Abstract
Streptococcus pneumoniae (the pneumococcus) is a major human pathogen and a leading cause of inflammatory infections such as pneumonia and otitis media. An important mechanism for host defense against S. pneumoniae is opsonophagocytic killing by neutrophils. To persist in the human host, the pneumococcus has developed strategies to evade opsonization and subsequent neutrophil-mediated killing. Utilizing a genomic approach, we identified NanA, the major pneumococcal neuraminidase, as a factor important for resistance to opsonophagocytic killing in ex vivo killing assays using human neutrophils. The effect of NanA was shown using both type 4 (TIGR4) and type 6A clinical isolates. NanA promotes this resistance by acting in conjunction with two other surface-associated exoglycosidases, BgaA, a β-galactosidase, and StrH, an N-acetylglucosaminidase. Experiments using human serum showed that these exoglycosidases reduced deposition of complement component C3 on the pneumococcal surface, providing a mechanism for this resistance. Additionally, we have shown that antibodies in human serum do not contribute to this phenotype. These results demonstrate that deglycosylation of a human serum glycoconjugate(s) by the combined effects of NanA, BgaA, and StrH, is important for resistance to complement deposition and subsequent phagocytic killing of S. pneumoniae.
Streptococcus pneumoniae (the pneumococcus) is a leading human pathogen that is responsible for over a million deaths annually (36). The organism resides in the nasopharynx, and although colonization is asymptomatic, it can spread from this site to cause diseases such as otitis media, pneumonia, and septicemia. During infection, the pneumococcus elicits an acute inflammatory response, characterized by an influx of phagocytic cells consisting primarily of neutrophils (58). Opsonophagocytic killing by neutrophils and other professional phagocytes is believed to be a major mechanism for host defense against pneumococcal infection. This is a multistep process in which bacteria must first be opsonized. A major mechanism for opsonization is via the complement system, which results in covalent deposition of C3b onto the bacterial surface (30, 41, 61). C3b can then be further cleaved to iC3b for recognition by complement receptor 3 (CR3). On neutrophils, this receptor binds complement-opsonized bacteria and stimulates phagocytosis, after which neutrophils efficiently kill the pneumococcus (8, 14, 31, 49).
Evading opsonophagocytosis is essential for persistence of this pathogen in the human host. This is evidenced by an increased prevalence of pneumococcal infection in patients with deficiencies in complement components (12, 45, 63). Also, mice that are rendered neutropenic are more susceptible to invasive pneumococcal infection (34). Recently it has been shown that during colonization there is a correlation between resistance to neutrophil-mediated killing and carriage of pneumococcal serotypes, where serotypes more resistant to killing have a higher prevalence (60). Like many successful extracellular pathogens, the pneumococcus is encapsulated by a thick coat of polysaccharide, which aids in evasion of phagocytic killing by masking underlying structures on the bacterial surface and reducing opsonization (18, 60). Capsular polysaccharide is the immunodominant antigen on the pneumococcus and is the basis for distinguishing strains, among 91 different serotypes. This antiphagocytic factor is crucial for pathogenesis, since unencapsulated strains rarely cause invasive disease and are severely attenuated in models of infection (35, 59). We have observed that even in the absence of capsule, however, S. pneumoniae retains some resistance to neutrophil killing. Therefore, we hypothesized that in addition to capsule, the pneumococcus expresses other factors that promote resistance to opsonophagocytic killing. To identify these factors, we took a whole-genome approach with a library of mutants generated with the mariner transposon and used ex vivo human neutrophil killing assays to screen for mutants that were more susceptible to neutrophil-mediated opsonophagocytic killing. One of the first genes identified by this screen encodes pneumococcal neuraminidase A (NanA), which catalyzes the release of terminally linked α2-3 and α2-6-linked sialic acid residues (7, 26).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. pneumoniae strains used in this study are described in Table 1. Strains were routinely grown at 37°C either in C medium supplemented with 5% yeast extract (C+Y medium) at pH 6.8 or in tryptic soy (TS) broth (Becton, Dickinson, & Co., Sparks, MD). Bacteria were also grown overnight at 37°C with 5% CO2 on TS plates containing 1.5% agar and 5,000 U of catalase (Worthington Biochemical Corporation, Freehold, NJ). When necessary, mutants were selected on TS that contained chloramphenicol (Cm) (2.5 μg/ml), spectinomycin (Sp) (200 μg/ml), kanamycin (Km) (500 μg/ml), or erythromycin (Erm) (1 μg/ml) as appropriate.
TABLE 1.
S. pneumoniae strains used in this study
| Strain | Serotype | Description | Reference or source |
|---|---|---|---|
| T4 | 4 | Clinical isolate TIGR4 | 51 |
| T4 cap | Una | Δcps locus (Kmr) | 55 |
| T4 nanA | 4 | ΔnanA (Cmr) | This study |
| T4 bgaA | 4 | ΔbgaA (Ermr) | This study |
| T4 strH | 4 | ΔstrH (Spr) | This study |
| T4 nanA(+nanA) | 4 | ΔnanA mutation replaced with WT copy of nanA from TIGR4 | This study |
| T4 nanA nanB | 4 | ΔnanA (Cmr) ΔnanB rpsL (K56T) (Smr) | 6 |
| T4 nanA::mar | 4 | nanA disrupted by the mariner Tn | This study |
| T4 nanA bgaA strH | 4 | ΔnanA (Cmr) ΔbgaA (Ermr) ΔstrH (Spr) | This study |
| 6Atr | 6A | Transparent variant of clinical isolate P303 | 25 |
| 6Atr nanA | 6A | ΔnanA (Cmr) | This study |
Un, unencapsulated.
Mutation of exoglycosidases and creation of the NanA revertant strain.
Insertion-deletion mutants were created for the genes encoding NanA, BgaA, and StrH using the constructs described by King et al. (26, 27). Mariner mutants of strain TIGR4 were created by in vitro transposon mutagenesis as previously described (16).
To create the nanA revertant strain, the nanA gene plus 1 kb of flanking genomic DNA from TIGR4 was PCR amplified using the primers nanAF3 (5′-ATG CTA CAG TTG TGG TAA CGA TTA C-3′) and nanAR2 (5′-CAT CAA CCA AAA AAT TGC TCA AAA G-3′). This product was then used to transform the T4 nanA strain of the pneumococcus to replace the insertion-deletion mutation with the wild-type (WT) copy of the gene. Transformation reaction mixtures were plated on TS without selection. The revertant strain was then negatively selected for by patching colonies onto TS plates with and without Cm to screen for transformants that have lost the insertion-deletion mutation. A Cm-sensitive transformant [T4 nanA(+nanA)] was then confirmed to have the WT copy of nanA by PCR and sequencing.
Isolation of neutrophils from human whole blood.
