Abstract
Microsporidia are obligate intracellular fungus-related parasites considered as emerging opportunistic human pathogens. Their extracellular infective and resistance stage is a spore surrounded by a unique plasma membrane protected by a thick cell wall consisting of two layers: the electron-lucent inner endospore which contains chitin and protein components and the outer-electron-dense and mainly proteinaceous exospore. We identified the whole sequences of two spore wall proteins in the microsporidian species Encephalitozoon hellem, designated EhSWP1a and EhSWP1b. Isolation of the genes encoding these SWP1-like proteins was performed using degenerate oligonucleotides based on the amino acid sequence alignment of the previously reported Encephalitozoon cuniculi and Encephalitozoon intestinalis SWP1s. Sequences lacking the 5′ and 3′ ends were then identified by PCR and reverse transcription (RT)-PCR amplifications. The swp1a and swp1b genes encode proteins of 509 and 533 amino acids, respectively, which present an identical N-terminal domain of 382 residues and a variable C-terminal extension mainly characterized by a 26-amino-acid (aa) deletion/insertion containing glutamate- and lysine-rich repeats. Using polyclonal antibodies raised against recombinant polypeptides, we showed that EhSWP1a and EhSWP1b appear as dithiothreitol (DTT)-soluble bands of 55 and 60 kDa in size, respectively. Immunolocalization experiments by IFA and transmission electron microscopy (TEM) indicated that both proteins are present at the onset of sporogony and are specifically located to the spore wall exospore in mature spores. Analysis of four E. hellem human isolates revealed that the C-terminal regions of both EhSWP1a and EhSWP1b are polymorphic, which is of interest for epidemiological studies.
Microsporidia constitute a phylum of small unicellular eukaryotes comprising more than 1,200 species, all obligate intracellular parasites able to form environmentally resistant spores. Although they are commonly found in arthropods and fishes, being responsible for important economic losses in the beekeeping and fish industries, some species are also of medical and veterinary significance (10). In humans, Microsporidia are now not only recognized as emerging pathogens in immunocompromised hosts, causing intestinal, ocular, muscular, and systemic diseases, but such infections are increasingly being described in immunocompetent people (1, 24, 34). Their zoonotic transmission is supported by the identification of human-infecting genotypes in a wide range of animals (23). In addition to their medical incidence, microsporidia have attracted attention as amitochondriate eukaryotes that were assumed to be among the earliest lineages of eukaryotes (32). However, data from molecular phylogenies based on various gene sequences argued against an early origin of Microsporidia and supported a placement of these organisms among Fungi (17, 19).
Although microsporidia differ greatly in host range and cell-type specificity, they share a similar mechanism for host cell invasion. The invasion process involves the sudden extrusion of the sporal polar tube, leading to the delivery of the infective sporoplasm inside the host cell, initiating the obligate intracellular development. In all Encephalitozoon species, the complete life cycle occurs inside a parasitophorous vacuole. First, a proliferative growth (merogony) is followed by sporogony, when meronts transform into sporonts characterized by the deposition of an electron-dense material on the plasma membrane. Then, sporonts divide into sporoblasts in which the formation of the thick wall is associated with the differentiation of the invasion apparatus. After completion of the maturation of sporoblasts, the rupture of host cells may lead to the release of resistant spores in the environment (11).
The microsporidian cell wall consists of two major layers surrounding the plasma membrane: (i) an electron-dense outer layer, the exospore, which is mainly proteinaceous, and (ii) an electron-lucent inner layer, the endospore, which contains chitin and protein components (4). The rigidity of the wall makes the spore resistant to various environmental stresses and maintains a high intrasporal hydrostatic pressure that could play a role in the polar tube extrusion (12). Recent reports showed that spore wall proteins are able to adhere to the host cell surface via sulfated glycosaminoglycans (GAGs) (16, 30). In addition, spore wall components have been shown to be useful tools for serological studies (31).
Knowledge regarding the microsporidian spore wall composition is still limited. Only a chitin deacetylase-like protein (7) and two proteins with unknown functions, named EnP1 and EnP2, have been shown to localize to the endospore in the mammal microsporidian Encephalitozoon cuniculi (27, 37). A major antigen (EcSWP1) has also been assigned to the exospore in E. cuniculi (5). Two homologous SWP1 proteins (EiSWP1 and EiSWP2) have been identified in Encephalitozooon intestinalis (15). SWP1 proteins are characterized by a conserved N-terminal part of 360 amino acids (aa) and possess highly divergent C-terminal regions. More recent proteomic-based approaches successfully identified several spore wall proteins in Nosema bombycis, the microsporidian species responsible for silkworm disease. These proteins have no sequence homology with ncephalitozoon SWP1 proteins (20, 35).
The purpose of the present study was to identify SWP1-like proteins in Encephalitozoon hellem, the third species of the Encephalitozoon genus known to cause infections from birds to mammals, including humans (23). Using PCR and reverse transcription (RT)-PCR approaches, we successfully identified the complete sequences of two swp1-like genes and demonstrated that proteins encoded by these genes localized to the spore wall exospore. We also showed that these swp1 genes are polymorphic, which is of high interest for epidemiological studies.
MATERIALS AND METHODS
Microsporidian spore production and DNA extraction.
Four Encephalitozoon hellem strains isolated from American AIDS patients were used in this study: EhD, donated by E. S. Didier; and V242, V257, and V213 (18), kindly given by G. S. Visvesvara and E. U. Canning. E. hellem, E. cuniculi, and E. intestinalis were maintained on human foreskin fibroblast (HFF) cells in 5% CO2 at 37°C in minimum essential medium (MEM) supplemented with 5% fetal calf serum, 2 mM glutamine (Invitrogen), and 20 μg/ml gentamicin. Supernatants containing microsporidian spores were collected every 3 days, harvested (5,000 × g for 10 min), washed, and stored at 4°C as described previously (9). Host cell debris was eliminated with 2% SDS at 50°C. DNA was released by boiling purified E. hellem spores for 10 min at 100°C in sterile distilled water.
Identification of Ehswp1 genes.
The sequences of the primers used for the different PCR amplification steps are listed in Table S1 in the supplemental material. Two degenerate primers (SWD1 and SWR1) were first designed in the conserved central region between EcSWP1 and EiSWP1 to amplify a 400-bp fragment. The lacking 5′ end was characterized using primers SWD2 and EhR1. The lacking 3′ ends of both Ehswp1a and Ehswp1b were determined from mRNA after reverse transcription using an oligo(dT) primer linked to an adapter sequence. PCR amplifications were then done using the primer corresponding to the adapter sequence and the primer EhD1. The whole sequences of Ehswp1a and Ehswp1b were confirmed with primers EhD2 and EhR2 for Ehswp1a and EhD2 and EhR3 for Ehswp1b. Genotyping studies were performed from four E. hellem isolates (EhD, V242, V257, and V213) with primers EhD3 and EhR4. All PCR amplifications were performed using a Perkin-Elmer DNA thermal cycler 2400 apparatus in a 50-μl reaction mixture according to standard conditions (Eurobio). After denaturing at 94°C for 3 min, 35 cycles were run with 30 s of denaturation at 94°C, 30 s of annealing at 50 to 55°C, and 1 min of extension at 72°C, followed by a 15-min final extension step at 72°C. PCR products were analyzed on 1% agarose gel and purified with QIAquick gel extraction kit (Qiagen). They were cloned in pGEM-T Easy vector (Promega) and sequenced.
