Abstract
Rickettsii rickettsii, the etiologic agent of Rocky Mountain spotted fever, replicates within the cytosol of infected cells and uses actin-based motility to spread inter- and intracellularly. Although the ultrastructure of the actin tail and host proteins associated with it are distinct from those of Listeria or Shigella, comparatively little is known regarding the rickettsial proteins involved in its organization. Here, we have used random transposon mutagenesis of R. rickettsii to generate a small-plaque mutant that is defective in actin-based motility and does not spread directly from cell to cell as is characteristic of spotted fever group rickettsiae. The transposon insertion site of this mutant strain was within Sca2, a member of a family of large autotransporter proteins. Sca2 exhibits several features suggestive of its apparent role in actin-based motility. It displays an N-terminal secretory signal peptide, a C-terminal predicted autotransporter domain, up to four predicted Wasp homology 2 (WH2) domains, and two proline-rich domains, one with similarity to eukaryotic formins. In a guinea pig model of infection, the Sca2 mutant did not elicit fever, suggesting that Sca2 and actin-based motility are virulence factors of spotted fever group rickettsiae.
Intracellular bacterial pathogens utilize various strategies to avoid detection and killing by the cells they infect. A fundamental strategy of intracellular survival involves the intracellular niche occupied by a pathogen. One approach is for bacteria to remain sequestered within a vacuole that dissociates from endosomal/lysosomal maturation pathways (20, 25, 31). An alternative strategy is for bacteria to escape from the endocytic vacuole in order to utilize the host cell cytosol as the site of replication. Listeria, Shigella, and Rickettsia are intracytoplasmic pathogens that not only replicate within the cytosol, but also exploit the host actin polymerization machinery to propel themselves into neighboring cells as a mechanism of cell-to-cell spread and immune evasion (14, 27).
Members of the genus Rickettsia are Gram-negative obligate intracellular bacteria transmitted to humans via arthropod vectors (5) and are the etiologic agents of a number of serious diseases in humans. Rickettsia spp. are historically divided into two groups, the spotted fever group and the typhus group (66). Rickettsia rickettsii, the causative agent of Rocky Mountain spotted fever, is the prototypic spotted fever group rickettsia and causes the most severe of spotted fever group rickettsioses. The spotted fever group of rickettsiae is comprised of over 20 closely related species that occur throughout the world. Many species are regionally important as disease agents, while others have never been associated with human disease (30, 43, 50, 63). Spotted fever group rickettsiae recruit actin at the surface of the bacteria to induce microfilament production and facilitate dissemination to adjacent cells (29, 33, 59). In contrast, the typhus group, which includes the etiologic agent of epidemic typhus, Rickettsia prowazekii, as well as Rickettsia typhi and Rickettsia canada, stimulate limited or no actin polymerization (33, 59).
Although Listeria, Shigella, and Rickettsia each use actin-based motility as a means of intra- and intercellular movement, the molecular mechanisms involved differ, suggesting that these diverse pathogens have independently acquired this capacity. Ultrastructurally, rickettsial actin tails are longer and are made up of linear actin filaments (29, 62). This is in contrast to Listeria monocytogenes and Shigella flexneri actin tails, which appear as highly cross-linked structures composed of multiple branched actin filaments (27, 29, 60). The branched actin structure of Listeria and Shigella actin tails is consistent with the presence of the host Arp2/3 complex, which is observed along the entirety of the Listeria and Shigella actin tails, but not rickettsial actin tails (21, 29, 65). The Arp 2/3 complex nucleates new actin filaments at a 70° angle from existing filaments (11, 46), and its absence from rickettsiae has been correlated with the unbranched filaments of rickettsial actin tails (29). A functional role for the Arp2/3 complex in the actin-based motility of Listeria and Shigella is demonstrated by the inhibition of their movement in cells expressing the verprolin, cofilin, and acidic (VCA) domain of neuronal-Wiscott-Aldrich syndrome protein (N-WASP) (32, 42), which acts as a dominant-negative inhibitor of the Arp2/3 complex (22, 45). Conversely, expression of the VCA domain had only a modest effect on the motility of R. rickettsii (32), suggesting that its actin-based motility may be independent of the Arp2/3 complex.
Most pathogens that parasitize the host actin nucleation machinery do so through the activation and recruitment of the host Arp2/3 complex (17). Arp2/3 requires activation by WASP family members to nucleate actin. In the case of L. monocytogenes, the protein ActA functionally mimics WASP family proteins in the recruitment and activation of Arp2/3 at one pole of the bacterial surface (34). S. flexneri expresses an autotransporter protein, IcsA, that recruits host WASP family proteins to activate the Arp2/3 complex (9, 38). Due to the limitations imposed by the obligate intracellular lifestyle of rickettsiae, identification of the bacterial gene products involved has been limited. The rickettsial protein RickA has been proposed as a molecule responsible for directing actin-based motility of Rickettsia, as it was found only in members of the spotted fever group yet was absent from the typhus group species (28). Indeed, RickA localizes to the surfaces of the bacteria, activates Arp2/3 complex proteins in vitro, and is sufficient to promote actin polymerization in vitro, as well as subsequent motility of RickA-coated beads in cell extracts (28, 34).
Here, we report the characterization of an R. rickettsii mutant generated via transposon mutagenesis that is unable to generate actin comet tails or spread directly from cell to cell, as does the congenic parental strain. Sca2 (surface cell antigen 2), a member of a family of large autotransporter proteins (10), was truncated by transposon insertion and appears to represent a rickettsial protein in addition to RickA that is necessary for the actin-based motility of rickettsiae. Importantly, in a guinea pig model, the Sca2 mutant does not induce fever as does the congenic wild-type strain, suggesting that Sca2 is a virulence determinant for spotted fever group rickettsiae.
MATERIALS AND METHODS
Bacteria and cell cultures.
R. rickettsii strain R with a clear-plaque morphology (R-clear) was propagated in Vero cells with Medium 199 (Invitrogen) plus 10% fetal bovine serum (HyClone Laboratories, Inc.) and purified by Renografin density gradient centrifugation as previously described (64).
Transposon mutagenesis.
R. rickettsii cells were transformed using the mariner-based Himar1 transposon system, as previously described (40), and directly plated onto Vero cell monolayers overlaid with 3 ml Medium 199 containing 0.05% agarose and 5% fetal bovine serum. After 24 h of incubation at 34°C, 2 ml of the overlay medium containing 400 ng/ml rifampin was added, and the infection was allowed to progress for 7 days at 34°C until visible plaques formed. Eight plaques were picked, recloned, and expanded by inoculation on fresh Vero cell monolayers incubated in the presence of rifampin.
Microscopy.
