Abstract
Brucella abortus is a facultative intracellular bacterial pathogen that causes abortion in domestic animals and undulant fever in humans. The mechanism of virulence of Brucella spp. is not yet fully understood. Therefore, it is crucial to identify new molecules that can function as virulence factors to better understand the host-pathogen interplay. Herein, we identified the gene encoding the phosphoglycerate kinase (PGK) of B. abortus strain 2308. To test the role of PGK in Brucella pathogenesis, a pgk deletion mutant was constructed. Replacement of the wild-type pgk by recombination was demonstrated by Southern and Western blot analyses. The B. abortus Δpgk mutant strain exhibited extreme attenuation in bone marrow-derived macrophages and in vivo in BALB/c, C57BL/6, 129/Sv, and interferon regulatory factor-1 knockout (IRF-1 KO) mice. Additionally, at 24 h postinfection the Δpgk mutant was not found within the same endoplasmic reticulum-derived compartment as the wild-type bacteria, but, instead, over 60% of Brucella-containing vacuoles (BCVs) retained the late endosomal/lysosomal marker LAMP1. Furthermore, the B. abortus Δpgk deletion mutant was used as a live vaccine. Challenge experiments revealed that the Δpgk mutant strain induced protective immunity in 129/Sv or IRF-1 KO mice that was superior to the protection conferred by commercial strain 19 or RB51. Finally, the results shown here demonstrated that Brucella PGK is critical for full bacterial virulence and that a Δpgk mutant may serve as a potential vaccine candidate in future studies.
Brucella spp. are responsible for a zoonosis that causes a serious economic impact worldwide, especially in developing countries, and a human disease that is difficult to treat (6). In animals, brucellosis is a major cause of abortions and infertility (25). In humans, infection can cause a serious debilitating disease manifested as undulant fever, endocarditis, arthritis, and osteomyelitis (30). Due to serious economic losses and public health risks, extensive efforts have been conducted to prevent the disease in animals through vaccination programs (28).
Brucella enters the host via the nasal, oral, and pharyngeal cavities; it proliferates within macrophages and prevents fusion of the phagosome with the lysosome by altering the intracellular traffic of the early phagosome vesicle (32, 41), and it is located in structures that resemble the endoplasmic reticulum (ER) (31). Therefore, Brucella is capable of establishing chronic infections due to its ability to avoid the killing mechanisms within macrophages and to escape the immune response, persisting in the host during its life span (10).
Despite the availability of live vaccine strains for cattle (S19 and RB51) and small ruminants (Rev-1), the vaccines have several drawbacks, including interference with diagnosis, resistance to antibiotics, and residual virulence, that prevent their use in humans (4, 5). Numerous attempts to develop safe and more effective vaccines, including the use of live vectors, DNA vaccine, or recombinant proteins, has had limited success (23, 29, 40). In the absence of defined protective immunogens, the use of attenuated vaccine strains offers the best approach.
Previous research in our laboratory has identified Brucella genes coding for metabolic enzymes (27, 36). Among them is pgk, which encodes the phosphoglycerate kinase (PGK). This enzyme catalyzes the reversible transfer of a phosphate group from 1,3-bisphosphoglycerate to ADP, resulting in phosphorylation of ATP and the formation of 3-phosphoglycerate. PGK is required both for glycolysis and gluconeogenesis (26). In the present study, a mutant of pgk was generated by gene replacement, and the effects of this mutation on intracellular bacterial survival were evaluated in vitro and in vivo.
MATERIALS AND METHODS
Mice.
A pair of interferon (IFN) regulatory factor-1 knockout (IRF-1−/− or IRF-1 KO) mice in a 129/Sv background was kindly donated by Luis F. Lima Reis from the Ludwig Institute for Cancer Research, São Paulo, Brazil, and the mice were bred and maintained in our animal facility at the Federal University of Minas Gerais (UFMG). BALB/c, C57BL/6, and 129/Sv mice were purchased from the UFMG and maintained at the Department of Biochemistry and Immunology animal care facility, and 6- to 9-week-old mice were used for experimental infection.
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Brucella abortus virulent strain S2308 and B. abortus vaccine strains RB51 and S19 were obtained from our own laboratory stock. They were grown in brucella broth medium (Becton Dickinson, Sparks, MD) for 3 days at 37°C on a rotary shaker (200 rpm), divided in aliquots, and frozen in 10% glycerol. Then, the number of CFU/ml was enumerated in one aliquot to determine the number of viable bacteria that we were inoculating in the animals. If necessary, the medium was supplemented with ampicillin or kanamycin (25 μg/ml) or chloramphenicol (20 μg/ml) and with 0.1% erythritol. Escherichia coli TOP 10F was cultured at 37°C in Luria-Bertani medium containing kanamycin (50 μg/ml) or ampicillin (100 μg/ml) as needed.
TABLE 1.
Bacterial strains and vectors used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli TOP 10F | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| B. abortus strains | ||
| S2308 | Wild type; smooth; virulent | Laboratory stock |
| S19 | Vaccine strain; smooth | Laboratory stock |
| RB51 | Rifr; rough mutant of S2308 | Laboratory stock |
| Δpgk strain | Kanr; Δpgk mutant of S2308 | This study |
| Plasmids | ||
| pUC4K | ColE1; Ampr Kanr | GE Healthcare |
| pBluescript KS | ColE1; bla | Stratagene |
| pBBR1MCS | Broad-host-range cloning vector; Cmr | 21 |
| pBBR1-pgk | 1,345-kb KpnI and BamHI fragment containing the complete B. abortus S2308 pgk gene cloned into pBBR1MCS | This study |
Isolation and DNA sequencing analysis of the B. abortus pgk gene.
The pgk gene was isolated through screening of the B. abortus S2308 genomic library, using the gap (glyceraldehyde-3-phosphate-dehydrogenase) gene fragment as a DNA probe (36). In this screening process a clone of approximately 22 kb was obtained and partially sequenced using primer walking to obtain the open reading frame (ORF) of the B. abortus pgk gene. Double-stranded DNA was sequenced by the dideoxy chain termination method (38) by using a MegaBACE 1000 system (GE Healthcare, São Paulo, Brazil). The clone was sequenced with a DYEnamic ET Dye Terminator kit (GE Healthcare); the primers used were M13 reverse sequence and M13 universal sequence from GE Healthcare, and specific primers were purchased commercially. The sequences of the specific primers used were as follows: F1, 5′-CGTGGTACGACAATGAATGG-3′; F2, 5′-CATTTTGCCGAAGACTGC-3′; F3, 5′-GGTCTTGATGTCGGCAAA-3′; F4, 5′-GACTTCACCTATATCTCA-3′; R1, 5′-ACCGGGCTTATGTGGATG-3′; R2, 5′-CTGGTGTGGCGTATCGGC-3′; and R3, 5′-CTGCACGAATTCAGGATC-3′. The sequence data were compiled and analyzed by using the sequence analysis program DNASIS, version 5.00 (Hitachi Software). Subsequent homology searches were performed by using BLAST programs.
