Abstract
Bacterial small, noncoding RNAs (sRNAs) participate in the posttranscriptional regulation of gene expression, often by affecting protein translation, transcript stability, and/or protein activity. For proper function, many sRNAs rely on the chaperone Hfq, which mediates the interaction of the sRNA with its target mRNA. Recent studies have demonstrated that Hfq contributes to the pathogenesis of a number of bacterial species, suggesting that sRNAs play an essential role in the regulation of virulence. The enteric pathogen Yersinia pseudotuberculosis causes the disease yersiniosis. Here we show that Hfq is required by Y. pseudotuberculosis to cause mortality in an intragastric mouse model of infection, and a strain lacking Hfq is attenuated 1,000-fold compared to the wild type. Hfq is also required for virulence through the intraperitoneal route of infection and for persistence of the bacterium in the Peyer's patches, mesenteric lymph nodes, and spleen, suggesting a role for Hfq in systemic infection. Furthermore, the Δhfq mutant of Y. pseudotuberculosis is hypermotile and displays increased production of a biosurfactant-like substance, reduced intracellular survival in macrophages, and decreased production of type III secretion effector proteins. Together, these data demonstrate that Hfq plays a critical role in the virulence of Y. pseudotuberculosis by participating in the regulation of multiple steps in the pathogenic process and further highlight the unique role of Hfq in the virulence of individual pathogens.
Small, noncoding RNAs (sRNAs) are integral components of posttranscriptionally based regulation of protein synthesis in prokaryotes and have been implicated in the control of quorum sensing, stress response, virulence factor production, and the regulation of outer membrane proteins (1, 7, 8, 21). Unlike microRNAs in eukaryotes, sRNAs are often encoded in intergenic regions, transcribed directly from their own promoters, and unprocessed and contain Rho-independent terminators (34). sRNAs directly bind to their target mRNAs, and these interactions can result in the up- or downregulation of protein synthesis (27). For example, an sRNA molecule can bind to a target mRNA and block the ribosome binding site or enhance RNase E-based degradation of transcripts to inhibit translation, such as is seen with MicA-based negative regulation of ompA in Escherichia coli (47). Conversely, an sRNA can bind in such a way as to relieve a hairpin structure in the 5′ untranslated region of an mRNA. This exposes the ribosome binding site to enhance translation, as has been demonstrated in the regulation of rpoS by the sRNA DsrA (27, 43).
The Hfq protein was first identified as a host bacterial factor required for the synthesis of bacteriophage Qβ RNA (13). It is now known that Hfq is a small (102 amino acids in E. coli, 101 amino acids in Yersinia spp.), conserved RNA chaperone protein present in many bacterial species that binds to and regulates the stability of bacterial mRNA transcripts (22, 46, 50). Furthermore, Hfq also binds to many sRNAs and enhances the RNA-RNA interaction between these sRNAs and their mRNA targets (35, 48, 54).
Recent studies have highlighted the contributions of Hfq and sRNAs to bacterial pathogenesis. It has been shown that Hfq is critical to the virulence of a number of pathogens, including Francisella tularensis, Listeria monocytogenes, Neisseria meningitidis, Salmonella enterica, and uropathogenic E. coli (9, 12, 26, 32, 40, 42). Given the pleiotropic nature of Hfq, it is not surprising that defects have been observed in growth under oxidative stress and high salt and in the presence of antimicrobial peptides; defects in quorum sensing, host cell invasion, and other virulence determinants have also been observed (12, 28, 42). Interestingly, the effects of Hfq seem to be unique in each bacterial species. For example, the growth and survival inside host cells of S. enterica and Brucella abortus, but not L. monocytogenes and F. tularensis, are reduced in the absence of Hfq (9, 32, 40, 42). In addition, regulation of species-specific factors is often dependent on Hfq, such as the heat-stable enterotoxin Yst of Yersinia enterocolitica and the SPI-1 regulator HilD of Salmonella (36, 39). One common feature of Hfq among most bacterial pathogens examined thus far is a reduction of virulence in the relevant animal model in the absence of Hfq, which supports its critical role in pathogenesis (9, 12, 26, 32, 40, 42).
The aim of the current work is to understand the contribution of Hfq to the pathogenesis of Yersinia pseudotuberculosis, which is a Gram-negative bacterium that causes yersiniosis, a generally mild gastrointestinal disease in humans. Y. pseudotuberculosis is very closely related to Yersinia pestis, the causative agent of plague (6, 53). Yersiniosis caused by Y. pseudotuberculosis is characterized by ileitis, mesenteric lymphadenitis, fever, and diarrhea (31, 37, 41), and the presence of previous medical conditions can increase the severity of the disease (49). Y. pseudotuberculosis is transmitted through the fecal-oral route, and it has been shown in mice that colonization of the Peyer's patches occurs shortly after the bacteria enter the intestinal lumen (30). From here dissemination to the blood, spleen, liver, and other organs can occur (3).
The potential for Hfq to interact with multiple mRNA targets suggests that this protein may play a role in a number of processes important to the virulence of Y. pseudotuberculosis. Recent work has demonstrated that Hfq contributes to the pathogenesis of Y. pestis in the mouse models of bubonic and septicemic plague. In the absence of Hfq, Y. pestis shows a reduced ability to colonize the spleen and liver and displays defects in both intracellular survival and growth in vitro under a number of stress-inducing conditions (14). Here we demonstrate that Y. pseudotuberculosis is also attenuated for virulence in the absence of Hfq and that Hfq participates in the regulation of motility, intracellular survival, and production of type III effectors in this bacterium.
MATERIALS AND METHODS
Reagents, bacterial strains, and growth conditions.
All reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless stated otherwise. Bacterial strains and plasmids used in this study are listed in Table 1. Oligonucleotide sequences are listed in Table 2. Y. pseudotuberculosis strain IP 32953 (designated PAN29) (6), its derivatives, and all other Yersinia strains were routinely grown at 26°C in liquid brain heart infusion (BHI) broth (Difco) or on BHI agar unless otherwise noted. E. coli strains were grown at 37°C in Luria-Bertani (LB) broth or on LB agar. When necessary, these media were supplemented with kanamycin (50 μg/ml), ampicillin (100 μg/ml), or Irgasan (2 μg/ml). Mutants were evaluated for the presence of key virulence loci (yopT, yopH, hmsR, and psn) by PCR.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Strain designation or plasmid marker(s) | Genotype and/or characteristics | Source or reference |
|---|---|---|---|
| Y. pseudotuberculosis strains | |||
| IP 32953 | PAN29 | Wild type | 6 |
| IP 32953 Δhfq | PAN38 | Δhfq | This study |
| IP 32953 ΔflgE | PAN154 | ΔflgE | This study |
| IP 32953 Δhfq ΔflgE | PAN155 | Δhfq ΔflgE | This study |
| IP 32953 pYV− | PAN100 | pYV−, lacks T3SS | This study |
| IP 32953 Δhfq pYV− | PAN101 | Δhfq pYV−, lacks T3SS | This study |
| IP 32953 Δhfq+phfq | PAN136 | Δhfq hfq complemented on multicopy plasmid | This study |
| IP 32953 Δhfq | PAN39 | Δhfq | This study |
| 32777 | PAN36 | Wild type | K. Satchell |
| 32777 Δhfq | PAN40 | Δhfq | This study |
| 32777 Δhfq+phfq | PAN137 | Δhfq hfq complemented on multicopy plasmid | This study |
| IP 32953 Δhfq+phfq | PAN181 | Δhfq hfq complemented on multicopy plasmid | This study |
| Plasmids | |||
| pSR47S | Kan | Homologous recombination vector, sacB counterselection | 33 |
| pWL302 | Kan | 500 bp up- and downstream of hfq coding region cloned into pSR47S | This study |
| pCS20 | Kan | 500 bp up- and downstream of flgE coding region cloned into pSR47S | This study |
| pET24a(+) | Kan | C-terminal His tag coding sequence, T7 promoter | Novagen |
| pWL226 | Kan | pET24a(+) with Yersinia hfq C-terminal His tag for overexpression | This study |
| pCR-Blunt II-TOPO | Kan, Amp | pUC-ori LacZα plac TOPO cloning site | Invitrogen |
| phfq | Kan | hfq gene and 1-kb upstream sequence inserted into pCR-Blunt II-TOPO | This study |
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide name | Oligonucleotide sequence |
|---|---|
| hfq 5′-493 Bam | CGGGATCCCGGGTGAAACCTTACCTTACCG |
| hfq 3′-1 Spe | GGACTAGTTCTATATTTTCCTTATTTGCTTG |
| hfq 5′+1 Spe | GGACTAGTAGCCCATTGCTGGTCGACCATG |
| hfq 3′+500 Not | ATAAGAATGCGGCCGCGGTCGCGATATGACGCAATTG |
| flgE 5′-501 Bam | GGATCCCATGGCAAAGCTGCTCAAGAGC |
| flgE 3′-37 Spe | ACTAGTGATATTGA CCGTGCGGGCTAG |
| flgE 5′+1 Spe | ACTAGTTGGATAAGCTTCTGTATACCGCC |
| flgE 3′+498 Not | GCGGCCGCGCGGGGATATCCACCAGTTTG |
| hfq 5′-1018 Bam | CGGGATCCTTATTATG GGGCCAACTGCTTC |
| hfq 3′306 Eco | GGAATTCTTATTCAGCGTCATCACTGTC CTGC |
| hfq 5′1 Nde | GGAATTCCATATGGCTAAGGGGCAATCTTTGC |
| hfq 3′303 Xho | CCGCTCGAGTTCAGCGTCATCACTGTCCTG |
| flgA 5′252 | GCGTTATGAGATAAGTTGTCCCGATGGTC |
| flgA 3′365 | TTTCTCCCGCGCTCAAGGGTCTG |
| flhC 5′253 | AAAGTGGCTATTGCACGGGTGTTG |
| flhC 3′112 | CCACTGTCAACGAAACGGACCAG |
| fliC 5′925 | TCTGCGGTCACCAACCTGAATAAC |
| fliC 3′1071 | AGCTTGAGACAACACAGAAGTCCC |
| yopE 5′510 | ACGCCTGTTTGTGGTATTCCCTTCTC |
| yopE 3′37 | AGCCCTTTGATCTCATTTGCTGCC |
| yopH 5′324 | GACACTACAAGACGCCAAAGTGCTG |
| yopH 3′423 | TGCGTGAAGGGCTGAATGTGAATG |
| yopJ 5′544 | AAACTTTACATCGAGAGAGATAGCCTG |
| yopJ 3′661 | TTACCGGGAGATACGGGTCCAAC |
| yopT 5′96 | CGCACACCGAGTGAAAGTGGAAAC |
| yopT 3′242 | AAGCTGCTGCGTTGGTTAGCTTTG |
| hflX 5′1100 | ACTTTGAATTGCGCTTGCCTCCTC |
| hflX 3′1215 | TCTAACCACCATACCGACATTCCC |
| gyrB.f.5′ | TCGCCGTGAAGGTAAAGTTC |
| gyrB.r.3′ | ATTGGTAAAGGTCTGGAAACTTGGCC |
Construction of mutant strains.