RT-PCR analysis.
Total RNA was extracted using the RNeasy minikit (Qiagen) from HFF cells 72 h postinfestation with E. hellem strains. mRNAs were reverse transcribed with an oligo(dT) primer linked to an adapter sequence using 15 U of the avian myeloblastosis virus (AMV) reverse transcriptase (Amersham). PCRs were then performed using a reverse primer corresponding to the adapter sequence and the forward primer EhD1 (see Table S1 in the supplemental material). PCR amplifications were performed as described above. PCR products were analyzed on 1.5% agarose gel, cloned in pGEM-T Easy vector (Promega), and sequenced.
Expression of recombinant EhSWP1s in Escherichia coli.
EhB1 and EhE1 primers (see Table S1 in the supplemental material) were designed to amplify a 945-bp fragment corresponding to the conserved N-terminal part (aa 20 to 334). EhB2 and EhR2 primers were used to amplify a 453-bp fragment representing the C-terminal part of EhSWP1a (EhSWP1a-C; aa 358 to 509). A third fragment, specific to EhSWP1b and encoding the 26-aa insertion (EhSWP1b-C; aa 388 to 423) was amplified with EhB3 and EhR3. Forward and reverse primers contained a BamHI restriction site and an EcoRI restriction site, respectively, at their 5′ ends. PCR amplifications were performed as described above with 30 s of annealing at 56°C and 1 min of extension at 72°C. PCR products were analyzed on 1% agarose gel and purified with QIA-quick gel extraction kit (Qiagen). After digestion with BamHI and EcoRI, the fragments were cloned in frame with glutathione S-transferase (GST) and an 8-histidine tag into the prokaryotic expression vector pGEX-4T1 (Pharmacia). The resulting recombinant plasmids (pGEX-4T1-SWP1-N, pGEX-4T1-SWP1a-C, and pGEX-4T1-SWP1b-C) were then used to transform the E. coli BL21+ strain, and protein expression was analyzed after induction by 2 mM IPTG (isopropyl-β-d-thiogalactoside; Sigma) for 4 h. Bacterial proteins were solubilized in 2.5% SDS-100 mM dithiothreitol (DTT) and analyzed by 10% SDS-PAGE. Recombinant proteins were purified on Ni-nitrilotriacetic acid (NTA) columns (Qiagen).
Antibody production.
Swiss mice were injected with purified recombinant proteins expressed in E. coli. The animal facility (agreement C63014.19) and the experimental staff (agreement 63-146) were approved by the French Veterinary Service. The experiments were conducted according to ethical rules. Mice were injected i.p. with samples homogenized with Freund's complete adjuvant for the first injection and Freund's incomplete adjuvant for the next injections (days 14, 21, and 28). Sera were collected 1 week after the last injection and stored at −20°C.
SDS-PAGE and Western blotting.
Proteins from healthy and E. hellem-, E. cuniculi-, or E. intestinalis-infected HFF cells were extracted in a lysis buffer containing 2.5% SDS and 100 mM DTT. Cells were destroyed by boiling for 10 min and repeated cycles of freezing-thawing in liquid nitrogen and sonications (10× for 30 s on ice). Protein samples were then analyzed by SDS-PAGE on 10% polyacrylamide gels. For immunological detection, proteins were first transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore). Membranes were saturated in PBS-5% skim milk and incubated for 3 h with appropriate dilutions of mouse antibodies (1:500 to 1:1,000). After washing in PBS-0.1% Triton, membranes were reacted for 1 h with alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (IgG) diluted at 1:10,000 (Promega) and revealed with the nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP) detection kit (Promega).
Indirect IFA.
For the indirect immunofluorescense assay (IFA), E. hellem, E. cuniculi, and E. intestinalis spores were washed in PBS and fixed with methanol for 20 min at −80°C. HFF cells growing on glass coverslips were infected with E. hellem, E. cuniculi, or E. intestinalis spores and fixed 96 h postinfestation. Slides were permeabilized with PBS-0.5% Triton X-100, saturated with PBS-5% skim milk, and incubated with antibodies diluted at 1:100 in PBS-0.1% Triton X-100. Primary antibodies were consequently detected by incubation for 1 h with Alexa Fluor 488-conjugated antimouse IgG diluted at 1:1,000 (Molecular Probes). Preparations were then examined with a Leica epifluorescence microscope.
TEM.
For transmission electron microscopy (TEM), E. hellem-infected HFF cells were fixed with 4% paraformaldehyde-0.1% glutaraldehyde in PBS. After infusion for 1 h at room temperature in a 25% glycerol-5% DMSO mixture, the samples were frozen in pasty nitrogen. After saturation for 1 h with 1% ovalbumin in PBS, grids were incubated for 3 h with various dilutions of spore wall antibodies then for another 1 h in the presence of a 1:100 dilution of goat anti-mouse IgG conjugated with 10-nm colloidal gold particles (Sigma). After washing, the grids were treated with a 4:1 (vol/vol) mixture of methylcellulose-4% uranyl acetate for 10 min, left to dry, and then observed with a JEOL 1200 X transmission electron microscope.
Serological tests.
The recombinant proteins EhSWP1-N, EhSWP1a-C, and EhSWP1b-C were purified on Ni-NTA columns, and 0.5 μg of each was spotted onto a PVDF membrane. The membranes were then treated as described in the section “SDS-PAGE and Western blotting.” Three different antibodies were hybridized: a monoclonal antihistidine antibody (Amersham) and two polyclonal antisera directed against E. hellem spores. The mouse antiserum was produced from purified and inactivated E. hellem spores; the rabbit antiserum was kindly provided by E. S. Didier.
Sequence analysis.
Gene and protein statistical analysis, molecular masses, isoelectric points, and potential O-glycosylation sites were determined using ExPASy Proteomics tools (http://www.expasy.org/tools/). Protein primary sequence properties were predicted using PSORT (http://psort.nibb.ac.jp). Search for homologous proteins in databases was done using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Nucleotide sequence accession numbers.
The sequence data for EhSWP1a and EhSWP1b have been submitted to the GenBank database under accession no. FJ870923 and FJ870924, respectively. The Ecswp1 (ECU10_1660), Eiswp1, and Ehswp2 nucleic sequence accession no. are AJ133745, AF355750, and AF355750, respectively. The predicted amino acid sequences for EcSWP1, EiSWP1, and EiSWP2 were obtained from accession no. CAD25887, AAL27283, and AAL27282, respectively.
RESULTS
Identification of two genes encoding SWP1-like proteins in Encephalitozoon hellem.
