Google Scholar]
- 27.Patton, T. G., K. C. Rice, M. K. Foster, and K. W. Bayles. 2005. The Staphylococcus aureus cidC gene encodes a pyruvate oxidase that affects acetate metabolism and cell death in stationary phase. Mol. Microbiol. 56:1664-1674. [DOI] [PubMed] [Google Scholar]
- 28.Pelz, A., K. P. Wieland, K. Putzbach, P. Hentschel, K. Albert, and F. Götz. 2005. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 280:32493-32498. [DOI] [PubMed] [Google Scholar]
- 29.Peters, G. 1984. Pathogenesis of staphylococcal infections of implanted plastics and intravascular catheters. Infection 12:235-239. [DOI] [PubMed] [Google Scholar]
- 30.Proctor, R. A., P. van Langevelde, M. Kristjansson, J. N. Maslow, and R. D. Arbeit. 1995. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 20:95-102. [DOI] [PubMed] [Google Scholar]
- 31.Proctor, R. A., C. von Eiff, B. C. Kahl, K. Becker, P. McNamara, M. Herrmann, and G. Peters. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4:295-305. [DOI] [PubMed] [Google Scholar]
- 32.Resch, A., S. Leicht, M. Saric, L. Pasztor, A. Jakob, F. Götz, and A. Nordheim. 2006. Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 6:1867-1877. [DOI] [PubMed] [Google Scholar]
- 33.Resch, A., R. Rosenstein, C. Nerz, and F. Götz. 2005. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 71:2663-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadykov, M. R., M. E. Olson, S. Halouska, Y. Zhu, P. D. Fey, R. Powers, and G. A. Somerville. 2008. Tricarboxylic acid cycle-dependent regulation of Staphylococcus epidermidis polysaccharide intercellular adhesin synthesis. J. Bacteriol. 190:7621-7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schägger, H., H. Aquila, and G. Von Jagow. 1988. Coomassie blue-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for direct visualization of polypeptides during electrophoresis. Anal. Biochem. 173:201-205. [DOI] [PubMed] [Google Scholar]
- 36.Seifert, H., D. Oltmanns, K. Becker, H. Wisplinghoff, and C. von Eiff. 2005. Staphylococcus lugdunensis pacemaker-related infection. Emerg. Infect. Dis. 11:1283-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seifert, H., H. Wisplinghoff, P. Schnabel, and C. von Eiff. 2003. Small colony variants of Staphylococcus aureus and pacemaker-related infection. Emerg. Infect. Dis. 9:1316-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Senn, M. M., M. Bischoff, C. von Eiff, and B. Berger-Bächi. 2005. σB activity in a Staphylococcus aureus hemB mutant. J. Bacteriol. 187:7397-7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Somerville, G. A., M. S. Chaussee, C. I. Morgan, J. R. Fitzgerald, D. W. Dorward, L. J. Reitzer, and J. M. Musser. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70:6373-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somerville, G. A., and R. A. Proctor. 2009. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol. Mol. Biol. Rev. 73:233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somerville, G. A., B. Said-Salim, J. M. Wickman, S. J. Raffel, B. N. Kreiswirth, and J. M. Musser. 2003. Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect. Immun. 71:4724-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinmetz, M., and R. Richter. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142:79-83. [DOI] [PubMed] [Google Scholar]
- 43.Stevens, D. L. 2003. Community-acquired Staphylococcus aureus infections: increasing virulence and emerging methicillin resistance in the new millennium. Curr. Opin. Infect. Dis. 16:189-191. [DOI] [PubMed] [Google Scholar]
- 44.Strasters, K. C., and K. C. Winkler. 1963. Carbohydrate metabolism of Staphylococcus aureus. J. Gen. Microbiol. 33:213-229. [DOI] [PubMed] [Google Scholar]
- 45.Tynecka, Z., Z. Szczesniak, A. Malm, and R. Los. 1999. Energy conservation in aerobically grown Staphylococcus aureus. Res. Microbiol. 150:555-566. [DOI] [PubMed] [Google Scholar]
- 46.von Eiff, C. 2008. Staphylococcus aureus small colony variants: a challenge to microbiologists and clinicians. Int. J. Antimicrob. Agents 31:507-510. [DOI] [PubMed] [Google Scholar]
- 47.von Eiff, C., D. Bettin, R. A. Proctor, B. Rolauffs, N. Lindner, W. Winkelmann, and G. Peters. 1997. Recovery of small colony variants of Staphylococcus aureus following gentamicin bead placement for osteomyelitis. Clin. Infect. Dis. 25:1250-1251. [DOI] [PubMed] [Google Scholar]
- 48.von Eiff, C., C. Heilmann, R. A. Proctor, C. Woltz, G. Peters, and F. Götz. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179:4706-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Eiff, C., P. McNamara, K. Becker, D. Bates, X. H. Lei, M. Ziman, B. R. Bochner, G. Peters, and R. A. Proctor. 2006. Phenotype microarray profiling of Staphylococcus aureus menD and hemB mutants with the small-colony-variant phenotype. J. Bacteriol. 188:687-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Eiff, C., P. Vaudaux, B. C. Kahl, D. Lew, S. Emler, A. Schmidt, G. Peters, and R. A. Proctor. 1999. Bloodstream infections caused by small-colony variants of coagulase-negative staphylococci following pacemaker implantation. Clin. Infect. Dis. 29:932-934. [DOI] [PubMed] [Google Scholar]
- 51.Vuong, C., J. B. Kidder, E. R. Jacobson, M. Otto, R. A. Proctor, and G. A. Somerville. 2005. Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J. Bacteriol. 187:2967-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]