Abstract
Acinetobacter baumannii is an emerging bacterial pathogen of considerable health care concern. Nonetheless, relatively little is known about the organism's virulence factors or their regulatory networks. Septicemia and ventilator-associated pneumonia are two of the more severe forms of A. baumannii disease. To identify virulence factors that may contribute to these disease processes, genetically diverse A. baumannii clinical isolates were evaluated for the ability to proliferate in human serum. A transposon mutant library was created in a strain background that propagated well in serum and screened for members with decreased serum growth. The results revealed that disruption of A. baumannii phospholipase D (PLD) caused a reduction in the organism's ability to thrive in serum, a deficiency in epithelial cell invasion, and diminished pathogenesis in a murine model of pneumonia. Collectively, these results suggest that PLD is an A. baumannii virulence factor.
Acinetobacter baumannii is rapidly emerging as an important opportunistic pathogen that can cause a wide range of infections, including ventilator-associated pneumonia, skin and soft tissue infections, secondary meningitis, and bacteremia (reviewed in reference 6). While community-associated A. baumannii infections have been reported, the organism is predominantly a nosocomial pathogen, and disease is generally limited to severely compromised individuals, such as intensive-care unit patients and military personnel suffering from traumatic injury (2, 21, 27, 31, 38). Due in part to the organism's widespread resistance to most antimicrobial agents, many infections can be quite severe, with mortality rates ranging from 26 to 68% (18, 32, 35). Moreover, pan-antibiotic-resistant strains have recently emerged both in the United States and elsewhere, accentuating the need for new therapeutic strategies for the treatment of Acinetobacter infections (7, 33).
One of the reasons that A. baumannii-associated disease is predominantly limited to hospitalized patients is that it is inherently poised to persist within health care settings, thereby providing reservoirs for transmission and infection. Indeed, the organism can readily colonize and survive on inanimate surfaces for extended periods of time and is resistant to desiccation and disinfectants (10, 12, 26, 42). A. baumannii is also capable of forming biofilms on abiotic surfaces, resulting in a physiological state that could, in part, contribute to its aforementioned ability to persist, although the causality of this relationship has yet to be determined. Mature A. baumannii biofilm production depends on the production of pili, which are assembled by the chaperone-usher secretion system (CsuABCDE), and at least one other factor, biofilm-associated protein (Bap) (20, 36).
Despite a worldwide increase in the incidence of A. baumannii infections, surprisingly little is known about the organism's virulence factors or their regulatory networks. Accordingly, the focus of our work is to characterize the bacterial factors that contribute to A. baumannii ventilator-associated pneumonia and septicemia, the two most severe forms of disease caused by the organism (6). Their characterization may lead to novel therapeutic strategies to limit A. baumannii-associated morbidity and mortality.
To date, in vitro studies have revealed that outer membrane protein A (OmpA) plays an important role in bronchial epithelial cell invasion, implying that it may contribute to the organism's ability to cause pneumonia (4). Nonetheless, little difference in lung bacterial burdens was observed between wild-type (WT) and ompA mutant cells in a murine-pneumonia model of infection, suggesting that other factors may play important roles in Acinetobacter-mediated pneumonia (4). It has also been shown that some A. baumannii strains are resistant to the killing action of human serum, suggesting that they harbor genetic components that augment the organism's ability to cause bacteremia. One proposed mechanism for the serum-resistant phenotype is the inhibition of host complement C3 protein cleavage and binding to the organism's cell surface, which may in turn inhibit host phagocytic cell recognition and result in a serum-resistant phenotype (16). However, the A. baumannii factor(s) responsible for this phenotype has not yet been identified. It has also been shown that A. baumannii penicillin-binding protein 7/8 may play a role in serum resistance, presumably by remodeling the bacterial cell surface in a manner that protects it from host defense components, such as antimicrobial peptides (30).
The current work describes our efforts to expand the characterization of A. baumannii factors that augment the organism's ability to survive in human serum. Accordingly, we assessed the ability of A. baumannii clinical isolates to proliferate in human serum. A transposon mutant library was generated in one strain that exhibited the ability to proliferate well in serum and was screened for members with diminished serum growth. One mutant harbored a transposon insertion within a putative A. baumannii phospholipase D (PLD) gene. Phospholipase D assays revealed that this gene (A1S_2989) does indeed affect phospholipase activity. Moreover, its mutation resulted in decreased A. baumannii epithelial cell invasion (in vitro). Consistent with the aforementioned decrease in the ability to proliferate in serum, pld mutant cells exhibited decreased bacteremia and colonization of visceral host organs in a murine infection model. Collectively, these results indicate that phospholipase D is an A. baumannii virulence factor and suggest that agents that inhibit the protein's activity may have considerable therapeutic potential.
MATERIALS AND METHODS
Bacterial strains and growth medium.
The bacterial strains and plasmids used in this study are listed in Table 1. All strains were grown in either Luria-Bertani (LB) medium (Becton Dickinson, Franklin Lakes, NJ), tryptic soy broth (TSB) (Becton Dickinson), or 100% human serum. For serum preparation, plasma (American Red Cross, Omaha, NE) was clotted by adding 20 mM calcium chloride (Sigma Aldrich, Saint Louis, MO) and thromboplastin (prepared from human brain following the methods in reference 28) and incubation at 37°C for 1 h. Serum was collected after centrifugation at 2,000 × g and stored at −80°C until it was needed. Where indicated, the medium was supplemented with the following antibiotics: kanamycin (50 μg ml−1; MP Biomedicals, Solon, OH), tetracycline (20 μg ml−1; Acros Organics, Morris Plains, NJ), ampicillin (50 μg ml−1; Thermo Fisher, Waltham, MA), or gentamicin (300 μg ml−1; Invitrogen, Carlsbad, CA).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Collection site or relevant genotype | PFGE typea | Serum growthb | Sourcec or reference |
|---|---|---|---|---|
| A. baumannii | ||||
| 510 | Sputum | A | I | UNMC (NE; 2006) |
| 2824 | Urine | A | UNMC (NE; 2008) | |
| 2898 | Ankle wound | A | UNMC (NE; 2008) | |
| 4564 | Urine | A | UNMC (NE; 2006) | |
| 4860 | Urine | A | UNMC (NE; 2007) | |
| 4884 | Urine | A | UNMC (NE; 2007) | |
| 4893 | Leg wound | A | UNMC (NE; 2007) | |
| 5191 | Urine | A | UNMC (NE; 2007) | |
| 5393 | Unknown | A | UNMC (NE; 2007) | |
| 5562 | Blood | A | I | UNMC (NE; 2007) |
| 5592 | Sputum | A | UNMC (NE; 2007) | |
| 5695 | Unknown | A | UNMC (NE; 2007) | |
| ATCC 17904 | Urine | B | R | ATCC (1963) |
| ATCC 17961 | Blood | C | R | ATCC (1968) |
| ATCC 17976 | Postoperative meningitis | D | I | ATCC (1951) |
| ATCC 17978 | Fetal meningitis | E | I | ATCC (1951) |
| 98-37-01 | Endotracheal tube | F | R | CDC (TX; 1998) |
| 98-37-02 | Sputum | G | R | CDC (TX; 1998) |
| 98-37-05 | Tracheal aspirate | H | I | CDC (TX; 1998) |
| 98-37-09 | Cerebrospinal fluid | I | R | CDC (TX; 1998) |
| ACJ1 | EZ-Tn5::A1S_3365 derivative of 98-37-09 | I | I | This work |
| ACJ2 | EZ-Tn5::A1S_2989 derivative of 98-37-09 | I | I | This work |
| ACJ3 | EZ-Tn5::A1S_0269 derivative of 98-37-09 | I | I | This work |
| ACJ4 | EZ-Tn5::A1S_3045 (promoter region) derivative of 98-37-09 | I | I | This work |
| ACJ5 | EZ-Tn5::A1S_3045 (promoter region) derivative of 98-37-10 | I | I | This work |
| ACJ6 | EZ-Tn5::A1S_3278 derivative of 98-37-09 | I | I | This work |
| 98-37-11 | Bronchial wash | J | R | CDC (TX; 1998) |
| 01-12-05 | Blood | K | R | CDC (IN; 2001) |
| 01-12-06 | Sputum | K | CDC (IN; 2001) | |
| 01-12-07 | Sputum | K | CDC (IN; 2001) | |
| 01-12-08 | Burn wound | K | CDC (IN; 2001) | |
| 01-12-09 | Catheter tip | K | CDC (IN; 2001) | |
| 07-09-54 | Unknown | L | I | CDC (KY; 2007) |
| 07-09-59 | Unknown | L | CDC (KY; 2007) | |
| 07-09-60 | Unknown | L | CDC (KY; 2007) | |
| 07-09-61 | Unknown | L | CDC (KY; 2007) | |
| 07-09-63 | Unknown | L | CDC (KY; 2007) | |
| E. coli | ||||
| INVαF′ | F′ endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 ϕlacZΔM15 Δ(lacZYA-argF)U169 λ− | Invitrogen | ||
| Plasmids | ||||
| pCR II-TOPO | Ampr Kanr | Invitrogen | ||
| pWH1266 | Ampr Tetr | 15 | ||
| pACJ01 | pWH1266/A1S_2989 Tetr | This work |
PFGE pattern of the isolate in comparison to all other isolates in the study.