A Y. pseudotuberculosis Δhfq strain was generated by homologous recombination. A 500-bp region upstream of the coding sequence for hfq was PCR amplified from Y. pseudotuberculosis IP 32953 using primers hfq 5′-493 Bam and hfq 3′-1 Spe, and a 500-bp region downstream of the coding sequence was PCR amplified using primers hfq 5′+1 Spe and hfq 3′+500 Not. These fragments were cloned into pSR47S, which carries a kanamycin resistance cassette and sacB (33). Clones were confirmed by DNA sequencing. The resulting plasmid pWL302 was introduced into Y. pseudotuberculosis IP 32953 by mating. Transconjugants were plated on BHI plus kanamycin plus Irgasan. Two kanamycin-sensitive Δhfq mutants were subsequently selected for by passage on BHI plus 5% sucrose agar and confirmed by PCR. These mutant strains are designated PAN38 and PAN39, respectively. The gene for Hfq was deleted from strain Y. pseudotuberculosis 32777 as above, with wild-type and Δhfq strains designated PAN36 and PAN40, respectively.
A deletion of the flgE gene was created in Y. pseudotuberculosis and the Y. pseudotuberculosis Δhfq strain by using the same homologous recombination technique as above. The up- and downstream sequences were PCR amplified from Y. pseudotuberculosis with primers flgE 5′-501 Bam and flgE 3′-37 Spe and primers flgE 5′+1 Spe and flgE 3′+498 Not, respectively. These strains are designated PAN154 and PAN155, respectively.
The pYV plasmid, which carries the genes for the type III secretion system (T3SS) and effectors, was cured from Y. pseudotuberculosis IP 32953 wild type and the Δhfq mutant by growth at 37°C on BHI plates containing magnesium chloride and sodium oxalate (MOX), as previously described (19). The loss of pYV was verified by PCR and by the loss of growth restriction at 37°C in the absence of calcium (15). These mutant strains are designated PAN100 and PAN101, respectively.
Construction of complementing plasmid phfq.
The coding region and 1,018 bp upstream of the transcriptional start site for hfq were PCR amplified from Y. pseudotuberculosis using primers hfq 5′-1018 Bam and hfq 3′306 Eco. This product was inserted into the plasmid vector pCR-Blunt II-TOPO (Invitrogen), and the sequence was verified. The plasmid, called phfq, was transformed into both Y. pseudotuberculosis IP 32953 Δhfq strains and the 32777 Δhfq strain by electroporation, and the strains were designated PAN136, PAN181, and PAN137, respectively.
Antibody production and immunoblot analysis.
The coding sequence for Hfq was PCR amplified from Yersinia using primers hfq 5′1 Nde and hfq 3′303 Xho and cloned into plasmid pET24a(+), which contains a C-terminal hexahistidine tag. The His-tagged Hfq protein was expressed in E. coli BL21(DE3) and purified under native conditions according to the Qiagen Expressionist protocol, and polyclonal anti-Hfq antibodies were raised in rabbits (Covance).
For immunoblot analysis, Y. pseudotuberculosis wild-type, Δhfq, and Δhfq+phfq strains (PAN29, PAN38, and PAN136, respectively) were grown overnight in BHI at 26°C; equivalent units of optical density at 620 nm (OD620) were taken from each culture, resuspended in sample buffer (10% glycerol, 100 mM Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate [SDS], 0.02 mg/ml bromophenol blue, 5% β-mercaptoethanol), and separated by SDS-PAGE. Proteins were transferred to nitrocellulose membranes for analysis of Hfq expression with the anti-Hfq antibody by immunoblotting.
Growth curves.
Y. pseudotuberculosis wild-type, Δhfq, and Δhfq+phfq strains (PAN29, PAN38, and PAN136, respectively) were cultured overnight in BHI at 26°C and then subcultured at an OD620 of 0.1 in 10 ml of BHI, BHI plus 2.5 mM CaCl2, M9, or M9 plus 2.5 mM CaCl2. Cultures were incubated with shaking at 250 rpm in 125-ml Erlenmeyer flasks at 26°C or 37°C for 12 h. Optical density was measured at 620 nm.
Animal infections.
All procedures involving animals were carried out in compliance with protocols approved by the Northwestern University institutional animal care and use committee. Eight-week-old female BALB/c mice were purchased from Harlan Laboratories and allowed to acclimate to the animal facility for 5 to 7 days prior to infection. To prepare the inocula, Y. pseudotuberculosis wild-type, Δhfq, and pYV− strains (PAN29, PAN38, and PAN100, respectively) were cultured overnight in BHI at 26°C, diluted to an OD620 of 0.1, and incubated at 26°C with shaking to an OD620 of 0.6. The cells were harvested by centrifugation, washed once with sterile phosphate-buffered saline (PBS), and diluted to the appropriate OD620 in PBS. Groups of 10 mice were inoculated intragastrically using a 22-gauge feeding needle with approximately 107 CFU of the Y. pseudotuberculosis wild-type, Δhfq, or pYV− strain and monitored for 21 days. The weights of individual mice were recorded every third day. In experiments examining the kinetics of infection, groups of 5 mice were infected as above and sacrificed at various times postinoculation. CFU per gram of tissue were determined in the spleen, visible Peyer's patches, tissue inclusive of mesenteric lymph nodes, and the small intestine by homogenizing organs in PBS and plating them on Yersinia selective agar (Difco). Mice with CFU counts in any organ or with any recorded weight loss were included in the analysis. For the dose-response survival curve, mice were anesthetized with a mixture of ketamine (100 mg/kg of body weight) and xylazine (10 mg/kg) in PBS given intraperitoneally immediately prior to intragastric inoculation with either 10-fold-increasing doses of the Y. pseudotuberculosis Δhfq strain (103 to 107 CFU) or 103 CFU of the wild-type strain and monitored for 21 days. For intraperitoneal infections, mice were injected using a 28-gauge needle with approximately 103 CFU of the Y. pseudotuberculosis wild-type, Δhfq, or pYV− strain and monitored for 21 days.
Gentamicin protection assays.