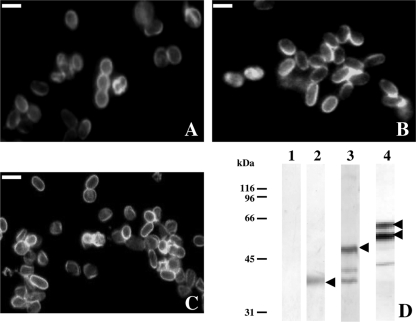
In a previous study, several monoclonal antibodies (MAbs) were generated against spores of Encephalitozoon intestinalis and Encephalitozoon hellem, most of these being specific to antigens from either the cell wall or the polar tube (21). Two MAbs, referred as 1E4 and 11B2, showed similar patterns of hybridization on Western blotting and reacted with the spore wall of E. intestinalis. MAb 11B2 was then used to identify a major glycoprotein from the exospore of E. intestinalis (EiSWP1) (15), a homologue to the first-identified E. cuniculi SWP1 (EcSWP1) (5). In the present study, these Mabs were used for IFA and Western blot analyses of HFF cells infected with each of the three Encephalitozoon species. As shown in Fig. 1, the 1E4 MAb cross-reacted with the cell wall of both E. cuniculi (Fig. 1B) and E. hellem (Fig. 1C). In the Western blot, it specifically recognized an ∼40-kDa band of E. intestinalis corresponding to the expected size for EiSWP1 (Fig. 1D, lane 2). A cross-reaction was also observed in E. cuniculi with a 50-kDa migrating band (expected size for EcSWP1) and some lower bands at 40 and 42 kDa (Fig. 1D, lane 3). In protein extracts from E. hellem-infected HFF cells, two major bands of 55 and 60 kDa in size were specifically recognized (Fig. 1D, lane 4), further suggesting that an swp1 or homologues also exist in E. hellem.
FIG. 1.
Immunolabeling in IFA and Western blotting with the monoclonal antibody 1E4. In IFA, MAb 1E4 reacts with the E. intestinalis spore wall (A). A cross-reaction is observed with the spore wall of both E. cuniculi (B) and E. hellem, strain EhD (C). (D) In the Western blot, a 40-kDa band, corresponding to the expected size of EiSWP1, is detected from E. intestinalis-infected HFF cells (lane 2). For E. cuniculi, a major band at 50 kDa (expected size for EcSWP1) is labeled (lane 3). The two bands at 40 and 42 kDa probably correspond to degradation products. For E. hellem, two major bands at 55 and 60 kDa are revealed (lane 4). Arrows indicate the major recognized proteins. Proteins were extracted from healthy and infected HFF cells in Laemmli buffer containing 2.5% SDS and 100 mM DTT and analyzed by 10% SDS-PAGE. Lane 1, healthy HFF cells; lanes 2, 3, and 4, HFF cells infected by E. intestinalis, E. cuniculi, and E. hellem (EhD), respectively. MAb 1E4 was diluted at 1:100 in IFA and 1:1,000 in Western blot. Bars, 1 μm.
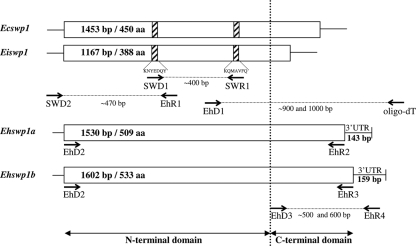
The strategy used for identifying the Ehswp1 gene(s) was based on different steps of PCR amplifications (Fig. 2). Two conserved peptides (KNYEDQY) and (KQMAVFQ) identified from the alignment between E. cuniculi and E. intestinalis SWP1s were first chosen to design degenerate oligonucleotide primers (see Fig. S1 in the supplemental material) that were used to amplify a unique ca. 400-bp-long product specific to E. hellem (Fig. 2). Amplification of a 470-bp DNA fragment corresponding to the sequence lacking the 5′ end was then performed using a primer (SWD2) designed in a conserved region deduced from the alignment of the 5′ regions downstream of the translation initiation codon in both Ecswp1 and Eiswp1. Because no conserved sequence was observed by comparing the 3′ regions upstream of the stop codon in swp1 genes, the sequence lacking the 3′ region was successfully amplified by RT-PCR using an oligo(dT) primer and RNA extracted from E. hellem-infected HFF cells. Surprisingly, two RT-PCR products of approximately 0.9 and 1 kbp in size were obtained. Sequencing of these products revealed that they may correspond to two divergent swp1 genes, with the amino acid alignment of their translated coding regions showing 71% of identity. The analysis of these sequences also showed A/T-rich 3′-untranslated regions (3′-UTRs) that are 143 and 159 bp long, respectively, and poly(A) tails beginning 8 nucleotides (nt) downstream of a putative polyadenylation signal (AATAAA). The whole encoding genes were then amplified from genomic DNA using a common primer designed at the ATG initiation codon (EhD2) and two primers determined at the divergent stop codon (EhR2 and EhR3). Sequencing confirmed the presence of two distinct open reading frames 1,530 bp and 1,602 bp in length (Fig. 2). We propose the designations Ehswp1a and Ehswp1b for these genes and EhSWP1a and EhSWP1b for the encoded proteins. The sequences of these two genes were submitted to the GenBank databases under accession no. FJ870923 and FJ870924.
FIG. 2.
PCR strategy used for the amplification of the genes encoding EhSWP1a and EhSWP1b. The first-round PCR was performed using degenerated primers SWD1 and SWR1 designed from conserved amino acid regions (hatched parts) in SWP1s of E. cuniculi (EcSWP1) and E. intestinalis (EiSWP1). The 5′-end region of swp1 genes was completed using SWD2 and EhR1. The 3′-end regions, including the 3′-untranslated regions (UTR), were then determined by RT-PCR with an oligo(dT) primer. After reverse transcription, the PCRs were performed using the primer corresponding to the adapter sequence linked to oligo(dT) and the forward primer EhD1 designed from the previously amplified 400-bp DNA sequence. The full open reading frames (ORFs) were finally validated through amplification with EhD2 and either EhR2 for Ehswp1a or EhR3 for Ehswp1b. The primer pair EhD3-EhR4 was used to analyze the polymorphism of the C-terminal part of SWP1s in four E. hellem isolates. EhD3 was designed from a conserved amino acid region between EhSWP1a and EhSWP1b (TTASGD) located upstream of their divergent C terminus. EhR4 was designed in a conserved part of the 3′-UTR of both EhSWP1a and EhSWP1b.
Properties of the primary sequences of EhSWP1a and EhSWP1b.