Vero cells grown on 12-mm coverslips in a 24-well plate were infected with R. rickettsii at a multiplicity of infection (MOI) of 5 for 30 min at room temperature. Fresh Medium 199 containing 2% fetal bovine serum (FBS) was added to each well, and the infection was allowed to progress for 24 h at 34°C. The monolayers were fixed in 3.7% paraformaldehyde, 25 mM NaPO4, 150 mM NaCl for 5 min and then washed in phosphate-buffered saline (25 mM NaPO4, 150 mM NaCl, pH 7.4) (PBS). The cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min and rinsed once with PBS. The rickettsiae were labeled with an anti-OmpB monoclonal antibody, 13-2 (1), and visualized with an anti-mouse immunoglobulin-Alexa Fluor 488 secondary antibody. F-actin was labeled with Alexa Fluor 598-phalloidin (2 U/ml). Coverslips were washed three times with PBS and once with distilled H2O (dH2O) and mounted with DakoCytomation Aqueous Mounting Medium (DakoCytomation). Images were acquired on a Nikon Microphot-FXA microscope with a 60× 1.4-numerical-aperture (NA) oil immersion objective (Nikon) and a Photometrics CoolSnap HQ camera.
Growth curves.
R. rickettsii R and the transposon mutants 9-7 and 9-3 were inoculated onto Vero cell monolayers grown in T25 flasks at an MOI of 0.05 bacteria/cell. Monolayers were harvested from a single flask daily for 8 days for plaquing. At the designated time, the monolayers were scraped into the medium and centrifuged at 12,000 rpm. The pellets were resuspended in 1 ml K36 buffer (0.1 M KCl, 0.015 M NaCl, 0.05 M K2HPO4, 0.05 M KH2PO4, pH 7.0) and lysed by disruption twice for 5 s each time with 1-mm glass beads using a Mini-Beadbeater (BioSpec, Inc.). The cell suspensions were frozen at −80°C until plaque assays were performed. The plaque assays were performed as previously described (16).
Genomic-DNA purification.
R. rickettsii R and the transposon mutants 9-7 and 9-3 were purified by Renografin density gradient and resuspended in 450 μl TE (50 mM Tris, 50 mM EDTA, pH 8.0). The suspension was then brought to 1% sodium dodecyl sulfate (SDS), 10 mM dithiothreitol, and 400 μg/ml RNase A (Invitrogen) for 10 min to achieve lysis, and proteinase K was added to 0.1 mg/ml and incubated at 60°C for 2 h. Proteinase K was supplemented at 20-min intervals. DNA was extracted three times with 1 volume of phenol-chloroform-isoamyl alcohol (25:24:1; Roche Applied Science), followed by two extractions with 1 volume of chloroform-isoamyl alcohol (24:1). The final aqueous phase was removed, and the DNA was precipitated with 0.1 volume of 5 M ammonium acetate plus 0.6 volume of isopropanol. The pellet was washed with 70% ethanol, dried, and resuspended in nuclease-free water to a concentration of 500 ng/μl.
Sequencing of transposon insertion sites.
The eight rifampin-resistant transformants were confirmed to display green fluorescent protein (GFPuv) expression by fluorescence microscopy, and the presence of the gfp gene was confirmed by PCR using the primer set GFPF (5′-GGTTGTCTGGTAAAAGGACAGGGCC-3′) and GFPR (5′-GCACAAATTTTCTGTCAGTGGAGAG-3′). All clones were positive for gfp. DNA was extracted from each strain as described above. Direct sequencing with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) was performed using 2 μg DNA and 2 μl BigDye Terminator Ready Reaction Mix. Primers complementary to regions within the transposon and directed outward into the rickettsial chromosome were designed (PMW3202, 5′-ATCCCCGGCAAGTTCATCCT-3′, and PMW5790, 5′-TAAAAGCAAGTATGTTCCTA-3′).
Southern blotting.
Genomic DNA was digested to completion at 37°C with HindIII (New England Biolabs), resolved by agarose gel electrophoresis, and stained with ethidium bromide. The migration of the DNA standards was recorded. The DNA was transferred onto a SensiBlot Plus nylon membrane (Fermentas Life Sciences, Canada) by capillary action with 20× SSC (3 M NaCl, 0.3 M sodium citrate, pH 7.0), after which the membrane was cross-linked by UV (302-nm) irradiation. The 524-bp gfp-specific probe was generated by PCR using primers GFPF and GFPR with the plasmid pMW1650 as a template. The PCR products were purified using QiaQuick columns (Qiagen) and labeled with [32P]dCTP by using Ready-to-Go DNA-labeling beads (GE Healthcare) according to the manufacturer's instructions. The nylon membrane was moistened with prehybridization buffer (1% SDS, 10% dextran sulfate, and 1 M NaCl) prior to the addition of a denatured gfp fragment probe (7 × 106 cpm) and salmon sperm DNA (0.1 mg/ml) (Invitrogen). Hybridization was performed overnight at 60°C in a hybridization oven with constant rotation. The membranes were washed twice in 2× SSC at room temperature for 5 min each time, twice in 2× SSC-1% SDS at 73°C for 30 min each time, and twice in 0.1× SSC at room temperature for 30 min each time. The membranes were air dried and exposed to CL-X Posure film (Thermo Scientific) at −80°C for 2 h.
Guinea pig infection.
Six-week-old female Hartley guinea pigs were inoculated intradermally with 100 PFU of each of the following: R. rickettsii strain R, R. rickettsii strain R mutant 9-7 (Sca2), R. rickettsii strain R mutant 9-3, formaldehyde-fixed R. rickettsii strain R, or an equivalent volume of K36 buffer alone. The remainder of the inoculum was plated immediately afterward to confirm the PFU. Temperatures were monitored rectally for 14 days beginning the day of infection. Blood was drawn from the guinea pigs 30 days postinfection, and antibody titers were determined by microimmunofluorescence.
RESULTS
Transposon mutagenesis of R. rickettsii.
The mariner-based transposon mutagenesis system developed for Rickettsia prowazekii (40) was used to generate a collection of R. rickettsii mutants. Eight clones were plaque purified and expanded for further characterization. Southern blot analysis revealed only a single predominant band in each of the mutants, demonstrating that transposition had occurred at a single site within the genome of each of the transformed strains (Fig. 1A). Direct sequencing of purified genomic DNA from each of the clones using primers from within the transposon delineated the precise insertion sites (Table 1). Transposition sites were randomly distributed throughout the rickettsial genome (Fig. 1B). Three of the clones contained transposon insertions in intergenic regions, while the remaining five clones contained a single insertion in the open reading frames specified in Table 1.
FIG. 1.
