Abstract
Chlamydiae replicate within a nonacidified vacuole, termed an inclusion. As obligate intracellular bacteria, chlamydiae actively modify their vacuole to exploit host signaling and trafficking pathways. Recently, we demonstrated that several Rab GTPases are actively targeted to the inclusion. To define the biological roles of inclusion localized Rab GTPases, we have begun to identify inclusion-localized Rab effectors. Here we demonstrate that oculocerebrorenal syndrome of Lowe protein 1 (OCRL1), a Golgi complex-localized phosphatidylinositol (PI)-5-phosphatase that binds to multiple Rab GTPases, localizes to chlamydial inclusions. By examining the intracellular localization of green fluorescent protein (GFP) fusion proteins that bind to unique phosphoinositide species, we also demonstrate that phosphatidylinositol-4-phosphate (PI4P), the product of OCRL1, is present at the inclusion membrane. Furthermore, two additional host proteins, Arf1, which together with PI4P mediates the recruitment of PI4P-binding proteins to the Golgi complex, and PI4KIIα, a major producer of Golgi complex-localized PI4P, also localize to chlamydial inclusions. Depletion of OCRL1, Arf1, or PI4KIIα by small interfering RNA (siRNA) decreases inclusion formation and the production of infectious progeny. Infectivity is further decreased in cells simultaneously depleted for all three host proteins, suggesting partially overlapping functions in infected cells. Collectively, these data demonstrate that Chlamydia species create a unique replication-competent vacuolar environment by modulating both the Rab GTPase and the PI composition of the chlamydial inclusion.
Chlamydia species are obligate intracellular bacteria that primarily infect ocular, urogenital, and pulmonary mucosal surfaces (48). Chlamydia trachomatis infections are the leading cause of both bacterially acquired sexually transmitted disease (11) and preventable blindness worldwide (59). Infections caused by Chlamydia pneumoniae primarily result in upper respiratory disease and pneumonia (48), but may also be an associated risk factor for the development of atherosclerosis (26).
Chlamydiae undergo a unique biphasic developmental cycle characterized by the interconversion of two morphologically and functionally distinct cell types: the infectious elementary body (EB) and the metabolically active reticulate body (RB). Active entry into nonphagocytic cells results in the enclosure of EBs within a membrane-bound vacuole known as the inclusion. Chlamydia-mediated remodeling of the nascent inclusion (53), results in avoidance of lysosomal fusion and trafficking of the inclusion to the perinuclear region or the host microtubule organizing center (MTOC) (reviewed in reference 21). Within the confines of the inclusion, chlamydiae target multiple host trafficking pathways resulting in the fusion of Golgi complex (28)- and multivesicular body (MVB)-derived vesicles (5) with the inclusion membrane. Additionally, lipid droplets (LD), which are neutral lipid storage organelles, accumulate at the periphery and within the lumen of the inclusion (14, 34). Direct exploitation of host trafficking pathways by chlamydiae is likely mediated via chlamydial proteins that are either secreted into the host cytosol (71) or localized to the inclusion membrane (46).
Although the mechanisms by which chlamydiae target host trafficking pathways are not completely understood, several host trafficking factors, such as microtubules and dynein (13, 27, 49), a microtubule-associated minus-end-directed motor protein (50), are required for the intracellular trafficking of the inclusion. In addition, Rab (47), Rho (9, 10, 57), and the Arf family of small GTPases (2) are also targeted by and/or recruited to chlamydial inclusions. Rho and Arf family members are required during chlamydial entry. While roles in maintenance of Golgi structure in infected cells have been described for several Rab GTPases (38), roles for the additional inclusion-localized Rab GTPases have not yet been defined. Rab GTPases are the largest family of small ras-like GTPases that are important regulators of vesicularly mediated trafficking pathways in eukaryotic cells (44, 60). Localized to distinct organelles or organellar domains (69), Rab GTPases cycle between an inactive GDP-bound cytosolic state and an active GTP-bound membrane-localized state. In the active GTP-bound state, Rab GTPases interact with a wide variety of downstream effectors to facilitate the formation of transport vesicles at donor membranes, transport and docking of vesicles, and fusion of vesicles at target membranes (44). Importantly, organelle identity is determined in part by the composition of active Rab GTPases as well as the specific phosphoinositide (PI) species that are present on each organelle (6, 42, 44, 55, 69).
Phosphoinositides are short-lived phosphorylated derivatives of phosphatidylinositol that play key roles in cellular signaling and vesicle-mediated trafficking by controlling the subcellular localization and activation of effector PI-binding proteins (15, 63). Phosphorylation at different positions of the inositol ring yields seven different PI species that differ both in function and intracellular localization. The intracellular localization of PIs is tightly regulated by PI-specific kinases and phosphatases that either add or remove specific phosphate groups, respectively (33, 63). The protein OCRL1 (oculocerebrorenal syndrome of Lowe protein 1) is a Golgi complex-localized PI-5′-phosphatase that can hydrolyze PI(4,5)P2 (PIP2) to produce PI4P. To a lesser extent, OCRL1 hydrolyzes PI(3,4,5)P3 (PIP3) as well as the soluble lipids Ins(1,4,5)P3 and Ins(1,3,4,5)P4 (17, 43, 58). OCRL1 is primarily localized to the trans-Golgi network (TGN) through the interaction with multiple Rab GTPases (22, 32) and also to clathrin-coated transport vesicles that traffic between early endosomes (EE) and the TGN (12). OCRL1 has multiple functions, including roles in clathrin-dependent EE-to-TGN trafficking (12, 61), actin dynamics at the plasma membrane (20, 58), homeostasis of Golgi complex-localized PI levels (39), and endocytosis (18).
PI4P, the product of OCRL1, is predominantly localized to the Golgi complex (36). The Golgi complex-localized PI-OH(4)-kinases (PI4K) and Arf1, also play important roles in PI4P metabolism and localization of PI4P-binding proteins at the Golgi complex. In mammals, there are two classes of PI4Ks: the wortmannin-sensitive type III enzymes (PI4KIIIα and PI4KIIIβ) and the wortmannin-insensitive type II enzymes (PI4KIIα and PI4KIIβ) (3). Two of these enzymes, PI4KIIIβ and PI4KIIα, are primarily localized to the Golgi complex and contribute to the Golgi complex distribution of PI4P (66, 67). Arf1 is a small GTPase of the Arf family of GTPases that localizes to the Golgi complex and functions in the production of transport vesicles through the recruitment of coat proteins (16). Arf1 also plays a role in PI4P homeostasis and membrane trafficking at the Golgi complex through the recruitment of PI4K enzymes and PI4P-binding proteins to the Golgi complex (24, 25, 36).
Here we demonstrate that three host proteins, OCRL1, PI4KIIα, and Arf1, all of which regulate PI4P levels or the intracellular localization of PI4P-binding proteins in the host cell, are recruited to chlamydial inclusions. Furthermore, PI4P, the product of both OCRL1 and PI4KIIα, localizes to the inclusion membranes of the different Chlamydia species. All three proteins are required for optimal chlamydial development. Collectively, these data suggest that chlamydiae target key host regulatory proteins to the inclusion to alter the PI composition of the inclusion, which may result in novel host-pathogen interactions important for chlamydial pathogenesis.
MATERIALS AND METHODS
Cell culture and organisms.
Monolayer cultures of HeLa 229 epithelial cells (CCL-1.2; American Type Culture Collection) were grown in RPMI 1640 (Mediatech) supplemented with 10% fetal bovine serum (Atlas Biologicals) and 10 μg/ml of gentamicin (Invitrogen) at 37°C in an atmosphere of 5% CO and 95% humidified air. Chlamydia trachomatis lymphogranuloma venereum strain LGV-434 (serotype L2), TW5/OT (serovar B), UW-3/CX (serotype D), Chlamydia muridarum, and C. pneumoniae (AR-39) were propagated in HeLa cells and purified by Hypaque-76 (Nycomed, Inc.) density centrifugation (8). Infections were carried out as follows. For C. trachomatis serovar L2 infection, 24 h posttransfection, the cells were infected at a multiplicity of infection (MOI) of approximately 1 and incubated for the indicated times in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and 10 μg/ml of gentamicin at 37°C in an atmosphere of 5% CO2 and 95% humidified air. For C. trachomatis serovars B and D, C. muridarum, and C. pneumoniae, the chlamydial inoculum was centrifuged onto transfected HeLa cell monolayers for 1 h at 20°C at 900 × g, and cycloheximide was then added to growth media at a final concentration of 1 μg/ml. Cells were infected for 18 to 24 h, except for C. pneumoniae infections, which were infected for 48 h. Infectivity was measured by enumerating inclusion-forming units (IFU) as described previously (23), except that inclusions were visualized by indirect immunofluorescent microscopy using polyclonal antiserum against formalin-fixed C. trachomatis serovar L2 EBs (antichlamydial antiserum) and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG.
Reagents and antibodies.
Mouse antichlamydial lipopolysaccharide (LPS) was generously provided by Harlan Caldwell (Rocky Mountain Laboratories [RML], National Institutes of Allergy and Infectious Diseases [NIAID], National Institutes of Health [NIH]), and rabbit anti-C. trachomatis EB antiserum (antichlamydial) was prepared as previously described (54). The following primary antibodies were used: anti-GM130 (BD Transduction Laboratories), anti-TGN46 (Abcam), antigiantin (Santa Cruz), and anti-PI4P (Echelon, Inc.). The following secondary antibodies were used: goat anti-mouse IgG or goat anti-rabbit IgG conjugated to Texas Red (Jackson ImmunoResearch Laboratories), goat anti-mouse and anti-rabbit IgG conjugated to Cy5 (Zymed Laboratories, Inc.), and goat anti-rabbit conjugated to FITC (Jackson ImmunoResearch Laboratories, Inc.). Calpain III inhibitor and MG-132 were purchased from EMD Chemicals, Inc., while WST-1 was purchased from Roche Applied Science.
Plasmid constructions.
The green fluorescent protein (GFP) plasmids utilized in this study were constructed by PCR amplification using standard molecular biology techniques. After cloning each PCR fragment into pGEM-T Easy (Promega), each gene was cloned into an enhanced GFP (EGFP) vector (Clontech) of the appropriate reading frame. GFP-OSBP-(PHR107ER108E)×2 was constructed in several steps. First site-directed mutagenesis was employed (QuikChange; Stratagene) to create GFP-OSBP-PHR107ER108E. DNA encoding OSBP-PHR107ER108E was then PCR amplified and cloned into pGEM-T Easy. The XhoI/EcoRI fragment containing the OSBP-PHR107ER108E with a 5′ linker sequence was subsequently cloned with the XhoI/EcoRI sites of GFP-OSBP-PHR107ER108E, creating a duplication of the mutated oxysterol binding protein-pleckstrin homology (OSBP-PH) domain. GFP-OSBP-(PH)×2 was constructed by PCR amplification and cloned in frame with GFP-OSBP-PH, creating a duplication of the OSBP-PH domain. GFP-OCRL1Δ237-539 was constructed by SOEing (splicing by overlap extension) PCR. First, amino acids 1 to 236 of OCRL1 and amino acids 539 to 8493 were independently PCR amplified. A second round of PCR amplification was performed using the PCR products described above as a template, and the product was cloned into EGFP-C2. GFP-OCRL1S564P was constructed by site-directed mutagenesis using QuikChange (Stratagene). All PCR amplifications were performed using HiFidelity Platinum Taq polymerase (Invitrogen), and each construct was confirmed by DNA sequencing (BioResource Center, Cornell University, Ithaca, NY). The plasmids (pCMV-OSBP, pCMV-PLCδ1, pCMV-PI4KIIα, and pCMV-PI4KIIβ) used as DNA templates were purchased from Origene, while pcDNA3.1-Arf1 was purchased from cDNA.org. The PH domain of Goodpasture antigen binding protein (GPBP) was PCR amplified from a cDNA HeLa library (Clontech). GFP-OCRL1 was a generous gift from M. Lowe (University of Manchester, Manchester, United Kingdom), and PI4KIIIβ-GFP was a generous gift from T. Balla (NICHD, NIH, Bethesda, MD).
