Abstract
Pneumococcal surface protein A (PspA) and PspC of Streptococcus pneumoniae are surface virulence proteins that interfere with complement deposition and elicit protective immune responses. The C-terminal halves of PspA and PspC have some structural similarity and contain highly cross-reactive proline-rich (PR) regions. In many PR regions of PspA and PspC, there exists an almost invariant nonproline block (NPB) of about 33 amino acids. Neither the PR regions nor their NPB exhibit the alpha-helical structure characteristic of much of the protection-eliciting N-terminal portions of PspA and PspC. Prior studies of PspA and PspC as immunogens focused primarily on the alpha-helical regions of these molecules that lack the PR and NPB regions. This report shows that immunization with recombinant PR (rPR) molecules and passive immunization with monoclonal antibodies reactive with either NPB or PR epitopes are protective against infection in mice. PR regions of both PspA and PspC were antibody accessible on the pneumococcal surface. Our results indicate that while PspA could serve as a target of these protective antibodies in invasive infections, PspC might not. When antibody responses to rPR immunogens were evaluated by using flow cytometry to measure antibody binding to live pneumococci, it was observed that the mice that survived subsequent challenge produced significantly higher levels of antibodies reactive with exposed PR epitopes than the mice that became moribund. Due to their conservation and cross-reactivity, the PR regions and NPB regions represent potential vaccine targets capable of eliciting cross-protection immunity against pneumococcal infection.
Pneumonia is the leading cause of mortality for children under the age of 5 years worldwide, and its most common etiology is Streptococcus pneumoniae (42). S. pneumoniae also cause otitis media and life-threatening meningitis. A 7-valent pneumococcal conjugate vaccine (PCV7) was introduced in the United States in 2000. PCV7 use reduced the number of cases of infections with vaccine capsular types in both immunized children (43) and nonimmunized individuals (18) in the same communities. But less than 5 years after the implementation of PCV7, reports of serotype replacement (increases in the number of invasive infections caused by strains of capsular serotypes not covered by the vaccine) began to appear (20, 22, 25, 40). The observation of this serotype replacement within a few years after vaccine implementation and the fact that there are at least 91 capsular types (36) raise concerns about the long-term effectiveness of capsule-based vaccines and stress the need for continued development of effective, noncapsular serotype-dependent pneumococcal vaccines (2, 39).
Surface proteins of pneumococci are important nonpolysaccharide vaccine candidates. Two of the more promising vaccine candidates are pneumococcal surface protein A (PspA) and pneumococcal surface protein C (PspC; also called CbpA). These two proteins have some similar structural features, and both proteins have been shown to elicit antibody-mediated protection against invasive pneumococcal infection (1, 8, 30, 31, 35). Antibodies to PspA generated in mice (28, 29) or humans (7, 34) are capable of passively protecting mice against infection. Strains of various capsule and PspA types can be protected against by immunizing with a single PspA (7). Recombinant alpha-helical regions of PspAs of different alpha-helical PspA families are cross-reactive and can be cross-protective (6, 7, 21, 24, 34), but the strongest protection in mice against some challenge strains is often observed when the immunizing and challenge PspAs are of the same alpha-helical PspA family (13, 38).
A gap in our knowledge of PspA and PspC immunogenicity exists, because with few exceptions, the published active and passive immunization experiments focused on immunity to the N-terminal alpha-helical regions of the protein or monoclonal antibodies (MAb) directed at the same alpha-helical regions. Although protection-eliciting sites exist within the N-terminal regions of PspA and PspC, these regions are diverse in their sequences and antigenic epitopes (8, 21, 23, 32). A proline-rich (PR) region, present in all PspAs and almost all PspCs, is not part of the alpha-helical regions of PspA or PspC molecules, and its immunogenicity has not been previously examined in detail. The PR region is remarkably similar within the paralogous PspA and PspC protein families and is much more conserved than the alpha-helical regions of either PspA or PspC proteins. The PR region consists of irregular repeats marked by the presence of a proline residue every two or three amino acids. The most common other amino acids are alanine and lysine. The most common sequence motif is PAPAP interrupted occasionally by PKP or, less commonly, by PEKP. About 56% of PspAs and 77% of PspCs are interrupted by a highly conserved block of amino acids termed the nonproline block (NPB) (8, 21-23, 45). The NPB is present in either PspA or PspC in about 90% of pneumococci. The NPB contains 33 amino acids, none of which are prolines (8, 21).
