Abstract
Chemotaxis is an important virulence factor for Helicobacter pylori colonization and infection. The chemotactic system of H. pylori is marked by the presence of multiple response regulators: CheY1, one CheY-like-containing CheA protein (CheAY2), and three CheV proteins. Recent studies have demonstrated that these molecules play unique roles in the chemotactic signal transduction mechanisms of H. pylori. Here we report the crystal structures of BeF3−-activated CheY1 from H. pylori resolved to 2.4 Å. Structural comparison of CheY1 with active-site residues of BeF3−-bound CheY from Escherichia coli and fluorescence quenching experiments revealed the importance of Thr84 in the phosphotransfer reaction. Complementation assays using various nonchemotactic E. coli mutants and pull-down experiments demonstrated that CheY1 displays differential association with the flagellar motor in E. coli. The structural rearrangement of helix 5 and the C-terminal loop in CheY1 provide a different interaction surface for FliM. On the other hand, interaction of the CheA-P2 domain with CheY1, but not with CheY2/CheV proteins, underlines the preferential recognition of CheY1 by CheA in the phosphotransfer reaction. Our results provide the first structural insight into the features of the H. pylori chemotactic system as a model for Epsilonproteobacteria.
Helicobacter pylori is a spiral-shaped, Gram-negative, microaerophilic bacterium associated with gastritis, duodenal ulcers, gastric adenocarcinoma and mucosa-associated lymphoid tumors (43). About 50% of the world population is infected with this pathogen (6). The bacteria colonize the mucus layer covering the epithelial cell surface of the stomach, and this colonization step is necessary to establish a long-term infection. A number of in vivo mutagenesis studies have demonstrated the importance of bacterial motility in colonization and long-term infection in mouse and gerbil stomach models (11, 17, 27, 41, 47).
Chemotaxis has been well studied in enteric bacteria, such as Escherichia coli and Salmonella enterica serovar Typhimurium that evolved a two-component system composed of histidine kinase CheA and response regulator CheY (39, 45). Chemoreceptors sense an increasing concentration of repellent and promote autophosphorylation of CheA through the adaptor protein CheW. Activated CheA phosphorylates CheY by phosphoryl transfer from its histidyl residue to an aspartyl residue of CheY. Phosphorylated CheY (CheY-P) has an increased affinity for the flagellar switch protein FliM and consequently switches the motor from counterclockwise rotation (running) to clockwise rotation (tumbling). To enable a quick response to environmental changes, signal transduction from CheY to FliM is terminated by autodephosphorylation of CheY-P or by dephosphorylation via phosphatase CheZ. To move along concentration gradients, bacteria have chemoreceptors that undergo adaptation that is regulated by a methylation and demethylation system: for example, CheR methyltransferase and CheB methylesterase in E. coli.
Efforts to sequence prokaryotic genomes allow for comparisons of the diverse chemotaxis systems (28). H. pylori is similar to E. coli in the sense that it carries genes for CheY (HP1067), CheA (HP0392), CheW (HP0391), and a remote CheZ homolog (HP0170) (2, 40). However, H. pylori differs from E. coli in that it carries genes for multiple CheY-like proteins—a bifunctional CheA with a CheY-like domain (CheAY2) (HP0392) fused to the C terminus and three CheV (CheV1 [HP0019], CheV3 [HP0393], and CheV2 [HP0616]) genes, each consisting of an N-terminal CheW-like domain and a C-terminal CheY-like domain. Taken together with the lack of CheB and CheR homologs in the H. pylori genome, the mechanisms of chemotactic signal transduction and adaptation in H. pylori remain to be elucidated.
Structural studies on inactive and active E. coli CheY (EcCheY) and their complexes with FliM have revealed the molecular mechanisms of CheY phosphorylation and activation (8, 20, 21). (In the following text, “Hp” plus the gene name denotes H. pylori.) Phosphorylation is initiated by nucleophilic attack of Asp57 by phosphoryl phosphorus. Bound phosphate that is hydrogen bonded with Thr87 and Lys109 causes a displacement of the β4-α4 and β5-α5 loops and restricted the inward positioning of Tyr106 (8). The activation of EcCheY requires a divalent metal ion (Mg2+/Mn2+), which is coordinated by Asp13, Asp57, the backbone carbonyl of Asn59, the phosphoryl oxygen and two water molecules in the active-site pocket (20). Activated EcCheY binds the N-terminal fragment of FliM through its α4-β5-α5 interface. Recent nuclear magnetic resonance (NMR) study suggests that the transient interaction between the surface residues around the active-site pocket of Thermotoga maritima CheY and the middle FliM domain causes a displacement of FliGC, which underlines the principle of motor switching (10).
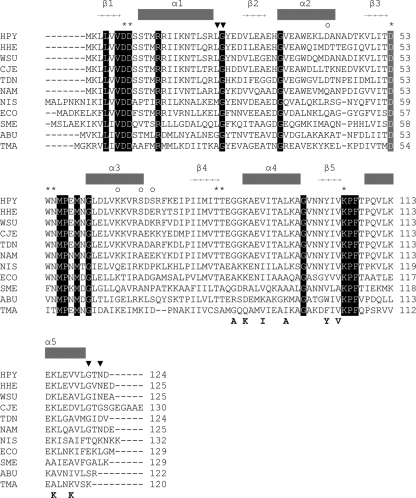
HpCheY1 shares a moderately high level of amino acid sequence identity (46%) with EcCheY (ClustalW2), and both of these proteins exhibit a dephosphorylation rate of 0.035 s−1 (15, 37). Residues involved in phosphorylation and activation of EcCheY are conserved in HpCheY1 (Fig. 1), suggesting that HpCheY1 shares a similar activation mechanism. The HpCheY1 mutant is smooth-swimming biased, which is in line with the E. coli model (24). In vitro experiments showed that HpCheY1 is phosphorylated by CheA and interacts with FliM upon activation by acetyl phosphate (23). These data support the hypothesis that the biological function of HpCheY1 is comparable to that of EcCheY. However, the chemotactic regulatory mechanisms of H. pylori are different from those of E. coli in that the H. pylori genome carries multiple response regulators. All response regulators can be phosphorylated by CheA, which shows a preference for HpCheY1 (15). Interestingly, HpCheY1-P is able to transfer the phosphate back to HpCheA, and the phosphate moiety is transferred to HpCheY2, suggesting that H. pylori may exhibit retrophosphorylation, although this observation is not yet supported by in vivo evidence. CheV is proposed to be involved in one of the three adaptation mechanisms of Bacillus subtilis (32). However, the biological significance of HpCheVs is not as well understood. HpCheV1 mutant is nonchemotactic, and heterologous overexpression of HpCheV2 and HpCheV3 in E. coli inhibits swarming of the bacteria (31), suggesting that all three HpCheVs are associated with chemotaxis. Using fixed-time diffusion analysis, a more recent study showed that HpCheV1 and HpCheV2 mutants are biased toward smooth swimming, while HpCheV3 shows the opposite bias (24). Given that HpCheVs can be phosphorylated in vitro to different levels by HpCheA (15) and that HpCheV mutants show different extents of deficiency in colonizing mouse stomachs (24), it is likely that HpCheVs have distinct and important roles in chemotaxis. Furthermore, the different extent of phosphotransfer observed in in vitro studies indicates that these proteins may display distinctive structural features (15, 31).