Eukaryotic cell transfection, siRNA, and infection.
For transient transfection experiments, HeLa 229 cells were grown on 12-mm-diameter glass coverslips (no. 1 thickness) in 24-well plates (Corning, Inc.) at 37°C in an atmosphere of 5% CO2 and 95% humidified air. Cells were washed once in serum-free RPMI 1640 and transfected with Lipofectamine (Invitrogen) using a total of 0.4 μg of DNA per well according to the manufacturer's protocol (Invitrogen). For small interfering RNA (siRNA) gene silencing experiments, cells were washed once in serum-free RMPI 1640 and transfected with 0.4 μl of a 50 μM stock of OCRL1 (Ambion; ID s9820), PI4KIIα (Ambion; ID 1170), Arf1 (Ambion; ID s1551), or negative control siRNA (Ambion; AM4611) for 72 h using Lipofectamine (Invitrogen). Cells were incubated with siRNA oligonucleotides for 72 h prior to infection.
Brefeldin A, nocodazole, and chloramphenicol treatment of cells.
For examination of association of GFP plasmids with the inclusion in the presence of nocodazole or brefeldin A (BFA), cells were infected with C. trachomatis for 18 h prior to treatment with 1 μg/ml BFA for 30 min at 37°C or with 20 μM nocodazole for 3 h at 37°C. For examination of trafficking of GFP-tagged plasmids to the inclusion, cells were pretreated with 20 μM nocodazole for 2 h at 37°C or with 1 μg/ml BFA for 30 min at 37°C prior to infection with C. trachomatis. Infected cells were incubated for an additional 18 h at 37°C in the presence of either BFA or nocodazole. To inhibit chlamydial protein synthesis, cells were treated with 25 μg/ml chloramphenicol immediately after infection with C. trachomatis and incubated for 18 h at 37°C.
Indirect immunofluorescence and LSCM.
Cells were fixed in 4% formaldehyde in PBS for 60 min or in ice-cold methanol for 10 min at room temperature. For antibody labeling of formaldehyde-fixed cells, cells were permeabilized in PBS containing 0.05% saponin and 0.2% bovine serum albumin (BSA) for 10 min at room temperature, and primary and secondary antibodies were incubated in permeabilization buffer sequentially for 60 min each at room temperature. Following incubation with each antibody, coverslips were washed three times with PBS containing 0.05% saponin and 0.2% BSA. For antibody labeling of methanol-fixed cells, cells were blocked in PBS containing 3% BSA for 10 min at room temperature, and primary and secondary antibodies were incubated in blocking buffer sequentially for 60 min each at room temperature. Following incubation with each antibody, coverslips were washed three times with PBS. Coverslips were mounted onto glass slides using Prolong Antifade (Molecular Probes) and viewed by laser-scanning confocal microscopy (LSCM). For LSCM, an Olympus Fluoview 500 confocal laser-scanning imaging system equipped with argon, krypton, and He-Ne lasers on an Olympus IX70 inverted microscope with a PLAPO 60× objective was used (Olympus America, Inc.). Images were processed using Adobe Photoshop 6.0 (Adobe Systems, Inc.).
RNA isolation and RT-PCR.
Total RNA from siRNA-treated HeLa cells was purified using Trizol reagent (Invitrogen) and the RNeasy mini kit according to manufacturer's protocol (Qiagen). Isolated RNA was treated with TURBO DNase (Ambion) prior to performing RT-PCR using the Access RT-PCR system (Promega).
RESULTS
OCRL1 is recruited to chlamydial inclusions in a species-independent manner.
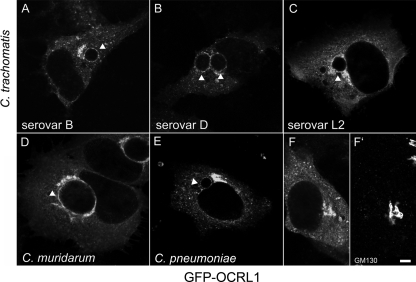
We have previously demonstrated that several Rab GTPases are recruited to the chlamydial inclusion in both a species-independent and -dependent manner (47). Since Rab GTPases function through their interaction with downstream effector molecules, Rab effectors are likely to interact with inclusion-localized Rab GTPases. Consistent with this hypothesis, BICD1, a Rab6 effector, is recruited to inclusions (41). In this study, we have focused on a second Rab effector, OCRL1, a PI-4,5-bisphosphate 5-phosphatase that interacts with multiple Rab GTPases (22, 32), of which several (e.g., Rab1, -6, -8, and -14) localize to chlamydial inclusions (47; data not shown). To determine whether OCRL1 localized to chlamydial inclusions, we examined the intracellular localization of GFP-OCRL1 in HeLa cells infected with C. trachomatis serovars L2, B, and D; Chlamydia muridarum; and C. pneumoniae. Cells were fixed and stained with anti-GM130, a marker for the cis-Golgi complex, to verify Golgi localization of GFP-OCRL1 (17, 32) (Fig. 1 F and F′) and with antichlamydial LPS to visualize the chlamydial inclusion (data not shown). Cells were viewed by laser-scanning confocal microscopy (LSCM). As shown in Fig. 1, GFP-OCRL1 is recruited to inclusions of all Chlamydia species examined, as demonstrated by the distinct rim-like immunofluorescent staining of GFP-OCRL1 that surrounds each of the inclusions. Although difficult to quantitate due to variable expression levels, the C. pneumonaie inclusion consistently recruits less GFP-OCRL1.
FIG. 1.
GFP-OCRL1 is recruited to chlamydial inclusions in a species-independent fashion. HeLa cells transiently expressing GFP-OCRL1 were infected with C. trachomatis serovar B (24 h) (A), C. trachomatis serovar D (18 h) (B), C. trachomatis serovar L2 (18 h) (C), C. muridarum (18 h) (D), and C. pneumoniae (48 h) (E). (F and F′) Uninfected HeLa cells transiently expressing GFP-OCRL1 were stained with GM130 antibody to confirm Golgi complex localization of GFP-OCRL1. Cells were fixed and viewed by LSCM. Arrowheads indicate inclusion. Scale bar, 5 μm.
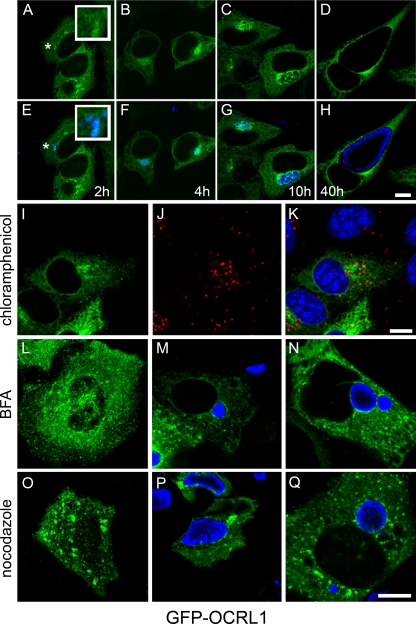
Since chlamydiae undergo a well-defined developmental cycle that is correlated with chlamydial differentiation and inclusion trafficking, we wanted to determine at what time during the chlamydial developmental cycle GFP-OCRL1 localized to the inclusion. To address this question, we examined the intracellular localization of GFP-OCRL1 in HeLa cells infected with C. trachomatis serovar L2 at high MOIs at different times postinfection (p.i.). Prior to 10 h p.i., chlamydiae remain in multiple unfused vacuoles that are aggregated at the peri-Golgi region in cells infected at high MOIs. Two hours p.i., EBs that have aggregated at the peri-Golgi region begin to accumulate GFP-OCRL1 (Fig. 2A and E). By 4 h p.i., the association with GFP-OCRL1 is much more prominent (Fig. 2B and F). These data suggest that GFP-OCRL1 is recruited to the chlamydial inclusion upon localization to the peri-Golgi region. GFP-OCRL1 remained associated with the inclusion during the remainder of the developmental cycle (Fig. 2D and H). Furthermore, GFP-OCRL1 failed to localize to chlamydia-containing vacuoles in the presence of chloramphenicol, a bacterial translation inhibitor (Fig. 2I to K). These data support the model that chlamydia-mediated remodeling of the inclusion is necessary for recruitment of OCRL1. Collectively, these data suggest that chlamydiae modify the inclusion early during the developmental cycle to recruit OCRL1 and that OCRL1 may play a role during chlamydial infection at both early and late times p.i.
FIG. 2.
Association of GFP-OCRL1 with the chlamydial inclusion. (A to H) HeLa cells transiently expressing GFP-OCRL1 were infected with C. trachomatis serovar L2 at an MOI of approximately 25 to 50. At the indicated time points p.i. (2 h, 4 h, 10 h, and 40 h), cells were fixed and stained with antichlamydial antiserum (blue) and viewed by LSCM. (E to H) Merged images of GFP-expressing cells (green) and chlamydia staining (blue). The insets are selected regions denoted by asterisks. Scale bar, 10 μm. (I to K) Inhibition of chlamydial gene expression with chloramphenicol prevents recruitment of GFP-OCRL1 to chlamydia-containing vacuoles. HeLa cells transiently expressing GFP-OCRL1 (I; green) were infected with C. trachomatis serovar L2 in the presence of 25 μg/ml chloramphenicol and incubated for 18 h at 37°C. Cells were fixed and stained with antichlamydial antisera (J; red) and viewed by LSCM. (K) Merged image of panels I and J. (L to N) Golgi complex-dependent trafficking is not essential for the trafficking and association of GFP-OCRL1 with the inclusion. (L) Uninfected HeLa cells transiently expressing GFP-OCRL1 (green) were treated with 1 μg/μl BFA for 30 min at 37°C. (M) HeLa cells transiently expressing GFP-OCRL1 were infected with C. trachomatis serovar D. Eighteen hours p.i., cells were treated with 1 μg/μl BFA for 30 min at 37°C. (N) HeLa cells transiently expressing GFP-OCRL1 were pretreated with 1 μg/μl BFA for 30 min prior to infection with C. trachomatis serovar D. Cells were incubated for an additional 18 h in the continued presence of BFA. Fixed cells were stained with antichlamydial antisera (blue) and viewed by LSCM. (O to Q) Microtubules are not essential for the trafficking or association of GFP-OCRL1 with inclusions. (O) Uninfected HeLa cells expressing GFP-OCRL1 were treated with 20 μM nocodazole for 3 h at 37°C. (P) HeLa cells transiently expressing GFP-OCRL1 were infected with C. trachomatis serovar D. Eighteen hours p.i., cells were treated with 20 μM nocodazole for 3 h at 37°C. (Q) HeLa cells transiently expressing GFP-OCRL1 were pretreated with 20 μm nocodazole for 2 h prior to infection with C. trachomatis serovar D. Cells were incubated for an additional 18 h in the presence of nocodazole. (O to Q) Fixed cells were stained with antichlamydial antisera (blue) and viewed by LSCM. Scale bar, 10 μm.