Human neutrophils were isolated as previously described (49). Briefly, heparinized whole blood from healthy human donors was run on a Polymorphprep gradient according to the manufacturer's instructions (Axis-Shield, Oslo, Norway). The neutrophil-enriched layer was collected and washed in Hanks buffer without Ca2+ and Mg2+ (GIBCO, Auckland, New Zealand) plus 0.1% (wt/vol) gelatin (−−+ buffer). Contaminating red blood cells (RBCs) were removed by hypotonic lysis in 0.83% NH4Cl. Cells were counted using trypan blue staining and adjusted to a concentration of 7 × 106 cells/ml in Hanks buffer containing Ca2+ and Mg2+ (GIBCO) plus 0.1% gelatin (+++ buffer) immediately before use.
Complement sources.
Three- to 4-week baby rabbit serum (BRS) was aliquoted and frozen at −80°C until use (Pel-Freez Biologicals, Rogers, AR). Normal human serum (NHS) was isolated from human whole blood. Blood was allowed to clot at 37°C for 30 min and then centrifuged at 1,000 × g for 20 min at 4°C. The serum layer was collected, aliquoted, and frozen at −80°C until use. Where indicated, serum was incubated at 56°C for 30 min to inactivate complement activity.
Opsonophagocytic killing assays.
Neutrophil killing assays were performed essentially as previously described by Davis et al. (10). Briefly, 103 PBS-washed late-log-phase bacteria (in 10 μl) were preopsonized in 20 μl of a complement source (66% BRS or 10% NHS), followed by incubation with 105 neutrophils (40 μl) in +++ buffer (130 μl). Reaction mixtures were then incubated at 37°C for 45 min with rotation. Opsonophagocytic killing assays were stopped by incubation in ice, and viable counts of bacteria were determined by dilution plating. Percent survival was determined relative to control reactions where no neutrophils were added. For experiments performed on different days, the absolute level of killing varied but relative differences between strains did not. Therefore, to compile data from different experiments, results are shown relative to killing of the WT strain in each independent experiment. Where indicated, data are shown as log10 CFU/ml for both the reactions where neutrophils were added and the control reactions where neutrophils were not added.
When specified, killing assays were modified. To complement the nanA mutant, 0.008 U of neuraminidase from Clostridium perfringens (Sigma Aldrich, St. Louis, MO) was added to serum immediately before addition of bacteria. To determine the role of complement receptor 3 in killing assays, a blocking antibody to this receptor (CBRM1/5) or an isotype control antibody (MOPC21) (Biolegend, San Diego, CA) was added to neutrophils at a concentration of 25 μg/106 neutrophils and incubated at 37°C for 30 min in +++ buffer prior to use in killing assays. To assess the role of NanA in the absence of IgG, opsonization reactions were performed with 20% IgG-depleted NHS. IgG was depleted from NHS using Hi-Trap protein G columns (GE Healthcare, Uppsala, Sweden), which resulted in removal of >95% of IgG. To assess the effect of NanA on the alternative pathway of complement activation, we performed opsonization reactions in the presence of MgEGTA to inhibit the classical pathway as previously described (13, 42). Bacteria were opsonized in 30% IgG-depleted NHS in gelatin veronal buffer containing MgEGTA (GVB-MgEGTA) (Boston Bioproducts, Worcester, MA). Since MgEGTA buffer would interfere with neutrophil-mediated killing, opsonized bacteria were then removed from serum and GVB-MgEGTA by centrifugation and resuspended in +++ buffer prior to addition of human neutrophils. IgG was depleted from NHS in these experiments using Hi-Trap protein G columns because agglutination of bacteria by IgG prevented centrifugation of cells from opsonization reactions.
Cell association assays.
Bacterial uptake assays were performed as previously described (32). Briefly, 107 PBS-washed bacteria grown to late log phase were labeled with fluorescein isothiocyanate (FITC) (0.2 mg/ml) for 30 min at 37°C. Unbound FITC was removed by washing in +++ buffer, and 4 × 105 FITC-labeled bacteria were then opsonized in 10% NHS and incubated with 2 × 105 neutrophils at 37°C for 30 min. Reactions were stopped by placing samples on ice and fixed by adding an equal volume of a freshly made paraformaldehyde solution (2% [wt/vol] in PBS). Uptake was assessed on a FACSCalibur flow cytometer, and at least 10,000 events were analyzed per sample. Percent cell association was determined relative to that in reactions where bacteria were preopsonized in 10% heat-inactivated NHS (HI NHS). Controls using bacteria that were not opsonized or FITC stained showed results similar to those when bacteria were opsonized in heat-inactivated serum.
Complement deposition assays.
C3 deposition was analyzed essentially as previously described by Brown et al. (5). Briefly, 106 PBS-washed bacteria grown to late log phase were incubated in 100 μl of 100% BRS or 10% NHS for 30 min at 37°C. Reactions were stopped by placing samples on ice. Opsonized cells were then washed and resuspended in Hanks balanced salt solution (HBSS) (GIBCO) containing 5% fetal calf serum (Sigma Aldrich). For samples opsonized in BRS, reaction mixtures were then incubated with a 1:100 dilution of an FITC-conjugated polyclonal goat anti-rabbit C3 antibody (MP Biomedical Cappel, Irvine, CA) for 30 min on ice. For samples opsonized in NHS, reaction mixtures were incubated with a 1:100 dilution of monoclonal antibody (MAb) 130.1, a mouse monoclonal antibody specific for human iC3b/C3b (50), for 30 min on ice, followed by incubation with an FITC anti-mouse IgG secondary antibody for 30 min on ice (Sigma Aldrich). After incubation with antibodies, samples were washed in PBS and fixed by resuspension in a freshly made paraformaldehyde solution (2% in PBS). C3 deposition was assessed on a FACSCalibur cell cytometer, and at least 10,000 events were analyzed per sample. C3 deposition was determined relative to that in reactions where bacteria were opsonized in heat-inactivated serum. As for opsonophagocytic killing assays, results were made relative to killing of the WT strain to compile data from different experiments. Controls where bacteria were not opsonized showed results similar to those for bacteria incubated in heat-inactivated serum.
Antibody binding assays.
Antibody binding assays were performed essentially as previously described (65). Briefly, 107 CFU of PBS-washed bacteria were opsonized in 1% NHS or 1% IgG-depleted NHS for 30 min on ice. Subsequently, bound IgG, IgA, and IgM were detected by incubating cells with FITC-conjugated secondary antibodies that recognized each antibody isotype, i.e., anti-human IgG whole molecule, anti-human IgA α-chain specific, and anti-human IgM μ-chain specific (Sigma Aldrich), for 1 h on ice. Cells were washed with HBSS containing 5% fetal calf serum, pelleted, and resuspended in a freshly made paraformaldehyde solution (2%). Antibody binding was analyzed on a FACSCalibur cell cytometer, and at least 10,000 events were analyzed per sample. Antibody binding was determined relative to that in reactions where unopsonized bacteria were incubated with the appropriate FITC-conjugated secondary antibodies.
Western blot analysis.