FIG. 1.
Multiple sequence alignment of CheY1 from Helicobacter pylori 26695 (HPY), Helicobacter hepaticus (HHE), Wolinella succinogenes (WSU), Campylobacter jejuni NCTC11168 (CJE), Sulfurimonas denitrificans (TDN), Nautilia profundicola (NAM), Nitratiruptor sp. strain SB155-2 (NIS), Arcobacter butzleri (ABU), Escherichia coli K-12 MG1655 (ECO), Sinorhizobium meliloti (SME), and Thermotoga maritima (TMA) (18). Secondary structure elements of HpCheY1 are shown above the sequence alignment. Totally conserved residues are shaded. Phosphorylation site Asp53 is shaded in gray. Asterisks represent residues of the active-site pocket. ▾, residues responsible for the upshift of α5 in HpCheY1; ○, residues that varied between strains of H. pylori. Residues (in EcCheY) involved in EcFliMN binding are marked below the alignment (21).
Despite the fact that the chemotactic system in H. pylori carries unique features and is critical for bacterial infection, none of the structures of the chemotactic proteins in H. pylori have been solved. In the present study, we solved the structure of BeF3−-activated HpCheY1. We demonstrated the binding of BeF3− to HpCheY1 and the interaction between BeF3−-HpCheY1 and HpFliM. Comparison of the BeF3−-HpCheY1 and BeF3−-EcCheY structures revealed a distinctive FliM binding surface. Further evidence of this distinctive binding surface was obtained by complementation assays using various nonchemotactic E. coli mutants. Our results have provided the first structural insight into the features of the H. pylori chemotactic system as a model for Epsilonproteobacteria. Comparison of the CheY structures from different bacteria has furthered our understanding of variations of chemotaxis pathways.
MATERIALS AND METHODS
Cloning, expression, and purification.
Genes encoding HpCheY1 (HP1067), HpCheY2 (HP0392; residues 677 to 803) (15), HpCheV1 (HP0019), HpCheV2 (HP0616), HpCheV3 (HP0393), the HpCheA-P2 domain (HP0392; residues 110 to 261), and the HpFliMNM domain (HP1031; residues 1 to 237) were amplified from H. pylori strain 26695 genomic DNA and cloned into various expression vectors in order to obtain optimal expression level and solubility. HpCheY1, HpCheV1, and HpCheV2 were cloned into the pGEX-6P-1 expression vector. HpCheY2 and HpCheV3 were cloned into pET28m-sumo1 vector. HpCheA-P2 and HpFliMNM were cloned into pT7-7 vector with a C-terminal 8-histidine tag. Mutants HpCheY1/D53A and HpCheY1/T84A were generated using QuikChange site-directed mutagenesis kit (Stratagene) and verified by a commercial sequencing service (1st BASE).
Recombinant proteins were expressed in Escherichia coli strain BL21 and purified according to standard protocols (48). After transformation, cells were grown at 37°C until the optical density at 600 nm (OD600) reached 0.4 to 0.6. Protein expression was induced by the addition of 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and the cells were further grown at 16°C or 25°C for 16 h. Cell pellet was resuspended in lysis buffer (10 mM Tris-HCl, pH 7.8, and 500 mM NaCl) and lysed by sonication. Cell lysate was clarified by centrifugation (40,000 × g, 1 h and 4°C) and the supernatant was incubated with preequilibrated affinity resin for 2 h at 4°C with gentle shaking. After washing the beads with the lysis buffer, the glutathione S-transferase (GST) tag was cleaved by Prescission protease, when necessary. For the His-sumo1 fusion protein, the tag was removed by incubation with sumo protease SENP1 in a mass ratio of 1:100 at 37°C for 30 min (48). The eluted proteins were then further purified by size exclusion chromatography using Superdex 75 or Superdex 200 equilibrated with buffer containing 10 mM HEPES (pH 7.0) and 150 mM NaCl.
Nickel pull-down assay. (i) CheAP2-CheY interaction assay.
An aliquot of 25 μl nickel-nitrilotriacetic acid (NTA) resin (Qiagen) was pipetted into a 1.5-ml Eppendorf tube and centrifuged (3,300 × g, 10 s, and 4°C). The beads were washed 3 times with binding buffer containing 20 mM imidazole (pH 7.0), 150 mM NaCl, and 2 mM MgCl2. It was followed by the addition of 1 ml purified HpCheA-P2-8×His protein solution in a concentration of 0.1 mg/ml. The mixture was incubated at 4°C for 1 h with gentle shaking. The beads were washed three times with the binding buffer followed by the addition of purified HpCheY1, HpCheY2, HpCheV1, HpCheV2, and HpCheV3 in a molar ratio of 1:1. The mixture was incubated for another 1 h at 4°C. The beads were washed three times with the buffer and then resuspended in 25 μl of 2× SDS-PAGE gel loading buffer and heated to 90°C for 8 min. The samples were subjected to SDS-PAGE analysis and Coomassie blue staining.
(ii) FliM-CheY interaction assay.
The pull-down experiment was performed as described previously with modifications (23). All experimental steps were performed at 25°C. HpFliMNM was immobilized onto nickel-NTA resin preequilibrated with buffer containing 10 mM HEPES (pH 7.6), 5 mM MgCl2, 250 mM KCl, 0.15% Tween 20, and 20 mM imidazole. After incubation for 1 h, the beads were washed for 3 times. HpCheY1 and HpCheY1/D53A were preincubated with 0.75 mM BeCl2 and 18 mM NaF for 10 min before being incubated with the immobilized HpFliMNM for 15 min. The beads were washed twice, boiled with gel loading buffer, and subjected to SDS-PAGE analysis.
Size exclusion chromatography.
Purified HpCheY1 and HpCheV3 were individually mixed with HpCheA-P2-8×His in a buffer containing 10 mM HEPES (pH 7.4), 150 mM NaCl, and 2 mM MgCl2. The mixture was incubated on ice for 30 min. Individual proteins or mixtures were applied to a Superdex 200 HR 10/30 gel filtration column (GE Healthcare Bio-Sciences Corp.) and eluted at 0.6 ml/min with the same buffer. Eluted proteins were subjected to SDS-PAGE analysis and Coomassie blue staining.
Fluorescence spectroscopy.