Generation of B. abortus pgk deletion mutant.
The B. abortus pgk gene was amplified by PCR from the 22-kb genomic clone and subcloned into pBluescript II KS+ (Stratagene, La Jolla, CA). Primers containing one artificial restriction site at each end were constructed according to the pgk nucleotide sequence. The primer sequences were (forward) 5′-GTAGGATCCATGATGTTCCGCACCCTT-3′ (containing a BamHI site) and (reverse) 5′-GGGGGTACCTCACTTCTTCAATACATC-3′ (containing a KpnI site). PCR was performed with a 10-μl volume containing assay buffer (200 mM Tris-HCl, pH 8.4, 500 mM KCl, 500 mM MgCl2), a mixture of the four deoxynucleoside triphosphates at 10 mM each, a 5 pmol/μl concentration of each primer, 10 ng of template DNA, and 2.5 U of Taq DNA polymerase (Promega). Amplification was performed with a PTC-100 Programmable Thermal Controller (MJ Research) at 95°C for 3 min, followed by 30 cycles of denaturation for 30 s at 95°C, annealing for 45 s at 68°C, and extension for 1 min at 72°C. The amplified gene fragment was digested with appropriate restriction endonucleases and cloned into pBluescript II SK+ (pBlue-pgk). To generate a pgk deletion mutant by homologous recombination, the recombinant plasmid (pBlue-pgk) was used to construct the target vector, pBlue-pgk-kan. pBlue-pgk was digested with EcoRI and ligated with an EcoRI 1.2-kb DNA fragment encoding the kanamycin resistance gene from plasmid pUC4K (GE Healthcare). Five micrograms of pBlue-pgk-kan plasmid DNA was added to 50 μl of B. abortus S2308 competent cells in sterile electroporation cuvettes with 0.2-cm electrode gaps (Bio-Rad Laboratories, Richmond, CA), and then electroporation was performed with a Gene Pulser II transfection apparatus (Bio-Rad Laboratories) at 25 μF, 2.5 kV, and 400 Ω. Then, colonies that were Kanr Amps and Kanr Ampr were selected as colonies in which double- or single-crossover events had occurred, respectively.
Characterization of the B. abortus pgk deletion mutant by Southern blotting.
To provide genetic evidence that the wild-type pgk gene was replaced by a pgk gene interrupted by the Kanr cassette, 10 μg of genomic DNA isolated from both the mutant strain and the wild-type strain (S2308) was digested with EcoRV and then loaded onto a 0.8% agarose gel for Southern blotting, performed as previously described (35).
Western blot analysis.
For analysis of pgk expression in B. abortus S2308, the Δpgk mutant strain and Δpgk strain complemented with pBBR1-pgk were grown in 10 ml of brucella broth (BD) overnight at 37°C with agitation (200 rpm). Then, 1 ml of each culture was pelleted and resuspended in SDS sample buffer at an optical density at 600 nm (OD600) of 1. The samples were boiled for 5 min, and 10 μl was loaded on a 12% SDS-PAGE gel. After the gel was run, the samples were transferred onto a nitrocellulose membrane (Hybond-ECL; GE Healthcare) for 1 h at 350 mA using a dry transfer system (Bio-Rad); the membrane was blocked overnight with 10% dry milk in TBST buffer (100 mM Tris-HCl, 150 mM NaCl, 0.05% Tween 20, pH 7.2). After the membranes were blocked, they were washed tree times with TBST buffer and probed with anti-PGK antibodies or naive mice serum diluted 1:100 in TBST buffer for 4 h at room temperature. The polyclonal anti-PGK antibodies were produced in our lab by immunizing mice with recombinant PGK (rPGK) protein. After reacting with the primary antibody, the blots were washed six times with TBST buffer and incubated for 1 h at room temperature with anti-mouse IgG conjugated to alkaline phosphatase (Promega) and diluted 1:2,000 in TBST buffer. After three washes with TBST buffer, the reaction was developed after the mixture was incubated at room temperature with NBT (nitroblue tetrazolium chloride) and BCIP (5-bromo-4-chloro-3-indolyl-1-phosphate).
Infection of bone-marrow derived macrophages (BMDM) with the Brucella mutant Δpgk.
Macrophages were derived from C57BL/6 mouse bone marrow as follows. Each femur and tibia was flushed with 5 ml of Hank's balanced salt solution (HBSS). The resulting cell suspension was centrifuged, and the cells were resuspended in Dulbecco's modified Eagle's medium (DMEM; Gibco) containing 10% fetal bovine Serum (FBS; Gibco) and 10% L929 cell-conditioned medium (LCCM) as a source of macrophage colony-stimulating factor (M-CSF) (24). The cells were distributed in 24-well plates and incubated at 37°C in a 5% CO2 atmosphere. Three days after seeding, another 0.1 ml of LCCM was added. On the seventh day, the medium was renewed. On the 10th day of culture, when cells were completely differentiated into macrophages, they were infected with B. abortus S2308 or Δpgk corresponding to a multiplicity of infection (MOI) of 50:1. The plates were then centrifuged at 600 × g for 10 min at 4°C in order to synchronize the infections. Phagocytosis was allowed to proceed for 30 min at 37°C. At this point, the culture medium was removed, and the monolayer was washed three times with phosphate-buffered saline (PBS). Cultures were incubated for 90 min at 37°C with medium containing 50 μg/ml gentamicin (Sigma) to kill extracellular bacteria. At each time point studied, the infected cells were washed three times with PBS and lysed in 1 ml of 0.1% Triton X-100 in double-distilled H2O (ddH2O). The number of viable intracellular Brucella bacteria recovered from lysed macrophages was determined at 2, 24, 48, 72, 120, and 168 h following infection. Tenfold serial dilutions of bacterial suspensions in PBS were plated in duplicate on brucella agar with or without kanamycin (50 μg/ml). The number of CFU was determined after 3 days of incubation at 37°C with 5% CO2. The experiments were performed in triplicate and repeated twice.
Confocal microscopy.