J774 murine macrophage-like cells were routinely cultured in Dulbecco's modified Eagle's medium (Cellgro; Mediatech) plus 10% heat-inactivated fetal calf serum (HyClone) and penicillin-streptomycin (100 μg/ml) (Cellgro; Mediatech) at 37°C in a 5% CO2 environment. Cells (5 × 105) were seeded into 24-well plates 16 to 18 h prior to use. Adherent cells were washed with PBS and incubated for 1 h with standard media lacking penicillin-streptomycin (binding buffer). Y. pseudotuberculosis wild-type, Δhfq, and Δhfq+phfq strains were cultured overnight in BHI at 26°C, washed with PBS, diluted to the appropriate CFU/ml in binding buffer (multiplicity of infection [MOI] of 10), and added to the J774 cells. The bacteria were incubated with host cells for 1 h. Cells were then washed five times in PBS and incubated for 30 min with binding buffer or binding buffer supplemented with 100 μg/ml gentamicin (Invitrogen). After 30 min cells were washed five times with PBS and lysed with 0.1% Triton X-100 in water, and serial dilutions were plated onto BHI plates to determine the number of host cell-associated bacterial cells. The inocula were also plated to calculate percent association relative to inocula. Alternatively, after 30 min of incubation with 100 μg/ml gentamicin, cells were washed once with PBS and incubated with binding buffer supplemented with 10 μg/ml gentamicin for 2 and 4 h. Cells were then washed and lysed as above. The experiments were performed in triplicate and with at least 3 independent biological replicates.
Hydrogen peroxide killing assay.
Y. pseudotuberculosis wild-type, Δhfq, and Δhfq+phfq strains (PAN29, PAN38, and PAN136, respectively) were grown overnight in BHI at 26°C and subcultured to an OD620 of 0.1. Bacteria were grown to mid-log phase and then diluted 1:10 in BHI. Hydrogen peroxide diluted in water or water alone was added to the bacteria to a concentration of 100 mM. All samples were incubated at 26°C in a roller drum, and aliquots were taken at 10 and 30 min posttreatment. Serial dilutions were plated onto BHI plates to determine the CFU/ml in the treated versus untreated samples. Experiments were performed in triplicate and with three independent biologic replicates.
Motility and biosurfactant assays.
Colonies of Y. pseudotuberculosis wild-type, Δhfq, Δhfq+phfq, ΔflgE, and Δhfq ΔflgE strains (PAN29, PAN38, PAN136, PAN154, and PAN155, respectively) were cultured overnight in BHI at 26°C. Aliquots (2 μl) of each were spotted onto soft agar motility plates (1.0% tryptone, 0.5% NaCl, 0.3% agar) and incubated at 22°C or 37°C, and at various times photographs were taken using the Gel Doc XR System (Bio-Rad). Aliquots (2 μl) of water were spotted inside and outside the border of the refractive compound to assess surface tension. Images of the released compound were taken using a Cannon 60D camera with a 100 mM F macrolens 2 days after the bacteria were spotted onto plates.
qRT-PCR.
Overnight cultures of Y. pseudotuberculosis wild-type and Δhfq strains (PAN29 and PAN38, respectively) were grown in triplicate and diluted to an OD620 of 0.1 in 22 ml BHI plus MOX. For the flhC, flgA, fliC, and hflX genes, cultures were grown at 26°C for 6 h. For the yopE, yopH, yopJ, and yopT genes, cultures were grown at 26°C for 1 h and then shifted to 37°C for 3 h. Five OD units were removed and added to RNAprotect bacterial reagent (Qiagen). RNA was isolated using the RiboPure bacterial kit (Ambion) and treated with DNase, and cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen). Quantitative reverse transcriptase PCR (qRT-PCR) was performed with the SYBR green dye in an iCycler thermocycler (Bio-Rad). The calculated threshold cycle (CT) was normalized to the CT of the gyrB gene from the same cDNA sample before calculation of the fold changes using the ΔΔCT method (2).
Type III secretion assays.
Y. pseudotuberculosis wild-type, Δhfq, and pYV− strains (PAN29, PAN38, and PAN100, respectively) were diluted to an OD620 of 0.1 in BHI plus MOX and cultured at 26°C for 1 h followed by 37°C for 3 h. Bacteria were centrifuged, and equivalent OD units of culture supernatants were harvested, filtered, and precipitated by the addition of trichloroacetic acid to 10%. Precipitated proteins were resuspended in equal volumes of 1 M Tris (pH 9.0) and sample buffer, separated by SDS-PAGE, and transferred to nitrocellulose or stained with Coomassie brilliant blue. For normalization purposes, 0.2 OD units of bacterial cultures was harvested and separated by SDS-PAGE. Bacterial cell pellets were washed three times with PBS, resuspended in PBS plus lysozyme (0.5 mg/ml), and incubated on ice for 30 min. Cells were then sonicated, and cellular debris was removed by centrifugation. Whole-cell lysates (50 μg) were separated by SDS-PAGE and transferred to nitrocellulose. Immunoblot analyses were performed using antibodies to YopE, YopH, YopJ, YopT, and RpoA (as a loading control).
Statistical analysis.
All experiments were performed two or more times, unless otherwise noted. Student's unpaired t test was used to compare wild-type, Δhfq, and Δhfq+phfq strains in growth curves, in the gentamicin protection assay, and in the hydrogen peroxide killing assay and to compare mouse weights during intragastric infection. For qRT-PCR experiments, significance was calculated by the Wilcoxon signed-rank test. For kinetics experiments, significant differences in CFU/organ were determined by the Mann-Whitney U test. In all cases, a P value of less than 0.05 was considered significant.
RESULTS
Deletion of hfq from Y. pseudotuberculosis and growth in vitro.
In order to examine the role of Hfq in the virulence of Y. pseudotuberculosis, we generated an unmarked isogenic mutant lacking the entire Hfq coding sequence. The loss of Hfq was verified by immunoblot analysis of overnight cultures of Y. pseudotuberculosis (data not shown). In addition, we confirmed the absence of polar effects on hflX, the gene immediately downstream of hfq, by qRT-PCR (data not shown). We then generated a plasmid-based complementing clone of hfq. Production of Hfq protein was detected in the complemented mutant, and Hfq appears to be noticeably overproduced in this strain compared to wild type, which is likely due to the multicopy nature of the plasmid (data not shown).
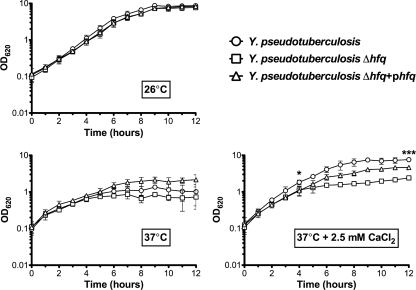
The absence of Hfq in some bacterial species has been associated with growth defects in various types of liquid media (12, 42, 50). In order to determine the effects of Hfq on the growth of Y. pseudotuberculosis, the wild-type, Δhfq, and Δhfq+phfq strains (PAN29, PAN38, and PAN136, respectively) were cultured in BHI (rich) broth at 26°C, 37°C, and 37°C plus 2.5 mM CaCl2. At 26°C there is no effect of Hfq on growth (Fig. 1), and overnight cultures of wild-type and Δhfq strains routinely reach the same density (data not shown). At 37°C, the loss of Hfq results in a slight, though not statistically significant, growth defect (Fig. 1). At 37°C in the presence of calcium, however, Δhfq bacteria exhibit a statistically significant growth defect during both exponential phase (P = 0.0419) and stationary phase (P = 0.0003) (Fig. 1). In particular, at 37°C with calcium Y. pseudotuberculosis has a slower doubling time in the absence of Hfq (1.35 h versus 3.85 h), and the Δhfq culture never reaches the optical density of the wild type. This defect is partly ameliorated in the complemented strain in stationary phase (P = 0.0061) (Fig. 1). The effects of Hfq on the growth of Y. pseudotuberculosis in a nutrient-limiting environment were also examined. Wild-type, Δhfq, and Δhfq+phfq bacteria were cultured in M9 medium as above (data not shown). As expected, all three strains reached a lower stationary-phase optical density than in the rich media; however, the observed trends remained the same for growth in M9 as in BHI. No significant growth defect was seen in the absence of Hfq at 26°C or at 37°C in the absence of CaCl2 (data not shown). A significant difference was seen during exponential phase when the Y. pseudotuberculosis Δhfq strain was cultured at 37°C with 2.5 mM CaCl2 (P = 0.0237) (data not shown).
FIG. 1.
Growth of Y. pseudotuberculosis Δhfq strain in rich media. Y. pseudotuberculosis wild-type, Δhfq, and Δhfq+phfq strains were cultured in BHI broth at 26°C, 37°C, and 37°C plus 2.5 mM CaCl2, and the OD620 of each culture was measured over the course of the growth curve. Each graph represents the mean of three independent biological replicates grown on three different days. The error bars represent the standard deviation of the optical density at each time point. Significance was calculated by Student's unpaired t test at 4 and 12 h (*, P = 0.0419; ***, P = 0.0003).
The Y. pseudotuberculosis Δhfq strain is attenuated for virulence in an intragastric mouse model of infection.
