Abstract
Cyclic diguanylic acid (c-di-GMP) is an intracellular signaling molecule involved in regulation of cellular functions such as motility, biofilm formation and virulence. Intracellular level of c-di-GMP is controlled through opposing diguanylate cyclase (DGC) and phosphodiesterase (PDE) activities of GGDEF and EAL domain proteins, respectively. We report the identification and characterization of cdpA, a gene encoding a protein containing an EAL domain in the Gram-negative soil bacillus and human pathogen Burkholderia pseudomallei KHW. Purified recombinant CdpA protein exhibited PDE activity in vitro. Evidence that CdpA is a major c-di-GMP-specific PDE in B. pseudomallei KHW was shown by an 8-fold-higher c-di-GMP level in the cdpA-null mutant as compared to the wild type and the complemented cdpA mutant. The presence of higher intracellular c-di-GMP levels in the cdpA-null mutant was associated with increased production of exopolysaccharides, increased cell-to-cell aggregation, absence of flagella and swimming motility, and increased biofilm formation. The relevance of CdpA in B. pseudomallei virulence was demonstrated by a 3-fold reduction in invasion of human lung epithelial cells and a 6-fold reduction in cytotoxicity on human macrophage cells infected with the cdpA mutant.
Burkholderia pseudomallei, a motile Gram-negative soil bacillus, is endemic to Southeast Asia and northern Australia. It causes melioidosis in humans, a potentially fatal septicemic infection which often manifests as acute pulmonary infection, localized skin infection, or acute septicemia (41). Its polar tuft of flagella is involved in swimming motility and is also a virulence determinant of B. pseudomallei during intranasal infection of mice (6). As an environmental saprophyte, it inhabits the soil at the root zone of plants where the bacteria readily form biofilm at the solid-liquid interface. B. pseudomallei can also be internalized within amoebic cysts or in the cytoplasm of arbuscular mycorrhizal fungi in order to survive hostile environmental conditions for a prolonged period (17, 21).
Pathogenicity of B. pseudomallei is dependent on a number of virulence factors and the expression of some virulence factors, such as phospholipase C and siderophores, is regulated by quorum sensing (29). B. pseudomallei mutants lacking components of the quorum-sensing systems exhibit reduced organ colonization of aerosolized BALB/c mice and increased time to death, while those lacking the BpsIR quorum sensing system were also impaired in biofilm formation and produced less MprA protease (29, 36, 37). Recent studies have linked the mechanisms of intercellular quorum sensing to intracellular signaling mediated by cyclic di-GMP (c-di-GMP) (39, 42, 43).
The sequenced genome of B. pseudomallei K96243 reveals several putative GGDEF-EAL domain proteins. These proteins use a combination of diguanylate cyclase (DGC) activities in the conserved GG(D/E)EF domains and phosphodiesterase (PDE) activities in the EAL domains to adjust intracellular c-di-GMP levels. Proteins containing both GGDEF and EAL domains usually display either DGC or PDE activity although an unusual GGDEF-EAL protein from Rhodobacter spheroides with both DGC and PDE activities was described recently (28, 33, 34). The intracellular c-di-GMP concentration positively regulates phenotypes such as sessility, biofilm formation, and expression of adhesive extracellular matrix (ECM) components and negatively influences phenotypes such as motility and virulence in bacteria (20, 28). The role of B. pseudomallei GGDEF and EAL proteins in motility, biofilm formation and virulence has not been studied. We describe an EAL domain protein encoded by cdpA, which functioned as a major c-di-GMP-specific phosphodiesterase in regulating intracellular levels of c-di-GMP in B. pseudomallei KHW. CdpA affects diverse phenotypes such as flagellum synthesis, bacterial motility, the production of exopolysaccharides, cell-to-cell aggregation, biofilm formation, cytotoxicity, and invasion of human cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. B. pseudomallei KHW is a virulent clinical isolate which was described previously (6). Bacteria were routinely cultured in Luria-Bertani (LB) medium (Becton Dickinson, Cockeysville, MD) at 37°C with shaking at 100 rpm. For nucleotide extraction and assays on motility, cell aggregation, and biofilm formation, B. pseudomallei was cultured AB medium (7) supplemented with 0.2% glucose and 0.5% Casamino Acids (CAA) instead of LB medium. The use of AB medium for these experiments was to minimize contamination with UV-absorbing compounds from LB medium that could interfere with the high-performance liquid chromatography (HPLC) analysis of nucleotides and also to provide a better correlation between levels of intracellular c-di-GMP nucleotide and bacterial motility, cell adhesiveness, and biofilm formation in bacteria cultured under identical conditions. Where appropriate, the antibiotics for E. coli cultures were added to the following concentrations: ampicillin, 100 μg/ml; gentamicin, 30 μg/ml; trimethoprim, 25 μg/ml; chloroamphenicol, 34 μg/ml; and tetracycline, 10 μg/ml. The concentrations for B. pseudomallei were as follows: kanamycin, 200 μg/ml; trimethoprim, 100 μg/ml; and tetracycline, 25 μg/ml. All antibiotics were purchased from Sigma (St. Louis, MO).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| B. pseudomallei | ||
| KHW | Virulent clinical isolate; Genr | E. H. Yap |
| KHWcdpA::Tet | Derivative of KHW with cdpA gene disrupted by insertion of 2.1-kb tetracycline cassette from pFTC1; Tetr Genr | This study |
| KHWcdpA::Tet/pUCP28T-cdpA | KHWcdpA::Tet complemented in trans with pUCP28TcdpA plasmid; Tetr Tmpr Genr | This study |
| KHW/pUCP28T-cdpA | KHW carrying pUCPT28T-cdpA plasmid; Tmpr Genr | This study |
| E. coli | ||
| DH5αλpir | DH5α with a λ prophage carrying the gene encoding the π protein; Kans Tmps Gens | 24 |
| HB101/pRK600 | Helper strain; containing pRK600 for triparental mating; Cmr | 9 |
| Plasmids | ||
| pFTC1 | Tetracycline resistance FRT vector; Tetr | 5 |
| pUCP28T | Broad-host-range vector; IncP OriT; pRO1600; ori Tmpr | 40 |
| pJQ200mp18 | Mobilizable suicide vector for allelic exchange; traJ sacB Genr | 26 |
| pJQ200mp18cdpA | pJQ200mp18 containing the 1.8-kb BamHI-digested cdpA PCR product; Genr | This study |
| pJQ200mp18cdpA::Tet | pJQ200mp18 carrying cdpA::Tet fragment; 2.1-kb Tetr cassette from pFTC1 inserted into the AccIII site within cdpA coding sequence; Genr Tetr | This study |
| pUCP28T-cdpA | pUCP28T carrying the full-length cdpA and its promoter; Tmpr | This study |
In silico sequence analysis.