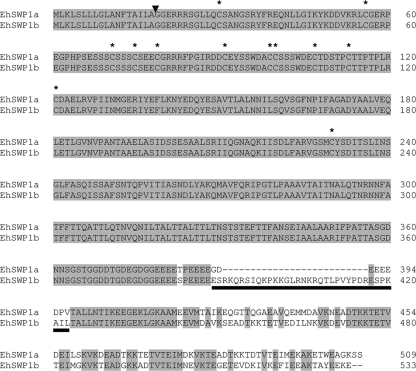
Ehswp1a and Ehswp1b, respectively, encode proteins of 509 and 533 aa. The N-terminal region of these proteins shows characteristics of a secretion signal peptide, as expected for spore wall proteins (Fig. 3). The predicted cleavage site of this signal peptide, between A18 and G19, would produce mature proteins with predicted molecular masses of 53,016 Da and 56,023 Da for EhSWP1a and EhSWP1b, respectively. Both proteins have acidic pIs (4.6 and 4.9, respectively) close to those of E. cuniculi and E. intestinalis SWPs and contain three major amino acids: threonine, glutamate, and alanine (Table 1). Although as a whole, 84.9% identity is found between EhSWP1a and EhSWP1b protein sequences, two domains can be distinguished: (i), a totally conserved region extending over the N-terminal 382 aa (100% of identity in amino acid and nucleotide sequences) and (ii) divergent C-terminal domains rich in charged residues (43.3 to 45%) of 127 aa in EhSWP1a and 151 aa in EhSWP1b, with only 54.7% identity (Fig. 3). Twelve cysteine residues are present at conserved positions in the N-terminal region of both sequences. The transition between the conserved N-terminal domain and the divergent C-terminal tail takes place between two tetraglutamate repeats. One main feature distinguishing the C-terminal tail of EhSWP1a and EhSWP1b is a 26-aa insertion/deletion located at position 390 (Fig. 3).
FIG. 3.
Amino acid sequence alignment of EhSWP1a and EhSWP1b. Identical residues are shaded. The arrow indicates the predicted cleavage site of the signal peptide. The 12 conserved cysteine residues are indicated by asterisks. The underlined sequence, including the insertion/deletion region of a 26-aa peptide, corresponds to the region that was selected for the expression of a recombinant polypeptide specific for EhSWP1b in Escherichia coli. Amino acids are numbered on the right. Eh, E. hellem.
TABLE 1.
Major characteristics of SWP1 proteins from E. cuniculi, E. intestinalis, and E. hellema
| Protein | Length (aa) |
pIc | % amino acid content shownc |
No. of cysteine residuesc | No. of potential O-glycosylation sitesc | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Precursor | Mature proteinb | S | G | A | T | E | ||||
| EcSWP1 | 450 | 432 | 5.2 | 18.9 | 14.1 | 6.5 | 7.6 | 3.9 | 11 | 41 |
| EiSWP1 | 388 | 370 | 5 | 14 | 6.2 | 8.6 | 8.9 | 5.6 | 10 | 19 |
| EiSWP2 | 1,002 | 984 | 4 | 5 | 17.2 | 3 | 7.8 | 25 | 10 | 3 |
| EhSWP1a | 509 | 491 | 4.6 | 8.9 | 6.3 | 10.1 | 11 | 10.6 | 12 | 2 |
| EhSWP1b | 533 | 515 | 4.9 | 8.9 | 6.0 | 9.3 | 9.9 | 10.3 | 12 | 1 |
Data for EcSWP1, EiSWP1, and EiSWP2 have been described previously (5, 15). The predicted amino acid sequences for EcSWP1, EiSWP1, and EiSWP2 were obtained from accession no. CAD25337, AAL27283, and AAL27282, respectively.
The mature proteins correspond to polypeptides devoid of predicted N-terminal signal peptide.
The pI, amino acid percentages, and number of O-glycosylation potential sites are deduced from the mature proteins.
When comparing the protein sequences of the SWP1 family members in the three Encephalitozoon species, two distinct regions can be found: a conserved N-terminal domain of 352 to 362 aa in length, with more than 60% of identity between EcSWP1, EiSWP1, EiSWP2, EhSWP1a, and EhSWP1b and a C-terminal tail that is highly variable both in length and in amino acid composition. In E. hellem, this region is 149 aa long in EhSWP1a and 173 aa long in EhSWP1b and mainly consists of degenerated repeats rich in three residues: glutamate, lysine and threonine (Fig. 3; see Fig. S1 in the supplemental material). A glutamate-rich C-terminal region is also found in EiSWP2 (15), whereas that of EcSWP1 is represented by glycine/serine-rich repeats (5). Among the 12 cysteine residues found in the N-terminal conserved parts of E. hellem SWP1s, five are present at the same positions in both E. cuniculi and E. intestinalis SWP proteins, suggesting an important role of these residues in the establishment of disulfide bridges. In contrast to EcSWP1 and EiSWP1, which contain 41 and 19 O-glycosylation sites, respectively, only two potential O-glycosylation sites are predicted for EhSWP1a and only one is predicted for EhSWP1b (Table 1).
Antisera raised against recombinant SWP1s reacted with one or two DTT-soluble protein electrophoretic bands and specifically labeled the E. hellem cell surface by IFA.
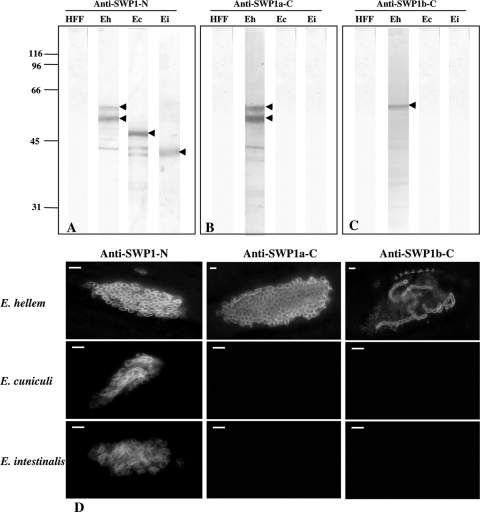
To assess EhSWP1a and EhSWP1b localization during the intracellular life cycle, we generated specific antibodies against three different recombinant proteins: one corresponding to the N-terminal conserved region (SWP1-N; aa 20 to 334), a second corresponding to the C-terminal part of EhSWP1a (SWP1a-C; aa 358 to 509), and a third covering the 26-aa insertion region specific to EhSWP1b (SWP1b-C; aa 389 to 423 [Fig. 3]). As expected, MAb 1E4, known to cross-react with the spore wall of the three Encephalitozoon species, recognized the recombinant protein SWP1-N (60% of identity is found between the five SWP1-like proteins identified in Encephalitozoon spp.; see Fig. S1 in the supplemental material) but not those corresponding to the C-terminal parts specific to E. hellem (not shown).