Recruitment of OCRL1 to chlamydial inclusions is mediated by its carboxy-terminal Rab-binding domain.
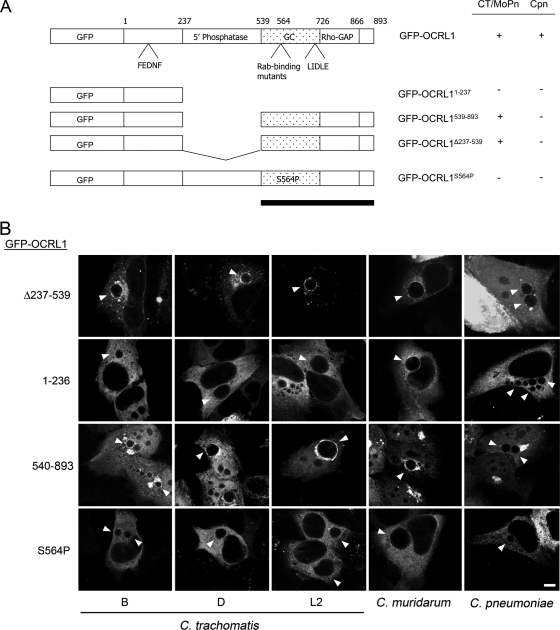
To delineate the region of OCRL1 that mediates its recruitment to the inclusion, we constructed GFP-tagged constructs encompassing different regions of OCRL1 as depicted in Fig. 3A. Amino acids 1 to 236 contain an AP-2 binding site (61), amino acids 237 to 539 encode the catalytic phosphatase domain (1, 12), and amino acids 540 to 893 are required for Golgi localization and contain separate Rab GTPase, clathrin heavy-chain binding domains, and a Rho-GAP-like domain (12, 61). HeLa cells transiently expressing GFP-OCRL1Δ237-539, GFP-OCRL11-236, or GFP-OCRL1540-893 were infected with C. trachomatis serovars B, D, and L2; C. muridarum; and C. pneumoniae (Fig. 3B). Cells were fixed, stained with anti-GM130 and anti-chlamydial LPS antisera (data not shown), and viewed by LSCM. Consistent with previously published data, GFP-OCRL11-236 was distributed throughout the cytosol, while GFP-OCRL1540-893 colocalized with GM130 at the Golgi complex (32). Both GFP-OCRL1540-893 and GFP-OCRL1Δ237-539 were recruited to all inclusions examined, except those of C. pneumoniae (Fig. 3B). In contrast, GFP-OCRL11-236 was not recruited to any chlamydial inclusion (Fig. 3B). These data demonstrate that amino acids 540 to 893 of OCRL1 are sufficient to localize GFP to C. trachomatis and C. muridarum but not to C. pneumoniae inclusions.
FIG. 3.
The carboxy-terminal Rab binding domain mediates the recruitment of OCRL1 to C. trachomatis and C. muridarum inclusions. (A) GFP-OCRL1 fusion proteins utilized in this study. Amino acids 1 to 236 contain an AP-2 binding site (FEDNF); amino acids 237 to 539 encode the catalytic 5-phosphatase domain; and amino acids 540 to 893 are required for Golgi complex localization (GC; stippled box) and contain separate Rab GTPase, clathrin heavy-chain binding domains (LIDLE), and the Rho-GAP-like domain. The black bar indicates the inclusion-targeting domain for C. trachomatis and C. muridarum. + indicates localization of GFP fusion protein to inclusion; − indicates no localization of the GFP fusion protein to the inclusion. CT, C. trachomatis; MoPn, C. muridarum; Cpn, C. pneumoniae. (B) HeLa cells transiently expressing GFP-OCRL1Δ237-539, GFP-OCRL11-236, GFP-OCRL1540-893, or GFP-OCRL1S564P were infected with C. trachomatis serovar B (24 h), C. trachomatis serovar D (18 h), C. trachomatis serovar L2 (18 h), C. muridarum (18 h), and C. pneumoniae (48 h). Cells were fixed and viewed by LSCM. Arrowheads indicate inclusions. Scale bar, 5 μm.
Since OCRL1-interacting Rab GTPases are localized to the inclusion (32, 47) and the minimal region required for inclusion localization to the C. trachomatis inclusion includes the Rab-binding domain, we wanted to determine whether the ability of OCRL1 to bind Rab GTPases is required for its association with the inclusion. To address this question, we constructed a single point mutant, GFP-OCRL1S564P, which is unable to bind Rab GTPases and fails to localize to the Golgi apparatus (32), and examined the ability of this mutant to localize to chlamydial inclusions. Yeast two-hybrid analysis confirmed that OCRL1S564P did not interact with Rab1 or Rab6 (data not shown), while LSCM confirmed that GFP-OCRL1S564P was localized to the cytosol in uninfected cells. In infected cells, GFP-OCRL1S564P failed to localize to the inclusions of all Chlamydia species examined (Fig. 3B). These data demonstrate that the ability of OCRL1 to interact with Rab GTPases is necessary for its localization to the chlamydial inclusion. The lack of recruitment of GFP-OCRL1Δ237-539 and GFP-OCRL1540-893 to the C. pneumoniae inclusion suggests that the Rab binding domain and amino acids localized to the central phosphatase region are each required for the localization of GFP-OCRL1 to the C. pneumoniae inclusion.
Since Rab binding has previously been shown to be necessary for the Golgi localization of OCRL1 (32), the failure of GFP-OCRL1S564P to be recruited to inclusions indicates that either OCRL1 must be targeted to the Golgi complex prior to trafficking to the inclusion or that interaction with Rab GTPases at the inclusion membrane is important for the ability of OCRL1 to localize to the inclusion. To examine whether transit through the Golgi complex was required, we examined the intracellular localization of GFP-OCRL1 in C. trachomatis serovar D-infected cells grown in the presence of BFA, a fungal metabolite that inhibits anterograde trafficking from the endoplasmic reticulum (ER) to the Golgi complex (51). To examine whether an intact Golgi complex is required for continued association of OCRL1 with the inclusion, at 18 h p.i., cells were treated with BFA for 30 min (Fig. 2M). In the presence of BFA, although GFP-OCRL1 no longer localized to the Golgi complex, it remained still associated with to the C. trachomatis inclusion, demonstrating that an intact Golgi apparatus is not required for the continued association of GFP-OCRL1 with the inclusion (Fig. 2M). Furthermore, GFP-OCRL1 still localized to inclusions in cells that were pretreated with BFA prior to infection, demonstrating that prior trafficking through the Golgi apparatus is not required for the initial trafficking to the inclusion (Fig. 2N). Since OCRL1 is also trafficked within the cell by microtubule-dependent mechanisms (12), we addressed whether the recruitment to and association of GFP-OCRL1 with the inclusion were microtubule dependent. Cells were either treated with nocodazole, a microtubule-depolymerizing drug, prior to infection (Fig. 2Q) or for 3 h at 18 h p.i. (Fig. 2P). In the absence of intact microtubules, GFP-OCRL1 still associated with the inclusion. Although GFP-OCRL1 is recruited to the chlamydial inclusion, in the presence of BFA and nocodazole, the recruitment is slightly diminished. Collectively, these data suggest that Golgi- or microtubule-dependent trafficking pathways may play minor roles, but they are not essential for the delivery to and/or maintenance of OCRL1 at the inclusion.
GFP-OSBP-PH and GFP-GPBP-PH localize to chlamydial inclusions.
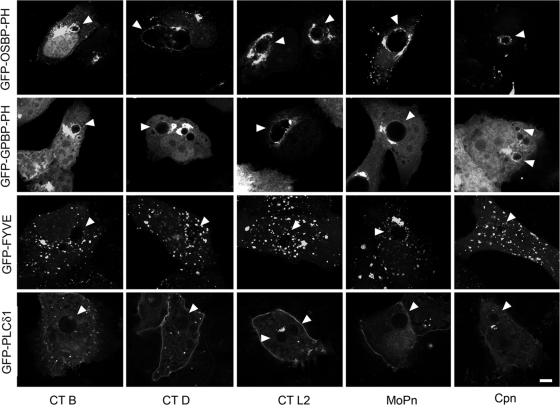
Since OCRL1 is a PI-5-phosphatase that produces PI4P (1, 39), we next examined whether PI4P was present at the inclusion. To test this, we examined the intracellular localization of GFP fusions containing the pleckstrin homology (PH) domains of the PI4P-binding proteins oxysterol-binding protein (OSBP) and Goodpasture antigen binding protein (GPBP) (37). The PH domains of OSBP and GPBP interact with PI4P and, to a lesser extent, PIP2 in vitro (37) and have been used to demonstrate the presence of PI4P at the Golgi complex (36). In addition, we also expressed GFP-FYVE, which localizes to EE and recognizes PI3P (40), and GFP-PLCδ1-PH, which localizes predominantly at the plasma membrane and recognizes PIP2 (68). As shown in Fig. 4, both GFP-OSBP-PH and GFP-GPBP-PH localized to the Golgi complex as well as to the inclusion membrane. In contrast, neither GFP-PLCδ1-PH nor GFP-FYVE localized to mature inclusions (Fig. 4). That lack of recruitment of GFP-PLCδ1-PH and GFP-FYVE confirms that the mature inclusion displays minimal to no characteristics of the plasma membrane or early endosomes. Since GFP-PLCδ1-PH recognizes PIP2 and is not localized to the inclusion, these data suggest that GFP-OSBP-PH and GFP-GPBP-PH are likely localized to the inclusion due to the presence of PI4P and not PIP2.
FIG. 4.
PI4P is present on the chlamydial inclusion membrane. HeLa cells transiently expressing GFP-OSBP-PH, GFP-GPBP-PH, GFP-PLCδ1-PH, or GFP-FYVE were infected with C. trachomatis serovar B (24 h), C. trachomatis serovar D (18 h), C. trachomatis serovar L2 (18 h), C. muridarum (18 h), and C. pneumoniae (48 h). Infected cells were fixed and viewed by LSCM. Arrowheads indicate inclusions. CT, C. trachomatis; MoPn, C. muridarum; Cpn, C. pneumoniae. Scale bar, 5 μm.