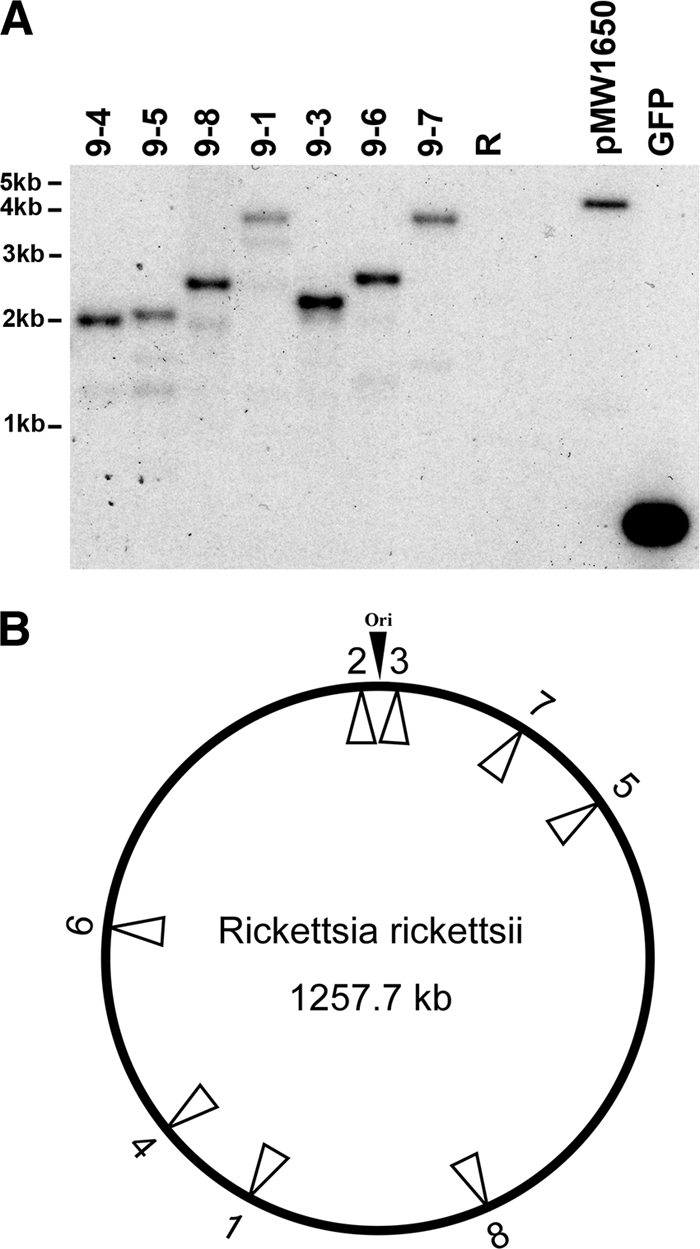
Transposon mutagenesis of R. rickettsii resulted in the generation of 8 unique clones. (A) Southern blot demonstrating a single transposition event that occurred in the generation of each mutant. Purified genomic DNA from mutant clones 9-1 through 9-8 and the congenic parental R. rickettsii strain (R) was digested with HindIII and subjected to Southern blotting with a 32P-labeled gfp-specific probe (GFP). Positive controls included pMW1650, as well as gfp-specific DNA used as the probe. Clone 9-2 did not appear to grow well and was not included in the Southern blot because of limited amounts of DNA recovered. (B) Diagram of the R. rickettsii chromosome depicting the relative locations of the insertion sites for each of the 8 mutants generated by transposon mutagenesis.
TABLE 1.
Precise transposon insertion site for each mutant
| Insertion site | Genome position | Sequence | NCBI protein ID | Function |
|---|---|---|---|---|
| 1 | 726453 | GTATTCTCTACAAGTAAA | ABV76354 | Hypothetical |
| 2 | 1246106 | TGTGTTATTATTTCATGA | Intergenic | |
| 3 | 14997 | ATAGTTCATAATTATAAC | Intergenic | |
| 4 | 807706 | TGACTCCATATTATCCTG | ABV76455 | Hypothetical |
| 5 | 191670 | TCAAATGCTATTTAATGA | Intergenic | |
| 6 | 966285 | GGAAGACGTAGTCTTGAC | ABV76633 | Hypothetical DNase |
| 7 | 112693 | AAACTTGATAAATCTGTT | ABV75720 | 190-kDa antigen precursor Sca2 |
| AAT79547.1 | ||||
| 8 | 546287 | CCGTCCTATAATAATACT | ABV76160 | Hypothetical RecB |
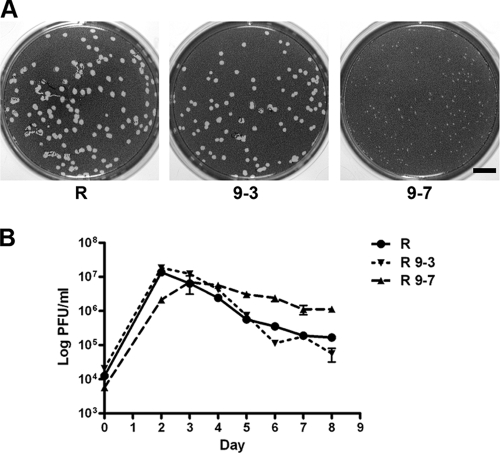
Small-plaque phenotype of the Sca2 mutant.
The clone in which Sca2 was disrupted (clone 9-7) displayed a unique plaque morphology in which the plaques were visibly smaller than those of the parental strain (Fig. 2A). To rule out the possibility that slower growth of the Sca2 mutant might be responsible for the small-plaque phenotype, growth rates were determined in Vero cell monolayers infected at an MOI of 0.05 with clones 9-7 and 9-3 and the parental R strain. One flask per group was harvested daily for 7 days, and rickettsiae were replated on fresh monolayers for quantitation of progeny PFU. The transposon mutants and wild-type rickettsiae replicated at equivalent rates, demonstrating that differences in intracellular multiplication cannot account for the differences in plaque size (Fig. 2B).
FIG. 2.
The Sca2 mutant displays a small-plaque phenotype but replicates at wild-type levels within Vero cells. (A) Plaques of the R. rickettsii parent strain R and mutants 9-3 and 9-7. Scale bar = 5 mm. (B) Growth curves of the R. rickettsii parent strain R and mutants 9-3 and 9-7 in Vero cells. PFU were enumerated in triplicate. The error bars represent standard errors of the mean.
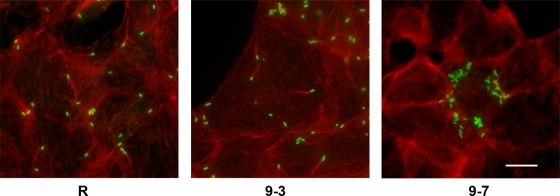
Sca2 mutant bacteria do not generate actin comet tails.