Infected C57BL/6 mouse BMDM grown on 12-mm glass coverslips in 24-well plates were fixed in 3% paraformaldehyde, pH 7.4, at 37°C for 15 min. Cells were labeled by inverting coverslips onto drops of primary antibodies diluted in 10% horse serum and 0.1% saponin in PBS and incubating the samples for 30 min at room temperature. The primary antibodies used for immunofluorescence microscopy were the following: cow anti-B. abortus polyclonal antibody, rat anti-mouse LAMP1 ID4B (Developmental Studies Hybridoma Bank, National Institute of Child Health and Human Development, University of Iowa), and rabbit anticalnexin polyclonal antibody (Stressgene Bioreagent Corp., British Columbia, Canada). Bound antibodies were detected by incubation with a 1:1,000 dilution of Alexa Fluor 488 goat anti-rat, a 1:100 dilution of Texas Red goat anti-cow, or a 1:500 dilution of donkey anti-rabbit Cyanin 3 antibody (Jackson ImmunoResearch Laboratories, Suffolk, United Kingdom) for 30 min at room temperature. Cells were washed twice with 0.1% saponin in PBS, once in PBS, and once in H2O and then mounted in Mowiol 4-88 mounting medium (Calbiochem, Darmstadt, Germany). Samples were examined on a Zeiss LSM 510 laser scanning confocal microscope for image acquisition. Images of 1,024 by 1,024 pixels were then assembled using Adobe Photoshop, version 7.0. Quantification was always done by counting intracellular bacteria in at least 50 cells in three independent experiments, as previously reported (37).
Persistence of Brucella Δpgk mutant in BALB/c, C57BL/6, and 129/Sv mice.
To assess the persistence of the Brucella Δpgk mutant in different mouse strains, eight animals from each group were examined at each sampling period. Female C57BL/6, 129/Sv, and BALB/c mice 6 to 8 weeks old were injected intraperitoneally (i.p.) with 1 × 106 CFU of either the B. abortus S2308 or Δpgk mutant strain in 0.1 ml of PBS. At 1, 2, 3, 4, and 6 weeks postinoculation, all mice in each group were killed, and bacterial counts were determined. Tenfold serial dilutions of the homogenized spleens were plated on brucella agar containing kanamycin to determine the number of Brucella Δpgk CFU per spleen compared to the number of the wild-type S2308 strain. Brucella colonies were counted after 3 days of incubation at 37°C with 5% CO2. Data are presented as log10 values of CFU per spleen. The experiment was repeated twice.
Virulence of Brucella Δpgk mutant in IRF-1 KO mice.
Four groups of eight IRF-1 KO mice were injected i.p. with 1 × 106 CFU of either B. abortus S2308, the Δpgk mutant, or vaccine strain S19 or RB51 in 0.1 ml. Mouse survival was observed during 30 days postinfection as previously demonstrated (18). The experiment was repeated twice.
Immunization of mice with the B. abortus Δpgk mutant.
Female BALB/c, 129/Sv, C57BL/6, and IRF-1 KO mice that were 6 to 9 weeks old were immunized i.p. with brucellae in 0.1 ml of PBS. Groups containing eight mice each were immunized with either B. abortus S19 or Δpgk at 1 × 105 CFU or with B. abortus RB51 at 1 × 107 CFU. Nonimmunized, control mice were injected i.p. with 0.1 ml of PBS. Twelve weeks after immunization, all mice in each group were challenged by i.p. injection of 1 × 106 CFU of B. abortus S2308. Experimentally infected BALB/c, 129/Sv, and C57BL/6 mice were killed 2 weeks later by cervical dislocation, and the spleens were collected and disrupted in 10 ml of PBS. A tenfold serial dilution was plated on brucella agar containing kanamycin or 0.1% erythritol for differentiation of B. abortus Δpgk, S19, and S2308. Additionally, we used the crystal violet method to differentiate between the RB51 and S2308 strains. After 3 days of incubation at 37°C, colonies were visualized, and the number of CFU of B. abortus S2308 per spleen was determined after the number of B. abortus S19 or B. abortus Δpgk CFU found by replica plating was subtracted. The degrees of protection in immunized animals and controls were expressed as the mean number of CFU of B. abortus S2308 for each mouse group obtained after challenge and log10 conversion. Log10 units of protection were obtained by subtracting the mean log10 CFU for the experimental group from the mean log10 CFU for the control group, as previously described (35). For IRF-1 KO mice, survival was observed during 30 days postinfection. The experiment was repeated twice.
Cytokine detection.
Six weeks after immunization, IRF-1 KO mice were sacrificed, and their spleens were removed under aseptic conditions. Splenocytes from naive or infected mice were obtained by mechanically disrupting the spleen and collecting the resulting single-cell suspension. Cells were suspended in RPMI 1640 medium (Gibco Laboratories, Grand Island, NY) supplemented with 2 mM l-glutamine, 25 mM HEPES, 10% heat-inactivated fetal bovine serum (Sigma), penicillin G sodium (100 U/ml), and streptomycin sulfate (100 μg/ml) (supplemented RPMI medium). Erythrocytes were eliminated with ACK lysis solution (150 mM NH4Cl, 1 mM Na2-EDTA [pH 7.3]). Splenocytes were cultured in 96-well microtiter plates with 1 × 106 cells/well in a volume of 0.2 ml to assess cytokine production. The culture was stimulated by the addition of 102 heat-killed bacteria (heat inactivation was performed at 80°C for 2 h) per cell or 2 μg/ml of concanavalin A. These cells were incubated at 37°C in a humidified chamber with 5% CO2. Aliquots of the supernatant were collected after 72 h of culture for IFN-γ measurements. Levels of IFN-γ in the supernatants were measured by a commercially available Duoset ELISA Development System kit (R&D Systems, Minneapolis, MN).
Statistical analysis.
Statistical analysis was performed with a Student's t test using the computer software package MINITAB (Minitab Inc., State College, PA).
Nucleotide sequence accession number.
The nucleotide sequence of the pgk gene of B. abortus was deposited in GenBank under accession number AF256214.
RESULTS
Characterization of the B. abortus Δpgk deletion mutant.
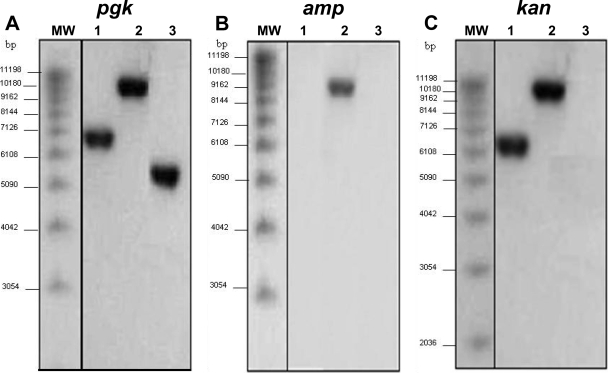
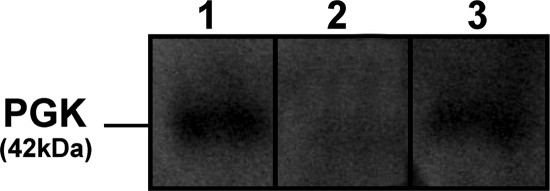
A defined Kanr Amps Δpgk deletion mutant of B. abortus S2308 was constructed by chromosomal gene replacement. Chromosomal DNA was isolated from these clones and from the parental strain for Southern blot analysis. The same hybridization profile was observed for all transformants selected from each different phenotype group, as shown in Fig. 1. DNA hybridization of EcoRV-digested chromosomal DNA by using the pgk probe produced one fragment at approximately 5.8 kb for wild-type B. abortus S2308 (Fig. 1A, lane 3) and a 7-kb band for the Kanr Amps Δpgk mutant (lane 1). A single recombination was confirmed when the Ampr probe hybridized to only one fragment corresponding to integration of the deletion plasmid (pBlue-pgk-kan) in the chromosome (Fig. 1B). When the kanamycin cassette was used as a probe, it hybridized to genomic DNA from the Kanr Amps or Kanr Ampr clone but not to genomic DNA of wild-type B. abortus S2308, which was used as a negative control (Fig. 1C). To determine whether pgk expression was taking place in Δpgk and if we would be able to complement the mutant strain with pBBR1-pgk plasmid, Western blot analysis was carried out using anti-PGK polyclonal antibodies. The wild-type and complemented Δpgk strains showed the presence of PGK protein of approximately 42 kDa, whereas the mutant strain lacked pgk expression (Fig. 2).