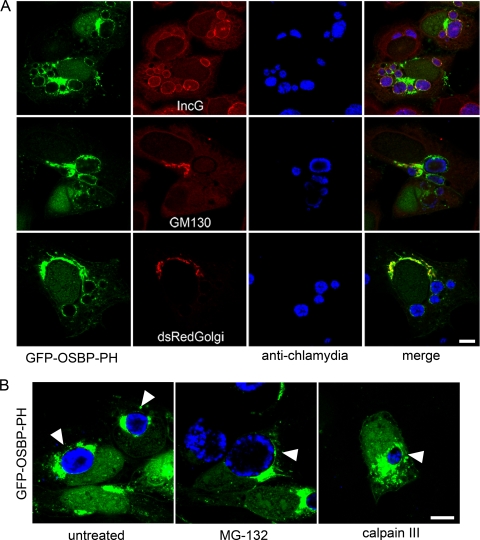
A recent report by Heuer and colleagues demonstrated that chlamydiae induce fragmentation of the Golgi apparatus, resulting in the close apposition of Golgi ministacks at the inclusion membrane (31). To determine whether the localization of PI4P to the inclusion resulted from association with Golgi ministacks, we compared the localization of GFP-OSBP-PH at the inclusion with several markers of the Golgi apparatus. Dual labeling of C. trachomatis serovar D-infected cells expressing GFP-OSBP-PH with the cis-Golgi marker, GM130, or coexpression with the trans-Golgi-localized dsRed-β1,4 galactosyltransferase (dsRed Golgi; Clontech) demonstrated that although there is some colocalization between GFP-OSBP-PH and these selected Golgi markers, the majority of the inclusion-localized GFP-OSBP-PH does not colocalize with markers of the Golgi complex (Fig. 5A). Furthermore, dual labeling of GFP-OSBP-PH with IncG demonstrated partial colocalization of GFP-OSBP-PH with IncG, thus providing further evidence that PI4P resides within the inclusion membrane (Fig. 5A). To confirm that PI4P does not localize to the inclusion as a result of chlamydia-mediated disruption of the Golgi apparatus, we treated cells with calpain III to prevent the formation of Golgi complex ministacks in infected cells. Inhibition of caspases with calpain III has previously been shown to prevent Chlamydia-mediated Golgi complex fragmentation (31). Cells were also treated with the proteosomal inhibitor MG132, which does not prevent Golgi complex fragmentation (31). Even though Golgi complex fragmentation was inhibited in cells treated with calpain III, as evidenced by the smaller inclusion size and the loss of golgin-84 cleavage (31; data not shown), PI4P still localized to the inclusion, as demonstrated by the localization of GFP-OSBP-PH (Fig. 5B).
FIG. 5.
GFP-OSBP-PH localizes to the inclusion membrane in the absence of Golgi complex markers. (A) HeLa cells transiently expressing GFP-OSBP-PH (green) were infected with C. trachomatis serovar D. Eighteen hours p.i., cells were fixed and dually labeled with either anti-IncG (red) and antichlamydial antisera (blue) or anti-GM130 (red) and antichlamydial antisera (blue). HeLa cells transiently coexpressing GFP-OSBP-PH (green) and dsRed-Golgi (red) (Clontech) were fixed and stained with antichlamydial antisera (blue). (B) HeLa cells transiently expressing GFP-OSBP-PH (green) were infected with C. trachomatis serovar L2. Eight hours p.i., cells were mock treated or treated with 2.5 μM MG132 or 100 μM calpain III and incubated for an additional 18 h at 37°C. Cells were fixed and stained with antichlamydial antisera (blue). Arrowheads indicated GFP-OSBP-PH localization on selected inclusions. Cells were viewed by LSCM. Scale bar, 10 μm.
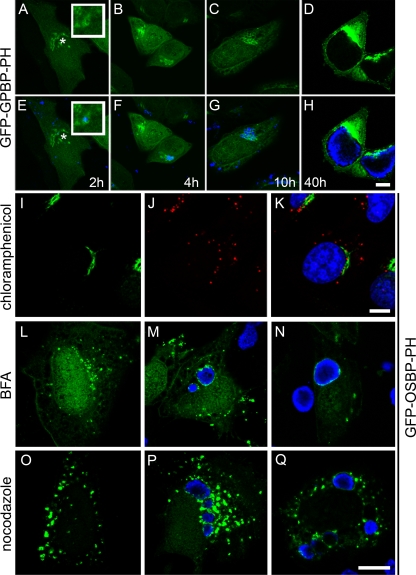
Since PI4P is enriched in the Golgi complex (36) and Golgi complex-derived vesicles are delivered to the inclusion (28, 29), we wanted to determine whether PI4P is delivered to the inclusion via a Golgi complex-derived-intermediate, or alternatively, whether it may be produced directly at the inclusion membrane. Since BFA induces the redistribution of GFP-OSBP-PH into the cytosol (4) and inhibits the delivery of Golgi complex-derived NBD-sphingomyelin to the inclusion (28), we examined whether GFP-OSBP-PH was localized to the inclusion in cells treated with BFA, either prior to infection (Fig. 6N) or postinfection (Fig. 6M). Although GFP-OSBP-PH no longer localized to the Golgi complex in the presence of BFA, it still localized to the inclusion (Fig. 6M and N). These data suggest that the PI4P present at the inclusion is not derived from Golgi complex-produced PI4P. Furthermore, similar to the recruitment of GFP-OCRL1, a time course analysis demonstrated that PI4P surrounds EBs aggregated at the peri-Golgi region between 2 and 4 h p.i., as determined by localization of GFP-GPBP-PH (Fig. 6A to H) and GFP-OSBP-PH (data not shown) to the chlamydia-containing vacuole. Similar to what was observed for OCRL1, the association of GFP-OSBP-PH to the inclusion was dependent on chlamydial gene expression (Fig. 6I-K). Although there was consistently less recruitment of GFP-OSBP-PH in the presence of nocodazole, it was still recruited, demonstrating that microtubules are also not essential (Fig. 6P and Q).
FIG. 6.
Association of the PI4P-binding PH domain with the chlamydial inclusion. (A to H) HeLa cells transiently expressing GFP-GPBP-PH were infected with C. trachomatis serovar L2 at an MOI of approximately 25 to 50. At the indicated time points p.i. (2 h, 4 h, 10 h, and 40 h), cells were fixed and stained with antichlamydial antiserum (blue) and viewed by LSCM. (E to H) merged images of GFP-expressing cells (green) and chlamydial staining (blue). The insets are selected regions denoted by asterisks. Scale bar, 10 μm. (I to K) Inhibition of chlamydial gene expression with chloramphenicol prevents recruitment of GFP-OSPB-PH to chlamydia-containing vacuoles. HeLa cells transiently expressing GFP-OSPB-PH (I; green) were infected with C. trachomatis serovar L2 in the presence of 25 μg/ml chloramphenicol and incubated for 18 h at 37°C. Cells were fixed and stained with antichlamydial antisera (J; red) and viewed by LSCM. (K) Merged image of panels I and J. (L to N) Golgi complex-dependent trafficking is not necessary for the trafficking and association of GFP-OSBP-PH with the inclusion. (L) Uninfected HeLa cells transiently expressing GFP-OSBP-PH (green) were treated with 1 μg/μl BFA for 30 min at 37°C. (M) HeLa cells transiently expressing GFP-OSBP-PH were infected with C. trachomatis serovar D. Eighteen hours p.i., cells were treated with 1 μg/μl BFA for 30 min at 37°C. (N) HeLa cells transiently expressing GFP-OSBP-PH were pretreated with 1 μg/μl BFA for 30 min prior to infection with C. trachomatis serovar D. Cells were incubated for an additional 18 h in the continued presence of BFA. (O to Q) Microtubules are not essential for the trafficking or association of GFP-OSBP-PH with inclusions. (O) Uninfected HeLa cells expressing GFP-OSBP-PH were treated with 20 μM nocodazole for 3 h at 37°C. (P) HeLa cells transiently expressing GFP-OSBP-PH were infected with C. trachomatis serovar D. Eighteen hours p.i., cells were treated with 20 μM nocodazole for 3 h at 37°C. (Q) HeLa cells transiently expressing GFP-OSBP-PH were pretreated with 20 μM nocodazole for 2 h prior to infection with C. trachomatis serovar D. Cells were incubated for an additional 18 h in the presence of nocodazole. (L to Q) Fixed cells were stained with antichlamydial antisera (blue) and viewed by LSCM. Scale bar, 10 μm.
Arf1 is recruited to chlamydial inclusions.
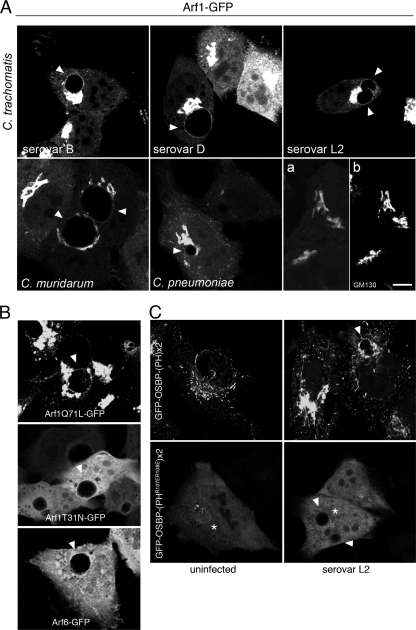
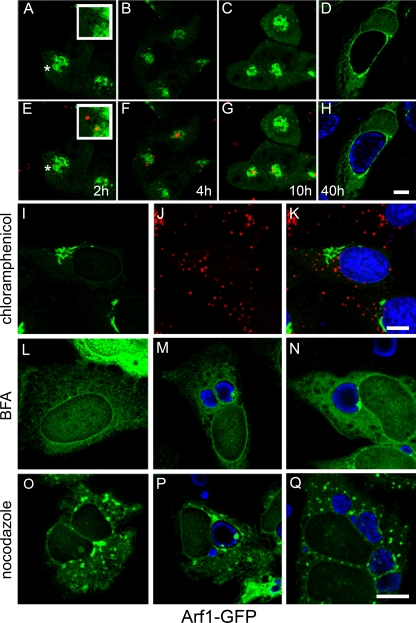
Although the majority of GFP-OSBP-PH and GFP-GPBP-PH is targeted to the Golgi complex by a PI4P-dependent mechanism, a small proportion is targeted by a PI4P-independent but Arf1-dependent mechanism (36). Arf1 is a small GTPase that functions in ER-to-Golgi complex trafficking and Golgi complex maintenance through recruitment of a wide variety of effector proteins (16). To determine whether Arf1 may also contribute to the recruitment of GFP-OBSP-PH and GFP-GPBP-PH to the inclusion, we first examined whether Arf1-GFP was present at the inclusion. In contrast to Arf6-GFP, which is recruited and activated at sites of chlamydial entry (2), Arf1-GFP is localized to mature C. trachomatis (serovars B, D, and L2) and C. muridarum inclusions, but not to C. pneumoniae inclusions (Fig. 7A and B). Furthermore, Arf1-GFP was localized to the inclusion in a guanine nucleotide-dependent manner with active GTP-Arf1 (Arf1Q71L-GFP) but not inactive GDP-Arf1 (Arf1T31N-GFP) localizing to the inclusion (Fig. 7B). Unlike GFP-OCRL1 and GFP-OSBP-PH, Arf1-1 was not clearly detected at chlamydia-containing vacuoles until at least 10 h p.i (Fig. 8A to H). Furthermore, although the recruitment of Arf1 to the inclusion was diminished slightly in the presence of nocodazole, Arf1-GFP was still recruited to the inclusion (Fig. 8P and Q). However, its association with the inclusion was partially sensitive to BFA (Fig. 8M and N). Since BFA inactivates Arf1 at the Golgi apparatus, these data suggest that a portion of Arf1 is regulated in a similar BFA-sensitive manner at the inclusion.