NHS or IgG-depleted NHS was diluted 1:10 in SDS sample buffer, and 20 μl per sample was loaded and separated by SDS-polyacrylamide gel electrophoresis (PAGE) on a 10% polyacrylamide gel. Proteins were then transferred to polyvinylidene difluoride (PVDF) transfer membranes (Thermo Scientific). Membranes were then probed with an alkaline phosphatase-conjugated anti-human IgG Fc-specific secondary antibody (Sigma Aldrich) diluted 1:10,000 for 1 h at room temperature. The presence of IgG in sera was detected with 4-nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolylphosphate.
Statistical analysis.
Statistical differences between groups were assessed by an unpaired Student's two-tailed t test (GraphPad PRISM 4; GraphPad Software).
RESULTS
NanA promotes resistance to opsonophagocytic killing.
To identify novel bacterial factors that promote resistance to opsonophagocytic killing, we screened mariner transposon mutants of the TIGR4 strain of S. pneumoniae (T4) for increased susceptibility to killing by human neutrophils. This was assessed using established ex vivo assays where bacteria were preopsonized in serum and then incubated with human neutrophils. During the initial screen, we identified that nanA, encoding the major pneumococcal neuraminidase, was important for promoting resistance to opsonophagocytic killing.
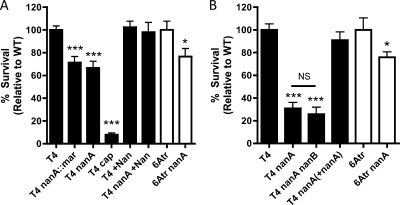
This phenotype was studied using two sources of serum to opsonize bacteria. The initial identification of the nanA mutant was performed using baby rabbit serum (BRS), as this was a source of complement without specific antipneumococcal antibodies (Fig. 1A). These results were then confirmed and further analyzed using normal human serum (NHS) to opsonize bacteria (Fig. 1B).
FIG. 1.
Survival of S. pneumoniae in neutrophil killing assays, showing comparisons of WT and nanA mutant strains in two independent backgrounds (TIGR4 and 6Atr). (A) Opsonophagocytic killing of S. pneumoniae by human neutrophils after bacteria were preopsonized in 66% BRS. Biochemical complementation (+Nan) was performed by addition of 0.008 U of C. perfringens neuraminidase per reaction. All results are relative to killing of the WT strain (T4 [black bars] or 6Atr [white bars]). (B) Opsonophagocytic killing of S. pneumoniae by human neutrophils after bacteria were preopsonized in 10% NHS. Data are the means from at least three independent experiments performed in duplicate ± standard errors of the means (SEM). *, P < 0.05; ***, P < 0.001; NS, not significant (compared to WT or as indicated).
The role of NanA in promoting resistance to opsonophagocytic killing was shown with the mariner mutant identified in the screen (T4 nanA::mar), as well as with a previously characterized insertion-deletion mutant (T4 nanA) (Fig. 1A) (6). The TIGR4 strain of the pneumococcus has an authentic frameshift resulting in secretion of NanA in this strain (51). To confirm that secretion of NanA was not required for this phenotype, a 6A strain that expresses NanA with an intact cell wall-anchoring domain was also studied (26). As in TIGR4, NanA promoted resistance to opsonophagocytic killing in the 6A strain (Fig. 1A and B, white bars).
The neuraminidase activity of NanA was required for this phenotype, because the addition of purified neuraminidase from Clostridium perfringens to killing assay mixtures complemented a nanA mutant (Fig. 1A, bars labeled +Nan). Additional evidence that the mutation in nanA is responsible for this phenotype was provided by testing a revertant strain [T4 nanA(+nanA)]. Correction of the mutation in nanA restored WT levels of resistance to opsonophagocytic killing (Fig. 1B). The phenotype of a nanA mutant is not due to differences in growth rate, as growth was similar to that of the WT in both serum and nutrient media (data not shown).
An unencapsulated mutant, T4 cap, is shown as a positive control for killing by human neutrophils to highlight the contribution of NanA to resistance relative to this major antiphagocytic factor (Fig. 1A). To ensure that differences in amounts of capsular polysaccharide/cell were not responsible for the phenotype seen in a nanA mutant, we performed a quantitative capture enzyme-linked immunosorbent assay (ELISA) and found equivalent levels of capsular polysaccharide in the nanA mutant and WT bacteria (data not shown) (25).
TIGR4 expresses two other neuraminidases, NanB and NanC (51). NanB is present in 96% of S. pneumoniae strains, while NanC is expressed in fewer than 51% of strains (6, 39). NanC is expressed by TIGR4 but not by the 6A strain, suggesting that NanC is not required for this phenotype, since a strain lacking it can still be resistant to opsonophagocytic killing (1A and B, white bars). Additionally, NanB does not significantly contribute to resistance to opsonophagocytic killing, since a nanA nanB double mutant is not more susceptible to neutrophil killing than a nanA mutant (Fig. 1B).
Killing of S. pneumoniae is dependent on opsonization by complement.
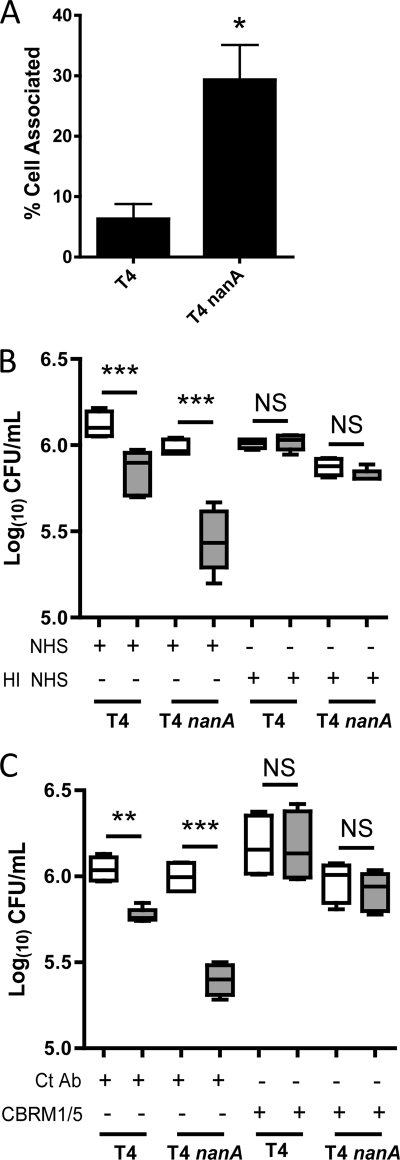
To further elucidate the mechanism for the effect on opsonophagocytic killing by NanA, we looked at bacterial uptake by neutrophils. Assaying for both adherent and ingested bacteria, we found that the nanA mutant was significantly more neutrophil associated than the WT strain (Fig. 2A). Therefore, NanA acts upstream of uptake by human neutrophils, possibly at the level of opsonization.
FIG. 2.