The three different sera were then used to detect EhSWP1a and EhSWP1b both in IFA and in whole protein extracts from E. hellem-infected cells separated by SDS-PAGE. As for MAb 1E4, anti-SWP1-N and anti-SWP1a-C antibodies consistently recognized two major protein bands (55 and 60 kDa in size) in cell lysates of E. hellem-infected HFF cells (Fig. 4A and B). These two bands were only detected when proteins were solubilized under reducing conditions, suggesting the presence of intra- and/or intermolecular disulfide bridges. In contrast, antisera produced against the specific part of EhSWP1b recognized the 60-kDa band, not the 55-kDa one (Fig. 4C). These results suggest that the 55-kDa migrating band corresponds to EhSWP1a, whereas the 60-kDa one corresponds to EhSWP1b. This is in agreement with the size predictions from the sequences: 53 kDa for EhSWP1a and 56 kDa for EhSWP1b. The fact that anti-SWP1a-C antisera also recognized EhSWP1b can be explained by the presence of conserved epitopes in the C-terminal tail of both proteins (see alignment in Fig. 3). The three antibodies were also applied to lysates of HFF cells infected with either E. cuniculi or E. intestinalis. As expected, the SWP1-N-specific antisera cross-reacted with SWP1s of both E. cuniculi (50 kDa) and E. intestinalis (40 kDa), while the antibodies specific to the C-terminal domains of EhSWP1a and EhSWP1b did not show any cross-reaction with E. cuniculi and E. intestinalis (Fig. 4B and C).
FIG. 4.
Immunodetection of SWP1-like proteins by Western blotting and IFA. (A to C) Western blot analyses were performed after SDS-PAGE on protein extracts from healthy human fibroblast cells (HFF) and HFF cells infected by E. cuniculi (Ec), E. intestinalis (Ei), and E. hellem (Eh) using antisera against SWP1-N (A), SWP1a-C (B), and SWP1b-C (C). Two major bands at 55 and 60 kDa (arrowheads) are detected for E. hellem with anti-SWP1-N (A) and anti-SWP1a-C (B), whereas sera raised against SWP1b-C only recognize the 60-kDa band (C; arrowhead). When using the anti-SWP1-N antibodies, cross-hybridizations are observed with an ∼50-kDa band for E. cuniculi and an ∼40-kDa band for E. intestinalis (A; arrowheads). (D) Indirect-immunofluorescence assay (IFA) with the three anti-SWP1 antisera. Antisera were applied to HFF cells 96 h postinfestation with E. hellem, E. cuniculi, or E. intestinalis. Cell surface labeling of most E. hellem intracellular stages is observed with anti-SWP1-N and anti-SWP1a-C, whereas only a few intracellular parasites are stained with anti-SWP1b-C. Antibodies specific to the N-terminal conserved part also recognize the cell surface of E. cuniculi and E. intestinalis. In contrast, sera raised against the C-terminal domains of E. hellem SWP1s do not cross-react with E. cuniculi and E. intestinalis. Mouse anti-SWP1 antisera were used at a 1:100 dilution. The secondary antibody was Alexa 488-conjugated goat anti-mouse IgG at a 1:1,000 dilution. Bars, 3 μm.
These results were further corroborated by immunofluorescence microscopy analyses of HFF cells infected with E. hellem, E. cuniculi, or E. intestinalis (Fig. 4D). A strong labeling associated with the cell surface of most E. hellem stages located inside the parasitophorous vacuole is observed using antisera directed against the conserved N-terminal domain of EhSWP1s. These antisera also react with the cell surface of extracellular mature spores (not shown). Similar results were obtained using antibodies raised against the C-terminal part of EhSWP1a. Interestingly, the antiserum directed against the EhSWP1b-specific epitope that includes the 26-aa insertion showed only a staining of the cell surface of some immature stages localized at the periphery of the parasitophorous vacuole. These results suggest that EhSWP1b may be only present in the first steps of the intracellular life cycle. Another explanation could be that the epitope corresponding to the 26-aa insertion is not accessible in most developmental stages including spores, as a result of some unknown conformational changes of EhSWP1b during spore wall formation. As expected, anti-SWP1-N antiserum cross-reacts with E. cuniculi and E. intestinalis, whereas antibodies produced against the C-terminal parts of EhSWP1a or EhSWP1b are highly specific for E. hellem.
Immunolabeling in electron microscopy confirms both proteins are present in most microsporidian differentiation stages and localize to the exospore of mature spores.
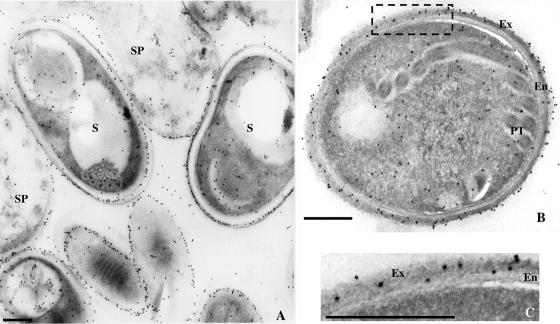
The microsporidian spore wall is composed by an inner layer called the endospore and an outer layer, the exospore. To better characterize the localization of both EhSWP1a and EhSWP1b within the spore wall, immunolabeling was done on ultrathin cryosections of E. hellem-infected HFF cells. The three antisera provided similar staining patterns, confirming the colocalization of EhSWP1a and EhSWP1b. Only the results obtained with the antibodies specific for the C-terminal part of EhSWP1b are presented in Fig. 5. The proteins seemed to be early expressed given that gold particles were found in meronts, the initial developmental stages of the microsporidian life cycle. In these stages that are devoid of cell wall, the detection of proteins in the cytoplasm probably reflects their transit through the endoplasmic reticulum (ER) and Golgi compartments (not shown). EhSWP1a and EhSWP1b are then targeted to the cell envelope in sporonts and sporoblasts that correspond to differentiation stages characterized by the spore wall biogenesis, including the endospore and the exospore (Fig. 5A). In immature and mature spores, the antibodies specifically decorated the external region of the cell wall designed as the exospore (Fig. 5B and C). Thus, immunolabeling data in TEM indicated that EhSWP1a and EhSWP1b are targeted to the exospore of the E. hellem spore wall and that both proteins have the same spatiotemporal localization, as suggested by IFA data.
FIG. 5.
Immunoelectron microscopy of different E. hellem developmental stages with antibodies raised against the C-terminal part of SWP1b. In sporonts (A) and sporoblasts, a strong labeling is observed at the periphery of the cells. In spores (A and B), gold particles are mainly associated with the electron-dense outer layer of the spore wall (i.e., the exospore). Panel C corresponds to a higher magnification of the exospore area (boxed in panel B). The antisera were used at a 1:20 dilution. The secondary antibody was goat anti-mouse IgG conjugated with 10-nm colloidal gold particles (Sigma). SP, sporont; S, spore; PT, polar tube; Ex, exospore; En, endospore. Bars, 200 nm.
Analysis of different E. hellem human isolates revealed an intraspecies variability of the C-terminal region for both EhSWP1a and EhSWP1b.