FIG. 7.
Arf1-GFP localizes to chlamydial inclusions but is not sufficient to mediate recruitment of GFP-OSBP-PH. (A) HeLa cells transiently expressing Arf1-GFP were infected with C. trachomatis serovar B (24 h), C. trachomatis serovar D (18 h), C. trachomatis serovar L2 (18 h), C. muridarum (18 h), and C. pneumoniae (48 h). Cells were fixed and viewed by LSCM. (a and b) Uninfected HeLa cells expressing Arf1-GFP (a) were fixed and stained with anti-GM130 (b) to confirm Golgi complex localization of the fusion protein. Scale bar, 10 μm. (B) HeLa cells transiently expressing constitutively active Arf1Q71L-GFP, dominant-negative Arf1T31N-GFP, or Arf6-GFP were infected with C. trachomatis serovar L2 for 18 h. Cells were fixed, stained with antichlamydial antisera (data not shown), and viewed by LSCM. (C) HeLa cells transiently expressing GFP-OSBP-(PH)×2 and GFP-OSBP-(PHR107ER108E)×2, a PI4P-binding-defective mutant, were mock infected or infected with C. trachomatis serovar L2 for 18 h. Cells were fixed and viewed by LSCM. Residual Golgi complex localization of the PI4P-binding deficient mutant is depicted by asterisks. Arrowheads indicate inclusions.
FIG. 8.
Association of Arf1-GFP with the chlamydial inclusion. (A to H) HeLa cells transiently expressing Arf1-GFP were infected with C. trachomatis serovar L2 at an MOI of approximately 25 to 50. At the indicated time points p.i. (2 h, 4 h, 10 h, and 40 h), cells were fixed and stained with antichlamydial antiserum (blue or red) and viewed by LSCM. (E to H) Merged images of GFP-expressing cells (green) and chlamydia staining (blue or red). The insets are selected regions denoted by asterisks. Scale bar, 10 μm. (I to K) Inhibition of chlamydial gene expression with chloramphenicol prevents recruitment of Arf1-GFP to chlamydia-containing vacuoles. HeLa cells transiently expressing Arf1-GFP (I; green) were infected with C. trachomatis serovar L2 in the presence of 25 μg/ml chloramphenicol and incubated for 18 h at 37°C. Cells were fixed and stained with antichlamydial antisera (J; red) and viewed by LSCM. (K) Merged image of I and J. (L to N) The trafficking and association of Arf1-GFP with the inclusion are sensitive to BFA. (L) Uninfected HeLa cells transiently expressing Arf1-GFP (green) were treated with 1 μg/μl BFA for 30 min at 37°C. (M) HeLa cells transiently expressing Arf1-GFP were infected with C. trachomatis serovar D. Eighteen hours p.i., cells were treated with 1 μg/μl BFA for 30 min at 37°C. (N) HeLa cells transiently expressing Arf1-GFP were pretreated with 1 μg/μl BFA for 30 min prior to infection with C. trachomatis serovar D. Cells were incubated for an additional 18 h in the continued presence of BFA. (O to Q) Microtubules are not essential for trafficking or association of Arf1-GFP with the inclusion. (O) Uninfected HeLa cells expressing Arf1-GFP were treated with 20 μM nocodazole for 3 h at 37°C. (P) HeLa cells transiently expressing Arf1-GFP were infected with C. trachomatis serovar D. Eighteen hours p.i., cells were treated with 20 μM nocodazole for 3 h at 37°C. (Q) HeLa cells transiently expressing Arf1-GFP were pretreated with 20 μM nocodazole for 2 h prior to infection with C. trachomatis serovar D. Cells were incubated for an additional 18 h in the presence of nocodazole. (L to Q) Fixed cells were stained with antichlamydial antisera (blue) and viewed by LSCM. Scale bar, 10 μm.
Since active Arf1-GFP is also localized to the inclusion and can recruit PI4P-binding PH domains to the Golgi complex in a PI4P-independent manner, we wanted to confirm that the association of GFP-OSBP-PH with the inclusion was indicative of an interaction with PI4P and not simply an interaction with the inclusion-localized Arf1-GFP. To test this, we constructed a mutant GFP derivative of the PH domain of OSBP, GFP-OSBP-(PHR107ER108E)×2, that does not interact with PI4P but still interacts with Arf1 (36). In mammalian cells, this mutant localizes to the Golgi complex in small amounts in an Arf1-dependent manner (36). Because the affinity for the Golgi apparatus is low, GFP-OSBP-(PHR107ER108E)×2 contains two copies of the mutated PH domain. As a control, we expressed GFP-OSBP-(PH)×2, which expresses two copies of the wild-type OSBP-PH domain. If GFP-OSBP-PH localizes to the inclusion due only to its interaction with Arf1, then similar amounts of the mutant compared to the wild type should localize to the inclusion. On the other hand, if GFP-OSBP-PH localizes to the inclusion due to its interaction with PI4P, then decreased or minimal amounts of the mutant should be recruited to the inclusion. As shown in Fig. 7C, although residual Golgi complex localization is observed, GFP-OSBP-(PHR107ER108E)×2 fails to localize to the inclusion in any detectable amount. Consistent with the lack of Arf1-GFP recruitment, together with the presence of GFP-OSBP-PH to C. pneumoniae inclusions, these data confirm that the localization of PI4P-binding PH domains to the inclusion reflects the presence of PI4P at the inclusion membrane.
PI4KIIα localizes to chlamydial inclusions.
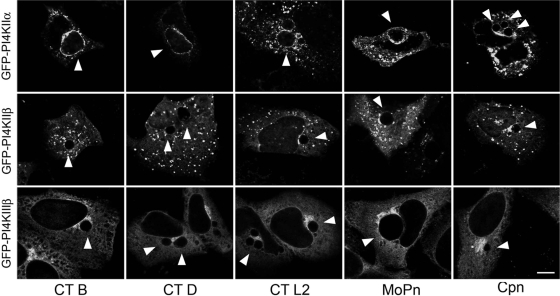
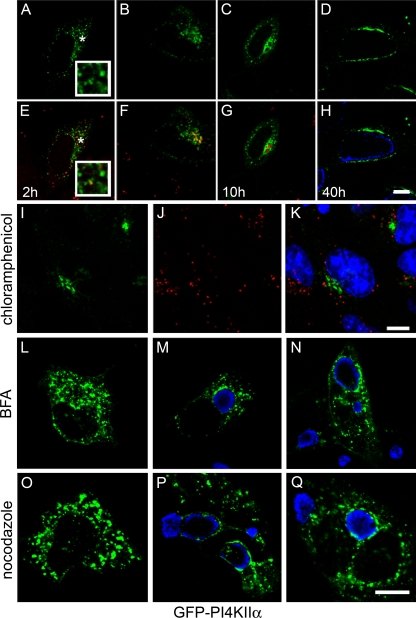
Although OCRL1 is thought to contribute to cellular levels of PI4P as loss of OCRL1 function can lead to an increase in cellular PIP2 levels in fibroblast cells from Lowe syndrome patients (70), the contribution of OCRL1 to PI4P levels at the Golgi complex is not entirely clear. At the Golgi complex, the major producers of PI4P are PI-OH(4)-kinases (PI4K). In mammals, there are 4 different PI4Ks, which belong to two different categories of enzymes, type II and type III, with one of each type localizing to the Golgi complex (3, 66). Since PI4P can be produced by either OCRL1 or PI4K, we wanted to determine other PI4P-producing host enzymes, in addition to OCRL1, were also localized to the inclusion. To test this, we analyzed the intracellular localization of three PI4K isoforms (PI4KIIα, PI4KIIβ, and PI4KIIIβ). As shown in Fig. 9, only GFP-PI4KIIα, which is normally localized to the Golgi complex and whose loss by siRNA depletion leads to loss of PI4P at the Golgi complex (36, 66), and to lesser extent, GFP-PI4KIIβ, localized to chlamydial inclusions. These data demonstrate that Chlamydia recruits at least two different PI4P-producing enzymes to the chlamydial inclusion. Similar to GFP-OCRL1, GFP-PI4KIIα is recruited to EBs aggregated at the peri-Golgi region between 2 and 4 h p.i. (Fig. 10A to H) and its recruitment is dependent on chlamydial gene expression (Fig. 10I to K). Finally, although the staining of GFP-PI4KIIα surrounding the inclusion is more punctate in the presence of either BFA (Fig. 10M and N) or nocodazole (Fig. 10P and Q), it is still recruited, suggesting that neither the Golgi apparatus or microtubules are essential for its trafficking to the inclusion.
FIG. 9.
PI4KIIα localizes to chlamydial inclusions. HeLa cells transiently expressing GFP-PI4KIIα, GFP-PI4KIIβ, or GFP-PI4KIIIβ were infected with C. trachomatis serovar B (24 h), C. trachomatis serovar D (18 h), C. trachomatis serovar L2 (18 h), C. muridarum (18 h), and C. pneumoniae (48 h). Cells were fixed and viewed by LSCM. CT, C. trachomatis; MoPn, C. muridarum; Cpn, C. pneumoniae. Arrowheads indicate inclusions. Scale bar, 10 μm.
FIG. 10.