The nucleotide sequences of GGDEF-EAL proteins of B. pseudomallei strain K96243 were obtained from the Sanger website. (http://www.sanger.ac.uk/Projects/B_pseudomallei). Additional sequences of GGDEF-EAL proteins were obtained from GenBank (http://www.ncbi.nlm.nih.gov/). In silico domain architecture analyses of these proteins was carried out using the Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/), and transmembrane domains were predicted using the TMpred program (http://www.ch.embnet.org/software/TMPRED_form.html) (13). Promoters were predicted using the Neural Network Promoter Prediction (NNPP) program for prokaryotes (http://www.fruitfly.org/seq_tools/promoter.html).
Construction of cdpA mutant and plasmid for complementation.
KHWcdpA::Tet mutants were generated by inserting a tetracycline resistance cassette from pFTC1 into the cdpA open reading frame. The full-length cdpA (corresponding to BPSL1263 in B. pseudomallei K96243) was amplified from B. pseudomallei KHW genomic DNA using the primers, pJQcdpA(BamHI) F (5′-CGGGATCCGAAGCCATCAGGAACA-3′) and pJQcdpA(BamHI) R (5′-ATGGATCCTCATGCGGTGGCGTG-3′) and inserted into the BamHI site of the suicide vector, pJQ200mp18 (26). A 2.1-kb tetracycline resistance (Tetr) cassette from pFTC1 (GenBank accession no. AY712950) was then inserted into the AccIII restriction site within cdpA. The pJQ200mp18cdpA::Tet plasmid was first introduced into Escherichia coli DH5αλpir (N. Judson, Gibco-BRL) and then into B. pseudomallei KHW by triparental conjugation using an E. coli HB101/pRK600 helper strain as described by de Lorenzo et al. (8). Exconjugants which had undergone allelic exchange were selected on LB agar containing 25 μg/ml of tetracycline, 100 μg/ml streptomycin, and 5% sucrose. Bacteria with the disrupted cdpA gene produced a 2.5-kb PCR product using cdpA::Tet(ver) F (5′-ACAAGTTCGCGGTGATGCTG-3′) and cdpA::Tet(ver) R (5′-TCGTGATCGGCTGGAAATGC-3′) primers instead of a 375-bp fragment in the parental B. pseudomallei KHW.
The cdpA-null mutation in the KHWcdpA::Tet mutant was confirmed by reverse transcription-PCR using total RNA prepared from the mutant and comparing it with the wild type. Briefly, 120 ng of total RNA was isolated from stationary-phase KHW cells and the KHWcdpA::Tet mutant cultured in AB medium containing 0.2% glucose (wt/vol) and 0.5% Casamino Acids, using the RNeasy minikit in combination with RNAprotect bacterial reagent (Qiagen). RT-PCR was carried out using Access RT-PCR kit (Promega) with the primer pair: cdpA(RT-PCR) F (5′-CGACGATTACCTGCGGATCAA-3′) and cdpA(RT-PCR) R (5′-CGAGATAGTTGATGAGGCCGA-3′). A 626-bp cdpA PCR product was observed in wild-type KHW but not in the KHWcdpA::Tet mutant. An RT-PCR of the 16S rRNA gene using primer pair 16SrDNA F (5′-GATGACGGTACCGGAAGAATAAGC-3′) and 16SrDNA R (5′-CCATGTCAAGGGTAGGTAAGGTTT-3′) was included as an internal control for the RNA template.
To construct a broad-host-range plasmid harboring the full-length cdpA, we used pUCP28TcdpA F (5′-CCGGAATTCACGAGCGCGGTGAAGTCGAG-3′) and pUCP28TcdpA R (5′-CCGGAATTCACGTCAGCCCCTCGCCTGGA-3′) to amplify a 4-kb DNA fragment containing the putative cdpA promoter using the Expand Long Template PCR system (Roche Diagnostics GmbH, Mannheim, Germany). The product was ligated into poly(T)-tailed, broad-host-range vector pUCP28T and introduced into E. coli DH5αλpir. E. coli DH5αλpir harboring the plasmid was then conjugated with the B. pseudomallei KHWcdpA::Tet mutant to create the complemented mutant. B. pseudomallei cells harboring the pUCP28T-cdpA plasmid were selected on LB agar containing 100 μg/ml trimethoprim and 100 μg/ml streptomycin. Expression of cdpA in the complemented mutant was verified by RT-PCR using the primer pair cdpA (RT-PCR) F and cdpA (RT-PCR) R.
Quantitative real-time PCR (qRT-PCR).
For quantitative real-time PCR (qRT-PCR), total RNA was extracted from triplicate 3-ml log-phase cultures (optical density at 600 nm [OD600] of ∼0.6) of wild-type and cdpA mutant cells in LB, using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Contaminating DNA was removed from the RNA samples using a Turbo DNase-free kit (Ambion, Applied Biosystems, Foster City, CA), and the amount of RNA in each sample was quantified by measuring absorbance at 260 nm, using a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies, Inc., Wilmington, DE). cDNA was synthesized from each RNA sample using TaqMan reverse transcription reagents (Applied Biosystems) containing 600 ng total RNA and 2.5 μM random hexamers in a final volume of 30 μl. Duplicate qRT-PCR was carried out for each RNA sample per pair of primers using 2× Fast SYBR green master mix (Applied Biosystems), 20 ng of cDNA, and 500 nM each gene-specific primer in a final volume of 20 μl. Thermal cycling was performed using an Applied Biosystems 7300 system and involved 40 cycles of 15 s at 95°C and 30 s at 60°C, followed by a melt curve analysis. Threshold cycle (CT) values were obtained using the default manual threshold setting of 0.2. The following gene-specific primer pairs were used: bsaN-3 (5′-TCTGAGCATCTGGAAATTCG-3′) and bsaN-4 (5′-GAGCCCGTAGCTCTCCTG-3′), bipB-F (5′-GCCTGATACTCGTCGGACTT-3′) and bipB-R (5′-TCGAAGCAGAAGCTCTTCAC-3′), fliC-1 (5′-CAGCAGATCTCGGAAGTGAA-3′) and fliC-2 (5′-AGGATGTTCTTGCCGTTGTA-3′), and recF-F (5′-CCTCACCGCACTCGCCAT-3′) and recF-R (5′-AGGCGCAGCGCACGATAC-3′). Expression levels of the genes in the wild-type and cdpA mutant were normalized to the expression level of its recombinase recF, an internal control. The normalized expression level of each gene in the cdpA mutant relative to the wild type was calculated using Livak's 2−ΔΔCT method (22).
Analysis of intracellular c-di-GMP levels using reversed-phase HPLC.