NanA acts upstream of phagocytosis, and complement is the predominant opsonin responsible for opsonophagocytic killing of S. pneumoniae. (A) Uptake of S. pneumoniae strains by human neutrophils. FITC-labeled bacteria were preopsonized in 10% NHS, followed by incubation with human neutrophils. The proportion of adherent and ingested pneumococci was assessed by flow cytometry. (B and C) The contribution of complement in the killing of S. pneumoniae was shown by performing opsonophagocytic killing assays using heat-inactivated NHS (HI NHS) to opsonize bacteria (B) and by performing assays in the presence of a blocking antibody to CR3 (CBRM1/5) or a control antibody (Ct Ab) (C). Gray boxes in panels B and C represent reactions where neutrophils were added, while white boxes represent control reactions where no neutrophils were added. Box-and-whiskers plots indicate high and low values, median, and interquartile ranges. Data are the results of at least two independent experiments performed in duplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant.
Opsonization by complement is a major mechanism for bacterial uptake. Complement components are heat labile, so the role of complement in opsonophagocytic killing of S. pneumoniae was first assessed using heat-inactivated human serum (HI NHS). When HI NHS was used to opsonize bacteria, killing of S. pneumoniae strains was almost completely abrogated (Fig. 2B). The role of complement in neutrophil-mediated opsonophagocytic killing of S. pneumoniae was also assessed using an antibody that blocks the major complement receptor responsible for phagocytosis, complement receptor 3 (CR3). As for heat-inactivated serum, killing of S. pneumoniae by human neutrophils was almost completely inhibited in the presence of the CR3-blocking antibody. This suggested that complement component C3 was the major opsonin in NHS responsible for killing by human neutrophils (Fig. 2C).
NanA promotes resistance to complement deposition.
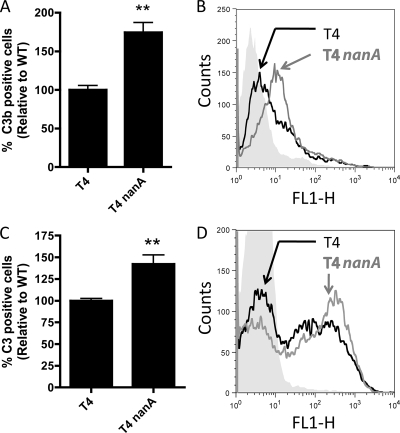
The level of C3b deposition on S. pneumoniae strains was assessed following opsonization by BRS or NHS using flow cytometry. As previously described, complement deposition on S. pneumoniae has a bimodal distribution (5), where cells either have C3 deposited or do not (Fig. 3A and C). In both BRS and NHS, a nanA mutant had a greater proportion of cells that were C3 positive than the WT (Fig. 3A to D).
FIG. 3.
C3 deposition on S. pneumoniae in NHS and BRS. Complement deposition was analyzed by flow cytometry. (A and B) Bacteria were preopsonized in 10% NHS, and C3 deposition was assessed using MAb 130.1, a monoclonal antibody specific for C3 breakdown products. All results are relative to bacteria preopsonized in 10% heat-inactivated NHS. (C and D) Bacteria were preopsonized in 20% BRS, and C3 deposition was assessed using a polyclonal antibody to C3. All results are relative to bacteria preopsonized in 20% heat-inactivated BRS. (A and C) Means from three independent experiments ± SEM. **, P < 0.01 compared to T4. (B and D) Representative histograms of the data comparing complement deposition on T4 (black lines), T4 nanA (gray lines), and T4 opsonized in heat-inactivated serum (gray fill).
NanA acts on the alternative pathway of complement activation.
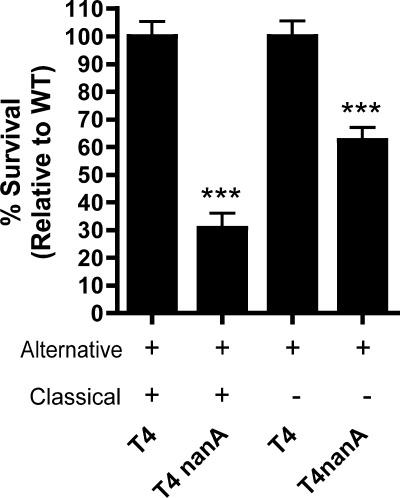
C3 deposition on the pneumococcus occurs through activation of both the classical and alternative pathways of complement (4, 5). To identify whether NanA acts on the alternative pathway, we performed opsonophagocytic killing assays using serum that was treated to prevent the activation of the classical pathway, as described in Materials and Methods. When the classical pathway was inhibited, there was still a significant effect of nanA, suggesting that NanA acts to prevent C3 deposition by the alternative pathway of complement activation (Fig. 4).
FIG. 4.
NanA has an effect on the alternative pathway of complement activation. To assess killing by the alternative pathway, opsonization was carried out with 30% IgG-depleted NHS in GVB-MgEGTA buffer. Data are the means from at least three independent experiments performed in duplicate ± SEM. ***, P < 0.001 compared to WT.
NanA does not act on antibodies in NHS to promote resistance to opsonophagocytic killing.
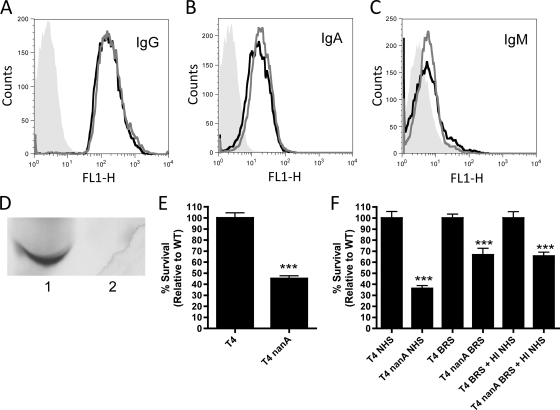
Interestingly, the phenotype of a nanA mutant in NHS is more dramatic than that seen in BRS. One of the major differences between these serum sources is the presence of specific antipneumococcal antibodies, which are potent activators of the complement system. NHS contains specific antipneumococcal IgG ≫ IgA > IgM, and bacterial recognition by these antibodies is not affected by NanA (Fig. 5A to C). Under similar conditions, no antipneumococcal immunoglobulins were detected in BRS (data not shown). As NanA displayed a significant effect on complement deposition in NHS (Fig. 3A and B), we hypothesized that one potential mechanism for this was via deglycosylation of antibodies. To test the contribution of IgG in our assays, we depleted IgG from NHS (Fig. 5D). However, even using IgG-depleted NHS we still saw a significant effect of NanA in opsonophagocytic killing assays (Fig. 5E). Next, we wanted to determine if any of the other antibody isotypes in NHS could account for the difference in phenotype observed in NHS versus BRS. This was assessed by adding heat-inactivated NHS as a source of antibodies to BRS. The phenotype of a nanA mutant in this mixture of heat-inactivated NHS and BRS was similar to that observed in BRS alone (Fig. 5F). This suggests that a heat-labile component of NHS is responsible for the increased phenotype of a nanA mutant in this serum source.