FIG. 1.
Characterization of the B. abortus Δpgk mutant by Southern blot analysis. EcoRV-digested genomic DNA was probed with the pgk (A), amp (B), or kan (C) DNA fragments. Lanes 1, B. abortus Δpgk mutant; lanes 2, clones in which a single recombination event took place; lanes 3, B. abortus S2308; lanes MW, molecular weight markers.
FIG. 2.
Western blot analysis of pgk expression in different B. abortus strains. B. abortus S2308 (lane 1), Δpgk mutant (lane 2), or Δpgk mutant complemented with pBBR1-pgk plasmid (lane 3) were subjected to SDS-PAGE, and Western blot analysis using polyclonal anti-PGK antibodies demonstrated the presence of a band of approximately 42 kDa in lanes 1 and 3, which indicated pgk expression.
B. abortus Δpgk mutant is attenuated in macrophages.
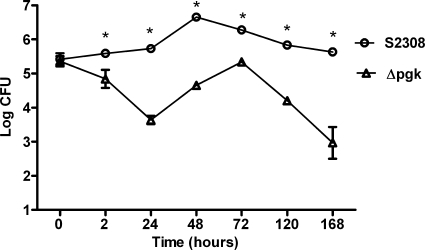
To investigate the role of the pgk gene in intracellular B. abortus survival, we evaluated the multiplication of B. abortus S2308 wild type and the B. abortus Δpgk mutant in bone-marrow derived macrophages. The number of viable bacteria was counted at 2, 24, 48, 120, and 168 h postinfection. To inhibit extracellular bacterial growth, gentamicin was added to the medium 30 min after infection at a concentration of 50 μg/ml. Shortly after infection (time zero) there was no difference (P > 0.05) between the wild type and the mutant in the number of bacteria infecting the macrophages (Fig. 3). In contrast, by 24 h postinfection there was a 2-log difference (P < 0.05) in the number of organisms surviving inside the macrophages. The Brucella Δpgk mutant displayed a lower rate of intracellular replication in macrophages than the wild-type strain S2308 at all times studied. These results demonstrate that the Δpgk mutant has a limited ability to replicate within macrophages.
FIG. 3.
Intracellular replication of B. abortus S2308 and Δpgk in BMDM. Adherent cells were infected at an MOI of 50 with B. abortus S2308 or the Δpgk mutant as described in Materials and Methods. At 2, 24, 48, 72, 120, and 168 h postinfection, macrophages were lysed and enumerated by serial dilutions plated in duplicate. The data points, presented as the log10 CFU per well, are the mean with standard error of the mean (SEM) of two independent experiments performed in triplicate. Statistically significant differences (P ≤ 0.05) between the Δpgk mutant value and that of the parental strain S2308 are indicated by an asterisk. This result is representative of two independent experiments.
Brucella Δpgk mutant does not recruit ER markers in BMDM.
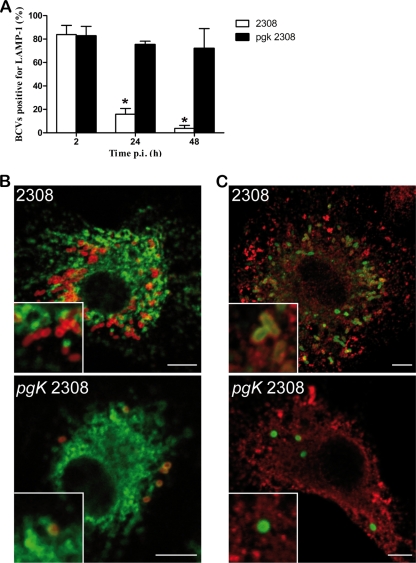
It has been well demonstrated that wild-type B. abortus establishes a replicative niche in macrophages, acquiring endoplasmic reticulum markers (9). At 24 h postinfection, wild-type bacteria is found in calnexin (endoplasmic reticulum marker)-positive and LAMP1 (late endosomal/lysosomal marker)-negative compartments. In contrast, immunofluorescence analysis of infected BMDM showed that, unlike wild-type bacteria, at 24 h postinfection the Δpgk mutant was not found in the replication vacuole containing ER markers (Fig. 4C), and instead over 60% of Brucella-containing vacuoles (BCVs) retained the late endosomal/lysosomal marker LAMP1 (Fig. 4A and B). Furthermore, at 48 h following infection, around 80% of BCVs containing Δpgk mutant bacteria retained LAMP1. These results are consistent with the attenuation of the Δpgk mutant compared to wild-type strain 2308.
FIG. 4.
Multiplication and intracellular localization of B. abortus Δpgk mutant and wild-type strain in BMDM. (A) Quantification of the percentage of wild-type or Δpgk mutant BCVs that contain LAMP1 by confocal immunofluorescence microscopy. The difference between the wild type and mutant was statistically significant at 24 and 48 h (P < 0.001) postinfection (p.i.). Data are means from three different experiments. (B) Representative confocal images of BMDM at 24 h postinfection with wild-type B. abortus or the Δpgk mutant. Brucella lipopolysaccharide (LPS) is labeled in red, and LAMP1 is in green. (C) Confocal images of BMDM at 24 h postinfection with wild-type B. abortus or the Δpgk mutant. Brucella LPS is labeled in green, and calnexin is shown in red. Scale bar, 5 μm.