Wild-type B. pseudomallei KHW, the cdpA mutant, and the complemented mutant were cultured for 24 h (OD600 of ∼1.8) in AB medium containing 0.2% glucose and 0.5% Casamino Acids at 37°C. Approximately 100 mg (wet weight) of cells (equivalent to 9 ml of culture with an OD600 of ∼1.8) was harvested by centrifugation at 4,000 × g for 10 min. The cells were washed twice in 0.9% NaCl, resuspended in 1 ml 0.9% NaCl, and boiled for 10 min at 100°C. The lysates were extracted twice using 1 ml ice-cold 65% (vol/vol) ethanol. The extracts were lyophilized using a vacuum dryer and then resuspended in 200 μl of water. Ten microliters was injected into a Hypersil C18 250- by 4.6-mm column (Phenomenex, CA) for RP-HPLC analysis (Agilent Series 1100) using a modification of the protocol described by Ryjenkov et al. (27). The run was carried out using a gradient profile of buffer A (100 mM KH2PO4, 4 mM tetrabutyl ammonium hydrogen sulfate [pH 5.9]) and buffer B (75% buffer A, 25% methanol) at a flow rate of 0.7 ml/min. The gradient profile of buffers A and B were as follows (values represent percentages of buffer B at the time shown): 0.0 min, 0%; 2.5 min, 0%; 5.0 min, 30%; 10.0 min, 60%; 14.0 min, 100%; 21.0 min, 100%; 22.0 min, 50%; and 23.0 min, 0%. Peaks corresponding to the respective nucleotides were detected at 254 nm using variable wavelength detector and identified by comparing the retention times with nucleotide standards (Sigma). Calibration curves for GTP, GMP, and c-di-GMP standards were used to quantify the level of intracellular nucleotides in the cell extracts. c-di-GMP was synthesized using a procedure reported by Hyodo et al. (15). The compound was purified by semipreparative HPLC using a COSMOSIL 5C18-AR-300 column (20 mm [diameter] by 250 mm [height]). Elution was carried out with the following gradient times and conditions (where A represents water with 1% TFA and B is a 20:80 mixture of water and acetonitrile with 1% TFA): 0 to 3 min, 100% A; 3 to 35 min (linear gradient), 100% A to 85% A-15% B; 35 to 45 min, 100% B; 45 to 55 min, 100% A. Detection was at 254 nm with a flow rate of 3 ml/min. Relevant fractions were collected, dried, and subsequently washed with acetonitrile. Centrifugation afforded a white solid of c-di-GMP. For c-di-GMP, the following analytical detection parameters were used: 1H NMR (500 MHz, D2O), δ 4.01 to 4.04 (m, 2H), 4.32 to 4.40 (m, 4H), 4.83 (s, 2H), 5.04 (m, 2H), 5.81 (s, 2H), 7.95 (s, 2H); 31P NMR (202.5 MHz, D2O) δ −1.05; HRMS (ESI−) C20H23O14N10P2− (M − H−) calculated m/z 689.0865, found m/z 689.0849.
Motility assay.
Two microliters of an overnight culture of the bacteria in AB medium containing 0.2% glucose and 0.5% Casamino Acids was inoculated into the center of an AB agar plate containing 0.3% agar, AB medium supplemented with 0.2% glucose (wt/vol), and 0.5% CAA. The plates were incubated at 37°C for 24 h, and swimming motility was observed as a circular swarm zone. Triplicate assays were performed for each strain.
Biofilm assay.
Biofilm formation was measured as described previously (4). For the 96-well format, 100 μl of a diluted (OD600 of ∼0.05) overnight bacterial culture in AB medium containing 0.2% glucose (wt/vol) and 0.5% CAA was added into each well of a 96-well microtiter plate. After 24 h at 37°C, the wells were washed twice with distilled water to remove planktonic cells, and the sessile bacteria were stained with 125 μl of 1% (wt/vol) crystal violet (CV) (Sigma) for 15 min at room temperature. The wells were washed carefully three times using 200 μl distilled water and air dried. Three hundred microliters of 95% (vol/vol) ethanol was added to each well to solubilize the stain, and the absorbance was measured at a wavelength of 570 nm. The experiment was performed twice, and for each the assays were performed in triplicates. Qualitative detection of adherent cells was performed by inoculating 3 ml of AB medium containing 0.2% glucose (wt/vol) and 0.5% CAA in a 15-ml sterile polyvinyl chloride (PVC) tube with 60 μl of an overnight bacteria culture and incubating the cultures at 30°C for 48 h. Planktonic bacteria were carefully removed, and the adherent cells were washed twice using distilled water. Adherent cells were detected by staining with 1% CV as described above. The tubes were air dried and photographed. Triplicate assays were performed for each strain.
Transmission electron microscopy.
Cells of the wild-type B. pseudomallei strain KHW, the cdpA mutant, and the complemented mutant were streaked onto agar containing AB medium with 0.2% glucose and 0.5% CAA and incubated at 37°C for 24 h. The bacteria colonies were resuspended in phosphate-buffered saline (PBS) by gentle rocking. A 300-mesh Formvar-coated copper grid (Agar Scientific, Stansted, United Kingdom) was placed on a drop of bacterial suspension for 5 min and then transferred to a drop of 2.5% glutaraldehyde in PBS (Agar Scientific) for 5 min to fix the bacteria. After being rinsed twice in sterile distilled water for 5 min each time, the grid was transferred on to a drop of 1% phosphotungstic acid (pH 6.0; BDH, Poole, United Kingdom) for 1 min to stain the bacteria and air dried. Duplicates were performed for each type of bacteria and the images were obtained using JEOL 1010 transmission electron microscope (JEOL, Ltd., Tokyo, Japan) at an acceleration voltage of 100 kV with calibrated magnification.
CR binding assay.
Overnight broth cultures of the bacteria were streaked on LB (without NaCl) agar plates containing Congo red (CR) (40 μg/ml) and Coomassie brilliant blue (20 μg/ml), and the extent of Congo red binding was observed after incubating the plates for 48 h at 37°C (28).
Cell aggregation assay.
Sixty microliters of an overnight culture of the bacteria was inoculated into 3 ml fresh LB medium and cultured under static conditions at 37°C for 24 h. The cultures were then examined for cell aggregation and photographed.
Cell invasion and cytotoxicity assays.
Cell invasion was assayed as described previously (4). Briefly, 105 human lung epithelial cells (A549) cultured in Dulbecco's modified Eagle's medium (Sigma) containing 10% fetal bovine serum (Sigma) were infected (multiplicity of infection [MOI] of 100:1) with mid-log-phase (OD600 of 0.6) B. pseudomallei cells for 2 h. The cells were then washed with PBS and resuspended in medium containing kanamycin (200 μg/ml) for 2 h to kill extracellular bacteria. After three rounds of washing with PBS, 1 ml of 0.1% Triton X-100 (Sigma) was added to lyse the A549 cells, and serial dilutions of the cell lysate were plated on LB agar to determine the numbers of intracellular bacteria. The experiments were performed at least three times in triplicate assays.