Human serum growth/resistance profile relative to all other PFGE types assessed; I and R indicate intermediate and robust growth, respectively.
Listed are the strain source with the location and/or year of isolation in parentheses. UNMC, University of Nebraska Medical Center.
PFGE.
The genetic diversity of all A. baumannii strains used in this study was assessed by pulsed-field gel electrophoresis (PFGE). Briefly, each bacterial strain was suspended in cell suspension buffer (100 mM EDTA, 100 mM Tris; Applied Biosystems, Austin, TX) to an optical density at 600 nm (OD600) of 1.15 and prepared in agarose plugs, and DNA was digested with 250 U ApaI enzyme (New England Biolabs, Ipswich, MA) as previously described (11). Samples were electrophoresed on a 1% SeaKem HGT Agarose Gel (Thermo Fisher) in 0.5× Tris-borate-EDTA (TBE) buffer. The initial switch time was 7 s, the final switch time was 20 s, and the run time was 18.5 h at 200 V. The agarose gel was stained with ethidium bromide to visualize and compare banding patterns.
Transposon mutagenesis.
An A. baumannii strain 98-37-09 transposon mutant library was created using the EZ-Tn5 <R6Kγori/KAN-2> Tnp transposome system, following the manufacturer's recommendations for prokaryotic cell transposition (Epicentre Biotechologies, Madison, WI). Briefly, EZ-Tn5 <R6Kγori/KAN-2> transposome (33 ng) was electroporated into 2 × 109 A. baumannii 98-37-09 electrocompetent cells in 0.1-cm cuvettes at 1.8 kV. Suspensions (100 μl) were then immediately added to 400 μl prewarmed SOC medium (Invitrogen) and incubated at 37°C and 225 rpm for 1 h. Transposon-containing cells were selected by plating them on LB agar containing kanamycin. In total, 6,000 transposon mutants were generated and subsequently inoculated into wells of 96-well round-bottom plates (Corning, Lowell, MA) containing 150 μl TSB medium supplemented with kanamycin. The plates were incubated at 37°C overnight to allow growth, and then 10 μl of each well suspension was transferred to individual wells of fresh 96-well round-bottom plates containing 100 μl of 5% bovine serum albumin (Sigma Aldrich)-5% l-glutamic acid (Sigma Aldrich) solution and sealed with plastic film (Thermo Fisher) for long-term storage at −80°C. A total of 65 transposon library members were randomly selected to assess the randomness and number of transposon insertions per isolate by Southern blotting. To do so, chromosomal DNA was digested with ApaI restriction enzyme, electrophoresed, and transferred to a nylon membrane (Hybond-N; GE Healthcare, Pittsburgh, PA). The membranes were probed with digoxigenin (DIG)-labeled probe specific for the transposon kanamycin resistance gene, which was generated using the PCR DIG Probe Synthesis Kit and primers (forward, 5′-GAAAACAGCATTCCAGGTATTAGAA-3′; reverse, 5′-CTTATGCATTTCTTTCCAGACTTGT-3′) as recommended by the manufacturer (Roche Applied Sciences, Indianapolis, IN). Anti-digoxigenin-AP Fab fragments were bound to the DIG-labeled probe by washing them and visualized with disodium 3-(4-methoxyspiro l,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan-4-yl) phenyl phosphate (CSPD) (Roche Applied Sciences).
Phospholipase D assay.
A. baumannii strains were assessed for PLD activity using the Amplex Red Phospholipase D Assay Kit according to the manufacturer's recommendations (Invitrogen). Briefly, A. baumannii strains 98-37-09 and ACJ2 were grown overnight in LB medium or LB medium supplemented with kanamycin, respectively. The strains were subsequently subcultured to an OD600 of 0.4. The cells were pelleted by centrifugation (2,000 × g; 4°C; 10 min), an aliquot of each supernatant was filter sterilized (0.22 μm), and PLD activity was measured. To normalize for differences in protein concentrations among samples tested, the protein concentration of the remainder of each supernatant was measured using the Bradford assay of trichloroacetic acid-precipitated material. The PLD assay results were normalized to the supernatant protein concentration.
Serum growth assay.
Representatives of each A. baumannii PFGE type listed in Table 1 and all members of an A. baumannii strain 98-37-09 transposon mutant library were assessed for the ability to proliferate in human serum. For PFGE type assays, strains were grown for 16 h in TSB medium at 37°C with aeration (225 rpm) and were then used to inoculate (1:100 dilution; approximately 1 × 107 CFU ml−1) 10 ml of human serum in 50-ml conical polypropylene tubes (Becton Dickinson). The tubes were incubated at 37°C at 225 rpm, and the OD600 or CFU ml−1 were measured at 15-h intervals over a 60-h period, as indicated in the text. For mutant library assays, library members were grown for 16 h in individual wells of 96-well round-bottom plates (Costar 3788; Corning) containing 150 μl TSB and kanamycin. Approximately 1 × 107 cells of each library member were subsequently transferred to new 96-well flat-bottom plates containing 200 μl human serum (Falcon 3072; Becton Dickinson). To prevent cross-contamination, the plates were then sealed with plastic film (Thermo Fisher) and incubated at 37°C for 60 h. The OD600 was determined initially and at 15-h intervals for the duration of the study.
Inverse PCR.
Inverse PCR was used to identify the transposon insertion sites of mutants exhibiting reduced cellular proliferation within human serum. To do so, total bacterial DNA was purified from each mutant of interest using a DNeasy Blood and Tissue Kit, following the manufacturer's recommendations (Qiagen, Valencia, CA). Two micrograms of purified DNA was then digested with the restriction enzyme AfeI (10 U; New England Biolabs) at 37°C for 1 h. The restriction fragments were circularized by ligation using 1.5 U of T4 DNA ligase (Invitrogen, Carlsbad, CA) at 16°C for 16 h. Following enzyme inactivation by heat treatment, 5 μl of each ligation mixture was subjected to inverse PCR using Platinum PCR Supermix High Fidelity (Invitrogen) and transposon-specific primers (forward, 5′-ACCTACAACAAAGCTCTCATCAACC-3′; reverse, 5′-CTACCCTGTGGAACACCTACATCT-3′) supplied in the EZ-Tn5 <R6Kγori/KAN-2> Tnp transposome kit (Epicentre Biotechnologies). Inverse PCR was performed in a GeneAmp PCR System 9700 thermocycler (Applied Biosystems) with the following parameters: 94°C for 10 min and 50 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 6 min, followed by an extension at 72°C for 10 min. The PCR products were electrophoresed in a 1% UltraPure Agarose gel (Invitrogen) at 75 V for 40 min and gel purified using a QIAquick Gel Extraction Kit (Qiagen). To determine the sequence of each PCR product, approximately 40 ng of gel-purified PCR products was ligated into 25 ng of pCRII-Topo vector and transformed into 50 μl Escherichia coli One Shot INVαF′ cells, following the manufacturer's recommendations for dual-promoter TA cloning (Invitrogen). Transformants were selected on LB agar containing kanamycin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (80 μg ml−1; Sigma Aldrich); the latter allowed an efficient screen of plasmids containing an insert within the vector lacZ gene. Following propagation, plasmid DNA was purified using QIAprep Spin Miniprep kits (Qiagen) and sequenced at the University of Nebraska Medical Center Sequencing Core Facility using vector-specific primers (forward, 5′-GTAAAACGACGGCCAG-3′; reverse, 5′-CAGGAAACAGCTATGAC-3′).