Because plaque size is correlated with the capacity for bacteria to spread to adjacent cells (9, 35, 49, 57), we used phalloidin staining of infected Vero cell monolayers to investigate whether the Sca2 mutant was able to generate actin comet tails. As has previously been shown (33), long actin tails were readily detected emanating from a single pole of wild-type rickettsiae (Fig. 3). In contrast, actin comet tails were not observed in cells infected with Sca2 mutant bacteria (clone 9-7), nor was actin visibly recruited to the surface of the microbe (Fig. 3). Clone 9-3 behaved like the parent rickettsiae. Notably, the Sca2 mutant did not appear to spread readily to adjacent cells, as did the parent rickettsiae or clone 9-3. The tendency to remain in an infected cell and replicate without spread to neighboring cells is characteristic of R. prowazekii (68), which lacks actin-based motility (33). Altogether, the data suggest that Sca2 is necessary for rickettsial actin-based motility and cell-to-cell spread.
FIG. 3.
The Sca2 mutant does not produce actin comet tails. Vero cells were infected with the parental R. rickettsii strain R, mutant strain 9-3, or Sca2 mutant 9-7. After 24 h, the cultures were fixed and stained for rickettsiae (green) and actin (red). Scale bar = 10 μm.
Sca2 is a rickettsial virulence factor.
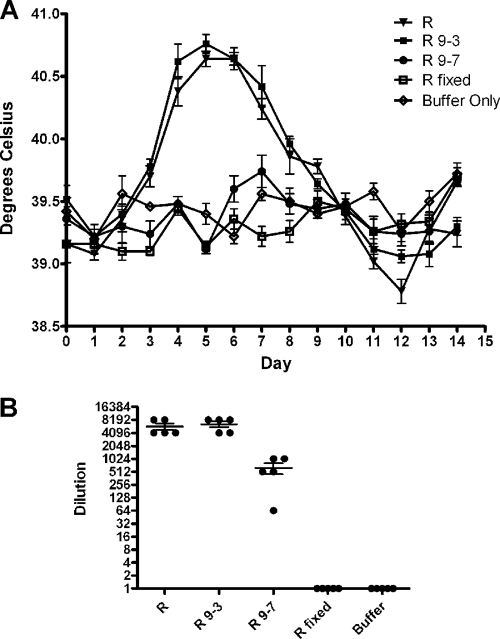
In animal models of virulence, Listeria ActA and Shigella IcsA have been shown to be important virulence determinants (9, 12, 36). Guinea pigs have been used as a model system for rickettsial pathogenesis, with fever induction providing the metric for virulence (2, 4, 8). In order to establish whether Sca2 is a rickettsial virulence factor, guinea pigs were inoculated intradermally with 100 PFU of each of the following strains: R. rickettsii strain R, R. rickettsii Sca2 mutant (clone 9-7), and R. rickettsii mutant (clone 9-3). Negative controls consisted of an equivalent number of formalin-fixed R. rickettsii cells, as well as animals sham-inoculated with K36 buffer. Rectal temperatures were monitored just prior to infection and every 24 h thereafter for 2 weeks. Wild-type R. rickettsii strain R induced a fever of >40°C by 2 days postinfection that peaked on day 5 (Fig. 4A). In contrast, infection with the Sca2 mutant did not result in fever induction. The Sca2 mutant, however, apparently replicated sufficiently to cause seroconversion in the guinea pigs (Fig. 4B). Animals inoculated with formalin-fixed rickettsiae, which do not replicate, did not show evidence of fever or seroconversion. Guinea pigs infected with the 9-3 mutant developed fevers with a peak temperature and duration similar to those seen in the infection with the wild-type strain. Thus, Sca2 is a virulence factor for R. rickettsii.
FIG. 4.
The Sca2 mutant displays reduced virulence in a guinea pig model. (A) Five guinea pigs were inoculated with either the congenic parental R. rickettsii strain (R), the mutant strain 9-3 (R 9-3), the Sca2 mutant 9-7 (R 9-7), or formalin-fixed R. rickettsii (R fixed) or were sham infected with K36 buffer alone (Buffer Only). Rectal temperatures were taken daily for 14 days beginning the day of inoculation. The error bars represent standard errors of the mean from 5 individual animals. (B) Sera collected at 30 days postinfection were used to determine anti-R. rickettsii titers by microimmunofluorescence. Bars indicate the means ± standard errors of the means.
The Sca2 amino acid sequence contains putative WH2 and formin homology domains.
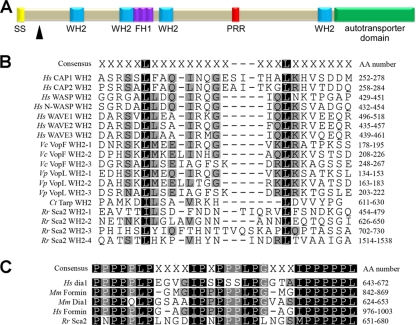
Sca2 was originally identified during analysis of nine completed rickettsial genomes as a member of a family of autotransporter proteins (10). It contains an N-terminal Sec-dependent signal sequence for protein translocation into the periplasmic space, along with the appropriate signal sequence cleavage site. Like all members of this family, it also bears a C-terminal autotransporter domain predicted to form a beta barrel in the outer membrane, facilitating translocation of the remainder of the protein to the outside of the bacterium (Fig. 5). In a search for amino acid sequences with similarity to the domains found in actin binding proteins, overlapping segments of the Sca2 protein sequence were aligned with known eukaryotic actin binding domains using ClustalV alignment. Four putative Wasp homology 2 (WH2) domains were identified within Sca2. Three were within the N-terminal half and the fourth just upstream of the autotransporter domain. A ClustalW alignment of the four putative rickettsial WH2 domains, as well as the WH2 domains from human CAP, WASP, and WAVE proteins; Vibrio VopF and VopL; and Chlamydia trachomatis Tarp, are shown in Fig. 5B. The putative WH2 domains from Sca2 maintain the characteristics needed for actin binding, including the hydrophobic residues found in the first half of the domain and the invariant leucine found in the LKKT motif in the second half of the region. Moreover, STRAP analysis (26) predicted coiled-coil-forming segments in the N-terminal portion of the first 3 rickettsial WH2 domains, a characteristic found in other, well-characterized WH2 domain-containing proteins (18). In addition, there are two proline-rich motifs within Sca2. One region consists of 8 consecutive proline residues in the C-terminal half of the protein, while the other region, found between the first putative WH2 domains, shares sequence homology with the formin homology 1-like (FH1) domain of formins, another class of eukaryotic proteins capable of nucleating actin (23, 52) (Fig. 5C). These predicted structures are consistent with a role for Sca2 in rickettsia-mediated actin dynamics. Sca2 in R. rickettsii 9-7 was disrupted at amino acid 95 of the predicted 1,873 amino acids of the intact protein and thus is lacking all of the features predicted to be involved in actin dynamics.