B. abortus Δpgk is highly attenuated in BALB/c, 129/Sv, C57BL/6, and IRF-1 KO mice.
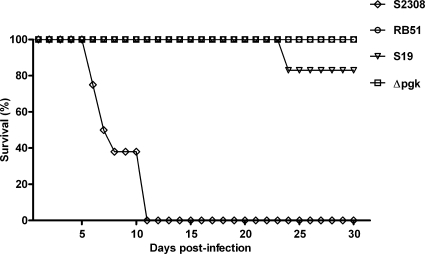
The ability of B. abortus to persist within BALB/c mice has been shown to correlate with virulence in the natural host (11). Groups of BALB/c, C57BL/6, and 129/Sv mice with different backgrounds were inoculated i.p. with the Δpgk mutant or B. abortus parental strain S2308 to determine differences in persistence. The number of Brucella CFU was evaluated at 1, 2, 3, 4, and 6 weeks postinfection in the spleen of each animal. The B. abortus Δpgk mutant strain displayed reduced virulence at all times tested in different mouse models compared to the virulence of the wild-type Brucella strain (Fig. 5). Additionally, IRF-1 KO mice were infected with B. abortus S2308, S19, RB51, or the Δpgk mutant strain. IRF-1 KO mice can detect different levels of Brucella virulence, providing a useful tool to screen B. abortus mutants regarding their level of intracellular survival (18). IRF-1 KO mice infected with S19, RB51, or Δpgk survived longer than mice infected with wild-type S2308 (P ≤ 0.005). All IRF-1 KO mice infected with strain S2308 died within 11 days postinfection. Eighty percent of IRF-1 KO mice injected with B. abortus S19 were alive at 30 days postinfection. Additionally, at 30 days postinfection all mice infected with attenuated B. abortus strain RB51 or the Δpgk mutant strain were alive, demonstrating that both strains are less virulent than S19 in this mouse model (Fig. 6) .
FIG. 5.
Persistence of B. abortus S2308 or Δpgk in BALB/c, C57BL/6, or 129/Sv mice. Eight mice were infected i.p. with a dose of 106 CFU of bacteria. Spleens were harvested at different times, and the number of CFU in disrupted tissue was determined by 10-fold serial dilution and plating. The values are means and standard deviations. The asterisks indicate statistically significant differences between the results obtained for the group that received the B. abortus Δpgk mutant compared to the animals injected with the B. abortus parental strain S2308 (P ≤ 0.05).
FIG. 6.
Virulence of B. abortus S2308, S19, RB51, or the Δpgk mutant strain in IRF-1 KO mice. Groups of 10 mice received intraperitoneally 106 CFU of each strain and were monitored daily for survival during 30 days.
Immunoprotection conferred by vaccination with the B. abortus Δpgk mutant strain in BALB/c, C57BL/6, 129/Sv, and IRF-1 KO mice.
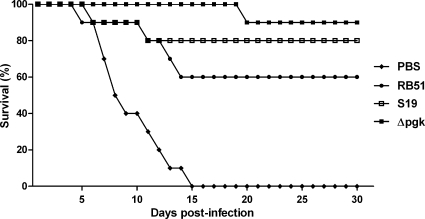
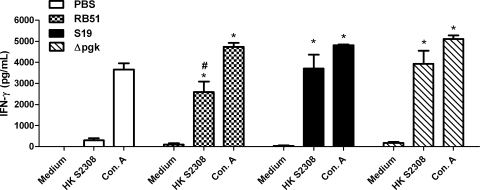
To determine if the B. abortus Δpgk mutant strain is able to induce protective immunity against infection, BALB/c, 129/Sv, C57BL/6, and IRF-1 KO mice immunized with the Δpgk mutant or with the S19 or RB51 vaccine strain were challenged with the B. abortus virulent S2308 strain. The numbers of bacterial CFU in the spleens were determined 12 weeks after challenge since Araya et al. (2) showed that nonspecific resistance to infection with unrelated bacteria is very low 6 weeks after immunization with Brucella. BALB/c, C57BL/6, and 129/Sv mice immunized with the B. abortus Δpgk mutant had significantly fewer splenic brucellae than nonimmunized animals (Table 2). Additionally, we observed similar log units of protection in BALB/c and C57BL/6 mice immunized with the Δpgk mutant strain (0.96 and 1.36 log units, respectively) compared to mice immunized with the commercial vaccine strain S19 (0.94 and 1.73 log units, respectively) and higher log units of protection in mice immunized with Δpgk than with the commercial vaccine strain RB51 (0.58 and 0.76 log units, respectively) following challenge. In 129/Sv mice, immunization with the B. abortus Δpgk mutant strain induced protection (3.28 log units) superior to that in animals immunized with the commercial vaccine strain S19 (2.46 log units) and higher log units of protection than in mice immunized with commercial vaccine strain RB51 (1.97 log units) following challenge. IRF-1 KO mice were immunized with the Brucella Δpgk mutant or the S19 or RB51 vaccine strain for 12 weeks and challenged with 1 × 106 CFU of virulent S2308. All IRF-1 KO mice immunized with any Brucella vaccine strain survived longer than nonimmunized IRF-1 KO mice, suggesting that immunological memory was activated in these animals and that it could provide protection. However, 90% of IRF-1 KO mice immunized with Δpgk were alive at 30 days postchallenge while 70% or 80% of animals vaccinated with RB51 or S19, respectively, survived during this period (Fig. 7). Therefore, the B. abortus Δpgk mutant significantly enhanced resistance to experimental infection compared to the S19 or RB51 commercially available vaccine strain. In order to determine if IFN-γ responses were altered in IRF-1 KO mice immunized with different vaccine strains, cytokine production in splenocyte culture was determined. Figure 8 demonstrates that the Δpgk mutant and S19 induced similar levels of IFN-γ in vaccinated IRF-1 KO mice. However, animals immunized with strain RB51 showed reduced production of IFN-γ compared to mice vaccinated with the Δpgk mutant and S19. This difference might be one of the reasons for the increased susceptibility to brucellosis in IRF-1 KO mice immunized with RB51 and challenged with virulent S2308.
TABLE 2.
Protective immunity induced by Δpgk mutant immunization
| Mouse strain and inoculum | Mean (SD) log10 CFU in mouse spleen | Log10 U of protectiona |
|---|---|---|
| BALB/c | ||
| PBS | 7.07 (0.12) | |
| B. abortus RB51 | 6.49 (0.42) | 0.58* |
| B. abortus S19 | 6.13 (0.59) | 0.94* |
| B. abortus Δpgk strain | 6.11 (0.41) | 0.96* |
| C57BL/6 | ||
| PBS | 6.66 (0.14) | |
| B. abortus RB51 | 5.90 (0.41) | 0.76* |
| B. abortus S19 | 4.93 (0.15) | 1.73*# |
| B. abortus Δpgk strain | 5.30 (0.15) | 1.36*# |
| 129/Sv | ||
| PBS | 7.38 (0.20) | |
| B. abortus RB51 | 5.42 (0.19) | 1.97* |
| B. abortus S19 | 4.93 (0.42) | 2.46*# |
| B. abortus Δpgk strain | 4.10 (0.11) | 3.28*#† |
Significance is indicated as follows: *, P ≤ 0.05 compared to PBS control group; #, P ≤ 0.05 compared to the RB51-vaccinated group; †, P ≤ 0.05 compared to the S19-vaccinated group.
FIG. 7.