FIG. 5.
Contribution of antibodies on the effect of NanA in NHS. (A to C) Levels of IgG (A), IgA (B), and IgM (C) in NHS were assessed by antibody binding assays on both T4 (black lines) and T4 nanA (gray lines) cells relative to unopsonized cells (gray fill). (D) Western blot detecting IgG heavy chain presence in NHS (1) and IgG-depleted NHS (2). (E) IgG-depleted serum was used to opsonize bacteria in opsonophagocytic killing assays. (F) Opsonophagocytic killing assays where bacteria were opsonized in either 10% NHS, 66% BRS, or a mixture of 10% BRS and 10% HI NHS (as a source of antibodies) to determine the role of antibodies in mediating the difference in phenotype observed in NHS versus BRS. Data are the means from at least two independent experiments performed in duplicate ± SEM. ***, P < 0.001 compared to WT.
NanA acts with two other exoglycosidases from S. pneumoniae to promote resistance to opsonophagocytic killing and complement deposition.
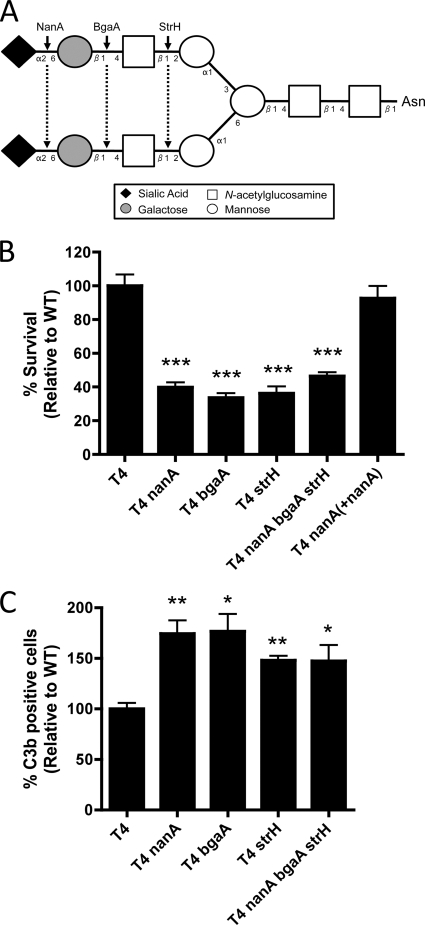
It has previously been demonstrated that NanA can act with a β-galactosidase, BgaA, and an N-acetylglucosaminidase, StrH, to sequentially remove sugars commonly found on the N-linked glycans of human glycoconjugates (the prototypic structure is shown in Fig. 6A) (6, 27). Therefore, we examined whether these other exoglycosidases also played a role in promoting resistance to opsonophagocytic killing. bgaA and strH mutants both showed a phenotype similar to that of a nanA mutant in opsonophagocytic killing and complement deposition assays (Fig. 6B and C). Additionally, a triple mutant with mutations in all three exoglycosidase genes showed a phenotype similar to that of each single mutant in both assays, supporting the hypothesis that these three exoglycosidases work on the same pathway/target (Fig. 6B and C). These three enzymes act exclusively on terminally linked substrates to sequentially remove sialic acid that is α2-3 or α2-6 linked to galactose (NanA), galactose that is β1-4 linked to N-acetylglucosamine (BgaA), and N-acetylglucosamine that is β1 linked to mannose (StrH) on human glycoconjugates (Fig. 6A) (27). Therefore, we conclude that the sequential activity of the three pneumococcal exoglycosidases promotes resistance to opsonophagocytic killing by deglycosylating a human glycoprotein(s) important for complement deposition.
FIG. 6.
Role of two other surface-exposed exoglycosidases of S. pneumoniae, BgaA and StrH, in resistance to opsonophagocytic killing by neutrophils. (A) Prototypic structure of complex N-linked biantennary glycans present on human glycoconjugates. Cleavage sites for the three pneumococcal exoglycosidases are indicated by arrows. (B) Neutrophil killing assays of S. pneumoniae, using 10% NHS to preopsonize bacteria. (C) C3 deposition assays of S. pneumoniae preopsonized in 10% NHS. Data are the means from three independent experiments ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared to T4).
DISCUSSION
Opsonophagocytic killing by neutrophils represents an important mechanism for clearance of pneumococcal infection (34). While some pneumococcal factors are known to inhibit opsonophagocytosis, a comprehensive search to identify all factors involved in this resistance has not been performed (20). Using a whole-genome approach, we identified that neuraminidase A (NanA) is important for resistance to opsonophagocytic killing by human neutrophils. Furthermore, we identified that NanA promotes resistance in conjunction with two other exoglycosidases from the pneumococcus, BgaA and StrH, by reducing complement deposition on the bacterial surface.
NanA is a well-characterized and ubiquitously expressed virulence factor in S. pneumoniae strains, and its role in the pathogenesis of the pneumococcus has been studied extensively. The effect of NanA on pathogenesis in vivo has been attributed to its various roles observed in vitro. NanA is able to remove sialic acid to expose receptors to aid pneumococcal adherence, directly bind epithelial cells via a lectin domain, aid in formation of biofilms, desialylate the surface of its competing nasopharyngeal flora, deglycosylate human glycoconjugates, and liberate carbohydrates to aid metabolic fitness of the organism (6, 27, 38, 48, 53, 54, 57). In vivo, a role for NanA in colonization and sepsis is less clear and is dependent upon the animal model employed (26, 52, 54). In contrast, during inflammatory diseases, such as pneumonia and otitis media, NanA has a more clear-cut role in the pathogenesis of this organism (33, 37, 52). These infections result in a robust influx of neutrophils and the serum components necessary for opsonization (2, 3, 15, 22, 23, 34, 58). Therefore, the effect of NanA in promoting resistance to opsonophagocytic killing demonstrated in this study could help explain the effect of this virulence factor observed in vivo.
NanA does not, however, appear to be acting alone to promote resistance to opsonophagocytic killing. NanA has previously been shown to act together with BgaA and StrH, two other surface-anchored exoglycosidases in the pneumococcus, which remove galactose that is β1-4 linked to N-acetylglucosamine (GlcNAc) and GlcNAc that is β1 linked to mannose, respectively (6, 9, 27, 28). All three enzymes act exclusively on terminally linked substrates, and recently it was shown that these exoglycosidases could sequentially remove sugars from complex N-linked glycans (27). Additionally, these enzymes are surface associated and have been shown to be more effective at deglycosylating a substrate when bound to the bacterial surface (7, 9, 24, 26, 27, 64). Glycosylation of host proteins can affect their stability, resistance to proteolysis, and functional activity (43). Therefore, it is possible that deglycosylation of a host glycoconjugate by the action of these three exoglycosidases is important for pneumococcal virulence in vivo. This is supported by the fact that these exoglycosidases deglycosylate human secretory component, immunoglobulin A, and lactoferrin, three human glycoconjugates thought to be important for pneumococcal clearance (27). The functional consequence of the deglycosylation of these host proteins or of other host proteins by these exoglycosidases, however, has not been examined.