Fluorescence measurement was carried out using a Perkin Elmer 50B spectrofluorimeter. Fluorescence was measured at an excitation wavelength of 293 nm and an emission wavelength of 341 nm with slit widths of 3 nm and 6 nm for excitation and emission, respectively. All reactions were carried out at 25°C. Equilibrium titrations of HpCheY1, HpCheY1/D53A, and HpCheY1/T84A by acetyl phosphate were carried out in a buffer containing 20 mM sodium phosphate (pH 7.5), 50 mM NaCl, and 2 mM MgCl2. The final concentrations were 1 μM for HpCheY1s and 0.3 to 10 mM for acetyl phosphate. Titration of HpCheY1 or HpCheY1/D53A with BeF3− was performed by sequential addition of NaF to the protein solution containing 10 μM BeCl2. The final concentration of NaF was set to 0 to 5 mM. Fluorescence changes upon addition of small molecules were monitored until the fluorescence signal stabilized. The fluorescence values were corrected for dilution. Km was determined as described previously (25, 31, 36). Acetyl phosphate concentration was plotted versus (Io − I)/(I − Iinf), where Io is initial fluorescence intensity, I is the intensity at the corresponding acetyl phosphate concentration, and Iinf is the intensity at the saturating concentration. From the plot, the reciprocal of the slope of the line corresponds to the Km value. According to proposed reaction scheme (25), shown as follows, Km = Ks k3/k2
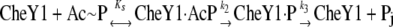 |
where Ks is the equilibrium dissociation constant between CheY1 and acetyl phosphate and k2 and k3 are the phosphorylation and dephosphorylation rate constants, respectively.
Swarming assay.
E. coli motility wild-type strain RP437 and chemotaxis mutant strains RP1616 (ΔcheZ) and RP5232 (ΔcheY) were gifts from J. S. Parkinson (29). The complementation assay was performed by individual transformation of pTrc99a, pTrc99a-HpCheY1, and pTrc99a-HpCheY1/D53A in strain RP437 and chemotaxis mutants RP1616 (ΔcheZ) and RP5232 (ΔcheY). An aliquot of a 1-μl overnight culture was spotted onto 0.4% TB soft agar (1% tryptone, 0.5% NaCl, and 0.5% glycerol, 0.4% agar). Ampicillin (100 μg/ml) and IPTG (isopropyl-β-d-thiogalactopyranoside; final concentration of 0.05, 0.1, or 0.5 mM) were added when necessary. The diameter of the chemotactic rings was measured after incubation at 30°C for 7 h.
Immunoblotting.
An aliquot of a 1-μl overnight culture of E. coli strains RP437 and RP1616 (ΔcheZ), RP9535 (ΔcheA), and RP5232 (ΔcheY) transformed with the recombinant plasmids were added to 3 ml of TB medium with 0, 0.05, 0.1, and 0.5 mM IPTG when necessary. Cells were grown overnight at 30°C with vigorous shaking. The cells were harvested and washed with a buffer containing 10 mM HEPES (pH 7.4) and 150 mM NaCl. The samples were subjected to SDS-PAGE analysis followed by immunoblotting with polyclonal anti-HpCheY1 antibody (Invitrogen).
Crystallization condition of HpCheY1.
Initial crystallization trials were performed by Crystal Screens I and II from Hampton Research (Hampton Research, Aliso Viejo, CA) using the sitting drop vapor diffusion method. Diffraction-quality crystals of HpCheY1 grown at 16°C were obtained from an optimized screening condition containing 0.1 M sodium acetate (pH 5.0), 0.2 M ammonium sulfate, 35% MPEG2000, and 1 mM MgCl2. Crystals of HpCheY1/D53A and T84A were grown under a similar condition. For BeF3−-HpCheY1, the crystals were grown in crystallization buffer containing 0.1 M sodium acetate (pH 5.0), 0.05 M ammonium sulfate, 1.6 mM BeCl2 (1:10 CheY1-BeCl2) and 16 mM NaF.
Data collection and processing.
A 1.8-Å X-ray data set for HpCheY1 crystal, 2.2-Å X-ray data set for BeF3−-HpCheY1 crystal, 2.2-Å X-ray data set for HpCheY1/D53A crystal, and 1.7-Å X-ray data set for HpCheY1/T84A crystal were collected at 100 K using a Rigaku MicroMax 007 X-ray generator at the Centre for Protein Science and Crystallography, The Chinese University of Hong Kong, and recorded on a RAXIS IV++ image plate. For each crystal, crystallization buffer containing 15% glycerol was used as a cryoprotectant. Images were processed using Mosflm (22) and scaled and reduced with SCALA from the CCP4 suite (5). All of the four protein structures were in spacegroup C2, and unit cell parameters and statistics for the data collected are summarized in Table 1 .
TABLE 1.
Diffraction data and refinement statistics of CheY
| Parameter | Result fora: |
|||
|---|---|---|---|---|
| CheY | CheY-BeF3− | CheY D53A | CheY T84A | |
| X-ray data statistics | ||||
| Spacegroup | C2 | C2 | C2 | C2 |
| Unit cell | ||||
| a, b, c (Å) | 70.3, 38.5, 38.9 | 70.3, 38.4, 38.7 | 70.3, 38.1, 38.6 | 70.4, 38.1, 39.0 |
| α, β, γ (Å) | 90.0, 107.0, 90.0 | 90.0, 107.0, 90.0 | 90.0, 107.4, 90.0 | 90.0, 107.6, 90.0 |
| Resolution range (Å) | 37.2-1.8 (1.9-1.8) | 33.3-2.2 (2.28-2.2) | 36.9-2.2 (2.28-2.2) | 23.08-1.7 (1.76-1.70) |
| No. of observations | 33,157 | 20,879 | 18,875 | 38,199 |
| No. of unique reflections | 9,175 | 4,888 | 4,993 | 10,972 |
| Redundancy | 3.6 (3.6) | 4.3 (4.4) | 3.8 (3.8) | 3.5 (3.3) |
| Completeness (%) | 98.1 (96.1) | 95.3 (91.8) | 98.5 (97.0) | 99.9 (100.0) |
| Rmerge | 0.031 (0.072) | 0.019 (0.029) | 0.082 (0.240) | 0.033 (0.199) |
| Mean I/σI | 30.3 (15.4) | 55.1 (39.0) | 10.6 (4.5) | 14.2 (5.2) |
| Refinement statistics | ||||
| Resolution | 37.16-1.8 | 33.46-2.4 | 36.89-2.2 | 23.08-1.7 |
| R value | 14.67 | 15.14 | 17.78 | 17.35 |
| Rfree (5% of data) | 18.61 | 22.01 | 23.11 | 20.28 |
| No. of atoms refined | ||||
| Protein | 966 | 966 | 963 | 964 |
| Water/SO4/Mg/BeF3 | 143/2/2/0 | 60/1/2/1 | 68/2/1/0 | 132/2/2/0 |
| RMSD from ideal geometry | ||||
| Bond lengths (Å) | 0.011 | 0.013 | 0.020 | 0.014 |
| Bond angles (°) | 1.37 | 1.40 | 1.87 | 1.42 |
| Ramachandran plot | 91.8/7.3/0.9 | 91.8/7.3/0.9 | 90.9/8.2/0.9 | 90.9/8.2/0.9 |
Values in parentheses are for the last resolution shell. Rmerge = (Σh Σj | <I(h)> − I (h)j|/Σh Σj <I(h)>), where <I(h)> is the mean intensity of symmetry-equivalent reflections. R value = Σ||Fobs| − |Fcalc||/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. The distribution of the residues in the most favored/additionally allowed regions of the Ramachandran plot was evaluated by PROCHECK (19).
Structure determination and refinement.