Construction of the A1S_2989 complementation plasmid.
The A. baumannii A1S_2989 (putative PLD) locus and 500 bp of upstream region were amplified from A. baumannii 98-37-09 DNA by PCR using Platinum PCR Supermix High Fidelity (Invitrogen) and primers (forward, 5′-CTGCAGATTATGGCACAATCCTTTCATTCCA-3′; reverse, 5′-CTGCAGGTAGAAGGCCATGATGTAAAAAGTT-3′), each containing a 5′-terminal PstI restriction site (underlined). The resulting 1.7-kb PCR product was ligated into a pCRII-Topo vector and transformed into E. coli INVαF′ cells for propagation. Plasmid DNA was subsequently purified using QIAprep Spin Miniprep kits (Qiagen) and then digested with PstI enzyme to liberate the plasmid insert. The PLD insert was gel purified using a QIAquick Gel Extraction Kit (Qiagen), ligated into a PstI-digested pWH1266 plasmid to generate plasmid pACJ01 and then electroporated into A. baumannii strain ACJ2. Transformants containing plasmid pACJ01 or vector alone were selected by growth on LB agar containing tetracycline. DNA sequencing confirmed the integrity of plasmid pACJ01.
Bacterial-invasion assay.
Human bronchial epithelial BEAS-2B cells and cervical cancer HeLa cells were infected with bacteria, as previously described (4). Briefly, cell lines were grown to confluence in 24-well plates (Becton Dickinson) and subsequently infected with 7.5 × 105 stationary-phase A. baumannii strain 98-37-09 or ACJ2 cells in 1 ml Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal calf serum (FCS) (Invitrogen) and, when applicable, tetracycline. The mixtures were incubated at 37°C and 6% CO2 for 4 h to allow invasion. The wells were then washed 3 times with Dulbecco's phosphate-buffered saline (DPBS) (Invitrogen). One milliliter DMEM-10% FCS with gentamicin was added to each well, and the plates were incubated for an additional 2 h to eliminate noninternalized A. baumannii cells. The supernatants were plated onto LB agar to ensure gentamicin had eliminated extracellular A. baumannii. The wells were washed 5 times with DPBS and then treated with 0.5 ml PBS containing 0.1% Triton X-100 (Invitrogen) for 20 min at 37°C to lyse the HeLa or BEAS-2B cells, allowing the release of internalized A. baumannii cells. The well contents were serially diluted and plated on LB medium for enumeration of CFU ml−1. The cytoskeleton inhibitor cytochalasin D was used to assess the extent to which gentamicin treatment measured host cell invasion, as previously described (4). Briefly, 2 μM cytochalasin D (Sigma Aldrich) was added to HeLa or BEAS-2B cells 30 min before the addition of A. baumannii strain 98-37-09 or ACJ2 cells and was maintained in the medium for the entire infection period. This concentration of cytochalasin D did not affect bacterial or host cell viability.
A. baumannii pneumonia mouse model.
A. baumannii strains 98-37-09 and ACJ2 were grown for 16 h in TSB and TSB supplemented with kanamycin, respectively, and then subcultured (1:100 dilution) into TSB without antibiotics and grown to stationary phase (10 h at 37°C, 225 rpm), corresponding to the optimal growth phase for pld transcription (data not shown). Bacteria were pelleted from 10 ml of culture, washed twice with endotoxin-free PBS (Invitrogen), and resuspended in endotoxin-free PBS to a final bacterial density of approximately 1.2 × 1010 CFU ml−1. Eight-week-old C57BL/6 mice were anesthetized by intraperitoneal injection of tribromoethanol. Pneumonia was induced by intranasal inoculation of 30 μl of bacterial suspension (3.8 × 108 CFU and 3.6 × 108 CFU of wild-type and pld mutant cells, respectively) prepared as described above. The mice were sacrificed at 24 or 48 h postinoculation by CO2 inhalation. For assessment of the bacterial burden, organs (lungs, hearts, and livers) were excised and homogenized in 1 ml sterile PBS (Invitrogen). The homogenized tissue was serially diluted and plated for colony formation on LB medium or LB medium with kanamycin for strains 98-37-09 and ACJ2, respectively. Statistical analyses were performed using a two-tailed, unpaired Student's t test, and the data are shown as the log CFU ml homogenized tissue−1. These experiments were approved by the Vanderbilt University Institutional Animal Care and Use Committee.
Histopathologic procedures.
Mouse hearts were removed from A. baumannii 98-37-09- or ACJ2-infected mice, rinsed in saline, and immersion fixed in 10% buffered neutral formalin. Transverse serial sections were stained with either hematoxylin and eosin or Gram stain, allowing microscopic examination of pulmonary, aortic, and atrioventricular valves.
RESULTS
Serum proliferation.
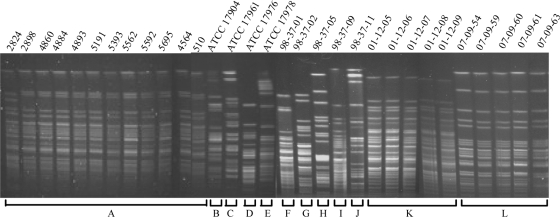
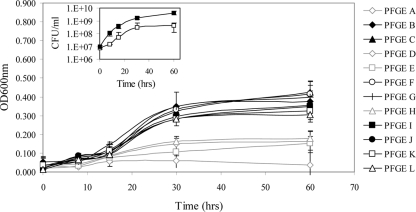
As a prerequisite to identifying A. baumannii genetic determinants that contribute to the organism's ability to survive in human serum, we acquired 31 clinical isolates from diverse U.S. geographic locations (Table 1). PFGE was used to evaluate the clonality of all isolates and established that they could be differentiated into 12 distinct genetic backgrounds (Fig. 1 and Table 1). A representative of each lineage was then assessed for the ability to proliferate in human serum over a course of 60 h. As shown in Fig. 2, most lineages exhibited little change in optical density during the first 7.5 h of incubation in human serum. By 15 h postinoculation, the optical density for most strains began to increase slightly. Growth increased further at 30 h postinoculation and plateaued by 60 h postinoculation. The results revealed that PFGE types C, F, and G (strains ATCC 17961, 98-37-01, and 98-37-02) consistently exhibited the most pronounced ability to thrive in human serum. Moreover, by comparing the growth characteristics of each strain at 60 h postinoculation to those of ATCC 17961, 98-37-01, and 98-37-02, all strains could be clearly differentiated into two classes: A, strains that readily propagated in human serum (i.e., that exhibited ≥50% growth in comparison), and B, strains that did not thrive in human serum (i.e., that exhibited <50% growth in comparison). We reasoned that strains such as 98-37-09 (PFGE type I) that exhibited a robust growth phenotype in serum harbored unique factors (or unique expression properties) that augmented their ability to proliferate in serum. Comparing the viable CFU of strain 98-37-09 (PFGE type I) to those of strain ATCC 17978 (PFGE type E), which exhibited reduced growth in serum in comparison to 98-37-09, verified that the growth phenotype of each strain mimicked its optical density measurement (Fig. 2, inset).
FIG. 1.
PFGE of clinical isolates used in this study. All clinical isolates were confirmed to be A. baumannii by Microscan Identification Panel assessment (data not shown) and were subjected to PFGE to assess their clonality. The results revealed that the 31 isolates obtained for the study represented 12 distinct genetic backgrounds (labeled A to L). Each strain's source, isolation site, and collection year are provided in Table 1.
FIG. 2.