In this study, we show that NanA, BgaA, and StrH work on the same pathway/target to promote resistance to opsonophagocytic killing by reducing complement deposition on the pneumococcus. As these enzymes function as exoglycosidases, we concluded that deglycosylation of a host glycoconjugate(s) promotes this resistance.
Using antibody binding assays and blocking antibodies to complement receptor 3, we showed that NHS is a source of both complement and antipneumococcal antibodies. Glycosylation of antibodies is known to be critical for their ability to fix complement (1, 21, 62); therefore, we wanted to test the hypothesis that deglycosylation of antibodies could help promote resistance to complement deposition. Using serum depleted of IgG, the predominant antibody isotype in NHS, we still saw a significant effect of NanA in opsonophagocytic killing assays. This suggests that IgG is not necessary for the effect of NanA in NHS. Additionally, using heat-inactivated NHS as a source of antibodies, we did not see an increase in phenotype for a nanA mutant when added to BRS. This suggests that a heat-labile component in NHS is responsible for promoting the greater effect of nanA in this serum source. Therefore, the difference between NHS and BRS could be due to increased exoglycosidase substrate specificity, since S. pneumoniae is a pathogen adapted to humans and glycosylation patterns can vary between species (17, 29, 40). Therefore, we believe that antibodies in NHS contribute to the deposition of complement on the pneumococcal surface but are not being acted upon by pneumococcal exoglycosidases. This is further supported by the fact that we do not see deglycosylation of human IgG using lectins that bind specifically to terminal mannose (data not shown). Thus, we conclude that deglycosylation of a serum component in NHS downstream of antibody binding and subsequent complement activation is important for resistance to complement deposition on the pneumococcus.
There has been a recent interest in studying glycosylation of complement components, and in some instances glycosylation is important for the function of these proteins (11, 19, 44, 46, 56). Most complement components are synthesized predominantly in the liver and contain complex biantennary glycans, as displayed in Fig. 6A (44). Therefore, deglycosylation of a complement component by NanA, BgaA, and StrH could provide a direct mechanism to promote resistance to complement deposition. Reducing the function of complement components, however, is only one mechanism whereby deglycosylation can directly affect complement deposition. Glycosylation can also be important for resistance to proteolysis; therefore, deglycosylation of a complement component(s) could increase its turnover by increasing its susceptibility to serum proteases (47). Opsonophagocytic killing assays looking at the effect of NanA on the alternative pathway suggest that deglycosylation affects at least this pathway of complement activation (Fig. 4). This does not, however, rule out a role for pneumococcal exoglycosidases in the classical pathway. In fact, since the effect of NanA on the alternative pathway was not as dramatic as that seen in complete NHS (containing both classical and alternative pathways intact) (Fig. 4), this suggests that NanA may have an effect on the classical pathway as well. Both the classical and alternative pathways of complement activation converge at the formation of the C3 convertase and deposition of C3 onto the bacterial surface. As C3 is shared by both complement pathways, it would seem to be a likely target for these exoglycosidases; however, this complement component does not have complex-type N-linked glycans (44). Therefore, the pneumococcal exoglycosidases could act upstream on a regulatory component of complement that affects C3 deposition. Alternatively, this resistance could require deglycosylation of multiple complement components or have an indirect role in promoting resistance to opsonophagocytic killing. With over 30 serum proteins involved in the complement cascade, however, pinpointing the factor reducing complement deposition is complex.
In summary, we have shown that NanA, BgaA, and StrH promote resistance to opsonophagocytic killing by increasing the ability of the pneumococcus to evade complement deposition and subsequent phagocytic killing. As complement contributes to both antibody-dependent and -independent clearance, this may be a mechanism to limit the effectiveness of both innate and adaptive immunity. Since glycoside hydrolases are not a unique feature of S. pneumoniae, deglycosylation may be a conserved strategy to evade the immune response.
Acknowledgments
We gratefully acknowledge Samantha King for providing the exoglycosidase mutants, John Lambris for providing MAb 130.1, and Ernesto Muñoz-Elías and Ramkumar Iyer for providing technical assistance and reagents for mariner transposon mutagenesis. We also thank Samantha King and John Lambris for helpful discussions.
This work was supported by Public Health Service grants AI44231 and AI38446 to J.N.W.
Editor: A. Camilli
Footnotes
Published ahead of print on 16 February 2010.
REFERENCES
- 1.Arnold, J. N., M. R. Wormald, R. B. Sim, P. M. Rudd, and R. A. Dwek. 2007. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 25:21-50. [DOI] [PubMed] [Google Scholar]
- 2.Bolger, M. S., D. S. Ross, H. Jiang, M. M. Frank, A. J. Ghio, D. A. Schwartz, and J. R. Wright. 2007. Complement levels and activity in the normal and LPS-injured lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 292:L748-L759. [DOI] [PubMed] [Google Scholar]
- 3.Broides, A., E. Leibovitz, R. Dagan, J. Press, S. Raiz, M. Kafka, A. Leiberman, and T. Yermiahu. 2002. Cytology of middle ear fluid during acute otitis media. Pediatr. Infect. Dis. J. 21:57-61. [DOI] [PubMed] [Google Scholar]
- 4.Brouwer, N., K. M. Dolman, M. van Houdt, M. Sta, D. Roos, and T. W. Kuijpers. 2008. Mannose-binding lectin (MBL) facilitates opsonophagocytosis of yeasts but not of bacteria despite MBL binding. J. Immunol. 180:4124-4132. [DOI] [PubMed] [Google Scholar]
- 5.Brown, J. S., T. Hussell, S. M. Gilliland, D. W. Holden, J. C. Paton, M. R. Ehrenstein, M. J. Walport, and M. Botto. 2002. The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. Proc. Natl. Acad. Sci. U. S. A. 99:16969-16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnaugh, A. M., L. J. Frantz, and S. J. King. 2008. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 190:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camara, M., G. J. Boulnois, P. W. Andrew, and T. J. Mitchell. 1994. A neuraminidase from Streptococcus pneumoniae has the features of a surface protein. Infect. Immun. 62:3688-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caron, E., and A. Hall. 1998. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science 282:1717-1721. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, V. A., N. Platt, and T. D. Butters. 1995. Cloning and expression of the beta-N-acetylglucosaminidase gene from Streptococcus pneumoniae. Generation of truncated enzymes with modified aglycon specificity. J. Biol. Chem. 270:8805-8814. [DOI] [PubMed] [Google Scholar]