FIG. 5.
Domain organization and sequence alignments of Sca2 WH2 and FH1-like domains. (A) Graphical illustration representing the domain organization of Sca2, a 1,821-amino-acid (aa) protein with a Sec-dependent secretion signal (SS) (aa 1 to 32), 4 putative WH2 domains (WH2-1, aa 454 to 479; WH2-2, aa 626 to 650; WH2-3, aa 702 to 730; and WH2-4, aa 1514 to 1538), an FH1 domain (aa 650 to 680), a proline-rich region (PRR) (aa 1057 to 1066), and an autotransporter domain (aa 1542 to 1821). The black arrowhead indicates the transposon insertion site in clone 9-7. (B) ClustalW amino acid sequence alignment of the four WH2 domains of Sca2 (R. rickettsii YP_001494228) and WH2-domain containing proteins from Homo sapiens, CAP1 (Q01518), CAP2 (P40123), WASP (P42768), N-WASP (O00401), WAVE1 (Q92558), WAVE2 (Q9Y6W5), and WAVE3 (Q9UPY6); from V. cholerae, VopF (AAZ32252); from V. parahaemolyticus, VopL (NP_800881); and from C. trachomatis, Tarp (YP_328278). (C) ClustalW amino acid sequence alignment of the Sca2 FH1-like domain and the portion of FH1 domain-containing proteins obtained through a BLASTp search. The accession numbers for proteins from humans are hDiap1, NP_001073280.1, and hFormin2, CAQ10135.1, and from mouse they are mDia1, AAH21396.1, and mFormin2, EDL13214.1. The consensus sequence was identified based upon a minimum of 75% amino acid identity at that position. Black shading indicates 100% similarity, and gray shading indicates >60% similarity.
Conservation of Sca2 in spotted fever group rickettsiae.
A BLAST search of Sca2 orthologs in the genus Rickettsia identified over 18 species with full-length Sca2 proteins and others with ostensible gene fragments (Table 2). These included the following members of the spotted fever group of rickettsiae: R. rickettsii, R. conorii, R. siberica, R. honei, R. japonica, R. parkeri, R. slovaca, R. massiliae, R. peacockii, R. montana, R. rhiphicephali, R. australis, R. akari, R. africae, R. aeschlimanni, and R. felis. An interesting observation is that R. peacockii encodes an intact Sca2 (24) but does not exhibit actin-based motility (6, 56). Interestingly, RickA of R. peacockii is disrupted by the presence of an insertion element, ISRpe1 (56). This observation suggests that actin-based motility may require the activities of at least two rickettsial proteins, RickA and Sca2, functioning in concert.
TABLE 2.
Presence or absence of Sca2 domains among various species of spotted fever group and typhus group rickettsiaea
| Group or species | SS | WH2-1 | WH2-2 | FH1 | WH2-3 | PRR | WH2-4 | ATD |
|---|---|---|---|---|---|---|---|---|
| Spotted fever groupb | + | + | + | + | + | + | + | + |
| R. honeiic | − | + | + | + | + | + | + | + |
| R. aeschlimannic | − | + | + | + | − | + | + | + |
| R. felisc | + | + | − | + | − | − | + | + |
| R. canadad | + | + | − | + | − | + | + | + |
| R. typhid | + | − | − | − | − | + | + | + |
| R. prowazekiid | − | − | − | − | − | − | + | + |
The presence of domains was determined by performing a BLASTp search using R. rickettsii Sca2 as the query, followed by amino acid sequence alignment in cobalt of the obtained matches. SS, Sec-dependent signal sequence; WH2, WH2 domain; PRR, proline-rich region; ATD, autotransporter domain. +, sequence retained; −, either the entire sequence is missing or the sequence maintained less than 50% identity to the consensus sequence.
Spotted fever group species that retained all identified domains: R. rickettsii strain Iowa (YP_001649468.1), R. rickettsii strain Sheila Smith (YP_001494228.1), R. conorii (NP_359747.1), R. siberica (ZP_00142412.1), R. japonica (AAT79540.1), R. parkeri (AAT79545.1), R. slovaca (AAT79549.1), R. massiliae (YP_001498970.1), R. peacockii (YP_002916397.1), R. montana (AAT79544.1), R. rhipacephali (AAT79546.1), R. australis (AAT79538.1), R. akari (YP_001492963.1), and R. africae (AAT79536.1).
Spotted fever group species that are missing 1 or more domains: R. honeii (AAT79535.1), R. aeschlimanni (AAT79542.1), and R. felis (YP_246083.1).
Typhus group Rickettsia species: R. canada (YP_001491816.1), R. typhi (YP_067021.1), and R. prowazekii (NP_220474.1).
Of the typhus group, R. prowazekii is predicted to encode only a truncated version of 341 amino acids encompassing the predicted fourth WH2 domain, as well as the autotransporter domain. R. prowazekii does not, however, encode the first three putative WH2 domains, the proline-rich domains, or secretory signals. Thus, R. prowazekii does not appear to express a functional Sca2 protein, which would be consistent with the lack of actin-based motility in the species. R. typhi is predicted to encode a protein of 1,483 amino acids whose highest degree of similarity overlaps the autotransporter domain and the fourth WH2 domain but also includes a region with limited similarity to the FH1 domain, as well as a secretory signal. This may explain the limited actin-based motility and short actin tails observed with R. typhi (29, 33, 59). R. canada has been reported not to exhibit actin-based motility (33) and has been described as encoding a split Sca2 (47, 48); however, the recently completed genome reportedly encodes an apparently intact Sca2 ortholog (YP_002916397), as well as a full-length RickA ortholog (YP_001492102).
DISCUSSION
Although intracellular movement of rickettsiae has been recognized for over half a century (37, 55, 67), the molecular mechanisms of actin-based motility have been better characterized in the more experimentally tractable facultative intracellular pathogens L. monocytogenes and S. flexneri (27, 29). These intracellular pathogens have provided important tools for the study of actin dynamics and the identification of cellular proteins regulating actin polymerization. Despite a more limited knowledge of bacterial factors involved in rickettsial actin-based motility, it is clear that distinct mechanisms are involved, based upon the unique ultrastructure of the actin tail itself, host accessory proteins associated with the tail, and the apparent lack of a requirement for the cellular actin nucleating Arp2/3 complex. Here, we have used transposon mutagenesis of R. rickettsii to identify an autotransporter protein (Sca2) that is required for actin-based motility. Compared to the parent strain or a transformant with a different insertion site, the Sca2 mutant displays reduced virulence in a guinea pig model of infection. Sca2 and actin-based motility, therefore, appear to be bone fide virulence determinants of spotted fever group rickettsiae.