Growth characteristics of A. baumannii PFGE types in human serum. Graphed are the growth characteristics of representative strains of 12 distinct A. baumannii PFGE types following 0, 7.5, 15, 30, and 60 h of incubation in human serum, as determined by the OD600 (starting inoculum, 1 × 107 CFU). The black lines and symbols indicate strains that exhibited the ability to proliferate well in serum, and the light-gray lines and symbols indicate strains that exhibited diminished serum growth in comparison to all other isolates analyzed. All OD600 values have been normalized to the optical density of serum alone, and standard deviations are shown (n = 6). (Inset) Growth curves of A. baumannii strain 98-37-09 (solid boxes) (PFGE type I; proliferates well in human serum) and ATCC 17978 (open boxes) (PFGE type E; reduced serum growth) grown in human serum; standard deviations are shown (n = 3).
Identification of A. baumannii serum growth factors.
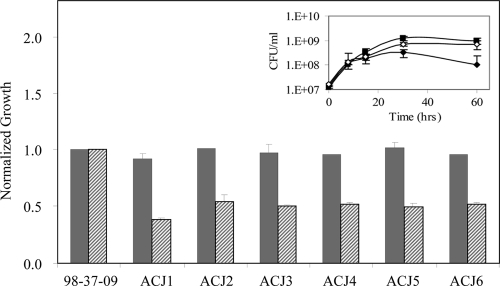
To identify A. baumannii genetic determinants that contribute to the organism's ability to survive and proliferate in human serum, a 98-37-09 transposon mutant library was created and subsequently screened for members with a reduced-serum-survival phenotype. We selected 98-37-09 as opposed to other strains that were determined to proliferate well in human serum because its antimicrobial susceptibility profile was amenable to the selection of commonly transferable antibiotic resistance markers (data not shown). Among the 6,000 A. baumannii 98-37-09 transposon mutants evaluated, 13 demonstrated a ≤1.6-fold increase in optical density when cultured for 60 h in human serum in comparison to parental 98-37-09 cells (data not shown). To distinguish whether these mutants harbored a mutation in a serum growth-dependent factor, as opposed to a general growth factor, we also assessed each mutant's ability to proliferate in laboratory LB medium. As shown in Fig. 3, six mutants exhibited a growth defect in human serum but exhibited wild-type growth rates in LB medium, suggesting that they harbored a loss-of-function mutation in a serum-specific growth factor. The transposon insertion sites for these mutants were determined to be located within the following loci by inverse PCR: disulfide bond formation protein (A1S_3365), putative general secretion pathway protein (A1S_0269), hydrolase isochorimatase family protein (A1S_3278), a putative promoter region of exoribonuclease R (A1S_3045; two mutants), and a putative phospholipase D (A1S_2989). While it is difficult to predict a priori how many of these factors affect the organism's ability to proliferate in serum, phospholipase D is a well-characterized virulence factor that has been suggested to play an important role in hematogenous dissemination of other bacterial pathogens within infected animals (23).
FIG. 3.
Growth characteristics of A. baumannii mutant library members with increased serum susceptibility. Shown are the growth characteristics of the wild type (strain 98-37-09) and isogenic mutants in LB medium (solid bars) and human serum (hatched bars), as measured by the OD600. All values have been plotted relative to wild-type growth (set at 1.0). Also shown are the growth curves of wild-type (solid boxes), ACJ2 (A1S_2989 mutant; solid diamonds), and ACJ2 harboring pACJ01 (wild-type copy of A1S_2989; open circles) in human serum (inset). Standard deviations are shown (n = 6).
A. baumannii locus A1S_2989 modulates phospholipase D activity.
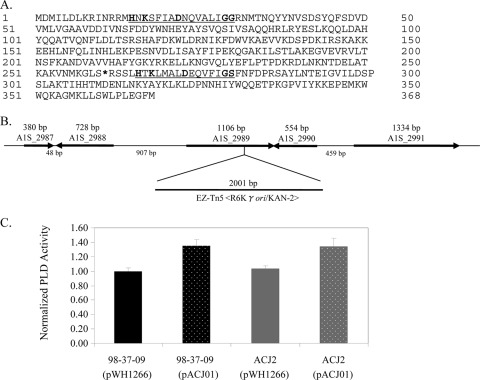
Most bacterial PLD proteins are secreted exoproteins containing two active-site HXKX4DX6G(G/S) motifs that catalyze hydrolysis of phosphatidylcholine, a major component of eukaryotic cell membranes, to phosphatidic acid (17, 29, 43). As shown in Fig. 4A, the A. baumannii A1S_2989 locus is predicted to encode a 368-amino-acid protein that contains two putative PLD active-site motifs, a perfect consensus motif spanning amino acid residues 15 to 30 and a nearly perfect match missing a single amino acid within the X6 region of the active-site motif spanning amino acid residues 265 to 279. The transposon insertion site within the serum-sensitive 98-37-09 derivative is located 4 amino acid residues upstream of the second putative PLD active site and, based on the predicted surrounding genome organization, is not likely to cause polar effects (Fig. 4A and B).
FIG. 4.
Characterization of the ATCC 17978 A1S_2989 locus. (A) Predicted amino acid sequence of ATCC 17978 locus A1S_2989-encoded protein with putative PLD active sites underlined; the boldface residues correspond to conserved HXKX4DX6G(G/S) amino acids. The asterisk indicates a transposon insertion site within strain ACJ2. (B) Chromosomal organization of A. baumannii strain ATCC 17978 locus A1S_2989 with the strain ACJ2 transposon EZ-Tn5 <R6Kγori/KAN-2> insertion site shown. (C) Graph of the phospholipase D activity assay results for wild-type (strain 98-37-09) and ACJ2 (A1S_2989 mutant) cells harboring either vector (pWH1266) or a plasmid containing a wild-type copy of A1S_2989 (pACJ01). The results were normalized to equal amounts of total starting protein, as described in Materials and Methods; standard deviations are shown (n = 6).
To assess whether A1S_2989 affects A. baumannii PLD activity, the phospholipase D activities of wild-type (98-37-09) and ACJ2 (A1S_2989 mutant) cell supernatants were compared. No significant difference in PLD activity was detected between wild-type and ACJ2 cells harboring plasmid vector alone (pWH1266), presumably reflecting the presence of additional phospholipase D genes within the strain background (Fig. 4C). Indeed, all publically available A. baumannii genomes contain multiple phospholipases, including at least two putative PLDs. Consistent with that possibility, microarray analysis of wild-type RNA revealed the expression of two PLD transcripts by strain 98-37-09 (data not shown). Importantly, both wild-type and ACJ2 cells harboring a plasmid-borne copy of A1S_2989 and the corresponding putative promoter region (pACJ01) exhibited a modest increase in phospholipase D activity in comparison to vector-containing cells (Fig. 4C). Collectively, these results suggest that A. baumannii A1S_2989 confers PLD activity. Moreover, it was found that ACJ2 cells harboring pACJ01 exhibited wild-type levels of growth in human serum (Fig. 3, inset).
A. baumannii pld mutant cells exhibit reduced epithelial cell invasion.
PLD activity has been proposed to play a role in Neisseria gonorrhoeae complement receptor type 3-mediated gonococcal invasion of cervical epithelial cells (8). Moreover, recent studies have revealed that A. baumannii invades nonphagocytic epithelial cells; it has been suggested that this could provide a means for the organism to circumvent host defenses by allowing bacterial persistence and penetration into deep tissues (4). Given the correlation between bacterial PLD and epithelial cell invasion observed for other organisms, we assessed whether A. baumannii PLD affects host cell invasion. To do so, the invasion properties of wild-type 98-37-09 and ACJ2 cells were compared.