Protection of IRF-1 KO mice vaccinated with B. abortus S19, RB51, or the Δpgk mutant strain. Animals received intraperitoneally 105 CFU of B. abortus S19, 105 CFU of Δpgk, or 107 CFU of RB51, and 12 weeks after immunization all mice were challenged by i.p. injection of 106 CFU of B. abortus S2308. Groups of 10 mice were monitored daily for survival for 30 days. Vaccination of mice with the Δpgk mutant induced protection superior to that of immunization with vaccine strain S19 or RB51.
FIG. 8.
IFN-γ production by spleen cells of IRF-1 KO mice vaccinated with S19, RB51, or the Δpgk mutant strain. IRF-1 KO mice were inoculated with 107 CFU of RB51, 105 CFU of S19, or 105 CFU of Δpgk. Six weeks after vaccination, splenocytes were recovered and stimulated with heat-inactivated B. abortus (HK 2308) or concanavalin A as a positive control. Splenocyte culture supernatants were harvested after 72 h of culture. Bars represent the mean ± standard deviation of quadruplicate sets of cells. Statistically significant differences are indicated by an asterisk (P ≤ 0.05) for the comparison to medium alone and by a number sign (#) for the comparison to the RB51-vaccinated group stimulated with HK 2308.
DISCUSSION
Intracellular bacteria require special features, such as adhesion, invasion, and replication, for interacting with the host and causing infection (15). Brucella behaves as a stealthy organism, disturbing the cell functions as little as possible (20). It is well established that upon entry the bacteria-lipid raft interaction in host cells allows Brucella to escape from the degradative endocytic pathway and further favors intracellular replication (22). Therefore, it is crucial to identify new molecules that can function as virulence factors to better understand this host-pathogen interplay.
The development of vaccines to control brucellosis has proven to be a challenge for years. The observation that the highest levels of protection are obtained when the host is immunized with live vaccines indicates that persistence and vaccine viability are key aspects required for an efficient vaccine (17, 43). Previous studies by other investigators and from our laboratory have identified genes related to survival and bacterial virulence (3, 7, 16, 27, 33, 35, 42). Among them, we have identified, sequenced, and disrupted the pgk gene of B. abortus. Furthermore, the Brucella pgk mutant has been evaluated for survival in macrophages and in different mouse strains to confirm attenuation and potential use as a vaccine candidate.
To address the role of Brucella PGK in bacterial virulence, a mutant was constructed. To confirm that inactivation of the Brucella pgk gene was achieved, Southern and Western blot analyses were performed. As shown in Fig. 2, Western blot analysis using polyclonal anti-PGK antibodies demonstrated the lack of pgk gene expression in the Δpgk mutant and showed that PGK production was restored when this mutant strain was complemented with the pBBR1-pgk plasmid. Further, the mutant was evaluated for survival and attenuation in bone marrow-derived macrophages. As shown in Fig. 3, Δpgk was defective for survival in macrophages compared to the wild-type strain. Additionally, to determine the fate of the Δpgk mutant intracellularly, confocal microscopy was performed in BMDM. As demonstrated in Fig. 4, at 24 h postinfection, wild-type bacteria was found in calnexin-positive and LAMP1-negative compartments. However, immunofluorescence analysis of infected BMDM showed that, unlike S2308, at 24 h postinfection the Δpgk mutant was not found within the ER-derived compartment and that, instead, over 60% of BCVs retained the late endosomal/lysosomal marker LAMP1.
In BALB/c, C57BL/6, and 129/Sv mice, the Δpgk mutant demonstrated different levels of spleen colonization from those of the wild-type S2308, indicating that virulence in vivo was altered by the absence of PGK (Fig. 5). The number of Brucella CFU was evaluated at 1, 2, 3, 4, and 6 weeks postinfection in the spleen of each animal. The B. abortus Δpgk mutant strain displayed reduced persistence at all times tested in different mouse models compared to the virulence of the wild-type Brucella strain. Additionally, IRF-1 KO mice were infected with B. abortus S2308, S19, RB51, or the Δpgk mutant strain. IRF-1 KO is an interesting animal model to analyze the contribution of an individual Brucella gene to bacterial virulence (18). IRF-1 KO mice infected with S19, RB51, or Δpgk survived longer than mice infected with wild-type S2308 (P ≤ 0.005). Eighty percent of IRF-1 KO mice injected with B. abortus S19 were alive at 30 days postinfection, and all mice injected with attenuated B. abortus strain RB51 or Δpgk mutant strain were alive.
Pathogenicity of brucellae and chronicity of brucellosis are due to the ability of the pathogen to adapt to the harsh environmental conditions found in host cells. The analysis of the intramacrophagic virulome of Brucella suis showed that low levels of nutrients and oxygen are major features of its replicative niche (19). A recent proteomic study has shown that Brucella adapts to low-oxygen conditions intracellularly by upregulating glycolysis and denitrification (1). Most of the upregulated proteins were involved in energy metabolism, such as transketolase, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and glycerol-3-phosphate ABC transporter. In the past, it was demonstrated that Brucella uses the pentose phosphate pathway via glycolysis for sugar degradation (34) and that a functional ribose kinase is essential for intramacrophagic growth (19). In parallel with these studies, we show here that PGK enzyme, involved in the downstream pathway of glycolysis, is critical for intramacrophagic growth and bacterial virulence in vivo. Recently, Fugier et al. (14) have shown that inhibition of host cell GAPDH and enolase, both involved in glycolysis, resulted in reduced intracellular replication of Brucella. It is possible that Brucella is also using the host cell glycolysis to its advantage, as a source of energy.
Although certain subunit vaccines have been demonstrated to be efficacious and useful (8, 12), one of the most promising of the vaccine approaches, based on published results and effectiveness in the field, is use of a live, attenuated agent (13). Vaccines currently in use are derived from spontaneously occurring attenuated forms that arise randomly and that therefore do not provide a means to control the combination of defects that attenuate survival (13). Therefore, to investigate whether the Δpgk mutant strain induces protection, the protective efficacy of this potential vaccine was assessed in BALB/c, 129/Sv, C57BL/6, and IRF-1 KO mice. We observed similar levels of protection in BALB/c and C57BL/6 mice immunized with the Δpgk mutant strain and in mice immunized with commercial vaccine strain S19 and higher log units of protection than with the commercial vaccine strain RB51. In 129/Sv mice, immunization with the B. abortus Δpgk mutant strain induced protection that was superior that achieved in animals immunized with commercial vaccine strains S19 and RB51. IRF-1 KO mice are defective in multiple immune components; however, they mount an adaptive immune response sufficient to protect against virulent challenge, and the protection is vaccine strain dependent. IRF-1 KO mice immunized with Δpgk were alive at 30 days postchallenge while 70% or 80% of animals vaccinated with RB51 or S19, respectively, survived during this period. Additionally, the number of Δpgk CFU in these mice at 6 weeks postimmunization decreased rapidly compared to the values for the S19 strain, showing the tendency of low residual vaccine load (data not shown). However, the Δpgk mutant still is more protective than the S19 strain. In conclusion, the B. abortus Δpgk mutant significantly enhanced resistance to experimental infection compared to the S19 or RB51 commercially available vaccine strain.