The HpCheY1 structure was solved by molecular replacement using a molecule of EcCheY (Protein Data Bank identification [PDB-ID] no. 3CHY) as a search model. Molecular replacement program PHASER (26) in CCP4 suite was performed with data in the resolution range 15 to 3.0 Å. For the other three structures, the refined structure of HpCheY1 was used as a search model. The randomly selected 5% of data was reserved for the Rfree calculation for all the three structures. Rounds of refinements and manual rebuilding were performed by using the programs REFMAC and COOT (5, 12), respectively. The electron density maps from 2Fo−Fc and Fo−Fc calculations were used for model building, and for all of the structures, strong electron density was found close to the active-site Asp 53. We modeled them as sulfate molecules for HpCheY1, HpCheY1/D53A, and HpCheY1/T84A structures. For the CheY1 crystal grown in the presence of beryllium and sodium, a beryllium fluoride molecule was modeled in the active site. The Ramachandran plots drawn by the program PROCHECK (19) showed that over 88% of all residues in the four CheY1 structures fall within the most favored region.
Protein structure accession numbers.
Coordinates have been deposited in the Protein Data Bank under PDB-ID no. 3GWG, 3H1E, 3H1F, and 3H1G.
RESULTS AND DISCUSSION
HpCheY1 is activated by beryllofluoride.
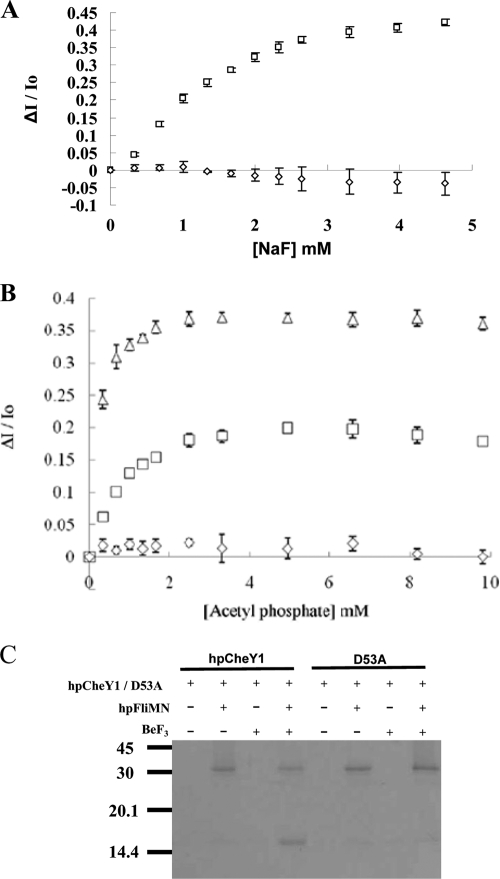
HpCheY1-P has a short half-life (20 s), precluding structural analysis of HpCheY1-P (15). BeF3− is known to form a persistent complex with response regulators by mimicking an acyl phosphate (4, 20). In prior to the structure-function analysis, the binding of BeF3− to HpCheY1 was examined by steady-state fluorescence quenching. A reaction mixture containing 1 μM HpCheY1 and 10 μM BeCl2 was titrated by sequential addition of 0.5 M NaF to a final concentration of 0 to 5 mM. HpCheY1 contains two tryptophan residues (Trp38 and Trp54). The conserved Trp54 residue in EcCheY has been used to report the binding of small molecules to the active-site pocket (25). The fluorescence of Trp38 is not affected by this binding because it is located at helix 2, and no conformational change in helix 2 was reported upon phosphorylation (20). As shown in Fig. 2A, the tryptophan fluorescence of HpCheY1 decreased upon addition of increasing amounts of NaF and was almost saturated at 4 mM NaF. The HpCheY1/D53A mutant, a nonphosphorylatable analog, showed no decrease in fluorescence, suggesting that HpCheY1 binds BeF3− and that an aspartic residue is necessary for the binding. As a control, phosphorylation of HpCheY1 by acetyl phosphate was monitored, and comparable decreases in fluorescence were observed for the wild type and the D53A mutant (Fig. 2B).
FIG. 2.
Activation of HpCheY1 by beryllofluoride. Fluorescence spectroscopy analysis of HpCheY1 (□), HpCheY1/D53A (⋄), and HpCheY1/T84A (▵) upon addition of sodium fluoride (A) and acetyl phosphate (B) is plotted. Fluorescence changes were plotted as ΔI/Io against concentration, where ΔI represents the cumulative changes in fluorescence intensity at the corresponding small molecule concentrations and Io is fluorescence intensity without addition of small molecules. (C) Pull-down study of HpCheY1 with HpFliM. Purified HpFliMNM in 0.1 mg/ml was immobilized on prewashed resin. HpCheY1 or the D53A mutant in a 2:1 molar ratio to HpFliMNM was incubated with immobilized HpFliMNM with or without BeF3− at 25°C for 15 min.
Activated CheY has an enhanced affinity for FliM (49), and a recent study demonstrated the binding of HpCheY1-P to HpFliM upon activation by acetyl phosphate (23). To further verify that HpCheY1-BeF3− mimics HpCheY1-P, the interaction of HpCheY1-BeF3− with HpFliM was examined by an in vitro pull-down assay. HpCheY1 and HpFliMNM (consisting of the N-terminal and middle domains fused to a C-terminal His8 tag) were purified to greater than 95% purity. HpFliMNM was immobilized on Ni-NTA resin, followed by incubation with wild-type HpCheY1 or the D53A mutant with or without BeF3−. As shown in Fig. 2C, only wild-type HpCheY1 complexed with BeF3− showed a stable interaction with HpFliMNM, suggesting that HpCheY1-BeF3− mimics the activated form of HpCheY1 in the interaction with HpFliMNM.
Description of the HpCheY1 structure.
The crystal structure of HpCheY1-BeF3− was solved at a resolution of 2.4 Å, with R = 15.14% and Rfree = 22.01%. The overall structure retained a (β/α)5 fold typical of response regulators. From a DALI search (13), HpCheY1 showed high structural homology to EcCheY/D13K/Y106W (PDB-ID 1U8T) (9) and to BeF3−-bound EcCheY/F14E/N59M/E89L (PDB-ID 3F7N) (30), with Cα root mean square deviation (RMSD) values of 0.776 Å and 0.684 Å, respectively. Alignment of the amino acid sequences of HpCheY1 proteins from different H. pylori strains revealed four variable residues: Asp42 positioned at α2 (Asn in Shi470), Lys66 at α3 (Ile in Shi470), Ser70 at α3 (Ala in P12/J99/Shi470), and Ser72 at the α3-β4 loop (Asn in J99 and Glu in Shi470). These residues are located on the distal side of the active site, which may not be directly involved in activation or FliM binding (Fig. 1).