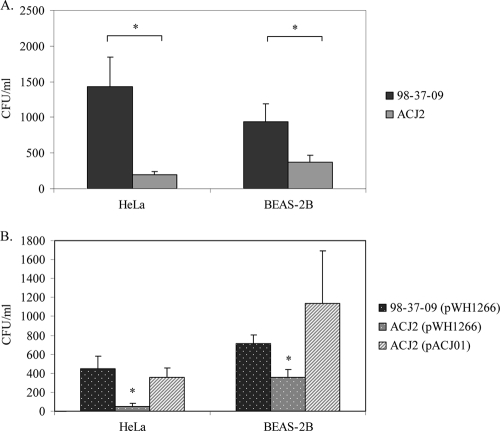
As shown in Fig. 5A, A. baumannii strain 98-37-09 demonstrated increased ability to invade HeLa cervical cancer cells in comparison to BEAS-2B bronchial epithelial cells. The reason for this disparity is unknown, but it is consistent with the results of Choi and colleagues, who observed differing cell line-specific invasion frequencies for A. baumannii strain ATCC 19606 (4). However, it should be noted that the previous study observed increased invasion of respiratory cells in comparison to other cell lines. Importantly, regardless of the cell line assessed, isogenic ACJ2 pld mutant cells exhibited a significant reduction (P < 0.01) in invasion. In comparison to wild-type cells, pld mutant (ACJ2) cell invasion decreased 7.4- and 3-fold for HeLa and BEAS-2B cells, respectively. Pretreatment of the cell lines with the cytoskeletal microfilament inhibitor cytochalasin D confirmed that A. baumannii invasion was dependent on the formation of epithelial cell actin microfilaments, as previously suggested (4). In the presence of cytochalasin D, cell invasion was significantly (P < 0.01) inhibited by 90% and 85% for wild-type and AJC2 cells, respectively (data not shown).
FIG. 5.
Invasion properties of A. baumannii 98-37-09, ACJ2, and complementation strains. (A) Internalization characteristics of wild-type (98-37-09) and ACJ2 (A1S_2989 mutant) cells following incubation with human cervical cancer (HeLa) or human bronchial epithelial (BEAS-2B) cells. Plotted are the numbers of CFU ml of A. baumannii−1 internalized following 4 h of incubation. (B) Internalization properties of wild-type and ACJ2 cells harboring plasmid vector (pWH1266) or an A1S_2989 complementation plasmid (pACJ01). The asterisks indicate statistically significant differences between wild-type and ACJ2 internalization as determined by Student's t test (A) or two-way analysis of variance (P < 0.05). Standard deviations are shown.
To verify that the pld-associated difference in invasion was due to disruption of the pld gene, as opposed to some other unrecognized feature of ACJ2 cells, the invasion properties of ACJ2 containing the A1S_2989 complementation plasmid, pACJ01, were measured. Interestingly, transformation of vector (pWH1226, a derivative of pBR322 containing the Acinetobacter origin of replication [15]) or vector containing pld (pACJ01) into both wild-type and ACJ2 cells caused a moderate bacterial growth defect when bacteria were grown in plasmid selection medium containing tetracycline. This same phenotype has been observed for other organisms harboring pBR322 derivatives (24, 25, 40, 41). Nonetheless, pACJ01 appeared to fully restore the invasion properties of ACJ2 cells to the levels of the wild type containing vector alone (Fig. 5B). Presumably the observed differences in invasion frequency between A. baumannii cells alone (Fig. 5A) and cells harboring plasmids (Fig. 5B) reflected the reduced growth properties of the latter in invasion medium, which contained tetracycline to ensure plasmid maintenance. Nonetheless, collectively, these results suggest that, in addition to contributing to serum survival, A. baumannii PLD plays an important role in epithelial cell invasion.
PLD affects bacteremia and extrapulmonary tissue bacterial burden.
Because pld-disrupted cells exhibited diminished serum survival and invasion and/or survival within lung epithelial cells, we used a murine-pneumonia model of infection to evaluate whether PLD affects lung pathogenesis, bacteremia, and visceral-organ bacterial burden. Accordingly, C57BL/6 mice were intranasally inoculated with 3.8 × 108 CFU or 3.6 × 108 CFU of 98-37-09 or ACJ2 (pld), respectively, and were then sacrificed at 24 or 48 h postinoculation. For assessment of the effects of PLD on bacteremia, the CFU per milliliter of blood were enumerated. To assess the effects of PLD on the bacterial burden in lungs, heart, and liver, the organs were excised and homogenized, and the CFU were enumerated. The infection results revealed that while pld mutant cells exhibited decreased tissue culture epithelial cell invasion, there was no significant difference between wild-type and pld mutant bacterial burdens in lung tissue. Presumably, this disparity reflects differences in the inocula and/or time of host cell exposure between the tissue culture assays (7 × 105 CFU; 4 h) and the murine-infection model (3.8 × 108 CFU; 24 h), either of which may mask the effects of PLD on lung invasion. Alternatively, PLD activity may play a minimal role in A. baumannii bronchial epithelial invasion in a multifactorial host environment. Histological examination of lung tissue revealed pathological changes consistent with pneumonia, including marked infiltration of immune cells, in both wild-type- and ACJ2-infected mice (data not shown).
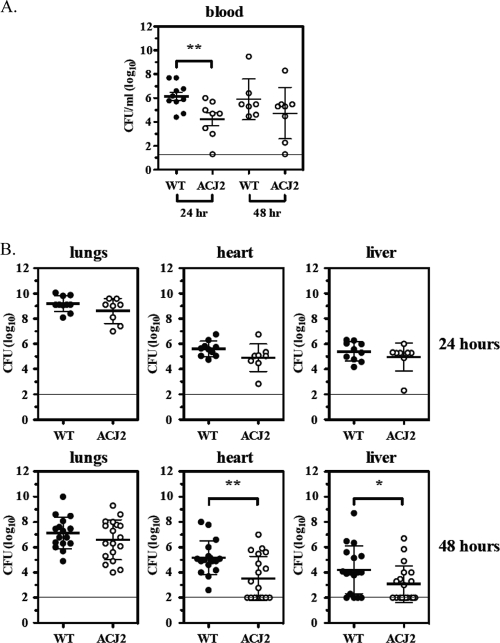
Although PLD did not seem to affect the lung bacterial burden, the pld mutant strain exhibited a striking reduction in the burden in blood, heart, and liver. More specifically, as shown in Fig. 6A, the mutant strain resulted in a significant decrease in bacteremia at 24 h postinoculation (1.5 × 106 CFU ml WT−1 versus 1.7 × 104 CFU ml ACJ2−1), which is consistent with the observation that pld mutant cells exhibit diminished serum proliferation. While not significant, this same trend was observed at 48 h postinoculation (7.8 × 105 CFU ml WT−1 versus 5.4 × 104 CFU ml ACJ2−1). Although few differences were observed at 24 h, by 48 h postinfection, pld mutant cells were observed to exhibit a diminished extrapulmonary tissue burden (Fig. 6B). More specifically, the mutant exhibited a significant decrease in heart burden (1.5 × 105 CFU ml WT−1 versus 3.3 × 103 CFU ml ACJ2−1) and a significant decrease in liver burden (1.6 × 104 CFU ml WT−1 versus 1.2 × 103 CFU ml ACJ2−1). A comparison of the bacterial burdens of heart and liver tissues from wild-type-infected mice at 24 and 48 h revealed a 0.5 to 1.0 log10 reduction in bacteria at the later time point. Conversely, pld mutant bacteria exhibited a 1.5 to 2.0 log10 reduction, suggesting that ACJ2 cells are more readily cleared within heart and liver tissues.
FIG. 6.
Pathogenic properties of A. baumannii strain 98-37-09 and ACJ2 cells in a murine-pneumonia model. (A) The averaged bacterial burdens of wild-type (98-37-09) and ACJ2 (A1S_2989 mutant) A. baumannii cells collected from the blood at 24 and 48 h after intranasal colonization (n ≥ 7); the horizontal line indicates the lower limit of detection. (B) The bacterial burdens of the wild type and ACJ2 obtained from lungs, hearts, and livers at 24 h (n ≥ 8) or 48 h (n ≥ 16) after intranasal colonization; the 48-h data points represent the results from two independent experiments; the horizontal lines indicate the lower limits of detection. The asterisks indicate statistically significant differences between the wild-type and ACJ2 bacterial burdens as determined by a two-tailed, unpaired Student t test (*, P < 0.05; **, P < 0.01). All data are shown as the log CFU ml blood or homogenized tissue−1.