IFN-γ is a critical cytokine in Brucella immunity and is required for macrophage bactericidal activity (39). Therefore, we decided to investigate the role of IFN-γ induced by the Δpgk mutant, RB51, or S19 vaccine strains in protection achieved in IRF-1 KO mice. As shown in Fig. 8, RB51-vaccinated IRF-1 KO mice produced less IFN-γ than S19- and Δpgk mutant-vaccinated mice, which paralleled the reduced protection induced by RB51 in this mouse model. Even though the IRF-1 KO mice have a dysregulation of interleukin-12 (IL-12) p40 induction, critical for Th1 cell development, these animals mounted a strong IFN-γ response when immunized with S19 or the Δpgk mutant strain. Additionally, the IRF-1 KO animals serve as an important model to rapidly assess vaccine efficacy of Brucella strains. The level of bacterial attenuation requires fine-tuning to avoid undesired effects on survival that may attenuate the organism so that the level of protective immunity provided is insufficient. Finally, the results reported here demonstrate that the Δpgk mutant possesses reduced persistence in macrophages and mice and that it induces protection superior to that of S19 and RB51; therefore, it should be considered a new potential vaccine candidate against brucellosis.
Acknowledgments
This study was supported by grants from CNPq, MAPA/CNPq, FAPEMIG, PNPD/CAPES, and INCT/Vacinas.
Editor: A. Camilli
Footnotes
Published ahead of print on 1 March 2010.
REFERENCES
- 1.Al Dahouk, S., S. Loisel-Meyer, H. C. Scholz, H. Tomaso, M. Kersten, A. Harder, H. Neubauer, S. Köhler, and V. Jubier-Maurin. 2009. Proteomic analysis of Brucella suis under oxygen deficiency reveals flexibility in adaptive expression of various pathways. Proteomics 9:3011-3021. [DOI] [PubMed] [Google Scholar]
- 2.Araya, L. N., P. H. Elzer, G. E. Rowe, F. M. Enright, and A. J. Winter. 1989. Temporal development of protective cell-mediated and humoral immunity in BALB/c mice infected with Brucella abortus. J. Immunol. 143:3330-3337. [PubMed] [Google Scholar]
- 3.Arenas-Gamboa, A. M., T. A. Ficht, M. M. Kahl-McDonagh, G. Gomez, and A. C. Rice-Ficht. 2009. The Brucella abortus S19 ΔvjbR live vaccine candidate is safer than S19 and confers protection against wild-type challenge in BALB/c mice when delivered in a sustained-release vehicle. Infect. Immun. 77:877-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashford, D. A., J. di Pietra, J. Lingappa, C. Woods, H. Noll, B. Neville, R. Weyant, S. L. Bragg, R. A. Spiegel, J. Tappero, and B. A. Perkins. 2004. Adverse events in humans associated with accidental exposure to the livestock brucellosis vaccine RB51. Vaccine 22:3435-3439. [DOI] [PubMed] [Google Scholar]
- 5.Berkelman, R. L. 2003. Human illness associated with use of veterinary vaccines. Clin. Infect. Dis. 37:407-414. [DOI] [PubMed] [Google Scholar]
- 6.Boschiroli, M. L., V. Foulongne, and D. O'Callaghan. 2001. Brucellosis: a worldwide zoonosis. Curr. Opin. Microbiol. 4:58-64. [DOI] [PubMed] [Google Scholar]
- 7.Burkhardt, S., M. P. Jiménez de Bagüés, J. P. Liautard, and S. Köhler. 2005. Analysis of the behavior of eryC mutants of Brucella suis attenuated in macrophages. Infect. Immun. 73:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassataro, J., K. A. Pasquevich, S. M. Estein, D. A. Laplagne, C. A. Velikovsky, S. de La Barrera, R. Bowden, C. A. Fossati, G. H. Giambartolomei, and F. A. Goldbaum. 2007. A recombinant subunit vaccine based on the insertion of 27 amino acids from Omp31 to the N terminus of BLS induced a similar degree of protection against B. ovis than Rev. 1 vaccination. Vaccine 25:4437-4446. [DOI] [PubMed] [Google Scholar]
- 9.Celli, J., C. de Chastellier, D. M. Franchini, J. Pizarro-Cerda, E. Moreno, and J. P. Gorvel. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbel, M. J. 1997. Brucellosis: an overview. Emerg. Infect. Dis. 3:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elzer, P. H., R. H. Jacobson, S. M. Jones, K. H. Nielsen, J. T. Douglas, and A. J. Winter. 1994. Antibody-mediated protection against Brucella abortus in BALB/c mice at successive periods after infection: variation between virulent strain 2308 and attenuated vaccine strain 19. Immunology 82:651-658. [PMC free article] [PubMed] [Google Scholar]
- 12.Estein, S. M., M. A. Fiorentino, F. A. Paolicchi, M. Clausse, J. Manazza, J. Cassataro, G. H. Giambartolomei, L. M. Coria, V. Zylberman, C. A. Fossati, R. Kjeken, and F. A. Goldbaum. 2009. The polymeric antigen BLSOmp31 confers protection against Brucella ovis infection in rams. Vaccine 27:6704-6711. [DOI] [PubMed] [Google Scholar]
- 13.Ficht, T. A., M. M. Kahl-McDonagh, A. M. Arenas-Gamboa, and A. C. Rice-Ficht. 2009. Brucellosis: the case for live, attenuated vaccines. Vaccine 27(Suppl. 4):D40-D43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fugier, E., S. P. Salcedo, C. de Chastellier, M. Pophillat, A. Muller, V. Arce-Gorvel, P. Fourquet, and J. P. Gorvel. 2009. The glyceraldehyde-3-phosphate dehydrogenase and the small GTPase Rab 2 are crucial for Brucella replication. PLoS Pathog. 5:e1000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzmán-Verri, C., E. Chaves-Olarte, C. Von Eichel-Streiber, I. Lópex-Goni, M. Thelestam, S. Arvidson, J. P. Gorvel, and E. Moreno. 2001. GTPases of the Rho subfamily are required for Brucella abortus internalization in nonprofessional phagocytes: direct activation of CDC42. J. Biol. Chem. 276:44435-44443. [DOI] [PubMed] [Google Scholar]