Cytotoxicity of B. pseudomallei on human macrophage THP-1 cells was evaluated by measuring the release of lactate dehydrogenase (LDH) using a cytotoxicity detection kit (Roche, Mannheim, Germany) as described previously (4). Briefly, 2 × 106 cells were cultured in 0.6 ml of RMPI 1640 (Sigma) containing 2% fetal bovine serum and 2 mM l-glutamine for 3 h. The cells were then infected (MOI of 100:1) with mid-log-phase B. pseudomallei cultures (OD600 of 0.6) for 1 h at 37°C in the presence of 5% CO2, and then kanamycin (200 μg/ml) was added for 4 h to inhibit growth of extracellular bacteria. One hundred microliters of the supernatant collected after centrifugation at 350 × g for 3 min was used for the LDH assay. Maximum release was achieved by lysing the cells with 1% Triton-X. LDH activity in supernatant of uninfected cells was taken as spontaneous release. Percentage cytotoxicity was calculated using the formula: % cytotoxicity = [(test LDH release − spontaneous release)/(maximal release − spontaneous release)].
The experiment was performed twice and triplicate assays were performed for each experiment.
Cloning, expression, and purification of recombinant CdpA protein.
cdpA (BPSL1263 homologue) was amplified from B. pseudomallei KHW genomic DNA using the primer pair pMALBPSL1263(XbaI)F (5′-CCGTCTAGAGAAGCCATCAGGAACA-3′) and pMALBPSL1263(HindIII)R (5′-AAGAAGCTTTCATGCGGTGGCGTG-3′). PCR was performed using BioTaq DNA polymerase (BioLine USA, Inc., Taunton, MA) and the following cycling conditions: 1 cycle at 94°C for 3 min; 30 cycles at 94°C for 1 min, 59°C for 1 min, and 72°C for 2 min; and 1 cycle at 72°C for 10 min. The resultant 1.8-kb PCR product and pMAL-c4x vector (New England Biolabs, Ltd., Hertsfordshire, United Kingdom) were digested with XbaI and HindIII and ligated using T4 DNA ligase to generate a CdpA-maltose binding protein (MBP) fusion protein. The ligation product was introduced into E. coli DH5α cells by electroporation (Bio-Rad MicroPulser, Bio-Rad Laboratories, Richmond, CA), and positive clones were confirmed by restriction digest using XbaI and HindIII and by DNA sequencing.
DH5α harboring pMALc4x-cdpA was cultured in 500 ml LB broth supplemented with 0.2% (wt/vol) glucose and ampicillin (100 μg/ml) at 37°C until the OD600 was 0.8. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM to induce the expression of MBP-CdpA at 18°C for 16 h. The cells were harvested by centrifugation at 4,000 × g for 20 min at 4°C, resuspended in 25 ml column buffer (20 mM Tris·HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol), and sonicated using an ultrasonic liquid processor XL-2020 (Misonix, Inc., Framingdale, NY). The cleared lysate obtained after centrifugation at 9,000 × g for 30 min at 4°C was passed through an amylose resin column, and the recombinant protein was purified according to the manufacturer's protocol (New England Biolabs). Fractions of the eluate containing the purified MBP-CdpA protein were pooled together and stored at −80°C. Recombinant MBP was also purified using DH5α harboring pMALc4x as described above.
In vitro phosphodiesterase assay.
The reaction was conducted in 20 μl assay buffer (50 mM Tris-HCl, pH 8.5) containing 1 mM MnCl2 and 5 mM bis-p-nitrophenylphosphate (bis-pNPP) (Sigma) as the substrate in the presence of either 1 μM, 2.5 μM, and 5 μM purified MBP-CdpA protein or purified MBP (1). The enzyme blank contained all other components except the MBP-CdpA protein and MBP. After incubation for 2 h at 37°C, 80 μl distilled water was added, and the absorbance at a wavelength of 410 nm was read against the enzyme blank. To test the requirement of divalent metal cations for the PDE activity, the reaction was performed in buffer containing 5 μM purified MBP-CdpA protein and 1 mM either MgCl2, CaCl2, or ZnCl2, or water in place of MnCl2. The experiments were performed twice, and triplicate assays were performed for each experiment.
Statistical evaluation.
The mean ± standard deviation (SD) was calculated for each sample. Unless otherwise stated, all assays were performed in triplicate, and the mean was taken as 1 data point. Significant differences between means were determined by analysis of variance (ANOVA) with post hoc Tukey's multiple comparison tests (InStat; GraphPad Software, San Diego, CA). P values of ≤0.05 were considered significant.
RESULTS
cdpA encodes a cyclic di-GMP phosphodiesterase.
Ten open reading frames (ORFs) encoding putative GGDEF-EAL proteins were identified in the genome sequence of B. pseudomallei K96243 (http://www.sanger.ac.uk/Projects/B_pseudomallei/). Of these, all except BPSL1263 encode proteins containing the conserved GG(D/E)EF domain. Five proteins contained only the GGEEF domain while the remaining four contained both GGDEF and EAL domains. BPSL1263 was chosen for this study as it encodes only a protein with the conserved EAL domain but not a GG(D/E)EF domain and is therefore likely to be a c-di-GMP phosphodiesterase. The BPSL1263 equivalent in B. pseudomallei KHW was named cdpA (for cyclic-di-GMP phosphodiesterase). CdpA has a predicted molecular mass of 67 kDa (611 residues) with one putative membrane-spanning domain (13).
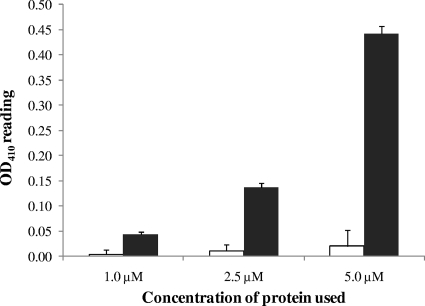
To investigate the phosphodiesterase (PDE) activity of CdpA in vitro, cdpA was overexpressed in E. coli DH5α using the pMALc4x vector and purified the recombinant maltose-binding protein (MBP)-CdpA fusion protein. The PDE activity of MBP-CdpA was measured using a colorimetric assay involving phosphodiesterase-mediated cleavage of the synthetic substrate, bis-pNPP and compared against activity due to MBP alone (Fig. 1). The PDE activity of MBP-CdpA showed a direct correlation with the concentration of enzyme used. Contamination by phosphodiesterase activity from the E. coli culture was very low and, unlike the PDE activity of MBP-CdpA, it did not increase with the concentration of the MBP used (Fig. 1). The PDE activity of MBP-CdpA showed an absolute requirement for manganese (Mn2+) and we could not detect any PDE activity in buffer without any divalent metal cations or in buffers containing Mg2+, Ca2+, Zn2+, or Co2+ instead of Mn2+ (data not shown).
FIG. 1.