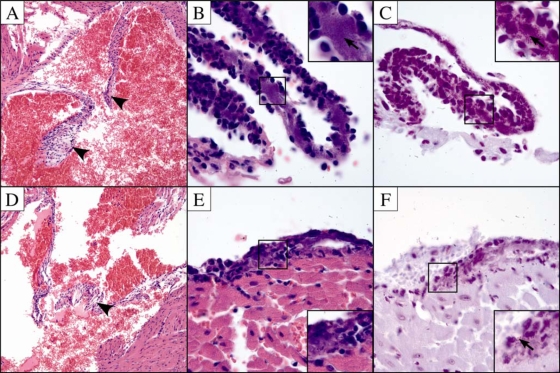
Histopathology was used, in part, to validate our murine-pneumonia results and expand our understanding of A. baumannii pathogenesis in this model. Accordingly, the hearts were removed from 98-37-09- and ACJ2-infected mice (48 h) and were subjected to microscopic evaluation. We did not observe inflammation or bacterial colonization of the myocardium or the heart valves in either experimental group (Fig. 7A and D). However, pericarditis was consistently observed in both wild-type- and ACJ2-infected animals (Fig. 7B and E). The inflammation consisted of neutrophils and macrophages expanding the pericardial tissue with minimal involvement of the epicardium. Bacteria were readily observed in the areas of inflammation on hematoxylin- and eosin-stained sections, and the presence of bacteria was confirmed by Gram staining (Fig. 7B and C). While bacteria were evident in both experimental groups, there appeared to be a reduction in pericardium-associated bacteria in ACJ2-infected animals in comparison to wild-type-infected mice, supporting the observation that PLD contributes to the heart bacterial burden (Fig. 7C and F). The finding that A. baumannii infection leads to pericarditis is not without precedent. Several reports have associated Acinetobacter with cardiac tamponade, a severe condition resulting from bacterial infection of the pericardial space that leads to increased pericardial fluid, restricted heart movement, and significant rates of morbidity and mortality, suggesting that the murine-pneumonia model described here is an appropriate system to study this disease (19, 37). In fact, to our knowledge, this represents the first description of A. baumannii-associated pericarditis in an animal model of infection. Histopathology results also revealed small foci of inflammation in the livers of mice infected with wild-type bacteria. However, bacteria were not identified in association with the inflammation on either hematoxylin- and eosin- or Gram-stained sections (data not shown). The livers of ACJ2-infected mice did not exhibit foci of inflammation. It is not clear why no bacteria were determined to be associated with infected mouse livers, in light of the fact that enumeration of CFU from homogenized livers of wild-type- and ACJ2-infected mice resulted in an average of 1.6 × 104 CFU ml−1 and 1.2 × 103 CFU ml−1, respectively. One possible explanation for this disparity may be differences between the limits of detectability of the two approaches. Histopathology allows a qualitative measure of the bacterial burden, whereas bacterial-viability measurements provide a more accurate and quantitative assessment. In that regard, microscopic evaluation may not allow the resolution needed to accurately detect low numbers of A. baumannii bacteria in liver tissue.
FIG. 7.
Histological sections of hearts from A. baumannii-infected C57BL/6 mice. (A to C) Histopathology results for mouse hearts from 98-37-09-infected mice (A) Section through the aortic valve (arrowheads) with no evidence of endocarditis (hematoxylin and eosin; magnification, ×40). (B) Section through pericardium that has lifted away from the heart surface demonstrating pericarditis with infiltration of neutrophils and macrophages (hematoxylin and eosin; magnification, ×100). Bacteria are easily observed in the pericardial tissue (arrow in the inset). (C) Section through pericardium (Gram stain; magnification, ×100). Bacteria are indicated by the arrow in the inset. (D to F) Histopathology results for ACJ2-infected mouse hearts. (D) Section through aortic valve (arrowhead) with no evidence of endocarditis (hematoxylin and eosin; magnification, ×20). (E) Section through pericardium demonstrating pericarditis with few bacteria, indicated by the arrow (hematoxylin and eosin; magnification, ×100). (F) Section through pericardium (Gram stain; magnification, ×100).
DISCUSSION
Although A. baumannii is an emerging bacterial pathogen of considerable health care interest, little is known about the factors that modulate the organism's ability to cause disease. One of the more severe forms of A. baumannii infection is bacteremia. King and colleagues recently established that A. baumannii strains exhibit different serum resistance, implying that some strain backgrounds may have a higher propensity to cause bacteremia than others (16). Characterizing the factors that modulate serum resistance and/or serum proliferation may delineate the mechanism(s) by which A. baumannii causes disease and provide novel strategies for therapeutic intervention in Acinetobacter infection. Accordingly, the focus of the current work was to expand our understanding of the A. baumannii genetic determinants that mediate serum growth. A collection of 31 clinical A. baumannii isolates was acquired, representing strains collected from diverse U.S. geographical regions over a course of 57 years. PFGE analyses determined that these isolates represented 12 distinct genetic lineages and that, in large part, outbreak strains or strains acquired from single sites tended to be clonal. One exception included 5 strains that were genetically distinct from one another but were isolated from a single outbreak in 1998 in Texas. The observed genetic diversity within the admittedly small strain collection used in these studies suggests that multiple lineages are capable of causing disease and that no single lineage predominates as the most problematic.
The A. baumannii PFGE types tested displayed differing abilities to proliferate in human serum, in agreement with studies by others (16). By exploiting the relatively robust serum growth phenotype of one strain, 98-37-09, we identified isogenic mutants that demonstrated diminished ability to proliferate in human serum. Six of these mutants exhibited parental growth phenotypes in laboratory culture medium, suggesting that they harbored a mutation in a serum-specific growth-dependent and/or resistance factor. One of these strains (A1S_2989) contained a mutation in phospholipase D.
Phospholipase D has been shown to play an important role in hematogenous dissemination of bacterial pathogens within infected animals and is considered a major virulence determinant of Corynebacterium pseudotuberculosis (1, 13, 34). Therefore, we hypothesized that PLD plays a role in A. baumannii pathogenesis and focused our effort on determining the in vitro and in vivo contributions of PLD to pathogenesis in the organism. As a first test of this possibility, we evaluated whether A. baumannii PLD affects the organism's ability to invade epithelial cells, in part because PLD plays an important role in N. gonorrhoeae host cell invasion (8). The results revealed that A. baumannii PLD is required for wild-type levels of epithelial cell invasion. Moreover, it is possible that PLD activity plays a more significant role during invasion than observed here. The reason for this is that all sequenced A. baumannii strains harbor at least two phospholipase D genes; thus, it is conceivable that the strain used in the current studies, 98-37-09, harbors multiple PLD alleles. Indeed, our preliminary microarray data indicate that 98-37-09 expresses an mRNA species that corresponds to a second phospholipase D gene (ATCC 17978 locus A1S_2891) (data not shown). Thus, wild-type levels of A. baumannii invasion may be attributable to the collective activities of multiple phospholipase D alleles. Regardless, our results indicate that the PLD product of locus A1S_2989 does affect the organism's ability to invade epithelial cells. This could be considered a means by which A. baumannii avoids the host's immune response and that contributes to antibiotic tolerance.
The finding that the PLD locus A1S_2989 affects A. baumannii's ability to survive in serum and invade lung epithelial cells provided compelling reasons to assess whether the locus codes for a bona fide virulence determinant. To directly test that possibility, wild-type and isogenic A1S_2989 mutant strains were assessed for the ability to establish infection in the lungs and to subsequently colonize other organs in a murine model of pneumonia. Those experiments revealed that A1S_2989 mutant cells did not exhibit aberrant lung bacterial burden in comparison to wild-type cells, a phenotype that was inconsistent with our in vitro lung epithelial cell invasion data. This disparity could be attributable to differences in inocula or times of host cell exposure between the two model systems. As stated above, the A. baumannii genome contains at least two pld alleles. Thus, it is also conceivable that in the context of the host cell milieu the second pld gene may overcome the loss of A1S_2989 or play a more important role in lung invasion and pathogenesis. Laboratory experiments are under way to characterize the second pld gene and to determine whether it affects A. baumannii pathogenesis.