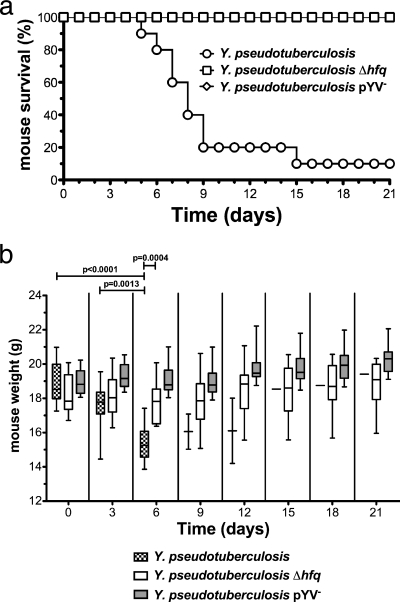
To determine if Hfq is required for the pathogenesis of Y. pseudotuberculosis in a model that mimics a natural route of infection, 9-week-old female BALB/c mice were infected intragastrically with 106 or 107 CFU of wild-type, Δhfq, or pYV− bacteria (PAN29, PAN38, and PAN100, respectively) and monitored for 21 days. When given 106 CFU, 100% of mice infected with the Δhfq strain survived, but only 30% survived when infected with the wild-type Y. pseudotuberculosis (data not shown). When inoculated with 107 CFU, 90% of mice infected with the wild-type strain died by day 15, while all mice infected with the Δhfq strain survived for the duration of the experiment (Fig. 2a). As expected, all mice infected with the pYV-cured strain survived, since this strain lacks the T3SS and effectors that are essential for Yersinia virulence. We also determined the weight of each mouse every 3 days, as the loss of body weight is indicative of a symptomatic infection. Mice infected with wild-type Y. pseudotuberculosis suffered significant weight loss prior to succumbing to the infection (Fig. 2b). Mice infected with the Δhfq strain initially displayed mild, but not statistically significant, weight loss but eventually recovered, while those infected with the pYV− strain never lost weight (Fig. 2b).
FIG. 2.
Survival of mice inoculated intragastrically with the Y. pseudotuberculosis Δhfq strain. Groups of 10 mice were inoculated via oral gavage with the Y. pseudotuberculosis wild-type, Δhfq, or pYV− strain (107 CFU) and monitored for 21 days. (a) Survival of infected mice over 21 days. (b) Body weight of infected mice over 21 days. The plot shows median weight, indicated by a solid line; a box represents the 25th and 75th percentiles, and whiskers represent the range. Significance was calculated by Student's unpaired t test.
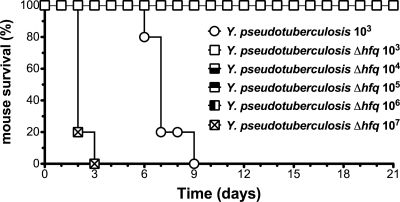
In order to determine if there is a dose at which Y. pseudotuberculosis deleted for hfq affects the survival of mice, we adapted a model in which the infectious dose can be lowered from 107 CFU to 103 CFU using a ketamine-xylazine cocktail to anesthetize the mice prior to intragastric inoculation. This model is useful because doses of bacteria can be administered many times higher than the 100% lethal dose (LD100) for wild-type Y. pseudotuberculosis without risking septic shock that would likely occur with the unanesthetized infection model (i.e., CFU approaching 1011) (24, 51). Ketamine-xylazine has been shown to induce a passing immunosuppressive state in rat gut mucosal homogenates by reducing inducible nitric oxide synthase (iNOS) and tumor necrosis factor alpha (TNF-α) production that lasts for 1 to 3 h after treatment (17, 45), which we hypothesize allows the bacteria to cause an infection at a lower dose. This hypothesis is supported by the fact that ketamine treatment allows for an intragastric Vibrio cholerae infection of adult mice, which is not possible in the absence of the anesthetic (38). Importantly, mice treated with ketamine-xylazine and infected with 103 CFU of avirulent, pYV− Y. pseudotuberculosis (lacking the T3SS plasmid) do not succumb to the infection and do not display any weight loss in this model (data not shown). Mice were injected intraperitoneally with a ketamine-xylazine cocktail immediately prior to infection with 103 CFU of wild-type Y. pseudotuberculosis or 10-fold-increasing doses of the Y. pseudotuberculosis Δhfq strain from 103 to 107 CFU. All ketamine-xylazine-treated mice infected with wild-type bacteria succumbed to the infection by 8 days postinoculation, while none of the ketamine-xylazine-treated mice infected with 103 to 106 CFU of the Δhfq strain died (Fig. 3). None of the ketamine-xylazine-treated mice infected with the highest dose of the Y. pseudotuberculosis Δhfq strain survived beyond day 3 (Fig. 3). These data indicate that the Y. pseudotuberculosis Δhfq strain is at least 1,000-fold less virulent than the wild-type strain under these conditions.
FIG. 3.
Survival of mice inoculated with increasing doses of the Y. pseudotuberculosis Δhfq strain. Groups of 5 mice were injected intraperitoneally with a ketamine-xylazine cocktail immediately prior to intragastric inoculation with Y. pseudotuberculosis (103 CFU) or 10-fold-increasing doses of the Y. pseudotuberculosis Δhfq strain (103 to 107 CFU). Data are representative of 2 independent experiments.
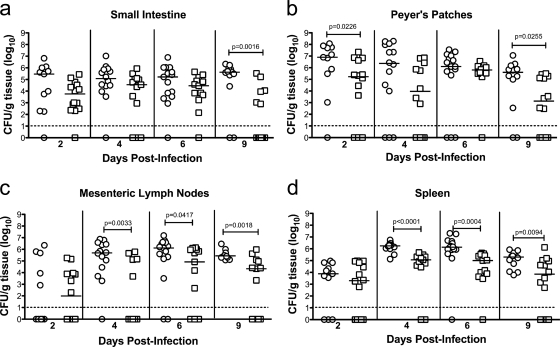
The defect in virulence of Y. pseudotuberculosis lacking Hfq may be due to a reduced ability of the bacteria to colonize the small intestine, to disseminate to other organs, or to persist in these organs. In order to evaluate the role of Hfq in bacterial colonization of these organs, unanesthetized mice were infected intragastrically with 107 CFU of the Y. pseudotuberculosis wild-type or Δhfq strain (PAN29 and PAN38, respectively). Mice were sacrificed 2, 4, 6, and 9 days postinoculation, and the small intestine, Peyer's patches, mesenteric lymph nodes, and spleen were removed, homogenized, and plated to determine the bacterial load in each organ. No significant difference was observed in the ability of the Y. pseudotuberculosis Δhfq strain to colonize the small intestine on day 2 postinoculation compared to the wild type, nor did the bacterial burden in the small intestine differ between wild-type- and Δhfq strain-infected mice on day 4 or 6 (Fig. 4a). However, a significant difference in CFU/g was seen on day 9 in the small intestine, with a trend toward clearance of the Δhfq strain occurring on or by this day (Fig. 4a). Similarly, there was no significant difference in the bacterial load in the Peyer's patches on day 4 or 6 between wild-type- and Δhfq strain-infected mice (Fig. 4b). A significant difference in bacterial load was seen in the Peyer's patches on days 2 and 9, however (Fig. 4B). In addition, the wild-type and Δhfq strains were able to disseminate to the mesenteric lymph nodes and spleen equally well on day 2 (Fig. 4c and d). A significant difference in the bacterial load in the mesenteric lymph nodes and spleen was observed on days 4, 6, and 9, indicating that in the absence of Hfq, Y. pseudotuberculosis may be diminished in its ability to survive or replicate in lymphoid organs (Fig. 4b to d).
FIG. 4.
Kinetics of infection with the Y. pseudotuberculosis Δhfq strain. Mice were inoculated intragastrically with wild-type or Δhfq Y. pseudotuberculosis (107 CFU), and after 2, 4, 6, and 9 days, CFU per gram of tissue in the spleen, visible Peyer's patches, mesenteric lymph nodes, and small intestine were determined. Graphs show bacterial counts from 3 combined experiments. Each point represents CFU/g recovered from a single animal (○, wild-type strain; □, Δhfq strain). A dashed line indicates the limit of detection. A solid line indicates the median of CFU recovered. Symbols below the limit of detection represent mice that survived but did not have detectable numbers of bacteria. Statistical significance was calculated by the Mann-Whitney U test.
Hfq is required for the virulence of Y. pseudotuberculosis in a systemic model of infection.