Analysis of PDE activity of CdpA in vitro. PDE activity of CdpA was assessed in vitro using 1 μM, 2.5 μM, and 5.0 μM purified MBP-CdpA protein or MBP and 5 mM bis-pNPP substrate. The chart showed a dose-dependent catalysis of the release of p-nitrophenol by increasing concentration of recombinant MBP-CdpA protein. Absorbance at 410 nm was read against an enzyme blank without any protein. Black bars represent p-nitrophenol release catalyzed the recombinant MBP-CdpA protein and white bars represent the contaminating PDE activity from E. coli purified MBP protein.
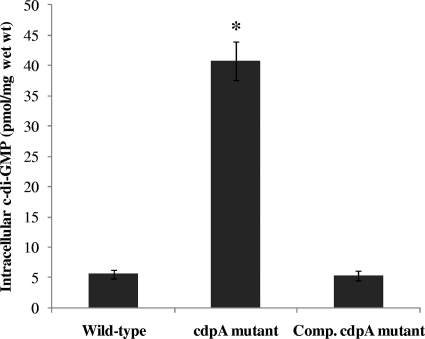
To verify the relevance of CdpA in regulation of intracellular c-di-GMP levels in B. pseudomallei KHW, we constructed a cdpA-null mutant by insertion mutagenesis. We also generated a broad-host-range plasmid, pUCP28T-cdpA, carrying full-length cdpA for trans complementation of the cdpA mutation in B. pseudomallei KHW. Both the cdpA-null mutation and trans complementation were verified by RT-PCR which showed the absence of cdpA transcript in the mutant and successful expression of the cdpA transcript in the complemented mutant (data not shown). Cyclic-di-GMP was extracted from stationary-phase wild-type bacteria, the cdpA mutant and the complemented mutant and measured using RP-HPLC (Fig. 2). The intracellular level of c-di-GMP in the cdpA mutant was 40.7 ± 3.2 pmol/mg wet weight, or at least 8-fold higher than the c-di-GMP levels in the wild-type B. pseudomallei KHW and complemented cdpA mutant, which were 5.6 ± 0.7 and 5.4 ± 0.8 pmol/mg wet weight, respectively. CdpA therefore plays a major role as a c-di-GMP phosphodiesterase regulating intracellular levels of the second messenger in B. pseudomallei KHW. There was no statistically significant difference in the levels of c-di-GMP in the cdpA complemented mutant and wild-type B. pseudomallei KHW (P > 0.05), thus demonstrating successful trans complementation of the cdpA mutation using the pUCP28T-cdpA construct (Fig. 2B).
FIG. 2.
Analysis of PDE activity of CdpA in vivo. Intracellular c-di-GMP content in wild-type B. pseudomallei KHW, the cdpA mutant and complemented (Comp.) cdpA mutant. Intracellular nucleotides were extracted from late-stationary-phase bacterial cultures and the c-di-GMP content measured using RP-HPLC. The amount of c-di-GMP was expressed per milligram (wet weight) of cells. Each bar is the mean of three independent experiments and any value which was statistically different (P < 0.05) from the wild type as assessed by the ANOVA test is denoted by an asterisk.
Inactivation of cdpA increased production of exopolysaccharides and autoaggregation.
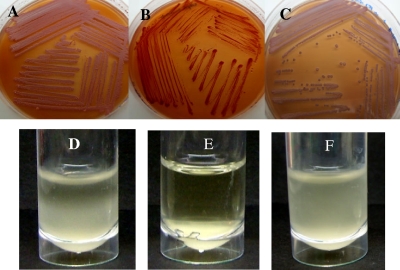
We used Congo red (CR) binding as a measure of biofilm/ECM production. CR binding shows a positive correlation with the presence of exopolysaccharides (EPS) in a number of bacterial species (10, 45). The cdpA mutant showed increased CR binding relative to the wild type and the complemented cdpA mutant, thus demonstrating increased production of exopolysaccharides in the cdpA mutant (Fig. 3A to C). We also compared cell-to-cell adhesion in wild-type bacteria and in the cdpA mutant using static broth cultures. Broth cultures of the cdpA mutant remained visibly clear after 24 h, with clumping of the bacterial cells at the bottom of the culture tube, while the broth cultures of the wild-type and complemented cdpA mutant showed bacterial cells in suspension (Fig. 3D to F). We conclude that the cells of the cdpA mutant were more adhesive and showed increased autoaggregation.
FIG. 3.
Effects of cdpA mutation on Congo red binding and formation of cell aggregates by B. pseudomallei. Increased production of exopolysaccharides was observed in the cdpA mutant which bound more CR (B) compared to the wild type (A) and complemented cdpA mutant (C). Bacterial cells of the cdpA mutant were also more adhesive, forming cell aggregates which sedimented to the bottom of the tube after 24 h static culture in LB broth (E). In comparison, the wild-type bacteria (D) and the complemented cdpA mutant (F) remained mostly in suspension in static broth cultures.
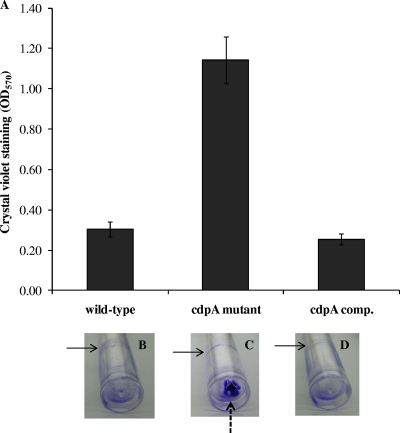
Next, we investigated the impact of increased exopolysaccharide production and adhesiveness in the cdpA mutant on biofilm formation under static conditions. Biofilm formation by bacteria adhering to the abiotic surface is measured by the intensity of crystal violet (CV) staining. The cdpA mutant stained 3.7-fold more CV compared to the wild type and the complemented cdpA mutant (Fig. 4A). We also noticed that cells of the cdpA mutant were attached mostly to the substratum rather than to the liquid-air interface as expected from its clumping behavior with qualitatively little increase in biofilm formation at the liquid-air interface in the cdpA mutant cells as compared to cdpA+ cells (Fig. 4B to D). Hence the biofilm assay using CV staining of bacteria adhering to the abiotic surface in a 96-well format does not distinguish between the two types of biofilm.
FIG. 4.
Effects of CdpA on biofilm formation. Biofilm formation was measured by CV staining assay using static broth cultures of wild-type B. pseudomallei KHW, the cdpA mutant and complemented (comp.) cdpA mutant in a 96-well PVC microtiter plate and in PVC tubes. (A) Bacteria were cultured in AB medium containing 0.2% glucose and 0.5% CAA in a 96-well PVC microplate format for 24 h at 30°C. Adherent bacteria were stained using 1% (wt/vol) crystal violet and measured spectroscopically at 570-nm wavelength. Panels B to D show CV-stained wild-type (B), cdpA mutant (C), and complemented cdpA mutant (D) cells. Solid arrows refer to biofilm formed at the liquid-air interface and the broken arrow refers to biofilm formed at the substratum.