Despite not affecting the lung bacterial load, the mutant exhibited a striking decrease in bacteremia (24 h) and both heart and liver bacterial burdens at 48 h postinoculation. These results imply that A1S_2989-encoded PLD may affect the organism's ability to thrive within blood early during infection; this observation is consistent with our finding that pld mutant cells exhibited decreased proliferation in human serum. It remains to be seen whether this, in turn, results in decreased opportunity for the organism to interact with and colonize other organs during infection. Despite differences in blood levels, we did not observe a corresponding decrease in pld mutant burden in visceral organs at 24 h postinoculation, suggesting that the levels of A. baumannii bacteremia are not directly correlated with the incidence of heart or liver tissue colonization. Alternatively, wild-type- and pld mutant-infected mice may have surpassed a bacteremia threshold above which the bacterial burden is no longer correlated directly with tissue colonization, or A1S_2989 may not play a role in A. baumannii colonization of host heart or liver tissue. Nonetheless, it was found that at 48 h postinoculation there was a significant decrease in both heart- and liver-associated pld mutant bacteria, implying that PLD affects the organism's ability to persist in these tissues. Indeed, while not significant, a comparison of tissue bacterial numbers for infected mice at 24 and 48 h revealed a trend in which pld mutant cells might have been more readily cleared than wild-type A. baumannii. This may reflect the fact that phosphatidylcholine has been suggested to pay a role in the clearance of other Gram-negative bacteria (22). In that regard, wild-type levels of A. baumannii phopholipase D could catalyze cleavage of host cell phosphatidylcholine and limit bacterial clearance, whereas pld mutant bacteria may have diminished ability to inactivate phosphatidylcholine and may be more readily cleared.
Histopathological examination of infected mice supported the observation that A. baumannii pld mutant cells exhibit diminished heart bacterial burden in comparison to wild-type cells. Moreover, it was found that the organism was localized to the pericardial membrane and correlated with pericarditis, a condition corresponding to clinical symptoms of Acinetobacter infection (19, 37). We believe that our results represent the first report establishing that the organism causes heart disease in an animal model of infection, suggesting that the murine-pneumonia model used here is an appropriate system to study Acinetobacter pathogenesis and Acinetobacter-associated pericarditis. Taken together, our results suggest that A1S_2989-encoded PLD is a previously unrecognized A. baumannii virulence factor that is required for wild-type levels of pathogenesis. It is worth noting that, similar to our observations, C. pseudotuberculosis disease is manifested through PLD-mediated host dissemination (1). Interestingly, C. pseudotuberculosis disease can be neutralized by passive immunization with rabbit anti-PLD serum and/or vaccination with inactive, purified PLD; extending those observations, A. baumannii PLD may represent a new target for therapeutic intervention in Acinetobacter infections (14, 44).
Another question that is currently under investigation in our laboratories is whether the remaining genes identified in this study contribute to A. baumannii pathogenesis. Of particular interest, A1S_0269 is expected to code for a putative type II secretion system protein (protein N) and is highly conserved across sequenced A. baumannii strains (≥99% amino acid identity). It is well recognized that type II secretion systems transfer proteases, toxins, and other enzymes from within bacteria to the extracellular milieu/host cells and contribute to bacterial pathogenesis (reviewed in reference 5). This raises the possibility that A. baumannii produces a type II secretion system, which presumably involves the product of A1S_0269 and could affect the organism's ability to cause disease. Likewise, exoribonuclease R (RNase R) is a 3′→5′ exoribonuclease that posttranscriptionally regulates the expression of virulence factors in Shigella flexneri, E. coli, Aeromonas hydrophila, and Helicobacter pylori (3, 9, 39). In some experimental systems, RNase R-deficient bacteria are attenuated in animal models of infection. Thus, while not assessed in the current study, it is intriguing to consider that other proteins identified here may contribute to A. baumannii disease processes.
Acknowledgments
This work was supported by University of Nebraska Medical Center (UNMC) research funds awarded to P.M.D. A.C.J. was supported by a UNMC Graduate Studies fellowship. I.H. was supported by Public Health Service award T32 GM07347 from the National Institute of General Medical Studies for the Vanderbilt Medical-Scientist Training Program.
We thank Judith Noble-Wang (Centers for Disease Control and Prevention) for providing many of the strains used in this work and Christelle Roux for thoughtful discussions.
Editor: F. C. Fang
Footnotes
Published ahead of print on 1 March 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Batey, R. G. 1986. Pathogenesis of caseous lymphadenitis in sheep and goats. Aust. Vet. J. 63:269-272. [DOI] [PubMed] [Google Scholar]
- 2.Chen, M. Z., P. R. Hsueh, L. N. Lee, C. J. Yu, P. C. Yang, and K. T. Luh. 2001. Severe community-acquired pneumonia due to Acinetobacter baumannii. Chest 120:1072-1077. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, Z. F., Y. Zuo, Z. Li, K. E. Rudd, and M. P. Deutscher. 1998. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 273:14077-14080. [DOI] [PubMed] [Google Scholar]
- 4.Choi, C. H., J. S. Lee, Y. C. Lee, T. I. Park, and J. C. Lee. 2008. Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol. 8:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cianciotto, N. P. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13:581-588. [DOI] [PubMed] [Google Scholar]
- 6.Dijkshoorn, L., A. Nemec, and H. Seifert. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939-951. [DOI] [PubMed] [Google Scholar]
- 7.Doi, Y., S. Husain, B. A. Potoski, K. R. McCurry, and D. L. Paterson. 2009. Extensively drug-resistant Acinetobacter baumannii. Emerg. Infect. Dis. 15:980-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards, J. L., D. D. Entz, and M. A. Apicella. 2003. Gonococcal phospholipase D modulates the expression and function of complement receptor 3 in primary cervical epithelial cells. Infect. Immun. 71:6381-6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erova, T. E., V. G. Kosykh, A. A. Fadl, J. Sha, A. J. Horneman, and A. K. Chopra. 2008. Cold shock exoribonuclease R (VacB) is involved in Aeromonas hydrophila pathogenesis. J. Bacteriol. 190:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Getchell-White, S. I., L. G. Donowitz, and D. H. Groschel. 1989. The inanimate environment of an intensive care unit as a potential source of nosocomial bacteria: evidence for long survival of Acinetobacter calcoaceticus. Infect. Control Hosp. Epidemiol. 10:402-407. [DOI] [PubMed] [Google Scholar]
- 11.Gouby, A., M. J. Carles-Nurit, N. Bouziges, G. Bourg, R. Mesnard, and P. J. Bouvet. 1992. Use of pulsed-field gel electrophoresis for investigation of hospital outbreaks of Acinetobacter baumannii. J. Clin. Microbiol. 30:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirai, Y. 1991. Survival of bacteria under dry conditions; from a viewpoint of nosocomial infection. J. Hosp. Infect. 19:191-200. [DOI] [PubMed] [Google Scholar]