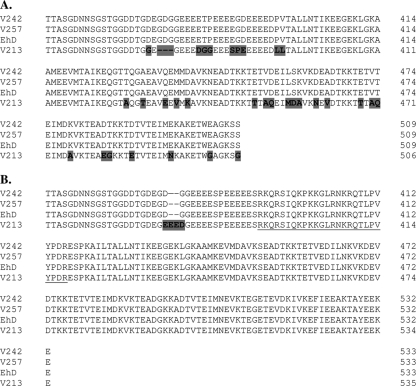
To determine whether the C-terminal end of EhSWP1a and EhSWP1b could be a good marker for genotyping studies, this region was analyzed in four E. hellem human isolates: V242, V257, V213, and EhD (used as a reference). Using primers EhD3 and EhR4, which encompass the variable K/E/T-rich C-terminal encoding region of both EhSWP1a and EhSWP1b (Fig. 2), two fragments of ∼500 and ∼600 bp were amplified from genomic DNA of each isolate and then cloned and sequenced. As previously described for the EhD isolate, the two fragments corresponded to Ehswp1a and Ehswp1b genes and the difference in size results from a 26-aa deletion/insertion encoding region. Alignment of protein sequences indicated 100% identity between isolates V242, V257, and EhD, for EhSWP1a as well as for EhSWP1b (Fig. 6), whereas some variations were found in isolate V213. As shown in Fig. 6B, the only difference in EhSWP1b between V213 and the three other isolates concerns a stretch of four residues. In contrast, some major differences were found in EhSWP1a: 32 residues were changed and a 3-residue stretch (GDG) was deleted in V213 compared to the three other isolates (Fig. 6A). In spite of this sequence polymorphism, no difference in SDS-PAGE and in Western blotting with anti-EhSWP1s antibodies could be detected between V213 and the three other isolates (not shown). In addition, amplification and sequencing of Ehswp1a and Ehswp1b whole sequences for the four isolates indicate that this polymorphism is only restricted to the C-terminal parts (not shown). Thus, the C-terminal end of SWP1 should be relevant for genotyping studies of E. hellem isolates.
FIG. 6.
Amino acid sequence alignment of the C-terminal domain of both EhSWP1a (A) and EhSWP1b (B) from four E. hellem human isolates. The sequence encoding the C-terminal region of EhSWP1a and EhSWP1b was amplified using primers EhD3 and EhR4 (Fig. 2), cloned in the pGEM-T Easy vector (Promega), and sequenced. Changes in amino acids for isolate V213 are shaded in gray. The 26 aa specific to EhSWP1b are underlined. Amino acids are numbered on the right.
Serological studies indicate that the C-terminal parts of EhSWP1a and EhSWP1b are not immunogenic.
Sera from rabbits and mice experimentally infected by E. hellem spores strongly reacted with the spore wall and the extruded polar tubes in IFA (not shown), showing that components of those structures are highly immunogenic as described previously (31). To evaluate the potential of recombinant SWP1s from E. hellem as diagnostic antigens for microsporidial infection, antisera from E. hellem-infected rabbit and mouse were tested against the three recombinant antigens produced in E. coli: SWP1-N, SWP1a-C, and SWP1b-C. Both antisera recognized the SWP1-N recombinant protein (see Fig. S2 in the supplemental material). In contrast, no reaction was obtained with the two recombinant antigens corresponding to the C-terminal domains specific to EhSWP1a and EhSWP1b, suggesting they are not immunogenic or not exposed to the immune system. We can thus conclude that the specific C-terminal parts of SWP1 proteins cannot be used as serological diagnostic tools to discriminate between Encephalitozoon species.
DISCUSSION
In the present study, using a combination of PCR and RT-PCR approaches, we identified in the human microsporidian Encephalitozoon hellem the whole sequences of two genes encoding closely related proteins that specifically localize to the exospore of mature spores. Both proteins belong to the SWP1 protein family, for which either one or two members have been previously identified in two other species of the Encephalitozoon genus: E. cuniculi and E. intestinalis (5, 15). All of these proteins are cysteine-rich antigens with a common N-terminal region and a highly divergent C-terminal end. Consistent with a translocation into the endoplasmic reticulum, the E. hellem SWP1 contains an N-terminal signal peptide that is highly conserved with those found in E. cuniculi and E. intestinalis SWP1/2 (see Fig. S1 in the supplemental material). The N-terminal domain of SWP1s is also characterized by the presence of cysteine residues in conserved positions between the three Encephalitozoon species. Consistent with the spore wall dissociation properties (soluble under reducing conditions), cysteine residues might be involved in inter- and/or intradisulfide bridges and potentially in EhSWP1a-EhSWP1b interaction. The existence of such interaction, already observed between EiSWP1 and EiSWP2 (15), still remains to be investigated.
No sequence homology has been found within public databases, and BLAST search failed to identify SWP1-like proteins in the genome sequence surveys of other Microsporidia, including the honeybee parasite Nosema ceranae (8), the grasshopper parasite Antonospora locustae (http://gmod.mbl.edu/perl/site/antonospora01), as well as the human pathogens Enterocytozoon bieneusi (2) and Anncaliia (ex Brachiola) algerae (personal data). These data suggest that SWP1-like proteins are specific to the Encephalitozoon genus or that orthologous proteins are highly divergent between nonphylogenetically related microsporidia. Such high divergence probably results from a rapid evolution of the sequence and has already been described for structural proteins such as polar tube proteins (28) and recently for other spore wall components (35). This evolution could suggest a role of these proteins in host cell interaction and consequently could reflect a host specificity. However, in contrast to highly divergent sequences, some spore wall components (e.g., N. bombycis NbSWP26 and EnP1) are highly conserved between distantly related microsporidia (20, 27).
The first immunological response developed by patients contaminated by E. cuniculi is targeted toward the spore wall component, showing that spore wall proteins are highly immunogenic and consequently useful for serodiagnostic (31). As shown here, the C-terminal-specific regions of SWP1 are not recognized by the host immune system despite their hydrophilic character, suggesting that SWP1 C-terminal ends are not exposed at the surface of the spore wall. In contrast, we showed the conserved N-terminal part is a good tool to detect Encephalitozoon genus infection. Because sera of humans infected by E. intestinalis and E. cuniculi recognize O mannosylation present on PTP1 (13, 25, 38), and because EiSWPs were shown to be glycosylated (15), the presence of this posttranslational modification on EhSWPs could be investigated to study their role in immune response, despite the reduced number of potential O-glycosylation sites.
Microsporidian cell wall protects the spore against environmental stresses and would maintain a high intrasporal hydrostatic pressure that should be of importance for polar tube extrusion and invasion. It has also been demonstrated that microsporidian spore adherence to the host cell surface via host cell sulfated glycosaminoglycans may be one initial event to mediate host cell invasion process (16). Southern et al. recently demonstrated that the spore wall protein EnP1 enables the spores to adhere to and to infect host cells in vitro (30). Further analyses are required to determine whether other components including SWP1 proteins can bind host cell receptors.