The cdpA mutant is aflagellate, nonmotile, and attenuated in cytotoxicity and invasion of mammalian cells.
Mutation in cdpA affected diverse phenotypes. Analyses of the bacterial morphology using TEM revealed that the wild-type B. pseudomallei strain KHW is a rod-shaped (1.5 μm by 0.8 μm) bacterium with a polar tuft of flagella (Fig. 5A). The cdpA mutant, which had a longer and narrower rod shape (2.3 μm by 0.5 μm), was aflagellate (Fig. 5B). Flagellum production was restored in the complemented cdpA mutant expressing wild-type cdpA (Fig. 5C). Therefore, absence of CdpA activity or the resultant high c-di-GMP levels prevented flagellum synthesis and assembly in B. pseudomallei. The aflagellate cdpA mutant also did not exhibit any swimming motility in semisolid agar (Fig. 5E). Swimming motility was observed only in the wild type and was restored in the complemented cdpA mutant (Fig. 5D and F). We further confirmed the absence of flagella in the cdpA mutant by qRT-PCR, which failed to detect any fliC expression in the cdpA mutant (Table 2).
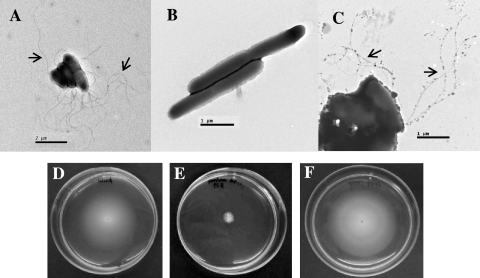
FIG. 5.
The cdpA mutant was aflagellate and did not exhibit swimming motility. Transmission electron micrographs of B. pseudomallei KHW (A), cdpA mutant (B), and the complemented cdpA mutant (C). The cdpA mutant was aflagellate and elongated compared to wild-type B. pseudomallei KHW. Flagella were present in the complemented cdpA mutant (C). Scale bars: panel A, 2 μm; panels B and C, 1 μm. Arrows indicate flagella. Wild-type B. pseudomallei KHW (D) and the complemented cdpA mutant (F) exhibited swimming motility in semisolid agar, but the cdpA mutant was nonmotile (E).
TABLE 2.
Comparison of bsaN and bipB expression in wild-type and cdpA mutant cells
| Strain | Normalized expression level relative to wild typea |
||
|---|---|---|---|
| bsaN | bipB | fliC | |
| Wild type | 1.112 ± 0.122 | 1.017 ± 0.234 | 1.001 ± 0.045 |
| cdpA mutant | 0.101 ± 0.027 | 0.003 ± 0.001 | 0.000 ± 0.000 |
The level of expression for each strain was first normalized to the expression level of recF in that strain. The values shown are the expression levels of the genes in the cdpA mutant relative to their average expression levels in the wild type (values of ∼1).
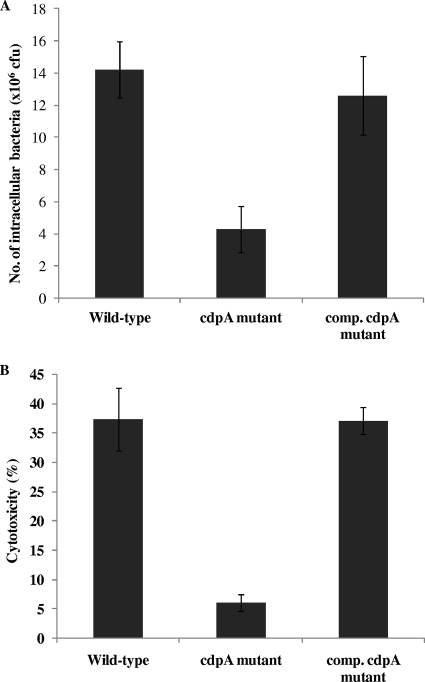
In view of the importance of flagella and swimming motility in B. pseudomallei virulence, we next determined whether the cdpA mutant would be attenuated in virulence in cell culture assays. We first compared the invasiveness of wild-type B. pseudomallei and the cdpA mutant on mammalian cells. When A549 human lung epithelial cells were cocultured with B. pseudomallei for 2 h, 3-fold less cdpA mutant bacteria were present in the A549 cells compared to cells cocultured with wild-type B. pseudomallei KHW (Fig. 6A). Although the reduction in cell invasion by the cdpA mutant was only 3-fold, we showed that the cdpA mutant was at least 6-fold less cytotoxic than wild-type bacteria on human macrophage cells (THP-1). B. pseudomallei has been shown to exhibit cytotoxcity on THP-1 cells via a caspase-1-induced mechanism and does not require prior stimulation with phorbol esters (32). Coculture of wild-type B. pseudomallei KHW with unactivated THP-1 cells for 4 h resulted in 37% killing of THP-1 cells, as compared to only 6% killing of the THP-1 cells by the cdpA mutant (Fig. 6B). In both cases, the invasiveness of the cdpA mutant and its cytotoxicity on THP-1 cells were restored to wild-type levels by trans complementation using a plasmid bearing wild-type cdpA, thus attributing the attenuation in virulence to the absence of CdpA. In order to ascertain how intracellular c-di-GMP affects cytotoxicity, we compared the expression of bsaN and bipB, encoding components of the B. pseudomallei type III secretion system (TTSS), in the wild-type and cdpA mutant using qRT-PCR. TTSSs are specialized transport machineries activated under specific conditions for the delivery of bacterial virulence proteins, called effectors, into the host cell cytoplasm. Once intracellular, these effectors function to alter host cellular processes in order to promote bacterial survival and colonization. BsaN (BPSS1546) is a homologue of the Salmonella enterica serovar Typhimurium invasion protein InvF and a member of the AraC family of transcriptional regulators, while BipB (BPSS1532) is a homologue of the Salmonella invasin protein B and a component of the translocon complex (31). The bsa (Burkholderia secretion apparatus) locus encodes at least 20 different subunits which enable B. pseudomallei to translocate effector proteins directly into the host cell and is required for its full virulence in a number of animal models (30, 38). In the cdpA mutant, expression of bsaN and bipB were downregulated by 11-fold and 340-fold, respectively, compared to the wild type (Table 2).
FIG. 6.
CdpA influences ability of B. pseudomallei to invade mammalian cells and its cytotoxicity. (A) The cdpA mutant showed reduced invasion of A549 human lung epithelial cells compared to the wild type and complemented (comp.) cdpA mutant. (B) Killing of THP-1 human macrophage cells was also reduced from 37% killing by wild-type B. pseudomallei strain KHW to 6% killing by the cdpA mutant. Cytotoxicity was restored to the wild-type level in the complemented cdpA mutant.