Association of GFP-PI4KIIα with the chlamydial inclusion. (A to H) HeLa cells transiently expressing GFP-PI4KIIα were infected with C. trachomatis serovar L2 at an MOI of approximately 25 to 50. At the indicated time points p.i. (2 h, 4 h, 10 h, and 40 h), cells were fixed and stained with antichlamydial antiserum (blue or red) and viewed by LSCM. (E to H) Merged images of GFP-expressing cells (green) and chlamydia staining (blue or red). The insets are selected regions denoted by asterisks. Scale bar, 10 μm. (I to K) Inhibition of chlamydial gene expression with chloramphenicol prevents recruitment of GFP-PI4KIIα to chlamydia-containing vacuoles. HeLa cells transiently expressing GFP-PI4KIIα (I; green) were infected with C. trachomatis serovar L2 in the presence of 25 μg/ml chloramphenicol and incubated for 18 h at 37°C. Cells were fixed and stained with antichlamydial antisera (J; red) and viewed by LSCM. (K) Merged image of panels I and J. (L to N) Golgi complex-dependent trafficking is not necessary for the trafficking and association of GFP-PI4KIIα with the inclusion. (L) Uninfected HeLa cells transiently expressing GFP-PI4KIIα (green) were treated with 1 μg/μl BFA for 30 min at 37°C. (M) HeLa cells transiently expressing GFP-PI4KIIα were infected with C. trachomatis serovar D. Eighteen hours p.i., cells were treated with 1 μg/μl BFA for 30 min at 37°C. (N) HeLa cells transiently expressing GFP-PI4KIIα were pretreated with 1 μg/μl BFA for 30 min prior to infection with C. trachomatis serovar D. Cells were incubated for an additional 18 h in the continued presence of BFA. (O to Q) Microtubules are not essential for the association or trafficking of GFP-PI4KIIα to the inclusion. (O) Uninfected HeLa cells expressing GFP-PI4KIIα were treated with 20 μM nocodazole for 3 h at 37°C. (P) HeLa cells transiently expressing GFP-PI4KIIα were infected with C. trachomatis serovar D. Eighteen hours p.i., cells were treated with 20 μM nocodazole for 3 h at 37°C. (Q) HeLa cells transiently expressing GFP-PI4KIIα were pretreated with 20 μM nocodazole for 2 h prior to infection with C. trachomatis serovar D. Cells were incubated for an additional 18 h in the presence of nocodazole. (L to Q) Fixed cells were stained with antichlamydial antisera (blue) and viewed by LSCM. Scale bar, 10 μm.
OCRL1, Arf1, and PI4KIIα are required for optimal chlamydial development.
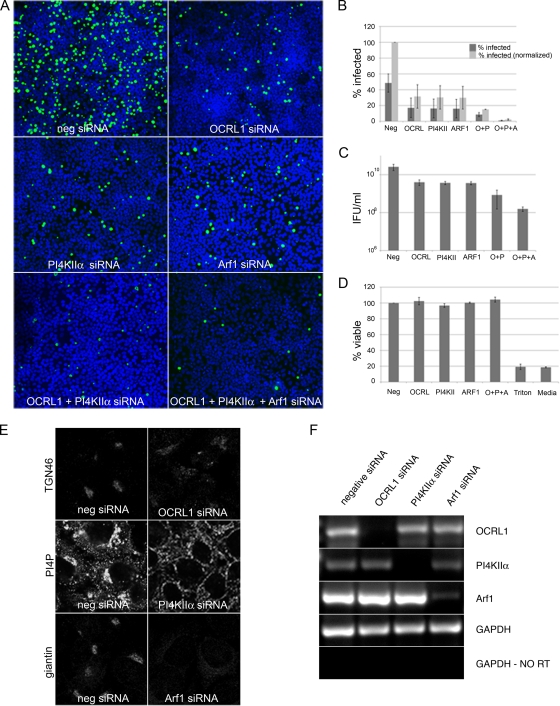
To examine whether OCRL1, Arf1, and PI4KIIα are required for chlamydial development, we knocked down expression of each gene in infected cells using siRNA-induced silencing. HeLa cells were exposed to gene-specific or negative control siRNAs for 72 h prior to infection with C. trachomatis serovar L2. Knockdown of each gene was confirmed by RT-PCR (Fig. 11F). In addition, knockdowns of OCRL1 and Arf1 were confirmed by mislocalization of TGN46 and giantin, respectively, and of PI4KIIα by loss of Golgi complex-localized PI4P (Fig. 11E). Cell viability was not affected by any of the siRNAs utilized (Fig. 11D). To measure infectivity, primary inclusion formation was enumerated by fixing and staining 18-h-infected cells with antichlamydial antisera (Fig. 11A and B). To measure chlamydial development, infectious progeny or inclusion-forming units (IFUs) were enumerated using a parallel set of samples that were infected for 44 h (Fig. 11C). In OCRL1, Arf1 and PI4KIIα siRNA-treated cells, inclusion formation was decreased by approximately 3-fold as compared to negative control siRNA-treated cells (Fig. 11A and B). Consistent with the decrease in inclusion formation, IFU formation was decreased approximately 6-fold in the absence of each of these proteins (Fig. 11C). Since both OCRL1 and PI4KIIα function to produce PI4P, we asked whether simultaneous depletion of both proteins caused a greater defect in infectivity. As shown in Fig. 11A and B, simultaneous depletion of both OCRL1 and PI4KIIa decreased primary inclusion by approximately 6-fold and IFU production by approximately 30-fold compared to negative siRNA-treated cells. Furthermore, depletion of all three proteins resulted in a much more dramatic decrease in infectivity (150-fold), suggesting OCRL1, PI4KIIα, and Arf1 perform partially overlapping functions that are important for chlamydial development (Fig. 11A to C). The decrease in primary inclusion formation suggests that either entry or early modification of the inclusion may be affected in the absence of each of these host proteins.
FIG. 11.
OCRL1, Arf1, and PI4KIIα are required for optimal chlamydial infection. siRNA-induced silencing was utilized to knock down OCRL1 (O), Arf1 (A), and PI4KIIα (P) expression in infected cells. HeLa cells were transfected with gene-specific or negative control siRNA oligonucleotide for 72 h. (F) RT-PCR was performed on total RNA isolated from OCRL1, PI4KIIα, Arf1, or negative (neg) siRNA-treated cells using gene-specific oligonucleotides as indicated. To control for equivalent amounts of total RNA, RT-PCR was performed with GAPDH-specific oligonucleotides. RT, reverse transcriptase. (E) Uninfected HeLa cells treated with indicated siRNAs for 72 h were fixed and stained as follows. OCRL1 siRNA-treated cells were stained with anti-TGN46, Arf1-siRNA-treated cells were stained with antigiantin, and PI4KIIα-siRNA-treated cells were stained with anti-PI4KIIα. (A, B, C, and D) Seventy-two hours posttransfection, siRNA-treated cells were infected with C. trachomatis serovar L2 for 18 h (A and B) or for 36 h (C). (A and B) To quantify primary inclusion formation, at 18 h p.i., cells were fixed and stained with antichlamydial antiserum. Inclusions were counted by indirect immunofluorescence microscopy. Percent infection was normalized such that the negative siRNA control cells were set to 100% infection. Experiments were performed in triplicate and repeated at least 3 times. Error bars indicate standard deviation. (C) To quantify infectivity, IFU were enumerated by lysing cells in distilled water (dH2O) at the indicated time points p.i. and titrating lysates on fresh HeLa cell monolayers. Experiments were performed in triplicate and repeated at least 3 times. Error bars indicate standard deviation. Statistics were performed with Microsoft Excel 2003 software using two-tailed t tests. (D) Cell viability was determined using WST-1 reagent. Cells were lysed with Triton X-100 as a control for inviable cells..
DISCUSSION
In eukaryotic cells, organelle identity is determined in part by the composition of active Rab GTPases and the specific PI species present on an organelle membrane. To alter the identity of their intracellular environment, many intracellular pathogens employ mechanisms that result in modulation of both the Rab GTPase and the PI composition of their parasitophorous vacuole (reviewed in reference 7). We have previously shown that the chlamydial inclusion is decorated with an unexpectedly large number of Rab GTPases that normally localize to multiple distinct host organelles (47) and at least one Golgi complex-localized Rab6 effector, BICD1 (41). In this article, we demonstrate the presence of a second Rab effector, OCRL1, at the chlamydial inclusion, whose depletion from host cells impairs chlamydial infection. Additionally, we demonstrate that PI4P, which is normally enriched in the Golgi complex and can be produced by the PI phosphatase activity of OCRL1, is localized to the inclusion. Finally, we show that two additional host proteins, Arf1 and PI4KIIα, which function to recruit PI4P-binding proteins and to produce PI4P at the Golgi complex, respectively, localize to chlamydial inclusions and that both proteins are also required for optimal chlamydial development. Collectively, these data suggest that chlamydiae modulate the identity of the inclusion by creating an organelle with a unique Rab GTPase and PI composition, which in turn may facilitate the exploitation of multiple host trafficking pathways.
To gain a better understanding of why chlamydiae target Rab GTPases, we initiated studies to determine whether specific Rab effectors were also localized to chlamydial inclusions. Utilizing a GFP-OCRL1 fusion protein that localizes to the TGN and EE in a manner similar to that of endogenous OCRL1, we demonstrated that OCRL1 is recruited to chlamydial inclusions in a species-independent manner. While full-length GFP-OCRL1 localized to inclusions of all Chlamydia species, the carboxy-terminal Rab-binding domain targeted GFP to C. trachomatis and C. muridarum but not C. pneumoniae inclusions. Species-specific recruitment of Rab GTPases may be one explanation for this observation. While the C. trachomatis inclusion is decorated with both Rab1 and Rab6, the C. pneumoniae inclusion is only decorated with Rab1 (47). Therefore, the interaction with the C. pneumoniae inclusion may be weaker, as evidenced by slightly diminished recruitment and require both the Rab-binding domain in addition to other regions of OCRL1, including the central phosphatase region. These data, together with the observation that Arf1-GFP is not recruited to C. pneumoniae inclusions, suggest that chlamydiae engage in multiple species-specific interactions that occur at the inclusion-host interface that may be important for the unique pathogenesis of each chlamydial species.
Since OCRL1 is capable of producing PI4P, we investigated whether PI4P was present at the inclusion membrane. Making use of GFP-tagged fusions that recognized different PI species, we demonstrated that PI4P, but not PIP2 or PI3P, was present at the inclusion membrane. Upon discovering that PI4P was present at the inclusion membrane, we examined whether additional host proteins that function to produce PI4P (PI4K) or interact with PI4P-binding proteins (Arf1) were also present. Using GFP-tagged fusion proteins, we confirmed the presence of PI4KIIα and Arf1 at the inclusion. Association was specific since neither PI4KIIIβ or Arf6 localized to mature inclusions. These data demonstrate that chlamydiae recruit at least three host proteins that function either to produce PI4P or regulate the cellular targeting of PI4P-binding proteins.
Since OCRL1, PI4KIIα, and Arf1 localize to the Golgi apparatus and Golgi complex-derived vesicles are known to traffic to the inclusion (28), they may be targeted to the inclusion via Golgi complex-dependent trafficking mechanisms. However, at least for OCRL1 and PI4KIIα, our experiments using BFA-treated cells would argue against this model. Although GFP-OCRL1, GFP-PI4KIIα, and GFP-OSBP-PH no longer localized to the Golgi apparatus in BFA-treated cells, each still localized to the inclusion, demonstrating that prior trafficking to the Golgi complex was not necessary for their localization to the inclusion. These data also suggest that the Rab-binding mutant, GFP-OCRL1S546P, failed to localize to the inclusion not because it could no longer localize to the Golgi complex, but rather because it no longer was able to bind to inclusion-localized Rab GTPases. However, OCRL1 is also localized to clathrin-coated vesicles that traffic between the EE and the TGN (12, 61) and Rab binding is also important for its endosomal role (32). Therefore, OCRL1 may traffic to the inclusion via the delivery of EEs containing OCRL1. Consistent with this model, EEs containing transferrin have been shown to be recruited to both nascent chlamydia-containing vacuoles and mature inclusions (52, 62). Furthermore, the recruitment of both EE-containing transferrin and GFP-OCRL1 to the chlamydia-containing vacuole occurs from 2 to 4 h p.i. (52). However, disruption of microtubule-dependent trafficking had only a minor impact on the recruitment of GFP-OCRL1 to the inclusion, suggesting that microtubule-dependent trafficking is also not essential. Although BFA diminished the association of Arf1 with the inclusion, especially with the C. trachomatis serovar L2 inclusion (data not shown), it is not clear whether Arf1 is trafficked via a Golgi complex-dependent mechanism or instead is released from the inclusion membrane as a result of its sensitivity to BFA.