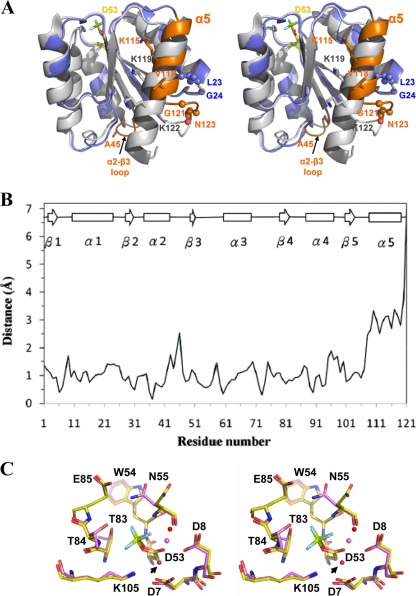
Structural alignment with BeF3−-EcCheY revealed that the conformation of most of the active site residues and of the hallmark residues for CheY activation, including Thr83 and Tyr106, aligned well when the structures of BeF3−-HpCheY1 and BeF3−-EcCheY were superimposed. This suggests that HpCheY1 shares a similar activation mechanism (8). On the other hand, a number of structural differences were noted. These differences were clustered at the α2-β3 loop, helix 5, and the C-terminal loop (Fig. 3A). The former one corresponded to an insertion of Ala45 in the HpCheY1 sequence; however, no distortion of the overall protein fold, especially the active site pocket, was observed. Interestingly, helix 5 of HpCheY1 was five residues shorter and was upshifted by approximately 3.4 Å. The movement of helix 5 may be related to a combination of effects from residues Gly121, which causes an early termination of the helical structure, and Asn123, which is hydrogen bonded to residues in the α1-β2 loop (backbone NH of Asn123 to the backbone carbonyl of Leu23 and Nδ of Asn123 to the backbone carbonyl of Gly24), leaving the C-terminal loop positioned in a rigid conformation. Gly121 is conserved in most Epsilonproteobacteria and in some other proteobacteria (Fig. 1). The corresponding Gly126 in the CheY2 structure from Sinorhizobium meliloti (PDB-ID no. 1P6U) also causes early termination of the helical structure, but the C-terminal loop flips to the other side (33). Sequence alignment of CheYs showed that the number and types of residues following Gly121 are variable. Residue Asn123 is only conserved in some related bacteria species. We speculate that the helical upshift of CheY1 is a unique feature in H. pylori and some closely related species. These structural discrepancies, which could not be predicted from the multiple sequence alignment, in fact provide insights into the molecular interaction with FliM. Analysis of the surface potential of HpCheY1 and EcCheY clearly demonstrated that although the hydrophobic surface of helix 4 is retained, the electropositive patch contributed by helix 5 for the FliM interaction was much weaker in the HpCheY1 structure (data not shown).
FIG. 3.
(A) Structural superimposition of BeF3−-HpCheY1 (blue) and BeF3−-EcCheY (white) (PDB-ID no. 1FQW) highlights the features of the HpCheY1 α2-β3 loop, α5, and the C-terminal loop (orange). Insertion at Ala45 and the major differences in residues involved in EcFliMN binding (Lys119 and Lys122 in EcCheY and Lys115 and Val118 in HpCheY1) are shown as sticks. Residues that contributed to α5 upshift are shown as spheres. Phosphorylation site Asp53 of HpCheY1 bonded with BeF3− is shown as a stick. (B) Differences in Cα positions between BeF3−-bound HpCheY1 and EcCheY. The least-square superposition program LSQKAB (CCP4) was used to superimpose HpCheY1 (residues 1 to 121) with EcCheY (residues 6 to 125) and to calculate individual Cα distances. Residue 45 of HpCheY1 was not aligned from the calculation and is not shown on the plot. Residues are numbered according to HpCheY1 sequence. Secondary structure is shown above the plot. (C) Stereo superimposition images of BeF3−-HpCheY1 (pink) and EcCheY (yellow) on the basis of structural alignment using Pymol. Residues in the active-site pocket are labeled. Metal ions (Mg2+ in HpCheY1) and two water molecules (red sphere) (from BeF3−-HpCheY1) involved in coordinating the metal ion (pink sphere) are shown. An arrow indicates the water molecule coordinated by Asp7.
We also attempted to solve the structure of apo-HpCheY1. To our surprise, a tetrahedral positive electron density was observed in the active-site pocket during model building (Fig. 4A). The electron density shown in the active-site pocket of the 1.8-Å “apo-HpCheY1” structure was interpreted as a sulfate ion because of the addition of 200 mM ammonium sulfate in the crystallization medium. The structure of the HpCheY1/D53A mutant was also solved to a resolution of 2.2 Å. However, the sulfate moiety was still found in the active-site pocket of HpCheY1/D53A. The overall structures of sulfate-bound HpCheY1 and HpCheY1/D53A were almost identical to the HpCheY1-BeF3− structure, including the α4-β4 loop and Tyr102, with Cα RMSD values of 0.147 Å and 0.199 Å, respectively, suggesting that sulfate-bound HpCheY1s may represent an activated form. We investigated whether the nonphosphorylatable D53A analog of HpCheY1 can be “activated” by high concentration of ammonium sulfate. We found that sulfate binds HpCheY1/D53A in vitro as the tryptophan fluorescence intensity of HpCheY1/D53A decreased by 30% upon titration with ammonium sulfate (data not shown). The titration was saturated when the concentration of ammonium sulfate reached around 360 mM, with a calculated Km of 176 ± 21 mM. To further investigate whether sulfate enhanced the ability of the D53A mutant to interact with HpFliM, an in vitro pull down assay was performed in the presence of 200 mM ammonium sulfate (without KCl). HpCheY1/D53A showed enhanced binding to HpFliMNM in the presence of ammonium sulfate (data not shown), but not in BeF3− (Fig. 2C), agreeing with the structural information of SO42−-bound HpCheY1/D53A in an activated configuration. Note that the interaction is of no known physiological relevance given the high concentration of ammonium sulfate.
FIG. 4.
Stereo view of the active site of HpCheY1 showing the 2Fo−Fc electron density around SO42− (A) and BeF3− (B) moieties contoured at 1.0 σ (7). Active-site residues are labeled and shown as sticks. Mg2+ is shown as a sphere.
A sulfate molecule positioned close to the active-site pocket has been reported in the crystal structure of apo-EcCheY (PDB-ID no. 3CHY), in which the SO42− was hydrogen bonded with Lys109 Nɛ and Asn59 Nδ (in the E. coli sequence) (44). However, our results show that the hydrogen bond networking of SO42− with the active-site residues in HpCheY1 was comparable to that found in BeF3−-HpCheY1. An “inward” orientation of the side chain of Asp53, in which the side chain is flipped toward the protein core, was noted in SO42−-bound HpCheY1 (Fig. 4). Such rearrangement is very likely induced by the charge-charge repulsion upon sulfate binding. The flipping of Asp53 is stabilized by hydrogen bond formation with its own peptide NH (2.85 Å), with the peptide NH of Trp54 (3.47 Å), with the peptide NH of Asp8 (3.41 Å), and with Mg2+ (2.44 Å).
Comparison of the active-site pockets.