DISCUSSION
We presented biochemical evidence that CdpA, an EAL domain-containing protein, is a phosphodiesterase which cleaves the synthetic substrate, bis-pNPP in vitro. The PDE activity has an absolute requirement for Mn2+, and this could not be substituted for by other divalent cations such as Mg2+, Ca2+, or Zn2+. This biochemical characteristic closely resembles that of HmsP c-di-GMP PDE of Yersinia pestis (1). Analysis of B. pseudomallei CdpA using the Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/) predicted it to be a membrane-bound signal transduction protein with a sensory (PAS) domain that is commonly associated with the sensing of oxygen and redox potential as part of the bacterial adaptive response (44). The presence of a transmembrane-spanning domain in CdpA explains the poor solubility and low yield of the recombinant MBP-CdpA. In E. coli, the DosCP complex comprising an oxygen-sensing DGC coupled to an oxygen-sensing PDE allows the bacterium to regulate its intracellular c-di-GMP concentration in response to O2 availability (35). As exemplified in Vibrio cholerae, other proteins containing the PilZ domain bind c-di-GMP and provide the link between oxygen-sensing and downstream processes involved in biofilm formation, motility, and virulence (25, 34). Although CdpA has a predicted DGC domain, this domain lacked the conserved GG(D/E)EF motif and is not expected to possess any DGC activity since structural analysis of PleD from C. crescentus revealed that the second glycine in this conserved motif is essential for the DGC catalytic function (3). A comparison of the intracellular c-di-GMP levels in the cdpA-null mutant and wild-type B. pseudomallei KHW verified that CdpA is a major c-di-GMP-specific PDE involved in the regulation of intracellular c-di-GMP levels in B. pseudomallei KHW. It would be interesting to determine if CdpA also interacts and respond to O2 availability.
Since high levels of c-di-GMP are associated with the production of adhesive matrix components, cell aggregation, and increased biofilm formation in several bacteria, the cdpA mutant which has high intracellular c-di-GMP levels also exhibited these phenotypes. CR staining showed that the cdpA mutant produced more exopolysaccharides than the wild type and was visibly more adhesive, forming aggregates in broth cultures. As expected, this resulted in increased adhesion of the cdpA mutant to the abiotic surface in the CV biofilm assay. This process is independent of swimming motility. Mutation in cdpA significantly increased in biofilm formation on the abiotic surface at the substratum but less so at the liquid-air interface in static broth cultures. It would be useful to further distinguish these two types of biofilms as the CV staining method is used widely for assessing biofilm formation. For example, with increased biofilm formation in the Vibrio cholerae vieA and Yersinia pestis hmsP mutants which are deficient in c-di-GMP PDEs, both showed increased CV staining (18, 34). In addition, the vieA mutant also showed increased CV staining at the liquid-air interface, while the hmsP mutant showed increased adherence to glass coverslips immersed in liquid broth.
Studies have shown that GGDEF or EAL proteins also control flagellar motility through gene expression, organelle assembly, or motor function. For instance, flagellar biosynthesis in P. aeruginosa is activated by FleQ, a c-di-GMP-responsive transcriptional regulator and the TipF c-diGMP phosphodiesterase homolog in Caulobacter crescentus functions as a flagellum assembly factor (12, 14). We therefore sought to find out if CdpA might have similar function to TipF in flagellum synthesis, especially since flagella are important virulence determinants in B. pseudomallei (6). The cdpA mutant, which was aflagellate, was indeed attenuated in cytotoxicity on THP-1 human macrophage cells, producing a 6-fold reduction in cytotoxicity compared to the wild type. This also agrees with our earlier observation that the aflagellate B. pseudomallei KHW ΔfliC mutant was avirulent (6). Flagella were also required by B. pseudomallei NCTC 13177 for adhesion and invasion during the early stage of infection of Acanthamoeba astronyxis cells (16). Swimming motility, which was absent in the cdpA mutant, is absolutely needed for B. pseudomallei to make contact with THP-1 cells in order to exert its cytotoxicity since a number of B. pseudomallei mutants defective in flagellum synthesis and swimming motility were also found to be attenuated in killing THP-1 cells. Cytotoxicity of these flagellar mutants was restored when the bacteria and THP-1 cells were centrifuged to allow contact to occur (Y. H. Gan, personal communication). Our results also showed that CdpA positively regulates expression of bsaN and bipB, components of the B. pseudomallei type III secretion system (TTSS). Downregulation of the expression of TTSS in the cdpA mutant would explain its attenuation in cytotoxicity. CdpA also positively regulates the expression of fliC, another virulence determinant for B. pseudomallei (7). The involvement of CdpA in biofilm formation, flagellum synthesis, motility, and invasion of host cells suggests a role for the protein as a regulator of virulence in response to environmental signals.
Bacterial aggregation into microcolonies is considered important for colonization of host cells in pathogenic bacteria such as V. cholera and in Neisseria meningitidis, but we did not observe an increase in invasion of A549 human lung epithelial cells by the cdpA mutant despite its more adhesive phenotype. N. meningitidis could manipulate host cell functions to reorganize the host cell surface such that microcolonies that formed were stabilized under dynamic conditions within blood vessels (19, 23). The B. pseudomallei cdpA mutant, which showed increased cell-to-cell adhesion, was in fact slightly less invasive on A549 cells than the wild type, possibly because formation of microcolonies might not be necessary for B. pseudomallei which normally infect lung epithelial cells via the respiratory route rather than the bloodstream. Data on the role of relevance of microcolony formation in colonization of eukaryotic cells by B. pseudomallei are also not clear. Although B. pseudomallei can form microcolonies to enhance its colonization of eukaryotic cells, the process was not required by all strains and was also temperature dependent, occurring at 27°C but not 37°C (2).
Given the pivotal roles of c-di-GMP-specific PDEs in the regulation of many bacterial behaviors, elucidation of the sensory and catalytic functions of CdpA in B. pseudomallei would contribute to deciphering the role of the cyclic di-GMP signaling in virulence and environmental adaptation of this important pathogen and also in the formation of biofilm, which is hypothesized to resemble the chronic form of melioidosis (11).
Acknowledgments
This study was supported by a grant from the Academic Research Fund, National University of Singapore (R183-000-194-112). H. S. Lee was supported by a graduate research scholarship from the National University of Singapore.
We thank A. M. Gamage and H. T. H. Saw for technical assistance and Y. H. Gan for primers for the qRT-PCR data.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 1 March 2010.