In order to further investigate if Y. pseudotuberculosis requires Hfq only in the initial stages of colonization or if it is also necessary for a systemic infection, mice were infected via the intraperitoneal route with 103 CFU of the Y. pseudotuberculosis wild-type, Δhfq, or pYV− strain (PAN29, PAN38, and PAN100, respectively) and survival was monitored for 21 days. This allowed the infection to bypass the step where Y. pseudotuberculosis colonizes the small intestine. Of the wild-type-infected mice, only 10% survived for the duration of the experiment, while in the Δhfq and pYV− strain-infected groups, all mice survived for 21 days (Fig. 5). This further suggests that Hfq is critical for the virulence of Y. pseudotuberculosis beyond the initial colonization steps and is required during a systemic infection.
FIG. 5.
Survival of mice inoculated intraperitoneally with the Y. pseudotuberculosis Δhfq strain. Groups of 10 mice were inoculated by intraperitoneal injection with wild-type, Δhfq, or pYV− Y. pseudotuberculosis (103 CFU) and monitored for 21 days. Data are representative of two independent experiments; in one of the experiments, a single mouse infected with the Δhfq strain succumbed to the infection on day 11 (not shown).
The Y. pseudotuberculosis Δhfq strain is hypermotile at 22°C.
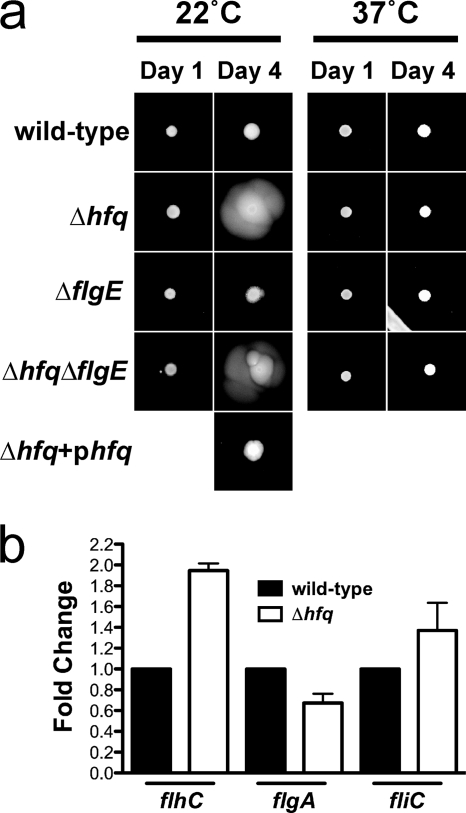
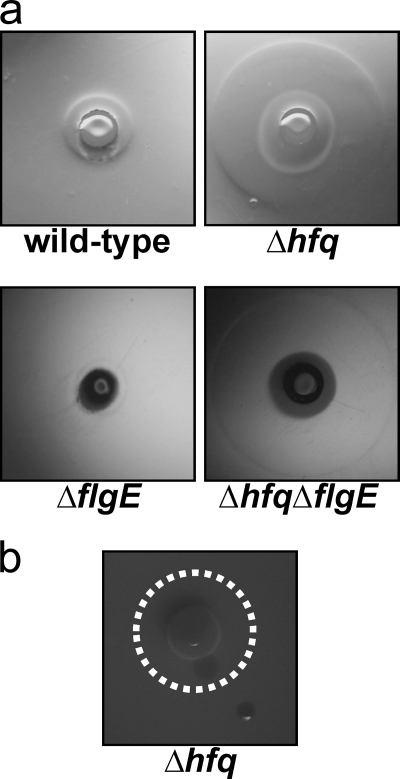
In order to determine how the absence of Hfq could lead to such a severe reduction in virulence, we examined key steps in the infectious process. As motility is a significant virulence determinant in many bacterial pathogens, we examined the effect of Hfq on the swarming motility of Y. pseudotuberculosis by growth on low-percentage agar plates. Overnight cultures of the Y. pseudotuberculosis wild-type, Δhfq, or Δhfq+phfq strain (PAN29, PAN38, and PAN136, respectively) were spotted onto swarm plates and grown at either 22°C or 37°C for 4 days. The absence of hfq results in increased motility at 22°C but not at 37°C, which indicates that Hfq is essential for the negative regulation of motility in Y. pseudotuberculosis at lower temperatures (Fig. 6a). To determine if hypermotility in the absence of Hfq is mediated through changes in flagellar synthesis or activity, we generated a deletion of the gene for the flagellar hook protein, flgE. The deletion of flgE in both the wild-type and Δhfq bacteria resulted in the same motility phenotype as that of the parental strains (Fig. 6a). Furthermore, qRT-PCR revealed that there is no significant difference in the transcript levels of early, middle, or late flagellar genes (flhC, flgA, and fliC) between the wild-type and mutant strains (Fig. 6b). Taken together, these data suggest that the hypermotility of the Y. pseudotuberculosis Δhfq strain is not mediated through changes in flagellar expression compared to wild type.
FIG. 6.
Motility of the Y. pseudotuberculosis Δhfq strain on semisolid agar. (a) Y. pseudotuberculosis wild-type, Δhfq, ΔflgE, Δhfq ΔflgE, and Δhfq+phfq strains were cultured on semisolid agar plates at 22°C or 37°C, and motility was monitored at 1 and 4 days postinoculation. Images are representative of several experiments. (b) qRT-PCR of flhC, flgA, and fliC transcripts. Bars represent the relative average fold change compared to wild type for each transcript of 3 independent experiments.
Enhanced production of a biosurfactant-like substance in the absence of Hfq.
Close examination of the low-percentage agar plates described above show a light-refractive compound visible in the agar surrounding the Y. pseudotuberculosis colony that is more evident in the absence of Hfq. The diameter of the refractive compound is larger in the absence of Hfq and is not affected by the presence or absence of flgE (Fig. 7a). Cultures of this material did not yield any bacterial growth, nor is this phenotype dependent on the T3SS (data not shown). Stewart et al. observed the production of a similar light-refractive compound by Legionella pneumophila, which they identified as a biosurfactant (44). Biosurfactants are characterized by their ability to reduce surface tension (11); therefore, in order to determine if the refractive compound observed here has properties of a biosurfactant, the Y. pseudotuberculosis Δhfq strain was plated on soft agar plates as above and allowed to grow for 2 days, after which time droplets of water were spotted inside and outside the area of refraction. The droplet of water inside collapsed immediately, while the droplet spotted outside maintained its integrity, which is consistent with the reduction in surface tension characteristic of a biosurfactant (Fig. 7b).
FIG. 7.
Production of a biosurfactant-like substance by the Y. pseudotuberculosis Δhfq strain. (a) Bacteria were prepared as described for Fig. 6 and cultured at 22°C for 2 days before photographs of a light-refractive compound, visible as a clear ring surrounding the bacterial colony, were taken. (b) Droplets of water were spotted inside and outside the compound on the motility plate 2 days after plating to demonstrate the reduced surface tension. A dotted white line delineates the border of the refractive compound.
The loss of Hfq reduces intracellular survival of Y. pseudotuberculosis in macrophage-like cells.
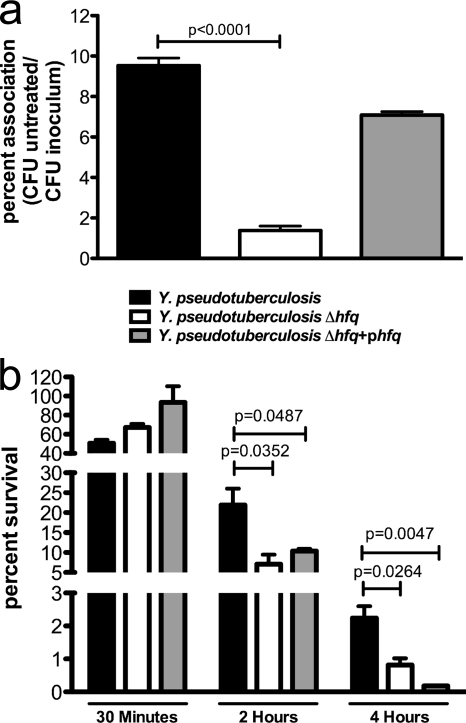
Previous work has shown that the intracellular survival of Y. pestis in cultured host macrophage cells is defective in the absence of Hfq (14). As this may be a critical step in pathogenesis, we examined the impact of Hfq on the ability of Y. pseudotuberculosis to survive within cultured macrophages. Y. pseudotuberculosis wild-type, Δhfq, and Δhfq+phfq strains (PAN29, PAN38, and PAN136, respectively) were incubated with J774 murine macrophage-like cells for 1 h (MOI of 10) and treated with gentamicin for 30 min, and CFU were determined. While there was a significant difference in the absolute numbers of cell-associated bacteria between the wild-type and Δhfq strains (Fig. 8a), there was no difference in the percentages of intracellular bacteria between the two strains (Fig. 8b). After 2 and 4 h of gentamicin treatment, however, the intracellular survival of the Y. pseudotuberculosis Δhfq strain was significantly decreased compared to that of the wild type (Fig. 8b). After 2 h and 4 h the phfq complementing strain is unable to restore survival to wild-type levels (Fig. 8b). We observed the same defect in a second, independently derived Δhfq mutant and complement of Y. pseudotuberculosis IP 32953 (PAN39 and PAN181, respectively), as well as in an Δhfq mutant and complement of a different isolate of Y. pseudotuberculosis, strain 32777 (PAN40 and PAN137, respectively) (data not shown). This suggests a defect with the plasmid-based complementation in this assay, rather than secondary, pleiotropic mutations in Δhfq bacteria. These data indicate that Hfq is necessary for adherence and intracellular survival of Y. pseudotuberculosis in host macrophages.