To begin to understand the roles of OCRL1, PI4KIIα, or Arf1 during chlamydial development, we performed gene knockdown experiments using gene-specific siRNAs. Depletion of OCRL1, PI4KIIα, or Arf1 resulted in a small but reproducible decrease in inclusion formation and infectious progeny production. Simultaneous depletion of both OCRL1 and PI4KIIα resulted in a greater defect with an approximate 10-fold decrease in infectivity, suggesting that the functions of OCRL1 and PI4KIIα in infected cells may partially overlap. Finally, simultaneous depletion of OCRL1, PI4KIIα, and Arf1 caused a dramatic decrease in infectivity, with a 100-fold decrease in infectivity and a similar decrease in the production of infectious organisms. Since fewer inclusions were formed in each case, depletion of each these proteins likely caused a defect in entry or in the initial remodeling and trafficking of the inclusion. Consistent with this idea, both OCRL1 and PI4KIIα have been shown to have roles in endocytosis (18) or in entry of Listeria monocytogenes (45), respectively.
Chlamydiae have recently been shown to induce fragmentation of the Golgi apparatus (31), and Rab6 and Rab11 play important roles in this process (38). Although OCRL1, PI4KIIα, and Arf1 are localized to the Golgi apparatus and OCRL1 interacts with Rab6 (32), our current evidence does not support a role for these proteins in destabilizing the Golgi apparatus. Since the infection burden is too low in our siRNA-treated cells to observe cleavage of Golgin-84 by immunoblot analysis, we cannot examine whether loss of each of these proteins affects the ability of chlamydiae to fragment the Golgi complex. However, depletion of each protein caused early defects, as demonstrated by the decrease in primary inclusion formation, whereas inhibition of Golgi complex fragmentation results in a late defect characterized by failure of RBs to differentiate into EBs (31). Conceivably these proteins may function at later stages during chlamydial development, but these roles would be masked by the earlier defect that we observed.
Based upon the presence of PI4P at the inclusion, we can envision several models to explain why each of these host proteins may be recruited to the inclusion. First, PIs are spatially localized within the host cell to help dictate organelle identity. PI4P is localized primarily to the Golgi complex (37) and has been shown to be essential for Golgi-to-PM trafficking in Saccharomyces cerevisiae (30). A similar role in mammalian cells has been proposed (24, 64). Therefore, chlamydiae may promote the production of PI4P at the inclusion to aid in recruiting host PI4P-binding proteins that would give the inclusion a Golgi complex-like identity or enable chlamydiae to exploit host nutrient-rich Golgi complex-dependent trafficking. However, no host-PI4P-binding proteins have been localized to the inclusion (data not shown) suggesting that if chlamydiae target host PI4P-binding proteins, they do so in a selective fashion. Alternatively, similar to Legionella pneumophila (65), inclusion-localized PI4P may instead act to promote the anchoring of secreted chlamydial PI4P-binding proteins. However, as of yet, we have no evidence to suggest that chlamydiae secrete PI4P-binding proteins into the host cell cytosol.
Second, PIP2 is predominantly localized to the plasma membrane, where it accumulates at chlamydial entry sites (2). Therefore, instead of functioning to produce PI4P, OCRL1, may function to remove its substrate PIP2 from the developing inclusion. This would effectively camouflage the inclusion from its plasma membrane origin by preventing PIP2-binding proteins from being recruited to the inclusion or by preventing the formation of PIP3, which would promote maturation along the endosomal pathway.
It is still unclear how PI4P is produced at the inclusion membrane. Both OCRL1 and PI4KIIα may contribute to the production of PI4P. Although OCRL1 can produce PI4P, OCRL1 may in fact function independent of its role in PI4P production. For example, OCRL1 functions in the intracellular trafficking of clathrin-coated vesicles between EE and the TGN. Overexpression or knockdown of OCRL1 induces the redistribution of clathrin- and the cation-independent mannose 6-phosphate receptor (CI-M6PR) to enlarged endosomal structures, which are defective in retrograde trafficking to the TGN (12). Since chlamydiae recruit transferrin-containing early endosomes (52), chlamydiae may target OCRL1 to exploit its role in EE-TGN trafficking. However, in cells overexpressing GFP-OCRL1, inclusion development was unaffected, which would argue against this model.
An early role for Arf1 is harder to imagine since Arf1-GFP is not detected on early chlamydia-containing vacuoles. However, a lack of association may be due to the sensitivity of our assay or due to a more transient association of Arf1 with the inclusion. At the Golgi complex, Arf1 acts in concert with PI4P to recruit PI4P-binding proteins (36, 56). Since the association of OSBP-PH with the C. trachomatis and C. pneumoniae inclusion occurs in the absence of Arf1, it is unlikely that Arf1 functions solely to recruit PI4P-binding proteins to the inclusion. Currently, the role of Arf1 during chlamydial infection is under investigation.
Future efforts are being directed toward understanding which enzyme, OCRL1 or PI4KIIα, is responsible for production of PI4P at the inclusion membrane and the specific role this lipid species may play during chlamydial infection. In recent years, it has become clear that the interactions between the inclusion and its host are much more extensive than previously believed with exploitation of multiple vesicle (5, 28, 34) and signaling pathways that lead to alterations in the host intermediate filament cytoskeleton (35), apoptosis (19), and immune recognition pathways (72). The discovery of three additional host proteins targeted by chlamydiae confirms the complexity of host-pathogen interactions and opens up the possibility of a multitude of protein interactions at the inclusion, all of which may be important for chlamydial pathogenesis.
Acknowledgments
We thank T. Balla and M. Lowe for generously providing plasmid constructs. We also thank Hélène Marquis and members of the Scidmore laboratory for critical reading of the manuscript and Fae Tompkins for technical assistance.
This work was supported by PHS grant RO1A1073831.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 15 March 2010.
REFERENCES
- 1.Attree, O., I. M. Olivos, I. Okabe, L. C. Bailey, D. L. Nelson, R. A. Lewis, R. R. McInnes, and R. L. Nussbaum. 1992. The Lowe's oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature 358:239-242. [DOI] [PubMed] [Google Scholar]
- 2.Balana, M. E., F. Niedergand, A. Subtil, A. Alcover, P. Chavrier, and A. Dautry-Varsat. 2005. Arf6 GTPase controls bacterial invasion by actin remodeling. J. Cell Sci. 118:2201-2210. [DOI] [PubMed] [Google Scholar]
- 3.Balla, A., and T. Balla. 2006. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 16:351-361. [DOI] [PubMed] [Google Scholar]
- 4.Balla, A., G. Tuymetova, A. Tsiomenko, P. Varnai, and T. Balla. 2005. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol. Biol. Cell 16:1282-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beatty, W. L. 2006. Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J. Cell.Sci. 119:350-359. [DOI] [PubMed] [Google Scholar]
- 6.Behnia, R., and S. Munro. 2005. Organelle identity and the signposts for membrane traffic. Nature 438:597-604. [DOI] [PubMed] [Google Scholar]
- 7.Brumell, J. H., and M. A. Scidmore. 2007. Manipulation of Rab GTPase function by intracellular bacterial pathogens. Microbiol. Mol. Biol. Rev. 71:636-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carabeo, R. A., C. A. Dooley, S. S. Grieshaber, and T. Hackstadt. 2007. Rac interacts with Abi-1 and WAVE2 to promote an Arp2/3-dependent actin recruitment during chlamydial invasion. Cell. Microbiol. 9:2278-2288. [DOI] [PubMed] [Google Scholar]
- 10.Carabeo, R. A., S. S. Grieshaber, A. Hasenkrug, C. Dooley, and T. Hackstadt. 2004. Requirement for the Rac GTPase in Chlamydia trachomatis invasion of non-phagocytic cells. Traffic 5:418-425. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 1997. Chlamydia trachomatis genital tract infections—United States, 1995. MMWR Morb. Mortal. Wkl. Rep. 46:193-198. [PubMed] [Google Scholar]
- 12.Choudhury, R., A. Diao, F. Zhang, E. Eisenberg, A. Saint-Pol, C. Williams, A. Konstantakopoulos, J. Lucocq, L. Johannes, C. Rabouilee, L. E. Greene, and M. Lowe. 2005. Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol. Biol. Cell 16:3467-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clausen, J. D., G. Christiansen, H. U. Holst, and S. Birklund. 1997. Chlamydia trachomatis utilizes the host cell microtubule network during early events of infection. Mol. Microbiol. 25:441-449. [DOI] [PubMed] [Google Scholar]
- 14.Cocchiaro, J. L., Y. Kumar, E. R. Fischer, T. Hackstadt, and R. H. Valdivia. 2008. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. U. S. A. 105:9379-9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Matteis, M. A., and A. Godi. 2004. PI-loting membrane traffic. Nat. Cell Biol. 6:487-492. [DOI] [PubMed] [Google Scholar]
- 16.Donaldson, J. G., A. Honda, and R. Weigert. 2005. Multiple activities for Arf1 at the Golgi complex. Biochim. Biophys. Acta 1744:364-373. [DOI] [PubMed] [Google Scholar]
- 17.Dressman, M. A., I. M. Olivos-Glander, R. L. Nussbaum, and S. F. Suchy. 2000. Ocrl1, a PtdIns(4,5)P(2) 5-phosphatase, is localized to the trans-Golgi network of fibroblasts and epithelial cells. J. Histochem. Cytochem. 48:179-190. [DOI] [PubMed] [Google Scholar]
- 18.Erdmann, K. S., Y. Mao, H. J. McCrea, R. Zoncu, S. Lee, S. Paradise, J. Modregger, D. Biemesderfer, D. Toomre, and P. De Camilli. 2007. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev. Cell 13:377-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan, T., H. Lu, L. Shi, G. A. McClarty, D. M. Nance, A. H. Greenberg, and G. Zhong. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faucherre, A., P. Desbois, F. Nagano, V. Satre, J. Junardi, G. Gacon, and O. Dorseuil. 2005. Lowe syndrome protein Ocrl1 is translocated to membrane ruffles upon Rac GTPase activation: a new perspective on Lowe syndrome pathology. Hum. Mol. Genet. 14:1441-1448. [DOI] [PubMed] [Google Scholar]