- 10.Davis, K. M., H. T. Akinbi, A. J. Standish, and J. N. Weiser. 2008. Resistance to mucosal lysozyme compensates for the fitness deficit of peptidoglycan modifications by Streptococcus pneumoniae. PLoS Pathog. 4:e1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenaille, F., M. Le Mignon, C. Groseil, C. Ramon, S. Riande, L. Siret, and N. Bihoreau. 2007. Site-specific N-glycan characterization of human complement factor H. Glycobiology 17:932-944. [DOI] [PubMed] [Google Scholar]
- 12.Figueroa, J. E., and P. Densen. 1991. Infectious diseases associated with complement deficiencies. Clin. Microbiol. Rev. 4:359-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fine, D. P. 1977. Comparison of ethyleneglycoltetraacetic acid and its magnesium salt as reagents for studying alternative complement pathway function. Infect. Immun. 16:124-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon, D. L., G. M. Johnson, and M. K. Hostetter. 1986. Ligand-receptor interactions in the phagocytosis of virulent Streptococcus pneumoniae by polymorphonuclear leukocytes. J. Infect. Dis. 154:619-626. [DOI] [PubMed] [Google Scholar]
- 15.Gross, G. N., S. R. Rehm, and A. K. Pierce. 1978. The effect of complement depletion on lung clearance of bacteria. J. Clin. Invest. 62:373-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 17.Hironaka, T., K. Furukawa, P. C. Esmon, T. Yokota, J. E. Brown, S. Sawada, M. A. Fournel, M. Kato, T. Minaga, and A. Kobata. 1993. Structural study of the sugar chains of porcine factor VIII: tissue- and species-specific glycosylation of factor VIII. Arch. Biochem. Biophys. 307:316-330. [DOI] [PubMed] [Google Scholar]
- 18.Hostetter, M. K. 1986. Serotypic variations among virulent pneumococci in deposition and degradation of covalently bound C3b: implications for phagocytosis and antibody production. J. Infect. Dis. 153:682-693. [DOI] [PubMed] [Google Scholar]
- 19.Inforzato, A., G. Peri, A. Doni, C. Garlanda, A. Mantovani, A. Bastone, A. Carpentieri, A. Amoresano, P. Pucci, A. Roos, M. R. Daha, S. Vincenti, G. Gallo, P. Carminati, R. De Santis, and G. Salvatori. 2006. Structure and function of the long pentraxin PTX3 glycosidic moiety: fine-tuning of the interaction with C1q and complement activation. Biochemistry 45:11540-11551. [DOI] [PubMed] [Google Scholar]
- 20.Jarva, H., T. S. Jokiranta, R. Wurzner, and S. Meri. 2003. Complement resistance mechanisms of streptococci. Mol. Immunol. 40:95-107. [DOI] [PubMed] [Google Scholar]
- 21.Jefferis, R., and J. Lund. 1997. Glycosylation of antibody molecules: structural and functional significance. Chem. Immunol. 65:111-128. [PubMed] [Google Scholar]
- 22.Kawana, M., C. Kawana, T. Yokoo, P. G. Quie, and G. S. Giebink. 1991. Oxidative metabolic products released from polymorphonuclear leukocytes in middle ear fluid during experimental pneumococcal otitis media. Infect. Immun. 59:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerr, A. R., G. K. Paterson, A. Riboldi-Tunnicliffe, and T. J. Mitchell. 2005. Innate immune defense against pneumococcal pneumonia requires pulmonary complement component C3. Infect. Immun. 73:4245-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kharat, A. S., and A. Tomasz. 2003. Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro. Infect. Immun. 71:2758-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, J. O., and J. N. Weiser. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177:368-377. [DOI] [PubMed] [Google Scholar]
- 26.King, S. J., K. R. Hippe, J. M. Gould, D. Bae, S. Peterson, R. T. Cline, C. Fasching, E. N. Janoff, and J. N. Weiser. 2004. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol. Microbiol. 54:159-171. [DOI] [PubMed] [Google Scholar]
- 27.King, S. J., K. R. Hippe, and J. N. Weiser. 2006. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol. Microbiol. 59:961-974. [DOI] [PubMed] [Google Scholar]
- 28.Kojima, K., M. Iwamori, S. Takasaki, K. Kubushiro, S. Nozawa, R. Iizuka, and Y. Nagai. 1987. Diplococcal beta-galactosidase with a specificity reacting to beta 1-4 linkage but not to beta 1-3 linkage as a useful exoglycosidase for the structural elucidation of glycolipids. Anal. Biochem. 165:465-469. [DOI] [PubMed] [Google Scholar]
- 29.Kuster, B., A. P. Hunter, S. F. Wheeler, R. A. Dwek, and D. J. Harvey. 1998. Structural determination of N-linked carbohydrates by matrix-assisted laser desorption/ionization-mass spectrometry following enzymatic release within sodium dodecyl sulphate-polyacrylamide electrophoresis gels: application to species-specific glycosylation of alpha1-acid glycoprotein. Electrophoresis 19:1950-1959. [DOI] [PubMed] [Google Scholar]
- 30.Lambris, J. D., D. Ricklin, and B. V. Geisbrecht. 2008. Complement evasion by human pathogens. Nat. Rev. Microbiol. 6:132-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Cabec, V., S. Carreno, A. Moisand, C. Bordier, and I. Maridonneau-Parini. 2002. Complement receptor 3 (CD11b/CD18) mediates type I and type II phagocytosis during nonopsonic and opsonic phagocytosis, respectively. J. Immunol. 169:2003-2009. [DOI] [PubMed] [Google Scholar]
- 32.Lu, L., Z. Ma, T. S. Jokiranta, A. R. Whitney, F. R. DeLeo, and J. R. Zhang. 2008. Species-specific interaction of Streptococcus pneumoniae with human complement factor H. J. Immunol. 181:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manco, S., F. Hernon, H. Yesilkaya, J. C. Paton, P. W. Andrew, and A. Kadioglu. 2006. Pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect. Immun. 74:4014-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthias, K. A., A. M. Roche, A. J. Standish, M. Shchepetov, and J. N. Weiser. 2008. Neutrophil-toxin interactions promote antigen delivery and mucosal clearance of Streptococcus pneumoniae. J. Immunol. 180:6246-6254. [DOI] [PubMed] [Google Scholar]
- 35.Nelson, A. L., A. M. Roche, J. M. Gould, K. Chim, A. J. Ratner, and J. N. Weiser. 2007. Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect. Immun. 75:83-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Brien, K. L., L. J. Wolfson, J. P. Watt, E. Henkle, M. Deloria-Knoll, N. McCall, E. Lee, K. Mulholland, O. S. Levine, and T. Cherian. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893-902. [DOI] [PubMed] [Google Scholar]
- 37.Orihuela, C. J., G. Gao, K. P. Francis, J. Yu, and E. I. Tuomanen. 2004. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J. Infect. Dis. 190:1661-1669. [DOI] [PubMed] [Google Scholar]