FIG. 8.
Association of the Y. pseudotuberculosis Δhfq strain with host cells. The Y. pseudotuberculosis wild-type, Δhfq, or Δhfq+phfq strain was incubated with J774 murine macrophage-like cells (MOI of 10). (a) Percentage of inoculum associated with host cells. (b) Percent intracellular bacteria after 30 min of treatment with gentamicin compared to untreated cells and percent intracellular bacteria after 2 and 4 h compared to CFU after 30 min. Bars represent the mean percentages, and error bars are standard errors of CFU from triplicate wells. Statistical analysis was performed with Student's unpaired t test. Data are representative of 3 independent experiments.
Hfq contributes to the resistance to oxidative stress.
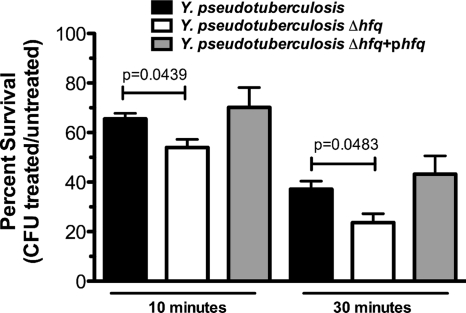
In order to determine if the defect in intracellular survival of Y. pseudotuberculosis in the absence of Hfq is due to a decreased ability of the bacteria to survive the oxidative burst, we exposed Y. pseudotuberculosis wild-type, Δhfq, and Δhfq+phfq (PAN29, PAN38, and PAN136, respectively) bacteria to H2O2. In the absence of Hfq, Y. pseudotuberculosis was significantly reduced in its ability to survive in the presence of H2O2 after 10 and 30 min of treatment (Fig. 9).
FIG. 9.
Survival in the presence of hydrogen peroxide. Y. pseudotuberculosis wild-type, Δhfq, and Δhfq+phfq strains were incubated for 10 or 30 min with 100 mM H2O2. Bars represent mean percent survival compared to untreated controls, and error bars represent standard errors of percent survival from 3 replicates. Statistical analysis was performed with Student's unpaired t test. Data are representative of 3 independent experiments.
Dysregulation of the T3SS in the absence of Hfq.
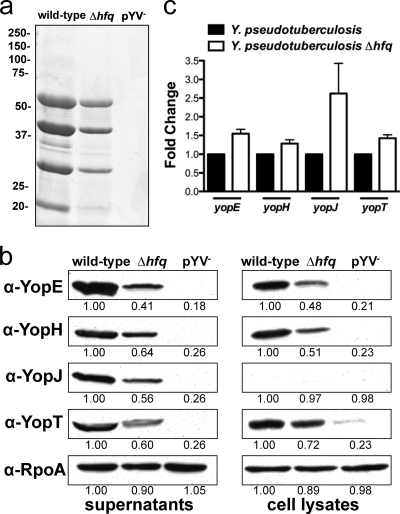
The type III secretion system (T3SS) is required by all pathogenic Yersinia species, including Y. pseudotuberculosis, for mammalian virulence (10). In order to determine if Hfq plays a role in the regulation of the T3SS in Y. pseudotuberculosis, bacteria were cultured for 3 h under secretion-inducing conditions (37°C, low Ca2+), and the culture supernatants and cell lysates from wild-type, Δhfq, and pYV− (PAN29, PAN38, and PAN100, respectively) bacteria were analyzed for the presence and abundance of Yop proteins. The overall protein profile was determined using Coomassie brilliant blue staining, and levels of the effector proteins YopE, YopH, YopJ, and YopT were assessed by immunoblotting with antibodies specific to each. The Coomassie blue-stained gel revealed that Hfq does not globally affect levels of secretion (supernatant) (Fig. 10a). In the absence of Hfq we observed decreased levels of all four Yop proteins tested in the cell pellets as well as the culture supernatants (Fig. 10b). To determine if the altered amounts of Yop proteins were due to changes in transcript abundance, we examined by qRT-PCR the relative mRNA levels of each Yop between the wild-type and Δhfq strains. There was no significant difference in the expression of any of the yop transcripts between wild-type and Δhfq bacteria (Fig. 10c). These data suggest that Hfq participates in the positive regulation of the T3SS, likely at a posttranscriptional level.
FIG. 10.
Production of type III secretion effector proteins in the absence of Hfq. Y. pseudotuberculosis wild-type, Δhfq, and pYV− strains were cultured in BHI broth for 3 h under Yop secretion-inducing conditions (37°C, low Ca2+). Cells were lysed, and culture supernatants were harvested, filtered, and precipitated with trichloroacetic acid. (a) The overall protein profile of the supernatant was determined by Coomassie brilliant blue staining. Molecular masses in kDa are indicated to the left. (b) Levels of YopE, YopH, YopJ, and YopT were assessed by immunoblotting in cells and culture supernatants. Blots are representative of 3 independent experiments. The relative density of each band compared to wild type is indicated. RpoA in whole-cell lysates was used as a loading control. (c) qRT-PCR of yopE, yopH, yopJ, and yopT transcripts. Bars represent the relative average fold change of each transcript compared to wild type for 3 independent experiments. There were no significant differences between wild-type and mutant transcript levels for all yop genes tested.
DISCUSSION
This study demonstrates that the small RNA chaperone Hfq plays a critical role in the pathogenesis of the enteric pathogen Y. pseudotuberculosis. A mouse model of infection shows that Hfq is required for the virulence of Y. pseudotuberculosis by a natural route of infection (Fig. 2a), and in the absence of Hfq, Y. pseudotuberculosis-infected mice do not succumb to the infection as their wild-type-infected counterparts do, even with a 1,000-fold-higher dose of bacteria (Fig. 3). This reduction in virulence may be due to the decreased ability of Hfq-deficient Y. pseudotuberculosis to survive and replicate in the Peyer's patches, mesenteric lymph nodes, and spleen (Fig. 4b to d).
The loss of Hfq does not completely abrogate the ability of Y. pseudotuberculosis to establish an infection in the mouse, however, as demonstrated by the moderate weight loss of the animals and the bacterial burden in the tissues of Δhfq strain-infected mice (Fig. 2b and 4a to d). In other pathogens, the loss of Hfq often results in a severe, multifold defect in the bacterial load in tissues and organs compared to a wild-type infection (14, 26, 42), whereas with Y. pseudotuberculosis, this does not appear to be the case. This suggests a unique contribution to virulence for Hfq in Y. pseudotuberculosis. While the attenuation in virulence attributed to Hfq may be due to the defect in growth at 37°C, it is also possible that Hfq contributes to the subversion of the host innate and adaptive immune response by Y. pseudotuberculosis. Hfq-dependent sRNAs may participate in the regulation of the expression of proteins that alter the immunogenicity of the bacterium or may affect the ability of Y. pseudotuberculosis to express virulence factors that prevent clearance by the host. Indeed, the Hfq homolog of F. tularensis was identified as an activator of the host immune system, supporting this possibility (16).
Alternatively, Hfq may have a role in regulating specific virulence factors that are essential for survival or replication in the lymphoid tissue. For instance, in the absence of Hfq the motility of Y. pseudotuberculosis is increased compared to wild-type bacteria (Fig. 6). While expression of the flagellar genes is also repressed at 37°C in Y. pseudotuberculosis, the observed hypermotility is independent of the flagellum (Fig. 6). Thus, we expect that this form of motility would not be overridden by the transcriptional regulation of flagellar expression (23). Non-flagellum-based motility could play a role in mammalian infection, particularly if the expression of the sRNA(s) that represses this phenomenon in wild-type bacteria in vitro is downregulated in vivo. Our results add an additional layer of sRNA-based regulation of motility beyond the CsrABC system that affects FlhDC expression in Y. pseudotuberculosis (18). This phenotype is in contrast to the decreased motility observed when Hfq is deleted from Salmonella enterica serovar Typhimurium and uropathogenic E. coli, which further demonstrates that the effects of Hfq are unique to each bacterial species (26, 42).