- 13.Hodgson, A. L., P. Bird, and I. T. Nisbet. 1990. Cloning, nucleotide sequence, and expression in Escherichia coli of the phospholipase D gene from Corynebacterium pseudotuberculosis. J. Bacteriol. 172:1256-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodgson, A. L., K. Carter, M. Tachedjian, J. Krywult, L. A. Corner, M. McColl, and A. Cameron. 1999. Efficacy of an ovine caseous lymphadenitis vaccine formulated using a genetically inactive form of the Corynebacterium pseudotuberculosis phospholipase D. Vaccine 17:802-808. [DOI] [PubMed] [Google Scholar]
- 15.Hunger, M., R. Schmucker, V. Kishan, and W. Hillen. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45-51. [DOI] [PubMed] [Google Scholar]
- 16.King, L. B., E. Swiatlo, A. Swiatlo, and L. S. McDaniel. 2009. Serum resistance and biofilm formation in clinical isolates of Acinetobacter baumannii. FEMS Immunol. Med. Microbiol. 55:414-421. [DOI] [PubMed] [Google Scholar]
- 17.Koonin, E. V. 1996. A duplicated catalytic motif in a new superfamily of phosphohydrolases and phospholipid synthases that includes poxvirus envelope proteins. Trends Biochem. Sci. 21:242-243. [PubMed] [Google Scholar]
- 18.Kwon, K. T., W. S. Oh, J. H. Song, H. H. Chang, S. I. Jung, S. W. Kim, S. Y. Ryu, S. T. Heo, D. S. Jung, J. Y. Rhee, S. Y. Shin, K. S. Ko, K. R. Peck, and N. Y. Lee. 2007. Impact of imipenem resistance on mortality in patients with Acinetobacter bacteraemia. J. Antimicrob. Chemother. 59:525-530. [DOI] [PubMed] [Google Scholar]
- 19.Lam, S. M., and T. Y. Huang. 1997. Acinetobacter pericarditis with tamponade in a patient with systemic lupus erythematosus. Lupus 6:480-483. [DOI] [PubMed] [Google Scholar]
- 20.Loehfelm, T. W., N. R. Luke, and A. A. Campagnari. 2008. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J. Bacteriol. 190:1036-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maegele, M., S. Gregor, E. Steinhausen, B. Bouillon, M. M. Heiss, W. Perbix, F. Wappler, D. Rixen, J. Geisen, B. Berger-Schreck, and R. Schwarz. 2005. The long-distance tertiary air transfer and care of tsunami victims: injury pattern and microbiological and psychological aspects. Crit. Care Med. 33:1136-1140. [DOI] [PubMed] [Google Scholar]
- 22.Mancilla-Ramirez, J., C. A. Dinarello, and J. I. Santos-Preciado. 1995. Phosphatidylcholine induces an increase in the production of interleukin-6 and improves survival of rats with neonatal sepsis caused by Klebsiella pneumoniae. Gac. Med. Mex. 131:14-22. [PubMed] [Google Scholar]
- 23.McKean, S. C., J. K. Davies, and R. J. Moore. 2007. Expression of phospholipase D, the major virulence factor of Corynebacterium pseudotuberculosis, is regulated by multiple environmental factors and plays a role in macrophage death. Microbiology 153:2203-2211. [DOI] [PubMed] [Google Scholar]
- 24.Moyed, H. S., and K. P. Bertrand. 1983. Mutations in multicopy Tn10 tet plasmids that confer resistance to inhibitory effects of inducers of tet gene expression. J. Bacteriol. 155:557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyed, H. S., T. T. Nguyen, and K. P. Bertrand. 1983. Multicopy Tn10 tet plasmids confer sensitivity to induction of tet gene expression. J. Bacteriol. 155:549-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musa, E. K., N. Desai, and M. W. Casewell. 1990. The survival of Acinetobacter calcoaceticus inoculated on fingertips and on formica. J. Hosp. Infect. 15:219-227. [DOI] [PubMed] [Google Scholar]
- 27.Oncul, O., O. Keskin, H. V. Acar, Y. Kucukardali, R. Evrenkaya, E. M. Atasoyu, C. Top., S. Nalbant, S. Ozkan, G. Emekdas, S. Cavuslu, M. H. Us, A. Pahsa, and M. Gokben. 2002. Hospital-acquired infections following the 1999 Marmara earthquake. J. Hosp. Infect. 51:47-51. [DOI] [PubMed] [Google Scholar]
- 28.Pitlick, F. A., and Y. Nemerson. 1976. Purification and characterization of tissue factor apoprotein. Methods Enzymol. 45:37-48. [DOI] [PubMed] [Google Scholar]
- 29.Ponting, C. P., and I. D. Kerr. 1996. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci. 5:914-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russo, T. A., U. MacDonald, J. M. Beanan, R. Olson, I. J. MacDonald, S. L. Sauberan, N. R. Luke, L. W. Schultz, and T. C. Umland. 2009. Penicillin-binding protein 7/8 contributes to the survival of Acinetobacter baumannii in vitro and in vivo. J. Infect. Dis. 199:513-521. [DOI] [PubMed] [Google Scholar]
- 31.Scott, P. T., K. Petersen, J. Fishbain, D. W. Craft, A. J. Ewell, K. Moran, D. C. Hack, G. A. Deye, S. Riddell, G. Christopher, J. D. Mancuso, B. P. Petruccelli, T. Endy, L. Lindler, K. Davis, E. G. Milstrey, L. Brosch, J. Pool, C. L. Blankenship, C. J. Witt, J. L. Malone, D. N. Tornberg, A. Srinivasan, and the Centers for Disease Control and Prevention. 2004. Acinetobacter baumannii infections among patients at military medical facilities treating injured U.S. service members, 2002-2004. Morb. Mortal. Wkly. Rep. 53:1063-1066. [PubMed] [Google Scholar]
- 32.Seifert, H., A. Strate, and G. Pulverer. 1995. Nosocomial bacteremia due to Acinetobacter baumannii. Clinical features, epidemiology, and predictors of mortality. Medicine (Baltimore) 74:340-349. [DOI] [PubMed] [Google Scholar]
- 33.Siegel, R. E. 2008. Emerging gram-negative antibiotic resistance: daunting challenges, declining sensitivities, and dire consequences. Respir. Care 53:471-479. [PubMed] [Google Scholar]
- 34.Soucek, A., C. Michalec, and A. Souckova. 1971. Identification and characterization of a new enzyme of the group “phospholipase D” isolated from Corynebacterium ovis. Biochim. Biophys. Acta 227:116-128. [DOI] [PubMed] [Google Scholar]
- 35.Sunenshine, R. H., M. O. Wright, L. L. Maragakis, A. D. Harris, X. Song, J. Hebden, S. E. Cosgrove, A. Anderson, J. Carnell, D. B. Jernigan, D. G. Kleinbaum, T. M. Perl, H. C. Standiford, and A. Srinivasan. 2007. Multidrug-resistant Acinetobacter infection mortality rate and length of hospitalization. Emerg. Infect. Dis. 13:97-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomaras, A. P., C. W. Dorsey, R. E. Edelmann, and L. A. Actis. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473-3484. [DOI] [PubMed] [Google Scholar]
- 37.Tomkowski, W. Z., P. Kuca, R. Gralec, J. Burakowski, P. Caban, T. Orlowski, and M. Kurzyna. 2003. Management of purulent pericarditis. Monaldi Arch. Chest Dis. 59:308-309. [PubMed] [Google Scholar]
- 38.Tong, M. J. 1972. Septic complications of war wounds. JAMA 219:1044-1047. [PubMed] [Google Scholar]
- 39.Tsao, M. Y., T. L. Lin, P. F. Hsieh, and J. T. Wang. 2009. The 3′-to-5′ exoribonuclease (encoded by HP1248) of Helicobacter pylori regulates motility and apoptosis-inducing genes. J. Bacteriol. 191:2691-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valenzuela, M. S., E. V. Ikpeazu, and K. A. Siddiqui. 1996. E. coli growth inhibition by a high copy number derivative of plasmid pBR322. Biochem. Biophys. Res. Commun. 219:876-883. [DOI] [PubMed] [Google Scholar]
- 41.Valenzuela, M. S., K. A. Siddiqui, and B. L. Sarkar. 1996. High expression of plasmid-encoded tetracycline resistance gene in E. coli causes a decrease in membrane-bound ATPase activity. Plasmid 36:19-25. [DOI] [PubMed] [Google Scholar]
- 42.Wendt, C., B. Dietze, E. Dietz, and H. Ruden. 1997. Survival of Acinetobacter baumannii on dry surfaces. J. Clin. Microbiol. 35:1394-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilderman, P. J., A. I. Vasil, Z. Johnson, and M. L. Vasil. 2001. Genetic and biochemical analyses of a eukaryotic-like phospholipase D of Pseudomonas aeruginosa suggest horizontal acquisition and a role for persistence in a chronic pulmonary infection model. Mol. Microbiol. 39:291-303. [DOI] [PubMed] [Google Scholar]
- 44.Zaki, M. M. 1976. Relation between the toxogenicity and pyogenicity of Corynebacterium ovis in experimentally infected mice. Res. Vet. Sci. 20:197-200. [PubMed] [Google Scholar]