As has been the case with other bacterial obligate intracellular parasites, genetic systems for manipulation of rickettsiae have been somewhat limited. The recent development of mariner-based transposon systems for rickettsiae has led to significant advances (19, 40, 53, 54). Challenges in efficiency of transformation and selection remain, but these systems have offered new promise for rickettsial research. Perhaps the most significant remaining obstacle is that, in the absence of growth on axenic media, disruption of genes essential for entry or other critical functions for intracellular growth and virulence would be lethal. The majority of the work in developing genetics for rickettsiae has been done with R. prowazekii (19, 40, 53, 54). Spotted fever group rickettsiae offer one advantage over the typhus group in that spotted fever rickettsiae readily form plaques while the typhus group does not (16, 44, 51); thus, direct plating and observation of plaque morphology are possible without the necessity of cloning by limiting dilution. In this manner, we were able to identify a plaque morphology consistent with impaired cell-to-cell spread and possible defects in actin-based motility.
Recent analysis of multiple rickettsial genomes of both the spotted fever and typhus groups has led to the identification of a large family of high-molecular-mass autotransporter proteins. From nine sequenced Rickettsia genomes, 17 members of this family were identified (10). Two members of the family, rickettsial OmpA (rOmpA) and rOmpB, are high-abundance antigenic proteins on the surfaces of rickettsiae and are targets of neutralizing antibody (3, 39). The functions of most members of the Sca family of autotransporters are unknown, but the proposed roles of rOmpA and rOmpB as adhesins (15, 39, 61) have led to the suggestion that many members of the Sca family may also be involved in the attachment process (10). Indeed, R. conorii Sca2 has recently been associated with adherence (13). While we did not formally measure the rates of attachment or entry of the Sca2 mutant described here, the growth rates and infectivity of Vero cells did not appear to be impaired. Effects on other cell types—arthropod cell lines, for example—were not investigated. Sca2, however, displays several features consistent with a role in actin dynamics. R. rickettsii Sca2 contains an N-terminal Sec-dependent translocation signal and cleavage site, as well as the predicted C-terminal autotransporter domain. Analysis of the Sca2 amino acid sequence revealed four putative WH2 domains, as well as 2 proline-rich regions, one of which bears similarity to the FH1 domain of formins. The presence of these domains is reminiscent of VopF and VopL proteins found in Vibrio cholerae and Vibrio parahaemolyticus, respectively (41, 58). VopF/VopL are substrates of the Vibrio type III secretion system and similarly contain FH1-like and WH2 domains, both of which are required for actin nucleation and polymerization. Indeed, recent studies have demonstrated actin nucleation activity of R. parkeri Sca2 in in vitro actin polymerization assays (C. Haglund and M. D. Welch, personal communication). The recognition of Sca2 as a protein essential for actin comet tail formation will provide new opportunities to study mechanisms of actin nucleation and/or cytoskeleton regulatory components in the rickettsial system.
RickA, which was initially identified bioinformatically based upon its absence from the genome of typhus group rickettsiae, activates the Arp2/3 complex in vitro and stimulated motility of RickA-coated beads in Xenopus extracts (28, 34); however, its role in the actin-based motility of rickettsiae has not yet been demonstrated on rickettsiae themselves. R. raoulti, a spotted fever group rickettsia that does not exhibit actin-based motility, expresses RickA and has led to a proposal that additional rickettsial proteins may be required (7). Conversely, R. peacockii expresses an apparently intact Sca2 ortholog, but RickA is disrupted, and R. peacockii does not exhibit actin-based motility (56). It is possible that RickA and Sca2 function together to promote actin-based motility. Genetic knockdown of RickA may confirm its role and aid in the identification of additional rickettsial proteins required for intracellular movement.
Actin comet tail formation has been shown to be an important virulence determinant for both Listeria and Shigella, as ActA and IcsA mutants are avirulent in their respective animal models (9, 12, 35). Because Sca2 is required for actin-based motility by R. rickettsii, it is perhaps not surprising that the Sca2 rickettsial mutant is unable to induce fever in a guinea pig model system. Sca2 and the capacity for actin-based motility are common features of most of the spotted fever group rickettsiae, although some members of the group have never been associated with human disease and do not cause fever in animal model systems (2). Clearly, multiple factors contribute to rickettsial virulence. The ongoing improvements in the ability to genetically modify rickettsiae provide an important new means to identify additional factors that contribute to rickettsial pathogenesis.
Acknowledgments
This work was supported by the Intramural Research Program of NIAID/NIH.
We thank D. O. Wood for providing plasmid pMW1650; M. Welch for sharing of preliminary data and advice on transformations; R. Heinzen, M. Welch, and C. Haglund for critical review of the manuscript; the RML Genomics Unit for DNA sequencing; and Anita Mora for graphic arts assistance.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 1 March 2010.