An unexpected result from this study is the discovery of a biosurfactant-like substance that is released by the Y. pseudotuberculosis Δhfq strain on a semisolid surface. This observation is intriguing, as biosurfactant production by Yersinia species has not been reported. Interestingly, Y. pestis does not produce this compound in either the presence or the absence of Hfq (data not shown). The biosurfactant-like substance is not produced at 37°C in our assay, which correlates with the presence of the hypermotility phenotype occurring only at lower temperatures and suggests that this substance could contribute to the hypermotility of the Y. pseudotuberculosis Δhfq strain at 22°C. Although the biosurfactant does not appear to be produced at physiologic temperatures in vitro, there may be stimuli that promote its synthesis under certain conditions during infection. Indeed, the in vivo function of this biosurfactant-like substance is unknown, but it could play a role during infection, as a biosurfactant as has been implicated in the virulence of Pseudomonas (5). The overproduction of the biosurfactant-like substance may contribute to the reduction in virulence in the mouse model caused by enhanced motility, a reduced ability to adhere to host cells, increased stimulation of the immune response, or another function, all of which may contribute to defects in the ability of Y. pseudotuberculosis to persist in lymphoid organs. A database search revealed putative glycosyltransferases in the genome of Y. pseudotuberculosis that are similar to the genes for rhlB and rhlC in Pseudomonas aeruginosa. YPTB1978, encoding a putative glycosyltransferase, with 39% similarity to rhlB, and speE, encoding a spermidine synthase, with 44% similarity to rhlC, may have functions that could be part of a biosurfactant synthesis pathway, and in other bacterial species, such genes have been implicated in changes in motility (4). This avenue requires further investigation to determine the biosynthetic pathway and biosurfactant material, as well as its potential contribution to virulence.
Appropriate host-pathogen interactions involving the macrophage are critical to the virulence of many bacterial species. It is possible, then, that the attenuation of the Δhfq strain may be explained by defects in the interaction of Y. pseudotuberculosis with host immune cells in the Peyer's patches, spleen, and mesenteric lymph nodes following dissemination from the small intestine. In the absence of Hfq, Y. pseudotuberculosis exhibits a significant defect in intracellular survival in macrophage-like cells (Fig. 8). This may be due to the reduced ability of the Y. pseudotuberculosis Δhfq strain to withstand the oxidative burst, simulated by the H2O2 killing assay (Fig. 9). Our experiments highlight the differences and unique role that Hfq and small RNAs can play in different pathogens. The loss of Hfq also reduces the growth and survival of S. enterica and B. abortus within the macrophage, suggesting that Hfq contributes to the regulation of factors that are involved in intracellular survival of a number of bacterial species. Indeed, the loss of Hfq results in an increase in phagocytosis of Y. pestis, as well as decreased intracellular survival (14). Conversely, there is no effect on the intracellular survival of L. monocytogenes and F. tularensis in the absence of Hfq (9, 32, 40, 42). Uropathogenic E. coli does not have a defect in adherence or invasion of cultured host cells, despite decreased colonization of the urinary tract and bladder tissue in a mouse model of infection (26). Given these results, it would be worthwhile to investigate if there is a common pathway for intracellular survival that is influenced by Hfq and its interactions with a particular set of conserved sRNAs or if the mechanism is unique to each organism.
Many species of pathogenic bacteria use the T3SS to inject effector proteins into host cells, and type III secretion is essential for the virulence of Yersinia species (10). Furthermore, Yersinia species have been shown to target immune cells for injection with type III effectors (25, 29); therefore, the defects in persistence of the Δhfq mutant that we see in the Peyer's patches, lymph nodes, and spleen could be related to dysregulation of the T3SS, as these organs are centers for lymphocytes. Our data show a coordinated decrease in the production of four Yop proteins (YopE, YopH, YopJ, and YopT) in the absence of Hfq, which indicates that Hfq may play a role in the regulation of Yop proteins directly or indirectly through interactions with a regulator of T3S effectors. For example, the protein LcrF coordinately regulates production of the effector Yops at the transcriptional level in response to temperature changes in an “all-or-none” fashion (20, 52). Furthermore, Rosenzweig et al. have shown that in Yersinia spp., polynucleotide phosphorylase (PNPase) is a negative regulator of the T3SS, while others have shown that the loss of Hfq in E. coli leads to increased PNPase activity, which together may indicate a role for Hfq on T3S via interactions with PNPase (34a, 40a).
It is possible that the decreased levels of T3S effectors in the Δhfq strain could account entirely for the inability of Y. pseudotuberculosis to cause death of the mouse through decreased fitness within the immune cell-containing lymphoid organs. Mice infected with the T3S mutant (pYV−) displayed no weight loss and showed no signs of illness in our intragastric model of infection, whereas mice infected with the Δhfq strain did lose weight and did show signs of illness, including huddling and decreased activity (Fig. 2b and data not shown). This suggests that Hfq may play a role in the regulation of other virulence factors beyond the T3SS. Furthermore, given the pleiotropic nature of Hfq, it is reasonable to anticipate that there are other targets of Hfq and sRNAs involved in virulence pathways. However, the slight production of Yop proteins seen in the Δhfq strain by immunoblotting may be sufficient enough to cause the level of illness seen in our mouse model (Fig. 10b); therefore, we cannot rule out changes to T3S as the sole contributor to the virulence defect.
We have demonstrated that the loss of Hfq significantly attenuates Y. pseudotuberculosis in a mouse model of infection and leads to defects in motility, intracellular survival, and type III secretion. Further analysis of these phenotypes will reveal the mechanisms by which Hfq and sRNAs mediate these effects in Y. pseudotuberculosis. Additionally, an analysis of the host response to Y. pseudotuberculosis in the presence and absence of Hfq may reveal if the disconnect between survival and colonization is based on an Hfq-dependent, host-mediated response. Finally, an understanding of the changes in protein expression in the absence of Hfq will reveal the targets of sRNAs that are regulated in an Hfq-dependent manner.
Acknowledgments
We thank James Bliska for his gift of the Yop antibodies and Melanie Marketon for the RpoA antibody, Karla Satchell for helpful discussions and advice, Carl Waltenbaugh for photography, and Jovanka Koo for help with animals.
This work was supported by the Searle Leadership Fund of Northwestern University, the Northwestern University Feinberg School of Medicine, and the NIH/NIAID Regional Center of Excellence for Bio-defense and Emerging Infectious Diseases Research (RCE) Program. We acknowledge membership within and support from the Region V “Great Lakes” RCE (NIH award 2-U54-AI-057153).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 15 March 2010.
REFERENCES
- 1.Altuvia, S., D. Weinstein-Fischer, A. Zhang, L. Postow, and G. Storz. 1997. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90:43-53. [DOI] [PubMed] [Google Scholar]
- 2.Applied Biosystems. 1997. ABI Prism 7700 sequence detection system user bulletin 2. Applied Biosystems, Foster City, CA.