- 38.Parker, D., G. Soong, P. Planet, J. Brower, A. J. Ratner, and A. Prince. 2009. The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect. Immun. 77:3722-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettigrew, M. M., K. P. Fennie, M. P. York, J. Daniels, and F. Ghaffar. 2006. Variation in the presence of neuraminidase genes among Streptococcus pneumoniae isolates with identical sequence types. Infect. Immun. 74:3360-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raju, T. S., J. B. Briggs, S. M. Borge, and A. J. Jones. 2000. Species-specific variation in glycosylation of IgG: evidence for the species-specific sialylation and branch-specific galactosylation and importance for engineering recombinant glycoprotein therapeutics. Glycobiology 10:477-486. [DOI] [PubMed] [Google Scholar]
- 41.Rambach, G., R. Wurzner, and C. Speth. 2008. Complement: an efficient sword of innate immunity. Contrib. Microbiol. 15:78-100. [DOI] [PubMed] [Google Scholar]
- 42.Ren, B., A. J. Szalai, S. K. Hollingshead, and D. E. Briles. 2004. Effects of PspA and antibodies to PspA on activation and deposition of complement on the pneumococcal surface. Infect. Immun. 72:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reuter, G., and H. J. Gabius. 1999. Eukaryotic glycosylation: whim of nature or multipurpose tool? Cell Mol. Life Sci. 55:368-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritchie, G. E., B. E. Moffatt, R. B. Sim, B. P. Morgan, R. A. Dwek, and P. M. Rudd. 2002. Glycosylation and the complement system. Chem. Rev. 102:305-320-319. [DOI] [PubMed] [Google Scholar]
- 45.Ross, S. C., and P. Densen. 1984. Complement deficiency states and infection: epidemiology, pathogenesis and consequences of neisserial and other infections in an immune deficiency. Medicine (Baltimore) 63:243-273. [PubMed] [Google Scholar]
- 46.Rudd, P. M., T. Elliott, P. Cresswell, I. A. Wilson, and R. A. Dwek. 2001. Glycosylation and the immune system. Science 291:2370-2376. [DOI] [PubMed] [Google Scholar]
- 47.Rudd, P. M., H. C. Joao, E. Coghill, P. Fiten, M. R. Saunders, G. Opdenakker, and R. A. Dwek. 1994. Glycoforms modify the dynamic stability and functional activity of an enzyme. Biochemistry 33:17-22. [DOI] [PubMed] [Google Scholar]
- 48.Shakhnovich, E. A., S. J. King, and J. N. Weiser. 2002. Neuraminidase expressed by Streptococcus pneumoniae desialylates the lipopolysaccharide of Neisseria meningitidis and Haemophilus influenzae: a paradigm for interbacterial competition among pathogens of the human respiratory tract. Infect. Immun. 70:7161-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Standish, A. J., and J. N. Weiser. 2009. Human neutrophils kill Streptococcus pneumoniae via serine proteases. J. Immunol. 183:2602-2609. [DOI] [PubMed] [Google Scholar]
- 50.Tamerius, J. D., M. K. Pangburn, and H. J. Muller-Eberhard. 1985. Detection of a neoantigen on human C3bi and C3d by monoclonal antibody. J. Immunol. 135:2015-2019. [PubMed] [Google Scholar]
- 51.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 52.Tong, H. H., L. E. Blue, M. A. James, and T. F. DeMaria. 2000. Evaluation of the virulence of a Streptococcus pneumoniae neuraminidase-deficient mutant in nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect. Immun. 68:921-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong, H. H., M. James, I. Grants, X. Liu, G. Shi, and T. F. DeMaria. 2001. Comparison of structural changes of cell surface carbohydrates in the eustachian tube epithelium of chinchillas infected with a Streptococcus pneumoniae neuraminidase-deficient mutant or its isogenic parent strain. Microb. Pathog. 31:309-317. [DOI] [PubMed] [Google Scholar]
- 54.Tong, H. H., X. Liu, Y. Chen, M. James, and T. Demaria. 2002. Effect of neuraminidase on receptor-mediated adherence of Streptococcus pneumoniae to chinchilla tracheal epithelium. Acta Otolaryngol. 122:413-419. [DOI] [PubMed] [Google Scholar]
- 55.Trzcinski, K., C. M. Thompson, and M. Lipsitch. 2003. Construction of otherwise isogenic serotype 6B, 7F, 14, and 19F capsular variants of Streptococcus pneumoniae strain TIGR4. Appl. Environ. Microbiol. 69:7364-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsiftsoglou, S. A., J. N. Arnold, P. Roversi, M. D. Crispin, C. Radcliffe, S. M. Lea, R. A. Dwek, P. M. Rudd, and R. B. Sim. 2006. Human complement factor I glycosylation: structural and functional characterisation of the N-linked oligosaccharides. Biochim. Biophys. Acta 1764:1757-1766. [DOI] [PubMed] [Google Scholar]
- 57.Uchiyama, S., A. F. Carlin, A. Khosravi, S. Weiman, A. Banerjee, D. Quach, G. Hightower, T. J. Mitchell, K. S. Doran, and V. Nizet. 2009. The surface-anchored NanA protein promotes pneumococcal brain endothelial cell invasion. J. Exp. Med. 206:1845-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Rossum, A. M., E. S. Lysenko, and J. N. Weiser. 2005. Host and bacterial factors contributing to the clearance of colonization by Streptococcus pneumoniae in a murine model. Infect. Immun. 73:7718-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watson, D. A., and D. M. Musher. 1990. Interruption of capsule production in Streptococcus pneumoniae serotype 3 by insertion of transposon Tn916. Infect. Immun. 58:3135-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinberger, D. M., K. Trzcinski, Y. J. Lu, D. Bogaert, A. Brandes, J. Galagan, P. W. Anderson, R. Malley, and M. Lipsitch. 2009. Pneumococcal capsular polysaccharide structure predicts serotype prevalence. PLoS Pathog. 5:e1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winkelstein, J. A., A. S. Abramovitz, and A. Tomasz. 1980. Activation of C3 via the alternative complement pathway results in fixation of C3b to the pneumococcal cell wall. J. Immunol. 124:2502-2506. [PubMed] [Google Scholar]
- 62.Wright, A., and S. L. Morrison. 1998. Effect of C2-associated carbohydrate structure on Ig effector function: studies with chimeric mouse-human IgG1 antibodies in glycosylation mutants of Chinese hamster ovary cells. J. Immunol. 160:3393-3402. [PubMed] [Google Scholar]
- 63.Yuste, J., A. Sen, L. Truedsson, G. Jonsson, L. S. Tay, C. Hyams, H. E. Baxendale, F. Goldblatt, M. Botto, and J. S. Brown. 2008. Impaired opsonization with C3b and phagocytosis of Streptococcus pneumoniae in sera from subjects with defects in the classical complement pathway. Infect. Immun. 76:3761-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zahner, D., and R. Hakenbeck. 2000. The Streptococcus pneumoniae beta-galactosidase is a surface protein. J. Bacteriol. 182:5919-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zola, T. A., E. S. Lysenko, and J. N. Weiser. 2009. Natural antibody to conserved targets of Haemophilus influenzae limits colonization of the murine nasopharynx. Infect. Immun. 77:3458-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]