REFERENCES
- 1.Anacker, R. L., R. E. Mann, and C. Gonzales. 1987. Reactivity of monoclonal antibodies to Rickettsia rickettsii with spotted fever and typhus group rickettsiae. J. Clin. Microbiol. 25:167-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anacker, R. L., T. F. McCaul, W. Burgdorfer, and R. K. Gerloff. 1980. Properties of selected rickettsiae of the spotted fever group. Infect. Immun. 27:468-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anacker, R. L., G. A. McDonald, R. H. List, and R. E. Mann. 1987. Neutralizing activity of monoclonal antibodies to heat-sensitive and heat-resistant epitopes of Rickettsia rickettsii surface proteins. Infect. Immun. 55:825-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anacker, R. L., R. N. Philip, J. C. Williams, R. H. List, and R. E. Mann. 1984. Biochemical and immunochemical analysis of Rickettsia rickettsii strains of various degrees of virulence. Infect. Immun. 44:559-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azad, A. F., and C. B. Beard. 1998. Rickettsial pathogens and their arthropod vectors. Emerg. Infect. Dis. 4:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldridge, G. D., N. Y. Burkhardt, J. A. Simser, T. J. Kurtti, and U. G. Munderloh. 2004. Sequence and expression analysis of the ompA gene of Rickettsia peacockii, an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni. Appl. Environ. Microbiol. 70:6628-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balraj, P., K. El Karkouri, G. Vestris, L. Espinosa, D. Raoult, and P. Renesto. 2008. RickA expression is not sufficient to promote actin-based motility of Rickettsia raoultii. PLoS One 3:e2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell, E. J., G. M. Kohls, H. G. Stoenner, and D. B. Lackman. 1963. Nonpathogenic rickettsias related to the spotted fever group isolated from ticks, Dermacentor variabilis and Dermacentor andersoni from Eastern Montana. J. Immunol. 90:770-781. [PubMed] [Google Scholar]
- 9.Bernardini, M. L., J. Mounier, H. d'Hauteville, M. Coquis-Rondon, and P. J. Sansonetti. 1989. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. U. S. A. 86:3867-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanc, G., M. Ngwamidiba, H. Ogata, P. E. Fournier, J. M. Claverie, and D. Raoult. 2005. Molecular evolution of Rickettsia surface antigens: evidence of positive selection. Mol. Biol. Evol. 22:2073-2083. [DOI] [PubMed] [Google Scholar]
- 11.Blanchoin, L., K. J. Amann, H. N. Higgs, J. B. Marchand, D. A. Kaiser, and T. D. Pollard. 2000. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404:1007-1011. [DOI] [PubMed] [Google Scholar]
- 12.Brundage, R. A., G. A. Smith, A. Camilli, J. A. Theriot, and D. A. Portnoy. 1993. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 90:11890-11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardwell, M. M., and J. J. Martinez. 2009. The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infect. Immun. 77:5272-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlsson, F., and E. J. Brown. 2006. Actin-based motility of intracellular bacteria, and polarized surface distribution of the bacterial effector molecules. J. Cell. Physiol. 209:288-296. [DOI] [PubMed] [Google Scholar]
- 15.Chan, G. Y., M. M. Cardwell, T. M. Hermanas, R. Uchiyama, and J. J. Martinez. 2009. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell Microbiol. 11:629-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cory, J., C. E. Yunker, R. A. Ormsbee, M. Peacock, H. Meibos, and G. Tallent. 1974. Plaque assay of rickettsiae in a mammalian cell line. Appl. Microbiol. 27:1157-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cossart, P. 2000. Actin-based motility of pathogens: the Arp2/3 complex is a central player. Cell Microbiol. 2:195-205. [DOI] [PubMed] [Google Scholar]
- 18.Dominguez, R. 2004. Actin-binding proteins—a unifying hypothesis. Trends Biochem. Sci. 29:572-578. [DOI] [PubMed] [Google Scholar]
- 19.Driskell, L. O., X. J. Yu, L. Zhang, Y. Liu, V. L. Popov, D. H. Walker, A. M. Tucker, and D. O. Wood. 2009. Directed mutagenesis of the Rickettsia prowazekii pld gene encoding phospholipase D. Infect. Immun. 77:3244-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duclos, S., and M. Desjardins. 2000. Subversion of a young phagosome: the survival strategies of intracellular pathogens. Cell Microbiol. 2:365-377. [DOI] [PubMed] [Google Scholar]
- 21.Egile, C., T. P. Loisel, V. Laurent, R. Li, D. Pantaloni, P. J. Sansonetti, and M. Carlier. 1999. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J. Cell Biol. 146:1319-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliott, D. A., D. J. Coleman, M. A. Lane, R. C. May, L. M. Machesky, and D. P. Clark. 2001. Cryptosporidium parvum infection requires host cell actin polymerization. Infect. Immun. 69:5940-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evangelista, M., D. Pruyne, D. C. Amberg, C. Boone, and A. Bretscher. 2002. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4:32-41. [DOI] [PubMed] [Google Scholar]
- 24.Felsheim, R. F., T. J. Kurtti, and U. G. Munderloh. 2009. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. PLoS One 4:e8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-del Portillo, F., and B. B. Finlay. 1995. The varied lifestyles of intracellular pathogens within eukaryotic vacuolar compartments. Trends Microbiol. 3:373-380. [DOI] [PubMed] [Google Scholar]
- 26.Gille, C., and C. Frommel. 2001. STRAP: editor for structural alignments of proteins. Bioinformatics 17:377-378. [DOI] [PubMed] [Google Scholar]
- 27.Goldberg, M. A. 2001. Actin-based motility of intracellular microbial pathogens. Microbiol. Mol. Biol. Rev. 65:595-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gouin, E., C. Egile, P. Dehoux, V. Villiers, J. Adams, F. Gertler, R. Li, and P. Cossart. 2004. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature 427:457-461. [DOI] [PubMed] [Google Scholar]
- 29.Gouin, E., H. Gantelet, C. Egile, I. Lasa, H. Ohayon, V. Villiers, P. Gounon, P. J. Sansonetti, and P. Cossart. 1999. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J. Cell Sci. 112:1697-1708. [DOI] [PubMed] [Google Scholar]
- 30.Hackstadt, T. 1996. The biology of rickettsiae. Infect. Agents Dis. 5:127-143. [PubMed] [Google Scholar]
- 31.Hackstadt, T. 2000. Redirection of host vesicle trafficking pathways by intracellular parasites. Traffic 1:93-99. [DOI] [PubMed] [Google Scholar]
- 32.Harlander, R. S., M. Way, Q. Ren, D. Howe, S. S. Grieshaber, and R. A. Heinzen. 2003. Effects of ectopically expressed neuronal Wiskott-Aldrich syndrome protein domains on Rickettsia rickettsii actin-based motility. Infect. Immun. 71:1551-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinzen, R. A., S. F. Hayes, M. G. Peacock, and T. Hackstadt. 1993. Directional actin polymerization associated with spotted fever group rickettsia infection of VERO cells. Infect. Immun. 61:1926-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeng, R. L., E. D. Goley, J. A. D'Alessio, O. Y. Chaga, T. M. Svitkina, G. G. Borisy, R. A. Heinzen, and M. D. Welch. 2004. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 6:761-769. [DOI] [PubMed] [Google Scholar]