Reliable genotyping tools are needed to improve our knowledge about the transmission, prevalence, and clinical significance of microsporidian-infecting humans, including Encephalitozoon species. The species E. hellem is commonly found in birds and in immunocompromised patients, suggesting a zoonotic transmission (23). The gene encoding the major polar tube protein PTP1 can be useful for E. cuniculi and E. hellem genotyping, based on their different numbers of central repeats (14, 26, 36). Three genotypes can be distinguished for each strain, with some intragenotyping variations in the case of E. cuniculi (36). In the present study, we have shown that the EhSWP1 C-terminal end presents an important inter- and intraspecies polymorphism establishing a more relevant genotypic tool for epidemiological studies than the internal transcribed spacer (ITS) or small subunit ribosomal DNA (ssu-rDNA) sequences (22, 29). In the four E. hellem isolates studied, two genotypes have been characterized, confirming the PTP1 data: genotype 1A comprises EhD, V242, and V257, whereas V213 belongs to genotype 1B (36). These results are consistent with molecular karyotypes showing strong polymorphism between these two genotypes (3). The genotype diversity and mechanisms involved in polymorphism appearance are still enigmatic but need to be investigated as they might have a physiological role in adaptation to their hosts (14).
Future development of new genotyping and serodiagnostic tools and study of the molecular architecture of the microsporidian spore wall should then include (i) the identification of other SWP proteins, for example, by proteomic-based approaches (6, 33); and (ii) the understanding of the interactions between these components that are required for cell wall biogenesis and host-parasite interface. Thus, further improvements of our knowledge concerning the role of cell wall proteins in the invasion process will help to elucidate the events of host cell attachment and penetration.
Supplementary Material
Acknowledgments
V.P. was supported by a grant from “Ministère de l'Éducation, de la Recherche et de la Technologie.”
We thank M. Diogon and P. Pino for useful criticism of the manuscript and for helpful discussion.
Editor: J. F. Urban, Jr.
Footnotes
Published ahead of print on 15 March 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abreu-Acosta, N., J. Lorenzo-Morales, Y. Leal-Guio, N. Coronado-Alvarez, P. Foronda, J. Alcoba-Florez, F. Izquierdo, N. Batista-Diaz, C. Del Aguila, and B. Valladares. 2005. Enterocytozoon bieneusi (microsporidia) in clinical samples from immunocompetent individuals in Tenerife, Canary Islands, Spain. Trans. R. Soc. Trop. Med. Hyg. 99:848-855. [DOI] [PubMed] [Google Scholar]
- 2.Akiyoshi, D. E., H. G. Morrison, S. Lei, X. Feng, Q. Zhang, N. Corradi, H. Mayanja, J. K. Tumwine, P. J. Keeling, L. M. Weiss, and S. Tzipori. 2009. Genomic survey of the non-cultivatable opportunistic human pathogen, Enterocytozoon bieneusi. PLoS Pathog. 5:e1000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biderre, C., A. Mathis, P. Deplazes, R. Weber, G. Metenier, and C. P. Vivares. 1999. Molecular karyotype diversity in the microsporidian Encephalitozoon cuniculi. Parasitology 118:439-445. [DOI] [PubMed] [Google Scholar]
- 4.Bigliardi, E., and L. Sacchi. 2001. Cell biology and invasion of the microsporidia. Microbes Infect. 3:373-379. [DOI] [PubMed] [Google Scholar]
- 5.Bohne, W., D. J. Ferguson, K. Kohler, and U. Gross. 2000. Developmental expression of a tandemly repeated, glycine- and serine-rich spore wall protein in the microsporidian pathogen Encephalitozoon cuniculi. Infect. Immun. 68:2268-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosson, D., L. Kuhn, F. Delbac, J. Garin, C. P. Vivarès, and C. Texier. 2006. Proteomic analysis of the eukaryotic parasite Encephalitozoon cuniculi (microsporidia): a reference map for proteins expressed in late sporogonial stages. Proteomics 6:3625-3635. [DOI] [PubMed] [Google Scholar]
- 7.Brosson, D., L. Kuhn, G. Prensier, C. P. Vivares, and C. Texier. 2005. The putative chitin deacetylase of Encephalitozoon cuniculi: a surface protein implicated in microsporidian spore-wall formation. FEMS Microbiol. Lett. 247:81-90. [DOI] [PubMed] [Google Scholar]
- 8.Cornman, R. S., Y. P. Chen, M. C. Schatz, C. Street, Y. Zhao, B. Desany, M. Egholm, S. Hutchison, J. S. Pettis, W. I. Lipkin, and J. D. Evans. 2009. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog. 5:e1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delbac, F., P. Peyret, G. Metenier, D. David, A. Danchin, and C. P. Vivares. 1998. On proteins of the microsporidian invasive apparatus: complete sequence of a polar tube protein of Encephalitozoon cuniculi. Mol. Microbiol. 29:825-834. [DOI] [PubMed] [Google Scholar]
- 10.Didier, E. S., L. M. Weiss, A. Cali, and F. Marciano-Cabral. 2009. Overview of the presentations on microsporidia and free-living amebae at the 10th International Workshops on Opportunistic Protists. Eukaryot. Cell 8:441-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franzen, C. 2004. Microsporidia: how can they invade other cells? Trends Parasitol. 20:275-279. [DOI] [PubMed] [Google Scholar]
- 12.Frixione, E., L. Ruiz, J. Cerbon, and A. H. Undeen. 1997. Germination of Nosema algerae (Microspora) spores: conditional inhibition by D2O, ethanol and Hg2+ suggests dependence of water influx upon membrane hydration and specific transmembrane pathways. J. Eukaryot. Microbiol. 44:109-116. [DOI] [PubMed] [Google Scholar]
- 13.Furuya, K., M. Omura, S. Kudo, W. Sugiura, and H. Azuma. 2008. Recognition profiles of microsporidian Encephalitozoon cuniculi polar tube protein 1 with human immunoglobulin M antibodies. Parasite Immunol. 30:13-21. [DOI] [PubMed] [Google Scholar]