- 16.Izadjoo, M. J., A. K. Bhattacharjee, C. M. Paranavitana, T. L. Hadfield, and D. L. Hoover. 2004. Oral vaccination with Brucella melitensis WR201 protects mice against intranasal challenge with virulent Brucella melitensis 16M. Infect. Immun. 72:4031-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahl-McDonagh, M. M., and T. A. Ficht. 2006. Evaluation of protection afforded by Brucella abortus and Brucella melitensis unmarked deletion mutants exhibiting different rates of clearance in BALB/c mice. Infect. Immun. 74:4048-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko, J., A. Gendron-Fitzpatrick, T. A. Ficht, and G. A. Splitter. 2002. Virulence criteria for Brucella abortus strains as determined by interferon regulatory factor 1-deficient mice. Infect. Immun. 70:7004-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Köhler, S., V. Foulongne, S. Ouahrani-Bettache, G. Bourg, J. Teyssier, M. Ramuz, and J. P. Liautard. 2002. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. U. S. A. 99:15711-15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohler, S., S. Michaux-Charachon, F. Porte, M. Ramuz, and J. P. Liautard. 2003. What is the nature of the replicative niche of a stealthy bug named Brucella? Trends Microbiol. 11:215-229. [DOI] [PubMed] [Google Scholar]
- 21.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800-802. [PubMed] [Google Scholar]
- 22.Lapaque, N., I. Moriyon, E. Moreno, and J. P. Gorvel. 2005. Brucella lipopolysaccharide acts as a virulence factor. Curr. Opin. Microbiol. 8:60-66. [DOI] [PubMed] [Google Scholar]
- 23.Leclerq, S., J. S. Harms, G. M. Rosinha, V. Azevedo, and S. C. Oliveira. 2002. Induction of a Th1-type of immune response but not protective immunity by intramuscular DNA immunization with Brucella abortus GroEL heat-shock gene. J. Med. Microbiol. 51:20-26. [DOI] [PubMed] [Google Scholar]
- 24.Macedo, G. C., D. M. Magnani, N. B. Carvalho, O. Bruna-Romero, R. T. Gazzinelli, and S. C. Oliveira. 2008. Central role of MyD88-dependent dendritic cell maturation and proinflammatory cytokine production to control Brucella abortus infection. J. Immunol. 180:1080-1087. [DOI] [PubMed] [Google Scholar]
- 25.Mahajan, N. K., R. C. Kulshrestha, and B. Vasudevan. 1986. Brucellosis—cause of abortion in sheep and its public health significance. Int. J. Zoonoses 13:174-179. [PubMed] [Google Scholar]
- 26.Meijer, W. G. 1994. The Calvin cycle enzyme phosphoglycerate kinase of Xanthobacter favus required for autotrophic CO2 fixation is not encoded by the cbb operon. J. Bacteriol. 176:6120-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyoshi, A., G. M. Rosinha, I. L. Camargo, C. M. Trant, F. C. Cardoso, V. Azevedo, and S. C. Oliveira. 2007. The role of the vacB gene in the pathogenesis of Brucella abortus. Microbes Infect. 9:375-381. [DOI] [PubMed] [Google Scholar]
- 28.Nicoletti, P. 1990. Vaccination against Brucella. Adv. Biotechnol. Processes 13:147-168. [PubMed] [Google Scholar]
- 29.Oliveira, S. C., and G. A. Splitter. 1996. Immunization of mice with recombinant L7/L12 ribosomal protein confers protection against Brucella abortus infection. Vaccine 14:959-962. [DOI] [PubMed] [Google Scholar]
- 30.Pappas, G., N. Akritidis, M. Bosilkovski, and E. Tsianos. 2005. Brucellosis. N. Engl. J. Med. 352:2325-2336. [DOI] [PubMed] [Google Scholar]
- 31.Pizarro-Cerda, J., E. Moreno, V. Sanguedolce, J. L. Mege, and J. P. Gorvel. 1998. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect. Immun. 66:2387-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pizarro-Cerda, J., S. Meresse, R. G. Parton, G. van der Goot, A. Sola-Landa, I. Lopez-Goni, E. Moreno, and J. P. Gorvel. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 66:5711-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajashekara, G., D. A. Glover, M. Banai, D. O'Callaghan, and G. A. Splitter. 2006. Attenuated bioluminescent Brucella melitensis mutants GR019 (virB4), GR024 (galE), and GR026 (BMEI1090-BMEI1091) confer protection in mice. Infect. Immun. 74:2925-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson, D. C., and W. G. McCullough. 1968. The glucose catabolism of the genus Brucella. I. Evaluation of pathways. Arch. Biochem. Biophys. 127:263-273. [DOI] [PubMed] [Google Scholar]
- 35.Rosinha, G. M., D. A. Freitas, A. Miyoshi, V. Azevedo, E. Campos, S. L. Cravero, O. Rossetti, G. Splitter, and S. C. Oliveira. 2002. Identification and characterization of a Brucella abortus ATP-binding cassette transporter homolog to Rhizobium meliloti ExsA and its role in virulence and protection in mice. Infect. Immun. 70:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosinha, G. M., A. Myioshi, V. Azevedo, G. A. Splitter, and S. C. Oliveira. 2002. Molecular and immunological characterisation of recombinant Brucella abortus glyceraldehyde-3-phosphate-dehydrogenase, a T- and B-cell reactive protein that induces partial protection when co-administered with an interleukin-12-expressing plasmid in a DNA vaccine formulation. J. Med. Microbiol. 51:661-671. [DOI] [PubMed] [Google Scholar]
- 37.Salcedo, S. P., M. I. Marchesini, H. Lelouard, E. Fugier, G. Jolly, S. Balor, A. Muller, N. Lapaque, O. Demaria, L. Alexopoulou, D. J. Comerci, R. A. Ugalde, P. Pierre, and J. P. Gorvel. 2008. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog. 4:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sathiyaseelan, J., R. Goenka, M. Parent, R. M. Benson, E. A. Murphy, D. M. Fernandes, A. S. Foulkes, and C. L. Baldwin. 2006. Treatment of Brucella-susceptible mice with IL-12 increases primary and secondary immunity. Cell Immunol. 243:1-9. [DOI] [PubMed] [Google Scholar]
- 40.Toth, T. E., J. A. Cobb, S. M. Boyle, R. M. Roop, and G. G. Schurig. 1995. Selective humoral immune response of BALB/c mice to Brucella abortus proteins expressed by vaccinia virus recombinants. Vet. Microbiol. 45:171-183. [DOI] [PubMed] [Google Scholar]
- 41.Ugalde, R. A. 1999. Intracellular lifestyle of Brucella spp. Common genes with other animal pathogens, plant pathogens, and endosymbionts. Microbes Infect. 1:1211-1219. [DOI] [PubMed] [Google Scholar]
- 42.Yang, X., T. Becker, N. Walters, and D. W. Pascual. 2006. Deletion of znuA virulence factor attenuates Brucella abortus and confers protection against wild-type challenge. Infect. Immun. 74:3874-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhan, Y., A. Kelso, and C. Cheers. 1995. Differential activation of Brucella-reactive CD4+ T cells by Brucella infection or immunization with antigenic extracts. Infect. Immun. 63:969-975. [DOI] [PMC free article] [PubMed] [Google Scholar]