REFERENCES
- 1.Bobrov, A. G., O. Kirillina, and R. D. Perry. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247:123-130. [DOI] [PubMed] [Google Scholar]
- 2.Boddey, J. A., C. P. Flegg, C. J. Day, I. R. Beacham, and I. R. Peak. 2006. Temperature-regulated microcolony formation by Burkholderia pseudomallei requires pilA and enhances association with cultured human cells. Infect. Immun. 74:5374-5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, C., R. Paul, D. Samoray, N. C. Amiot, B. Giese, U. Jenal, and T. Schirmer. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. U. S. A. 101:17084-17089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, Y. Y., and K. L. Chua. 2005. The Burkholderia pseudomallei BpeAB-OprB efflux pump: expression and impact on quorum sensing and virulence. J. Bacteriol. 187:4707-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, K. H., J. B. Gaynor, K. G. White, C. Lopez, C. M. Bosio, R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443-448. [DOI] [PubMed] [Google Scholar]
- 6.Chua, K. L., Y. Y. Chan, and Y. H. Gan. 2003. Flagella are virulence determinants of Burkholderia pseudomallei. Infect. Immun. 71:1622-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, D., and O. Maaloe. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99-112. [Google Scholar]
- 8.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 10.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furukawa, S., S. L. Kuchma, and G. A. O'Toole. 2006. Keeping their options open: acute versus persistent infections. J. Bacteriol. 188:1211-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hickman, J. W., and C. S. Harwood. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69:376-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmann, K., and W. Stoffel. 1993. TMBASE—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 14.Huitema, E., S. Pritchard, D. Matteson, S. K. Radhakrishnan, and P. H. Viollier. 2006. Bacterial birth scar proteins mark future flagellum assembly site. Cell 124:1025-1037. [DOI] [PubMed] [Google Scholar]
- 15.Hyodo, M., and Y. Hayakawa. 2004. An improved method for synthesizing cyclic bis(3′-5′)diguanylic acid (c-di-GMP). Bull. Chem. Soc. Jpn. 77:2089-2093. [Google Scholar]
- 16.Inglis, T. J., T. Robertson, D. E. Woods, N. Dutton, and B. J. Chang. 2003. Flagellum-mediated adhesion by Burkholderia pseudomallei precedes invasion of Acanthamoeba astronyxis. Infect. Immun. 71:2280-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inglis, T. J., and J. L. Sagripanti. 2006. Environmental factors that affect the survival and persistence of Burkholderia pseudomallei. Appl. Environ. Microbiol. 72:6865-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirillina, O., J. D. Fetherston, A. G. Bobrov, J. Abney, and R. D. Perry. 2004. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 54:75-88. [DOI] [PubMed] [Google Scholar]
- 19.Kirn, T. J., M. J. Lafferty, C. M. Sandoe, and R. K. Taylor. 2000. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35:896-910. [DOI] [PubMed] [Google Scholar]
- 20.Kulasakara, H., V. Lee, A. Brencic, N. Liberati, J. Urbach, S. Miyata, D. G. Lee, A. N. Neely, M. Hyodo, Y. Hayakawa, F. M. Ausubel, and S. Lory. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U. S. A. 103:2839-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy, A., B. J. Chang, L. K. Abbott, J. Kuo, G. Harnett, and T. J. Inglis. 2003. Invasion of spores of the arbuscular mycorrhizal fungus Gigaspora decipiens by Burkholderia spp. Appl. Environ. Microbiol. 69:6250-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 23.Mikaty, G., M. Soyer, E. Mairey, N. Henry, D. Dyer, K. T. Forest, P. Morand, S. Guadagnini, M. C. Prevost, X. Nassif, and G. Dumenil. 2009. Extracellular bacterial pathogen induces host cell surface reorganization to resist shear stress. PLoS Pathog. 5:e1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pratt, J. T., R. Tamayo, A. D. Tischler, and A. Camilli. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 282:12860-12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 27.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 29.Song, Y., C. Xie, Y. M. Ong, Y. H. Gan, and K. L. Chua. 2005. The BpsIR quorum-sensing system of Burkholderia pseudomallei. J. Bacteriol. 187:785-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens, M. P., A. Haque, T. Atkins, J. Hill, M. W. Wood, A. Easton, M. Nelson, C. Underwood-Fowler, R. W. Titball, G. J. Bancroft, and E. E. Galyov. 2004. Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis. Microbiology 150:2669-2676. [DOI] [PubMed] [Google Scholar]
- 31.Stevens, M. P., M. W. Wood, L. A. Taylor, P. Monaghan, P. Hawes, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2002. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46:649-659. [DOI] [PubMed] [Google Scholar]
- 32.Sun, G. W., J. Lu, S. Pervaiz, W. P. Cao, and Y. H. Gan. 2005. Caspase-1 dependent macrophage death induced by Burkholderia pseudomallei. Cell. Microbiol. 7:1447-1458. [DOI] [PubMed] [Google Scholar]
- 33.Tarutina, M., D. A. Ryjenkov, and M. Gomelsky. 2006. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J. Biol. Chem. 281:34751-34758. [DOI] [PubMed] [Google Scholar]
- 34.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuckerman, J. R., G. Gonzalez, E. H. Sousa, X. Wan, J. A. Saito, M. Alam, and M. A. Gilles-Gonzalez. 2009. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 48:9764-9774. [DOI] [PubMed] [Google Scholar]
- 36.Ulrich, R. L., D. DeShazer, E. E. Brueggemann, H. B. Hines, P. C. Oyston, and J. A. Jeddeloh. 2004. Role of quorum sensing in the pathogenicity of Burkholderia pseudomallei. J. Med. Microbiol. 53:1053-1064. [DOI] [PubMed] [Google Scholar]
- 37.Valade, E., F. M. Thibault, Y. P. Gauthier, M. Palencia, M. Y. Popoff, and D. R. Vidal. 2004. The PmlI-PmlR quorum-sensing system in Burkholderia pseudomallei plays a key role in virulence and modulates production of the MprA protease. J. Bacteriol. 186:2288-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warawa, J., and D. E. Woods. 2005. Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. FEMS Microbiol. Lett. 242:101-108. [DOI] [PubMed] [Google Scholar]
- 39.Waters, C. M., W. Lu, J. D. Rabinowitz, and B. L. Bassler. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190:2527-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 41.White, N. J. 2003. Melioidosis. Lancet 361:1715-1722. [DOI] [PubMed] [Google Scholar]
- 42.Williamson, N. R., P. C. Fineran, W. Ogawa, L. R. Woodley, and G. P. Salmond. 2008. Integrated regulation involving quorum sensing, a two-component system, a GGDEF/EAL domain protein and a post-transcriptional regulator controls swarming and RhlA-dependent surfactant biosynthesis in Serratia. Environ. Microbiol. 10:1202-1217. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, X., X. Meng, and B. Sun. 2008. An EAL domain protein and cyclic AMP contribute to the interaction between the two quorum sensing systems in Escherichia coli. Cell Res. 18:937-948. [DOI] [PubMed] [Google Scholar]
- 44.Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331-333. [DOI] [PubMed] [Google Scholar]
- 45.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]