- 14.Haro, M., C. Del Aguila, S. Fenoy, and N. Henriques-Gil. 2003. Intraspecies genotype variability of the microsporidian parasite Encephalitozoon hellem. J. Clin. Microbiol. 41:4166-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayman, J. R., S. F. Hayes, J. Amon, and T. E. Nash. 2001. Developmental expression of two spore wall proteins during maturation of the microsporidian Encephalitozoon intestinalis. Infect. Immun. 69:7057-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayman, J. R., T. R. Southern, and T. E. Nash. 2005. Role of sulfated glycans in adherence of the microsporidian Encephalitozoon intestinalis to host cells in vitro. Infect. Immun. 73:841-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hibbett, D. S., M. Binder, J. F. Bischoff, M. Blackwell, P. F. Cannon, O. E. Eriksson, S. Huhndorf, T. James, P. M. Kirk, R. Lucking, H. Thorsten Lumbsch, F. Lutzoni, P. B. Matheny, D. J. McLaughlin, M. J. Powell, S. Redhead, C. L. Schoch, J. W. Spatafora, J. A. Stalpers, R. Vilgalys, M. C. Aime, A. Aptroot, R. Bauer, D. Begerow, G. L. Benny, L. A. Castlebury, P. W. Crous, Y. C. Dai, W. Gams, D. M. Geiser, G. W. Griffith, C. Gueidan, D. L. Hawksworth, G. Hestmark, K. Hosaka, R. A. Humber, K. D. Hyde, J. E. Ironside, U. Koljalg, C. P. Kurtzman, K. H. Larsson, R. Lichtwardt, J. Longcore, J. Miadlikowska, A. Miller, J. M. Moncalvo, S. Mozley-Standridge, F. Oberwinkler, E. Parmasto, V. Reeb, J. D. Rogers, C. Roux, L. Ryvarden, J. P. Sampaio, A. Schussler, J. Sugiyama, R. G. Thorn, L. Tibell, W. A. Untereiner, C. Walker, Z. Wang, A. Weir, M. Weiss, M. M. White, K. Winka, Y. J. Yao, and N. Zhang. 2007. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 111:509-547. [DOI] [PubMed] [Google Scholar]
- 18.Hollister, W. S., E. U. Canning, and N. I. Colbourn. 1993. A species of Encephalitozoon isolated from an AIDS patient: criteria for species differentiation. Folia Parasitol. (Praha) 40:293-295. [PubMed] [Google Scholar]
- 19.Katinka, M. D., S. Duprat, E. Cornillot, G. Metenier, F. Thomarat, G. Prensier, V. Barbe, E. Peyretaillade, P. Brottier, P. Wincker, F. Delbac, H. El Alaoui, P. Peyret, W. Saurin, M. Gouy, J. Weissenbach, and C. P. Vivares. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414:450-453. [DOI] [PubMed] [Google Scholar]
- 20.Li, Y., Z. Wu, G. Pan, W. He, R. Zhang, J. Hu, and Z. Zhou. 2009. Identification of a novel spore wall protein (SWP26) from microsporidia Nosema bombycis. Int. J. Parasitol. 39:391-398. [DOI] [PubMed] [Google Scholar]
- 21.Lujan, H. D., J. T. Conrad, C. G. Clark, M. C. Touz, F. Delbac, C. P. Vivares, and T. E. Nash. 1998. Detection of microsporidia spore-specific antigens by monoclonal antibodies. Hybridoma 17:237-243. [DOI] [PubMed] [Google Scholar]
- 22.Mathis, A., I. Tanner, R. Weber, and P. Deplazes. 1999. Genetic and phenotypic intraspecific variation in the microsporidian Encephalitozoon hellem. Int. J. Parasitol. 29:767-770. [DOI] [PubMed] [Google Scholar]
- 23.Mathis, A., R. Weber, and P. Deplazes. 2005. Zoonotic potential of the microsporidia. Clin. Microbiol. Rev. 18:423-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omura, M., K. Furuya, S. Kudo, W. Sugiura, and H. Azuma. 2007. Detecting immunoglobulin M antibodies against microsporidian Encephalitozoon cuniculi polar tubes in sera from healthy and human immunodeficiency virus-infected persons in Japan. Clin. Vaccine Immunol. 14:168-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peek, R., F. Delbac, D. Speijer, V. Polonais, S. Greve, E. Wentink-Bonnema, J. Ringrose, and T. van Gool. 2005. Carbohydrate moieties of microsporidian polar tube proteins are targeted by immunoglobulin G in immunocompetent individuals. Infect. Immun. 73:7906-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peuvel, I., F. Delbac, G. Metenier, P. Peyret, and C. P. Vivares. 2000. Polymorphism of the gene encoding a major polar tube protein PTP1 in two microsporidia of the genus Encephalitozoon. Parasitology 121:581-587. [DOI] [PubMed] [Google Scholar]
- 27.Peuvel-Fanget, I., V. Polonais, D. Brosson, C. Texier, L. Kuhn, P. Peyret, C. Vivares, and F. Delbac. 2006. EnP1 and EnP2, two proteins associated with the Encephalitozoon cuniculi endospore, the chitin-rich inner layer of the microsporidian spore wall. Int. J. Parasitol. 36:309-318. [DOI] [PubMed] [Google Scholar]
- 28.Polonais, V., G. Prensier, G. Metenier, C. P. Vivares, and F. Delbac. 2005. Microsporidian polar tube proteins: highly divergent but closely linked genes encode PTP1 and PTP2 in members of the evolutionarily distant Antonospora and Encephalitozoon groups. Fungal Genet. Biol. 42:791-803. [DOI] [PubMed] [Google Scholar]
- 29.Snowden, K. F., K. Logan, and D. N. Phalen. 2000. Isolation and characterization of an avian isolate of Encephalitozoon hellem. Parasitology 121:9-14. [DOI] [PubMed] [Google Scholar]
- 30.Southern, T. R., C. E. Jolly, M. E. Lester, and J. R. Hayman. 2007. EnP1, a microsporidian spore wall protein that enables spores to adhere to and infect host cells in vitro. Eukaryot. Cell 6:1354-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Gool, T., C. Biderre, F. Delbac, E. Wentink-Bonnema, R. Peek, and C. P. Vivares. 2004. Serodiagnostic studies in an immunocompetent individual infected with Encephalitozoon cuniculi. J. Infect. Dis. 189:2243-2249. [DOI] [PubMed] [Google Scholar]
- 32.Vossbrinck, C. R., J. V. Maddox, S. Friedman, B. A. Debrunner-Vossbrinck, and C. R. Woese. 1987. Ribosomal RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature 326:411-414. [DOI] [PubMed] [Google Scholar]
- 33.Wang, J. Y., C. Chambon, C. D. Lu, K. W. Huang, C. P. Vivares, and C. Texier. 2007. A proteomic-based approach for the characterization of some major structural proteins involved in host-parasite relationships from the silkworm parasite Nosema bombycis (Microsporidia). Proteomics 7:1461-1472. [DOI] [PubMed] [Google Scholar]
- 34.Wichro, E., D. Hoelzl, R. Krause, G. Bertha, F. Reinthaler, and C. Wenisch. 2005. Microsporidiosis in travel-associated chronic diarrhea in immune-competent patients. Am. J. Trop. Med. Hyg. 73:285-287. [PubMed] [Google Scholar]
- 35.Wu, Z., Y. Li, G. Pan, X. Tan, J. Hu, Z. Zhou, and Z. Xiang. 2008. Proteomic analysis of spore wall proteins and identification of two spore wall proteins from Nosema bombycis (Microsporidia). Proteomics 8:2447-2461. [DOI] [PubMed] [Google Scholar]
- 36.Xiao, L., L. Li, H. Moura, I. Sulaiman, A. A. Lal, S. Gatti, M. Scaglia, E. S. Didier, and G. S. Visvesvara. 2001. Genotyping Encephalitozoon hellem isolates by analysis of the polar tube protein gene. J. Clin. Microbiol. 39:2191-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, Y., P. Takvorian, A. Cali, F. Wang, H. Zhang, G. Orr, and L. M. Weiss. 2006. Identification of a new spore wall protein from Encephalitozoon cuniculi. Infect. Immun. 74:239-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, Y., P. M. Takvorian, A. Cali, G. Orr, and L. M. Weiss. 2004. Glycosylation of the major polar tube protein of Encephalitozoon hellem, a microsporidian parasite that infects humans. Infect. Immun. 72:6341-6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.