The active-site residues of HpCheY1 aligned well with those in EcCheY, except for the flipping of the Asn55 side chain and the replacement of Thr84 with an alanine in the EcCheY sequence (equivalent to Ala88 in EcCheY). The side chain of the conserved Asn55 in HpCheY1 was flipped to an alternative conformation and was hydrogen bonded with the carboxylate Oɛ of Glu85 (2.9 Å). The coupling of Asn55 and Glu85 has been implicated in controlling autodephosphorylation (42). Sequence alignment of CheYs showed that Thr84 is conserved within strains of H. pylori and in several species of Epsilonproteobacteria (Fig. 1). Thr84 Oγ in the β4-α4 loop is hydrogen bonded with BeF3− (3.50 Å), which may affect the phosphotransfer reaction. The role of Thr84 in HpCheY1 phosphorylation was investigated by introducing a Thr-to-Ala mutation, and phosphorylation of HpCheY1 by acetyl phosphate was measured by equilibrium titration (up to 10 mM). The T84A mutant had an approximately 4- to 5-fold (0.22 ± 0.055 mM) lower Km value than that of the wild type (1.07 ± 0.31 mM) (Fig. 2B). The Km value determined for HpCheY1 is comparable to the previously determined Km value of EcCheY (3.2 ± 0.4 mM) (36). The lower Km value for HpCheY1 may be due to the differences in ionic strength in the experiment performed (200 mM buffer salt used in the previous experiment, compared to 50 mM used in our experiment).
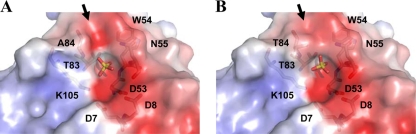
The Km value is derived from the binding constant and rate constant (Km = Ks·k3/k2). A lower Km value could be due to an increase in the binding affinity between CheY and the phosphodonor (smaller Ks), a faster rate of phosphorylation of bound CheY (larger k2) or a slower rate of autodephosphorylation (smaller k3) (36). It has been reported that Ala88 in EcCheY cannot be substituted for by amino acid residues with long side chains (38). Multiple sequence alignment from various response regulators showed that Thr84 is most frequently replaced by small residues, including Ser, Ala, and Gly, and more rarely by hydrophobic Val and Ile (42). The structure of HpCheY1/T84A with SO42− bound was almost identical to the wild type (CαRMSD = 0.1 Å), suggesting that the mutation would have no effect on backbone orientation. No significant difference around the active-site pocket was observed when comparing the surface electrostatic potential of wild-type HpCheY1 and the T84A mutant. A more electronegative surface was observed in the T84A mutant due to the exposed negative charge of Oγ of Thr83 (Fig. 5). Future study of the T84V mutant would provide insight into the role of the hydroxyl group in phosphorylation.
FIG. 5.
Electrostatic surface representation with the contour level ± 5 kT/e showing the active-site pocket of SO42−-bound HpCheY1/T84A (A) and SO42−-bound HpCheY1 (B). The position of SO42− is shown as a stick. Active-site residues are labeled. The arrow indicates the difference on the electrostatic surface between the two structures (see text). Electrostatic surfaces were calculated using the software APBS (3). Ligands were not included in the calculation.
Comparison of the motor binding surfaces.
Although HpCheY1 and EcCheY share high sequence identity and structural homology, we have noted a major difference at the FliM binding surface. The HpCheY1 structure differs from the EcCheY structure because of the upshift and shortening of helix 5 (Fig. 3A). We hypothesized that this structural difference would lead to a different CheY-FliM interaction in H. pylori. We examined the biological function of wild-type and mutant HpCheY1 in E. coli using a swarming assay. Bacterial swarming ability was assessed by transforming pTrc99a vectors encoding HpCheY1 and HpCheY1/D53A into wild-type E. coli (RP437) and into cheY (RP5232)- and cheZ (RP1616)-null mutants (Fig. 6). A control experiment using the empty pTrc99a vector was also conducted. Our results show that expression of HpCheY1 did not restore swarming of the cheY-null E. coli mutant. On the other hand, HpCheY1 inhibited swarming of wild-type E. coli in an IPTG concentration-dependent manner, suggesting that HpCheY1 associates with the E. coli chemotaxis system. Surprisingly, the expression of the HpCheY1/D53A mutant, a nonphosphorylatable mutant, inhibited E. coli swarming but did not affect cell growth. This result is different from that obtained by Alexandre et al. (1), who demonstrated that the heterologous expression of a wild-type CheY homolog from Azospirillum brasilense, but not the active-site mutant, inhibited the swarming of E. coli. Our results suggest that HpCheY1/D53A may interact with the chemotactic system of E. coli. Heterologous expression of CheY homologs from Rhodobacter sphaeroides partially restored cheZ-null mutant swarming, suggesting that CheY, with no motor binding affinity, competes with EcCheY for phosphate (35). In our study, HpCheY1, but not HpCheY1/D53A, restored the swarming ability of the cheZ-null mutant. It is possible that phosphorylation ability is necessary to bring the run-to-tumble ratio of the cheZ-null mutant close to that of the wild type. Wild-type HpCheY1, but not the D53A mutant, was able to receive a phosphate from EcCheA to modulate the concentration of EcCheY-P, therefore controlling the run-to-tumble ratio. Similar effects have also been observed in heterologous gene expression experiments using HpCheV2 and HpCheV3 (31). HpCheY1 failed to restore swarming of the cheY-null mutant, suggesting that HpCheY1 does not interact with EcFliM, even though HpCheY1 can be phosphorylated by EcCheA.
FIG. 6.
Swarming motility assay of heterologous expression of HpCheY1 and HpCheY1-D53A in the E. coli wild type (RP437) and cheZ (RP1616)- and cheY (RP5232)-null mutants. Overnight cultures of cells carrying plasmids were inoculated onto TB-0.4% semisolid agar plates that were incubated at 30°C for 7 h. (A) Representative images of the swarming rings produced from the expression of pTrc99a (control), HpCheY1, or HpCheY1/D53A in 0.5 mM IPTG are shown. (B) Mean diameters of the rings are plotted against IPTG concentrations. The experiment was replicated 3 times. ⋄, pTrc99a; ▪, HpCheY1; ▵, HpCheY1-D53A. (C) Protein expression levels of HpCheYs in E. coli wild-type and mutant strains were probed by anti-HpCheY1 antibody.
Sequence alignment between HpCheY1 and EcCheY suggested that the hydrophobic residues in α4 and β5 that are involved in FliMNM binding are either identical in the two proteins or are replaced by residues with similar amino acid properties (Fig. 1). However, salt bridge formation between the residues in helix 5 and the EcFliM peptide was disrupted (between the Asp12 Oδ of EcFliM-Lys119 and Nζ of EcCheY and between the Asn16 carboxylate of EcFliM-Lys122 and Nζ of EcCheY) (21). Although Lys119 of EcCheY is conserved in HpCheY1 (Lys115), the upshift of helix 5 caused displacement of Lys by a distance of approximately 4.4 Å (the distance between the Cα atom of Lys 115 in HpCheY and that of the equivalent Cα in EcCheY). Additionally, it was noted that the FliM-interacting α4/β5/α5 surface is more hydrophobic in HpCheY1. Specifically, the corresponding position of Lys122 in EcCheY is occupied by Val118 in HpCheY1 (Fig. 3A). Results of pull-down experiments from us and from Lowenthal et al. (23) show that HpCheY1-HpFliM interaction could be detected only if the concentration of KCl in the binding buffer was higher than 250 mM, while EcCheY1-EcFliM interaction was detected with no NaCl/KCl added (46). We speculated that the HpCheY1-HpFliM interaction would involve more extensive hydrophobic interactions. Sequence alignment of HpFliM and EcFliM revealed that the N-terminal fragments responsible for the interactions with CheY differ by four residues (G2A, S4-del, A9E, and N16E). It is likely that the interaction in H. pylori would be different from that in E. coli.