- 21.Fields, K. A., and T. Hackstadt. 2002. The chlamydial inclusion: escape from the endocytic pathway. Annu. Rev. Cell Dev. Biol. 18:221-245. [DOI] [PubMed] [Google Scholar]
- 22.Fukuda, M., E. Kanno, K. Ishibashi, and T. Itoh. 2008. Large scale screening for novel Rab effectors reveals unexpected broad Rab-binding specificity. Mol. Cell. Proteomics 6:1031-1042. [DOI] [PubMed] [Google Scholar]
- 23.Furness, G., D. M. Graham, and P. Reeve. 1960. The titration of trachoma and inclusion blennorrhoea viruses in cell cultures. J. Gen. Microbiol. 23:613-619. [DOI] [PubMed] [Google Scholar]
- 24.Godi, A., A. Di Campli, A. Konstantakopoulos, G. Di Tullio, D. R. Alessi, G. S. Kular, T. Daniele, P. Marra, J. Lucocq, and M. A. De Matteis. 2004. FAPPs control Golgi-to-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 6:393-404. [DOI] [PubMed] [Google Scholar]
- 25.Godi, A., P. Pertile, R. Meyers, P. Marra, G. Di Tullio, C. Iurisci, A. Luini, D. Corda, and M. A. De Matteis. 1999. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1:280-287. [DOI] [PubMed] [Google Scholar]
- 26.Grayston, J. T. 1992. Infections caused by C. pneumoniae strain TWAR. Clin. Infect. Dis. 15:757-761. [DOI] [PubMed] [Google Scholar]
- 27.Grieshaber, S. S., N. A. Grieshaber, and T. Hackstadt. 2003. Chlamydia trachomatis utilizes host cell dynein to traffic to the microtubule-organizing center in a p50 dynamitin-independent process. J. Cell. Sci. 116:3793-3802. [DOI] [PubMed] [Google Scholar]
- 28.Hackstadt, T., D. D. Rockey, R. A. Heinzen, and M. A. Scidmore. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 15:964-977. [PMC free article] [PubMed] [Google Scholar]
- 29.Hackstadt, T., M. A. Scidmore, and D. D. Rockey. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. U. S. A. 92:4877-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hama, H., E. A. Schnieders, J. Thorner, J. Y. Takemoto, and D. B. DeWald. 1999. Direct involvement of phosphatidylinositol-4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274:34294-34300. [DOI] [PubMed] [Google Scholar]
- 31.Heuer, D., A. R. Lipinski, N. Machuy, A. Karlas, A. Wehrens, F. Siedler, V. Brinkmann, and T. F. Meyer. 2009. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature 457:731-735. [DOI] [PubMed] [Google Scholar]
- 32.Hyvola, N., A. Diao, E. McKenzie, A. Skippen, S. Cockcroft, and M. Lowe. 2006. Membrane targeting and activation of the Lowe syndrome OCRL1 by rab GTPases. EMBO J. 25:3750-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krauß, M., and V. Haucke. 2007. Phosphoinositide-metabolizing enzymes at the interface between membrane traffic and cell signaling. EMBO Rep. 8:241-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar, Y., J. Cocchiaro, and R. H. Valdivia. 2006. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr. Biol. 16:1646-1651. [DOI] [PubMed] [Google Scholar]
- 35.Kumar, Y., and R. H. Valdivia. 2008. Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe 4:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine, T. P., and S. Munro. 2002. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 12:695-704. [DOI] [PubMed] [Google Scholar]
- 37.Levine, T. P., and S. Munro. 1998. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr. Biol. 8:729-739. [DOI] [PubMed] [Google Scholar]
- 38.Lipinski, A. R., J. Heymann, C. Meissner, A. Karlas, V. Brinkmann, T. F. Meyer, and D. Heuer. 2009. Rab6 and Rab11 regulate Chlamydia trachomatis development and golgin-84-dependent Golgi fragmentation. PLoS Pathog. 5:e1000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowe, M. 2005. Structure and function of the Lowe syndrome protein OCRL1. Traffic 6:711-719. [DOI] [PubMed] [Google Scholar]
- 40.Misra, S., G. J. Miller, and J. H. Hurley. 2001. Recognizing phosphatidylinsoitol 3-phosphate. Cell 107:559-562. [DOI] [PubMed] [Google Scholar]
- 41.Moorhead, A. R., K. A. Rzomp, and M. A. Scidmore. 2007. The Rab6 effector Bicaudal D1 associates with Chlamydia trachomatis inclusions in a biovar-specific manner. Infect. Immun. 74:5632-5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munro, S. 2004. Organelle identity and the organization of membrane traffic. Nat. Cell Biol. 6:469-472. [DOI] [PubMed] [Google Scholar]
- 43.Olivos-Glander, I. M., P. A. Janne, and R. L. Nussbaum. 1995. The oculocerebrorenal syndrome gene product is a 105-kD protein localized to the Golgi complex. Am. J. Hum. Genet. 57:817-823. [PMC free article] [PubMed] [Google Scholar]
- 44.Pfeffer, S. R. 2001. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 11:487-491. [DOI] [PubMed] [Google Scholar]
- 45.Pizarro-Cerda, J., B. Payrastre, Y. J. Wang, E. Veiga, H. L. Yin, and P. Cossart. 2007. Type II phosphatidylinositol 4-kinases promote Listeria monocytogenes entry into target cells. Cell. Microbiol. 9:2381-2390. [DOI] [PubMed] [Google Scholar]
- 46.Rockey, D. D., M. A. Scidmore, J. P. Bannantine, and W. J. Brown. 2002. Proteins in the chlamydial inclusion membrane. Microbes Infect. 4:333-340. [DOI] [PubMed] [Google Scholar]
- 47.Rzomp, K. A., L. D. Scholtes, B. J. Briggs, G. R. Whittaker, and M. A. Scidmore. 2003. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect. Immun. 71:5855-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schachter, J. 1999. Infection and disease epidemiology, p. 139-169. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, DC.
- 49.Schramm, N., and P. B. Wyrick. 1995. Cytoskeletal requirements in Chlamydia trachomatis infection in host cells. Infect. Immun. 63:324-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schroer, T. A. 2004. Dynactin. Annu. Rev. Cell Dev. Biol. 20:759-799. [DOI] [PubMed] [Google Scholar]
- 51.Sciaky, N., J. F. Presley, C. Smith, K. J. Zaal, N. Cole, J. E. Moreira, M. Terasaki, E. Siggia, and J. Lippincott-Schwartz. 1997. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J. Cell Biol. 139:1137-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scidmore, M. A., E. R. Fischer, and T. Hackstadt. 1996. Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J. Cell Biol. 134:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scidmore, M. A., D. D. Rockey, E. R. Fischer, R. A. Heinzen, and T. Hackstadt. 1996. Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect. Immun. 64:5366-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scidmore-Carlson, M. A., E. I. Shaw, C. A. Dooley, E. R. Fischer, and T. Hackstadt. 1999. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol. Microbiol. 33:753-765. [DOI] [PubMed] [Google Scholar]
- 55.Seabra, M. C., and C. Wasmeier. 2004. Controlling the location and activation of Rab GTPases. Curr. Opin. Cell Biol. 16:451-457. [DOI] [PubMed] [Google Scholar]
- 56.Shin, H. W., and K. Nakayama. 2004. Dual control of membrane targeting by PtdIns(4)P and ARF. Trends Biochem. Sci. 29:513-515. [DOI] [PubMed] [Google Scholar]
- 57.Subtil, A., B. Wyplosz, M. E. Balana, and A. Dautry-Varsat. 2004. Analysis of Chlamydia caviae entry sites and involvement of Cdc42 and Rac activity. J. Cell Sci. 117:3923-3933. [DOI] [PubMed] [Google Scholar]
- 58.Suchy, S. F., I. M. Olivos-Glander, and R. L. Nussbaum. 1995. Lowe syndrome protein, a deficiency of a phosphatidylinositol 4,5 bisphosphate 5-phosphatase in the Golgi apparatus. Hum. Mol. Genet. 4:2245-2250. [DOI] [PubMed] [Google Scholar]
- 59.Thylefors, B., A. D. Negrel, R. Pararajasegaram, and K. Y. Dadzie. 1995. Global data on blindness. Bull. World Health Organ. 73:115-121. [PMC free article] [PubMed] [Google Scholar]
- 60.Tuvim, M. J., R. Adachi, S. Hoffenberg, and B. F. Dickey. 2001. Traffic control: Rab GTPases and the regulation of interorganellar transport. News Physiol. Sci. 16:56-61. [DOI] [PubMed] [Google Scholar]
- 61.Ungewickell, A., M. E. Ward, E. Ungewickell, and P. W. Majerus. 2004. The inositol polyphosphate 5-phosphatase Ocrl1 associates with endosomes that are partially coated with clathrin. Proc. Natl. Acad. Sci. U. S. A. 101:13501-13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Ooij, C., G. Apodaca, and J. Engel. 1997. Characterization of the Chlamydia trachomatis vacuole and its interaction with the host endocytic pathway in HeLa cells. Infect. Immun. 65:758-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vicinanza, M., G. D'Angelo, A. Di Campli, and M. A. De Matteis. 2008. Function and dysfunction of the PI system in membrane trafficking. EMBO J. 27:2457-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, Y. J., J. Wang, H. Q. Sun, M. Martinez, Y. X. Sun, E. Macia, T. Kirchhausen, J. P. Albanesi, M. G. Roth, and H. L. Yin. 2003. Phosphatidylinositol 4 phosphate regulates the targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell 114:299-310. [DOI] [PubMed] [Google Scholar]
- 65.Weber, S. S., C. Ragaz, K. Reus, Y. Nyfeler, and H. Hilbi. 2006. Legionella pneumophila exploits PI(4)P to anchor secreted effectors proteins to the replicative vacuole. PLoS Pathog. 2:418-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weixel, K. M., A. Blenental-Perry, S. C. Watkins, M. Aridor, and O. A. Weisz. 2005. Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J. Biol. Chem. 280:10501-10508. [DOI] [PubMed] [Google Scholar]
- 67.Wong, K., D. Meyers, and L. C. Cantley. 1997. Subcellular locations of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 272:13236-13241. [DOI] [PubMed] [Google Scholar]
- 68.Yagisawa, H., M. Hirata, T. Kanematsu, Y. Watanabe, S. Ozaki, K. Sakuma, H. Tanaka, N. Yabuta, H. Kamata, H. Hirata, and H. Nojima. 1994. Expression and characterization of an inositol 1,4,5-trisphosphate binding domain of phosphatidylinositol-specific phospholipase C-δ1. J. Biol. Chem. 269:20179-20188. [PubMed] [Google Scholar]
- 69.Zerial, M., and H. McBride. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107-117. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, X., P. A. Hartz, E. Philip, L. C. Racusen, and P. W. Majerus. 1998. Cell lines from kidney proximal tubules of a patient with Lowe syndrome lack OCRL1 inositol polyphosphate 5-phosphatase and accumulate phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 273:1574-1582. [DOI] [PubMed] [Google Scholar]
- 71.Zhong, G., P. Fan, H. Ji, F. Dong, and Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193:935-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong, G., T. Fan, and L. Liu. 1999. Chlamydia inhibits interferon γ-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J. Exp. Med. 189:1931-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]