- 35.Kocks, C., E. Gouin, M. Tabouret, P. Berche, H. Ohayon, and P. Cossart. 1992. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68:521-531. [DOI] [PubMed] [Google Scholar]
- 36.Kocks, C., R. Hellio, P. Gounon, H. Ohayon, and P. Cossart. 1993. Polarized distribution of Listeria monocytogenes surface protein ActA at the site of directional actin assembly. J. Cell Sci. 105:699-710. [DOI] [PubMed] [Google Scholar]
- 37.Kokorin, I. N., E. D. Miskarova, O. S. Gudima, E. A. Kabanova, and C. D. Kiet. 1977. Intracellular development of Rickettsia. Zh. Mikrobiol. Epidemiol. Immunobiol. 1977:26-29. [PubMed] [Google Scholar]
- 38.Lett, M., C. Sasakawa, N. Okada, T. Sakai, S. Makino, M. Yamada, K. Komatsu, and M. Yoshikawa. 1989. virG, a plasmid-coded virulence gene of Shigella flexneri: identification of the virG protein and determination of the complete coding sequence. J. Bacteriol. 171:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, H., B. Lenz, and D. H. Walker. 1988. Protective monoclonal antibodies recognize heat-labile epitopes on surface proteins of spotted fever group rickettsiae. Infect. Immun. 56:2587-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, Z. M., A. M. Tucker, L. O. Driskell, and D. O. Wood. 2007. Mariner-based transposon mutagenesis of Rickettsia prowazekii. Appl. Environ. Microbiol. 73:6644-6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liverman, A. D. B., H. Cheng, J. E. Trosky, D. W. Leung, M. L. Yarbrough, D. L. Burdette, M. K. Rosen, and K. Orth. 2007. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc. Natl. Acad. Sci. U. S. A. 104:17117-17122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.May, R. C., M. E. Hall, H. N. Higgs, T. D. Pollard, T. Chakraborty, J. Wehland, L. M. Machesky, and A. S. Sechi. 1999. The Arp2/3 complex is essential for the actin-based motility of Listeria monocytogenes. Curr. Biol. 9:759-762. [DOI] [PubMed] [Google Scholar]
- 43.McDade, J. E., and V. F. Newhouse. 1986. Natural history of Rickettsia rickettsii. Annu. Rev. Microbiol. 40:287-309. [DOI] [PubMed] [Google Scholar]
- 44.McDade, J. E., J. R. Stakebake, and P. J. Gerone. 1969. Plaque assay system for several species of Rickettsia. J. Bacteriol. 99:910-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreau, V., F. Frischknecht, I. Reckmann, R. Vincentelli, G. Rabut, D. Stewart, and M. Way. 2000. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat. Cell Biol. 2:441-448. [DOI] [PubMed] [Google Scholar]
- 46.Mullins, R. D., J. A. Heuser, and T. D. Pollard. 1998. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U. S. A. 95:6181-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ngwamidiba, M., G. Blanc, H. Ogata, D. Raoult, and P. E. Fournier. 2005. Phylogenetic study of Rickettsia species using sequences of the autotransporter protein-encoding gene sca2. Ann. N. Y. Acad. Sci. 1063:94-99. [DOI] [PubMed] [Google Scholar]
- 48.Ngwamidiba, M., G. Blanc, D. Raoult, and P. E. Fournier. 2006. Sca1, a previously undescribed paralog from autotransporter protein-encoding genes in Rickettsia species. BMC Microbiol. 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oaks, E. V., M. E. Wingfield, and S. B. Formal. 1985. Plaque formation by virulent Shigella flexneri. Infect. Immun. 48:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parola, P., C. D. Paddock, and D. Raoult. 2005. Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clin. Microbiol. Rev. 18:719-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Policastro, P. F., M. G. Peacock, and T. Hackstadt. 1996. Improved plaque assays for Rickettsia prowazekii in Vero76 cells. J. Clin. Micriobiol. 34:1944-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pruyne, D., M. Evangelista, C. Yang, E. Bi, S. Zigmond, A. Bretscher, and C. Boone. 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science 297:531-532. [DOI] [PubMed] [Google Scholar]
- 53.Qin, A., A. M. Tucker, A. Hines, and D. O. Wood. 2004. Transposon mutagenesis of the obligate intracellular pathogen Rickettsia prowazekii. Appl. Environ. Microbiol. 70:2816-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rachek, L. I., A. Hines, A. M. Tucker, H. H. Winkler, and D. O. Wood. 2000. Transformation of Rickettsia prowazekii to erythromycin resistance encoded by the Escherichia coli ereB gene. J. Bacteriol. 182:3289-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaechter, M. J., F. M. Bozeman, and J. E. Smadel. 1957. Study on the growth of rickettsiae. II. Morphologic observations of living rickettsiae in tissue culture cells. Virology 3:160-172. [DOI] [PubMed] [Google Scholar]
- 56.Simser, J. A., M. A. Rahman, S. M. Dreher-Lesnick, and A. F. Azad. 2005. A novel and naturally occurring transposon, ISRpe1 in the Rickettsia peacockii genome disrupting the rickA gene involved in actin-based motility. Mol. Microbiol. 58:71-79. [DOI] [PubMed] [Google Scholar]
- 57.Sun, A. N., A. Camilli, and D. A. Portnoy. 1990. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect. Immun. 58:3770-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tam, V. C., D. Serruto, M. Dziejman, W. Brieher, and J. J. Mekalanos. 2007. A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe 1:95-107. [DOI] [PubMed] [Google Scholar]
- 59.Teyssiere, N., C. Chiche-Portiche, and D. Raoult. 1992. Intracellular movements of Rickettsia conorii and R. typhi based on actin polymerization. Res. Microbiol. 143:821-829. [DOI] [PubMed] [Google Scholar]
- 60.Tilney, L. G., and D. A. Portnoy. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109:1597-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uchiyama, R. 2003. Adherence to and invasion of Vero cells by recombinant Escherichia coli expressing the outer membrane protein rOmpB of Rickettsia japonica. Ann. N. Y. Acad. Sci. 900:585-590. [DOI] [PubMed] [Google Scholar]
- 62.Van Kirk, L. S., S. F. Hayes, and R. A. Heinzen. 2000. Ultrastructure of Rickettsia rickettsii actin tails and localization of cytoskeletal proteins. Infect. Immun. 68:4706-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiss, E. 1982. The biology of rickettsiae. Annu. Rev. Microbiol. 36:345-370. [DOI] [PubMed] [Google Scholar]
- 64.Weiss, E., J. C. Coolbaugh, and J. C. Williams. 1975. Separation of viable Rickettsia typhi from yolk sac and L cell host components by renografin density gradient centrifugation. Appl. Microbiol. 30:456-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Welch, M. D., A. Iwamatsu, and T. J. Mitchison. 1997. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature 385:265-269. [DOI] [PubMed] [Google Scholar]
- 66.Winkler, H. H. 1990. Rickettsia species (as organisms). Annu. Rev. Microbiol. 44:131-153. [DOI] [PubMed] [Google Scholar]
- 67.Wisseman, C. L., Jr., E. A. Edlinger, A. D. Waddell, and M. R. Jones. 1976. Infection cycle of Rickettsia rickettsii in chicken embryo and L-929 cells in culture. Infect. Immun. 14:1052-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wisseman, C. L., Jr., and A. D. Waddell. 1975. In vitro studies on Rickettsia-host cell interactions: intracellular growth cycle of virulent and attenuated Rickettsia prowazeki in chicken embryo cells in slide chamber cultures. Infect. Immun. 11:1391-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]