HpCheA-P2 preferentially interacts with HpCheY1.
One of the distinctive features of the H. pylori chemotactic pathway is the presence of a CheY-like domain fused to CheA (HpCheAY2); another distinctive feature is the presence of three CheV proteins. An in vitro experiment showed that HpCheA displayed a higher preference for HpCheY1 than HpCheY2; HpCheA had the lowest preference for the HpCheV proteins (15). The P2 domain in CheA consists of a docking site for response regulators and serves to increase the rate of phosphotransfer by concentrating CheY near P1 (14). We suspected that the binding affinities of response regulators for HpCheA-P2 would contribute to the differential rate of phosphotransfer in H. pylori. Interaction of HpCheA-P2 with the HpCheY and HpCheV proteins was investigated by a pull-down experiment and by gel filtration analysis.
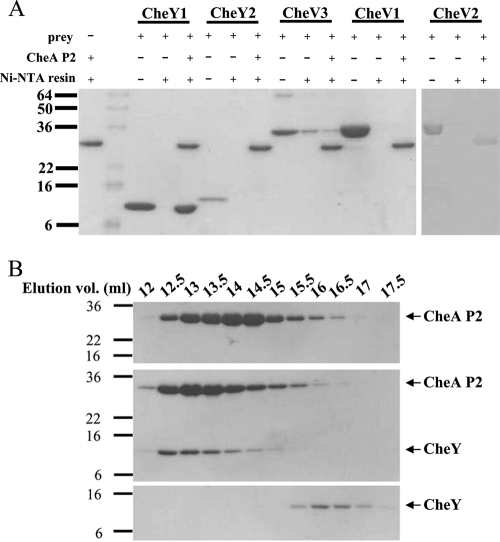
Recombinant HpCheA-P2-His8, HpCheY1, HpCheY2, HpCheV1, HpCheV2, and HpCheV3 were purified to greater than 95% purity. HpCheA-P2 was immobilized on Ni-NTA resin, followed by incubation with HpCheYs or HpCheVs. As shown in Fig. 7A, only HpCheY1 was shown to interact with HpCheA-P2, suggesting that HpCheY1 is the sole interacting partner of HpCheA-P2. The interaction between HpCheY1 and HpCheA-P2 was further investigated by gel filtration analysis. HpCheA-P2 and HpCheY1 were eluted at 14.57 ml and 16.05 ml, respectively, when run individually. When the two proteins were mixed and then subjected to the gel filtration analysis, the elution profile was shifted to 12.89 ml. Complex formation was further verified by SDS-PAGE analysis (Fig. 7B). Our results suggest that CheY1 formed a stable complex with CheA-P2. When the experiment was repeated with CheV3 and CheA-P2, the elution profile of the CheV3-CheA-P2 mixture did not change as compared to the elution profiles of the individual proteins (data not shown). We attempted to identify the surface of CheA-P2 that interacts with CheVs by homology modeling using Modeler (34). CheV3 was chosen as the representative CheV because it shares the highest sequence identity with HpCheY1. The P2 interaction patch on the α4/β5/α5 surface of HpCheV3 was found to be more electronegative than those of EcCheY and HpCheY1 (data not shown).
FIG. 7.
Interaction study of HpCheA-P2 and HpCheYs/CheVs. (A) Purified CheA-P2 in 0.1 mg/ml was immobilized. Response regulators in a 1:1 molar ratio to CheA-P2 were then added and incubated at 4°C for 1 h. After washing, the beads were boiled at 90°C and loaded for SDS-PAGE. (B) Elution profiles of CheY1 (lower panel), P2 (upper panel), and the CheY1-P2 complex (middle) separated by 10/30 Superdex 200 size exclusion chromatography. The elution volume is indicated above the graph. The elution volumes of P2, CheY1, and the CheY1-P2 complex are 14.57 ml, 16.05 ml, and 12.89 ml, respectively.
The high binding affinity of HpCheY1 for HpCheA-P2 observed in the present study is consistent with previous results showing that HpCheA has a greater phosphotransfer efficiency to HpCheY1 (15). Our data further suggest that the differential phosphotransfer efficiency is regulated by the interaction between the HpCheY/HpCheV proteins and the CheA-P2 domain. The P2 domain stands out because of its low sequence conservation among other regions of CheA, and HpCheA-P2 only shares 15% sequence identity with EcCheA-P2. In fact, the regulation would be complicated in HpCheV proteins, as the N-terminal CheW-like domain may interact with the P5 regulatory domain of CheA and affect the phosphotransfer activity. How this interaction affects the phosphotransfer reaction remains unclear. Structure determination studies of other CheY-like domains and of their complexes with CheA would help us unravel the complex regulatory mechanisms underlying the chemotactic network in H. pylori.
Acknowledgments
We thank John S. Parkinson for providing the E. coli mutants.
This work was supported by a Competitive Earmarked Research Grant (CUHK 4592/06 M) from the Research Grants Council of Hong Kong.
Footnotes
Published ahead of print on 5 March 2010.
REFERENCES
- 1.Alexandre, G., and I. B. Zhulin. 2003. Different evolutionary constraints on chemotaxis proteins CheW and CheY revealed by heterologous expression studies and protein sequence analysis. J. Bacteriol. 185:544-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, L. D. T. Moir, et al. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho, H. S., S. Y. Lee, D. Yan, X. Pan, J. S. Parkinson, S. Kustu, D. E. Wemmer, and J. G. Pelton. 2000. NMR structure of activated cheY. J. Mol. Biol. 297:543-551. [DOI] [PubMed] [Google Scholar]
- 5.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 6.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 7.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 8.Dyer, C. M., and F. W. Dahlquist. 2006. Switched or not? The structure of unphosphorylated CheY bound to the N terminus of FliM. J. Bacteriol. 188:7354-7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyer, C. M., M. L. Quillin, A. Campos, J. Lu, M. M. McEvoy, A. C. Hausrath, E. M. Westbrook, P. Matsumura, B. W. Matthews, and F. W. Dahlquist. 2004. Structure of the constitutively active double mutant CheYD13KY106W alone and in complex with a FliM Peptide. J. Mol. Biol. 342:1325-1335. [DOI] [PubMed] [Google Scholar]
- 10.Dyer, C. M., A. S. Vartanian, H. Zhou, and F. M. Dahlquist. 2009. A molecular mechanism of bacterial flagellar motor switching. J. Mol. Biol. 338:71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 13.Holm, L., S. Kaariainen, P. Rosenstrom, and A. Schenkel. 2008. Searching protein structure databases with DaliLite v. 3. Bioinformatics 24:2780-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahreis, K., T. B. Morrison, A. Garzón, and J. S. Parkinson. 2004. Chemotactic signaling by an Escherichia coli CheA mutant that lacks the binding domain for phosphoacceptor partners. J. Bacteriol. 186:2664-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiménez-Pearson, M. A., I. Delany, V. Scarlato, and D. Beier. 2005. Phosphate flow in the chemotactic response system of Helicobacter pylori. Microbiology 151:3299-3311. [DOI] [PubMed] [Google Scholar]
- 16.Reference deleted.