- 3.Barnes, P. D., M. A. Bergman, J. Mecsas, and R. R. Isberg. 2006. Yersinia pseudotuberculosis disseminates directly from a replicating bacterial pool in the intestine. J. Exp. Med. 203:1591-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caiazza, N. C., R. M. Shanks, and G. A. O'Toole. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 187:7351-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calfee, M. W., J. G. Shelton, J. A. McCubrey, and E. C. Pesci. 2005. Solubility and bioactivity of the Pseudomonas quinolone signal are increased by a Pseudomonas aeruginosa-produced surfactant. Infect. Immun. 73:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee, A., Y. Cui, and A. K. Chatterjee. 2002. RsmA and the quorum-sensing signal, N-[3-oxohexanoyl]-L-homoserine lactone, control the levels of rsmB RNA in Erwinia carotovora subsp. carotovora by affecting its stability. J. Bacteriol. 184:4089-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, S., A. Zhang, L. B. Blyn, and G. Storz. 2004. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 186:6689-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christiansen, J. K., M. H. Larsen, H. Ingmer, L. Sogaard-Andersen, and B. H. Kallipolitis. 2004. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 186:3355-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M. P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai, J. D., and I. M. Banat. 1997. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 61:47-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fantappie, L., M. M. Metruccio, K. L. Seib, F. Oriente, E. Cartocci, F. Ferlicca, M. M. Giuliani, V. Scarlato, and I. Delany. 2009. The RNA chaperone Hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect. Immun. 77:1842-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franze de Fernandez, M. T., L. Eoyang, and J. T. August. 1968. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature 219:588-590. [DOI] [PubMed] [Google Scholar]
- 14.Geng, J., Y. Song, L. Yang, Y. Feng, Y. Qiu, G. Li, J. Guo, Y. Bi, Y. Qu, W. Wang, X. Wang, Z. Guo, R. Yang, and Y. Han. 2009. Involvement of the post-transcriptional regulator Hfq in Yersinia pestis virulence. PloS One 4:e6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goguen, J. D., J. Yother, and S. C. Straley. 1984. Genetic analysis of the low calcium response in Yersinia pestis mud1(Ap lac) insertion mutants. J. Bacteriol. 160:842-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havlasova, J., L. Hernychova, M. Brychta, M. Hubalek, J. Lenco, P. Larsson, M. Lundqvist, M. Forsman, Z. Krocova, J. Stulik, and A. Macela. 2005. Proteomic analysis of anti-Francisella tularensis LVS antibody response in murine model of tularemia. Proteomics 5:2090-2103. [DOI] [PubMed] [Google Scholar]
- 17.Helmer, K. S., Y. Cui, L. Chang, A. Dewan, and D. W. Mercer. 2003. Effects of ketamine/xylazine on expression of tumor necrosis factor-alpha, inducible nitric oxide synthase, and cyclo-oxygenase-2 in rat gastric mucosa during endotoxemia. Shock 20:63-69. [DOI] [PubMed] [Google Scholar]
- 18.Heroven, A. K., K. Bohme, M. Rohde, and P. Dersch. 2008. A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol. Microbiol. 68:1179-1195. [DOI] [PubMed] [Google Scholar]
- 19.Higuchi, K., and J. L. Smith. 1961. Studies on the nutrition and physiology of Pasteurella pestis. VI. A differential plating medium for the estimation of the mutation rate to avirulence. J. Bacteriol. 81:605-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoe, N. P., and J. D. Goguen. 1993. Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J. Bacteriol. 175:7901-7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551-561. [DOI] [PubMed] [Google Scholar]
- 22.Kajitani, M., and A. Ishihama. 1991. Identification and sequence determination of the host factor gene for bacteriophage Q beta. Nucleic Acids Res. 19:1063-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapatral, V., J. W. Olson, J. C. Pepe, V. L. Miller, and S. A. Minnich. 1996. Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol. Microbiol. 19:1061-1071. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi, K., R. Hasunuma, S. Kikuchi, R. Ryll, K. Morikawa, and Y. Kumazawa. 2002. Time- and dose-dependent effect of fosfomycin on suppression of infection-induced endotoxin shock in mice. Biol. Pharm. Bull. 25:1658-1661. [DOI] [PubMed] [Google Scholar]
- 25.Koberle, M., A. Klein-Gunther, M. Schutz, M. Fritz, S. Berchtold, E. Tolosa, I. B. Autenrieth, and E. Bohn. 2009. Yersinia enterocolitica targets cells of the innate and adaptive immune system by injection of Yops in a mouse infection model. PLoS Pathog. 5:e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulesus, R. R., K. Diaz-Perez, E. S. Slechta, D. S. Eto, and M. A. Mulvey. 2008. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect. Immun. 76:3019-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lease, R. A., M. E. Cusick, and M. Belfort. 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. U. S. A. 95:12456-12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 29.Marketon, M. M., R. W. DePaolo, K. L. DeBord, B. Jabri, and O. Schneewind. 2005. Plague bacteria target immune cells during infection. Science 309:1739-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marra, A., and R. R. Isberg. 1997. Invasin-dependent and invasin-independent pathways for translocation of Yersinia pseudotuberculosis across the Peyer's patch intestinal epithelium. Infect. Immun. 65:3412-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayer, L., and A. J. Greenstein. 1976. Acute yersinial ileitis: a distinct entity. Am. J. Gastroenterol. 65:548-551. [PubMed] [Google Scholar]
- 32.Meibom, K. L., A. L. Forslund, K. Kuoppa, K. Alkhuder, I. Dubail, M. Dupuis, A. Forsberg, and A. Charbit. 2009. Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect. Immun. 77:1866-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merriam, J. J., R. Mathur, R. Maxfield-Boumil, and R. R. Isberg. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizuno. T, M.-Y. Chou, and M. Inouye. 1983. Regulation of gene expression by a small RNA transcript (micRNA) in Escherichia coli K-12. Proc. Jpn. Acad. Ser. B 59:335-338. [Google Scholar]
- 34a.Mohanty, B. K., V. F. Maples, and S. R. Kushner. 2004. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 54:905-920. [DOI] [PubMed] [Google Scholar]
- 35.Moller, T., T. Franch, P. Hojrup, D. R. Keene, H. P. Bachinger, R. G. Brennan, and P. Valentin-Hansen. 2002. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9:23-30. [DOI] [PubMed] [Google Scholar]
- 36.Nakao, H., H. Watanabe, S. Nakayama, and T. Takeda. 1995. yst gene expression in Yersinia enterocolitica is positively regulated by a chromosomal region that is highly homologous to Escherichia coli host factor 1 gene (hfq). Mol. Microbiol. 18:859-865. [DOI] [PubMed] [Google Scholar]
- 37.Nuorti, J. P., T. Niskanen, S. Hallanvuo, J. Mikkola, E. Kela, M. Hatakka, M. Fredriksson-Ahomaa, O. Lyytikainen, A. Siitonen, H. Korkeala, and P. Ruutu. 2004. A widespread outbreak of Yersinia pseudotuberculosis O:3 infection from iceberg lettuce. J. Infect. Dis. 189:766-774. [DOI] [PubMed] [Google Scholar]
- 38.Olivier, V., J. Queen, and K. J. Satchell. 2009. Successful small intestine colonization of adult mice by Vibrio cholerae requires ketamine anesthesia and accessory toxins. PloS One 4:e7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfeiffer, V., A. Sittka, R. Tomer, K. Tedin, V. Brinkmann, and J. Vogel. 2007. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol. Microbiol. 66:1174-1191. [DOI] [PubMed] [Google Scholar]
- 40.Robertson, G. T., and R. M. Roop, Jr. 1999. The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 34:690-700. [DOI] [PubMed] [Google Scholar]
- 40a.Rosenzweig, J. A., B. Chromy, A. Echeverry, J. Yang, B. Adkins, G. V. Plano, S. McCutchen-Maloney, and K. Schesser. 2007. Polynucleotide phosphorylase independently controls virulence factor expression levels and export in Yersinia spp. FEMS Microbiol. Lett. 270:255-264. [DOI] [PubMed] [Google Scholar]
- 41.Rosso, M. L., S. Chauvaux, R. Dessein, C. Laurans, L. Frangeul, C. Lacroix, A. Schiavo, M. A. Dillies, J. Foulon, J. Y. Coppee, C. Medigue, E. Carniel, M. Simonet, and M. Marceau. 2008. Growth of Yersinia pseudotuberculosis in human plasma: impacts on virulence and metabolic gene expression. BMC Microbiol. 8:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sittka, A., V. Pfeiffer, K. Tedin, and J. Vogel. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63:193-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sledjeski, D. D., A. Gupta, and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart, C. R., O. Rossier, and N. P. Cianciotto. 2009. Surface translocation by Legionella pneumophila: a form of sliding motility that is dependent upon type II protein secretion. J. Bacteriol. 191:1537-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suliburk, J. W., E. A. Gonzalez, S. D. Moore-Olufemi, N. Weisbrodt, F. A. Moore, and D. W. Mercer. 2005. Ketamine inhibits lipopolysacharide (LPS) induced gastric luminal fluid accumulation. J. Surg. Res. 127:203-207. [DOI] [PubMed] [Google Scholar]
- 46.Tsui, H. C., G. Feng, and M. E. Winkler. 1997. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 179:7476-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Udekwu, K. I., F. Darfeuille, J. Vogel, J. Reimegard, E. Holmqvist, and E. G. Wagner. 2005. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 19:2355-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vecerek, B., I. Moll, T. Afonyushkin, V. Kaberdin, and U. Blasi. 2003. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol. Microbiol. 50:897-909. [DOI] [PubMed] [Google Scholar]
- 49.Vincent, P., A. Leclercq, L. Martin, J. M. Duez, M. Simonet, and E. Carniel. 2008. Sudden onset of pseudotuberculosis in humans, France, 2004-05. Emerg. Infect. Dis. 14:1119-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vytvytska, O., J. S. Jakobsen, G. Balcunaite, J. S. Andersen, M. Baccarini, and A. von Gabain. 1998. Host factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth rate-dependent fashion and regulates its stability. Proc. Natl. Acad. Sci. U. S. A. 95:14118-14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wayte, J., A. T. Silva, T. Krausz, and J. Cohen. 1993. Observations on the role of tumor necrosis factor-alpha in a murine model of shock due to Streptococcus pyogenes. Crit. Care Med. 21:1207-1212. [DOI] [PubMed] [Google Scholar]
- 52.Wiley, D. J., R. Rosqvist, and K. Schesser. 2007. Induction of the Yersinia type 3 secretion system as an all-or-none phenomenon. J. Mol. Bio. 373:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wren, B. W. 2003. The yersiniae—a model genus to study the rapid evolution of bacterial pathogens. Nat. Rev. Microbiol. 1:55-64. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9:11-22. [DOI] [PubMed] [Google Scholar]