- 17.Kavermann, H., B. P. Burns, K. Angermuller, S. Odenbreit, W. Fischer, K. Melchers, and R. Haas. 2003. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197:813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. ClustalW and ClustalX version 2. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski, R. A., M. W. MacArthur, and J. M. Thornton. 1998. Validation of protein models derived from experiment. Curr. Opin. Struct. Biol. 8:631-639. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S. Y., H. S. Cho, J. G. Pelton, D. Yan, E. A. Berry, and D. E. Wemmer. 2001. Crystal structure of activated CheY. Comparison with other activated receiver domains. J. Biol. Chem. 276:16425-16431. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S. Y., H. S. Cho, J. G. Pelton, D. Yan, R. K. Henderson, D. S. King, L. Huang, S. Kustu, E. A. Berry, and D. E. Wemmer. 2001. Crystal structure of an activated response regulator bound to its target. Nat. Struct. Biol. 8:52-56. [DOI] [PubMed] [Google Scholar]
- 22.Leslie, A. 1995. MOSFLM users guide. MRC Laboratory of Moelcular Biology Cambridge, United Kingdom.
- 23.Lowenthal, A. C., M. Hill, L. K. Sycuro, K. Mehmood, N. R. Salama, and K. M. Ottemann. 2009. Functional analysis of the Helicobacter pylori flagellar switch proteins. J. Bacteriol. 191:7147-7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowenthal, A. C., C. Simon, A. S. Fair, K. Mehmood, K. Terry, S. Anastasia, and K. M. Ottemann. 2009. A fixed-time diffusion analysis method determines that the three cheV genes of Helicobacter pylori differentially affect motility. Microbiology 55:1181-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukat, G. S., W. R. McCleary, A. M. Stock, and J. B. Stock. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. U. S. A. 89:718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L.C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGee, D. J., M. L. Langford, E. L. Watson, J. E. Carter, Y. T. Chen, and K. M. Ottemann. 2005. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect. Immun. 73:1820-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, L. D., M. H. Russell, and G. Alexandre. 2009. Diversity in bacterial chemotactic responses and niche adaptation. Adv. Appl. Microbiol. 66:53-75. [DOI] [PubMed] [Google Scholar]
- 29.Parkinson, J. P. 1978. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J. Bacteriol. 135:45-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pazy, Y., A. C. Wollish, S. A. Thomas, P. J. Miller, E. J. Collins, R. B. Bourret, and R. E. Silversmith. 2009. Matching biochemical reaction kinetics to the timescales of life: structural determinants that influence the autodephosphorylation rate of response regulator proteins. J. Mol. Biol. 392:1205-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pittman, M. S., M. Goodwin, and D. J. Kelly. 2001. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147:2493-2504. [DOI] [PubMed] [Google Scholar]
- 32.Rao, C. V., G. D. Glekas, and G. W. Ordal. 2008. The three adaptation systems of Bacillus subtilis chemotaxis. Trends Microbiol. 16:480-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riepl, H., B. Scharf, R. Schmitt, H. R. Kalbitzer, and T. Maurer. 2004. Solution structures of the inactive and BeF3-activated response regulator CheY2. J. Mol. Biol. 338:287-297. [DOI] [PubMed] [Google Scholar]
- 34.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 35.Shah, D. S., S. L. Porter, D. C. Harris, G. H. Wadhams, P. A. Hamblin, and J. P. Armitage. 2000. Identification of a fourth cheY gene in Rhodobacter sphaeroides and interspecies interaction within the bacterial chemotaxis signal transduction pathway. Mol. Microbiol. 35:101-112. [DOI] [PubMed] [Google Scholar]
- 36.Silversmith, R. E., J. L. Appleby, and R. B. Bourret. 1997. Catalytic mechanism of phosphorylation and dephosphorylation of CheY: kinetic characterization of imidazole phosphates as phosphodonors and the role of acid catalysis. Biochemistry 36:14965-14974. [DOI] [PubMed] [Google Scholar]
- 37.Silversmith, R. E., J. G. Smith, G. P. Guanga, J. T. Les, and R. B. Bourret. 2001. Alteration of a nonconserved active site residue in the chemotaxis response regulator cheY affects phosphorylation and interaction with cheZ. J. Biol. Chem. 276:18478-18484. [DOI] [PubMed] [Google Scholar]
- 38.Smith, J. G., J. A. Latiolais, G. P. Guanga, S. Citineni, R. E. Silversmith, and R. B. Bourret. 2003. Investigation of the role of electrostatic charge in activation of the Escherichia coli response regulator CheY. J. Bacteriol. 185:6385-6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szurmant, H., and G. W. Ordal. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68:301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terry, K., A. C. Go, and K. M. Ottemann. 2006. Proteomic mapping of a suppressor of non-chemotactic cheW mutants reveals that Helicobacter pylori contains a new chemotaxis protein. Mol. Microbiol. 61:871-882. [DOI] [PubMed] [Google Scholar]
- 41.Terry, K., S. M. Williams, L. Connolly, and K. M. Ottemann. 2005. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect. Immun. 73:803-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas, S. A., J. A. Brewster, and R. B. Bourret. 2008. Two variable active site residues modulate response regulator phosphoryl group stability. Mol. Microbiol. 69:453-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uemura, N., S. Okamoto, S. Yamamoto, N. Matsumura, S. Yamaguchi, M. Yamakido, K. Taniyama, N. Sasaki, and R. J. Schlemper. 2001. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345:784-789. [DOI] [PubMed] [Google Scholar]
- 44.Volz, K., and P. Matsumura. 1991. Crystal structure of Escherichia coli CheY refined at 1.7-Å resolution. J. Biol. Chem. 266:15511-15519. [DOI] [PubMed] [Google Scholar]
- 45.Wadhams, G. H., and J. P. Armitage. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5:1024-1037. [DOI] [PubMed] [Google Scholar]
- 46.Welch, M., K. Oosawa, S. Aizawa, and M. Eisenbach. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:8787-8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams, S. M., Y. T. Chen, T. M. Andermann, J. E. Carter, D. J. McGee, and K. M. Ottemann. 2007. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect. Immun. 75:3747-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, Z., and S. W. Au. 2005. Mapping residues of SUMO precursors essential in differential maturation by SUMO-specific protease, SENP1. Biochem. J. 386:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan, D., H. S. Cho, C. A. Hastings, M. M. Igo, S. Y. Lee, J. G. Pelton, V. Stewart, D. E. Wemmer, and S. Kustu. 1999. Beryllofluoride mimics phosphorylation of NtrC and other bacterial response regulators. Proc. Natl. Acad. Sci. U. S. A. 96:14789-14794. [DOI] [PMC free article] [PubMed] [Google Scholar]