In 1999, Brooks-Walter et al. found that immunization with a PspC containing a PR region could protect mice from lethal infection by a pneumococcal strain lacking a pspC gene (8). This surprising cross-protection was hypothesized to occur via reactivity of antibodies elicited by PspC with the conserved PR region of PspA on the infecting strain. This cross-protection had been unexpected, since the PR domain of PspA and PspC was anticipated to be in close proximity to the cell wall (29, 47). The hypothesis was supported, however, by the finding that antibodies from the PspC-immunized mice bound to recombinant PspA (rPspA) containing a PR region, but not to rPspA lacking the PR region (8). Additional support for a protection-eliciting role of the PR and NPB regions comes from an analysis of prior studies in which mice were immunized with rPspC with alpha-helical domains containing or lacking a PR region. Xin et al. immunized mice with an rPspC containing a PR region with an NPB and elicited protection against a challenge strain containing PR regions with NPBs in both PspA and PspC (44). However, Ferreira et al. were not able to elicit protection with an rPspC antigen lacking a PR region (15). Recent reports have provided further evidence that the human immune system frequently sees and responds to the PR region. In an epitopic screening of human patient sera, among the peptides most reactive with patient sera were ones that contain PR regions from both PspA and PspC of type 4 strain TIGR4 (16, 33).
Given the hypothesis that immunization with PspC or PspA might elicit cross-protective antibodies to the shared PR region and also given that human serum is frequently found to contain antibodies to the PR region (16, 33), we used active immunization and passive transfer of monoclonal antibodies to examine the epitopic sites in the PR region in greater detail, to determine whether the antibodies elicited by PR region immunogens are protective against pneumococcal infection, and to explore properties of the protective antibodies. Our findings validate the hypothesis that the PR regions of PspA and PspC are capable of eliciting antibody-mediated protection against pneumococcal infection. Abstracts of some of the data contained in this paper were published in 2006 and 2007 (10, 11).
MATERIALS AND METHODS
Bacterial strains.
Strains used in this experiment are listed and referenced in Table 1. All strains were grown in Todd-Hewitt broth with 5% yeast extract (THY; Becton Dickinson & Co., Sparks, MD) at 37°C. Infection stocks were collected at an optical density at 600 nm (OD600) of 0.40, washed, resuspended in fresh THY with 8% glycerol, and stored at −80°C until use. The presence of the NPB and other potential epitopes in pspA or pspC was determined for each of the strains used in this study by the binding of antibodies to choline eluates of pneumococcal strains by Western blot analysis and by examining both published and unpublished sequences of the pspA and pspC genes.
TABLE 1.
Strains used in this studya
| Strain | Sequence reference | Wild type | Capsule | NPB |
PspA |
||
|---|---|---|---|---|---|---|---|
| PspA | PspC | Family | Clade | ||||
| D39 | 8, 21 | NA | 2 | + | + | 1 | 2 |
| JY53 | 46 | D39 | 2 | Tru | + | NA | NA |
| TRE118 | 19 | D39 | 2 | + | Ins/dup | 1 | 2 |
| TRE121.3 | 19 | D39 | 2 | Ins/dup | Ins/dup | NA | NA |
| 3JYP2670 | This paper | NA | 3 | + | − | 2 | 4 |
| WU2 | 21 | NA | 3 | + | − | 1 | 2 |
| BG12730 | This paper | NA | 6A | − | + | 2 | 3 |
| AC094 | 21 | NA | + | Unknown | 1 | 1 | |
| TIGR4 | 41 | NA | 4 | − | + | 2 | 3 |
| BG9739 | 21 | NA | 1 | 1 | |||
NA, not applicable; Tru, a truncated version of PspA is expressed that does not contain choline-binding repeats and is secreted from the bacterial surface; Ins/dup, insertion duplication to interrupt protein production; +, presence of the NPB in the molecule; −, absence of the NPB in the molecule.
Cloning and expression of the proline-rich regions in plasmid constructs.
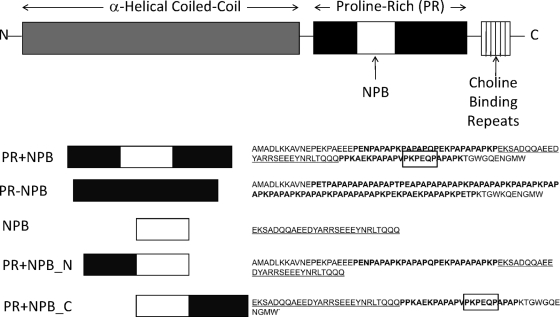
For details of the plasmids and recombinant proteins that were constructed for this work, see Table S1 in the supplemental material. In each case, an internal gene fragment of the pspA gene from the indicated strain was amplified by PCR using the primers given in Table 1. PCRs were carried out for 30 cycles in a total volume of 50 μl by following the manufacturer's recommendations. For the pET-32a constructs, the amplified gene fragments were then digested with NcoI and SalI, and the ∼300-bp pspA gene fragments were incorporated between NcoI and SalI sites of a pET32a vector (Novagen) to create pPAC001 expressing PR protein containing the NPB (PR+NPB) or pPAC003 expressing PR protein lacking the NPB (PR-NPB). For the pET-41Ek/LIC (ligation-independent cloning) or the pET-46Ek/LIC constructs, the amplified gene fragments were digested with T4 DNA polymerase (Novagen), and the ∼100-bp pspA gene fragment was then incorporated into predigested vectors (Novagen). DNA sequencing confirmed that all the recombinant plasmids contained the expected pspA gene fragments as shown in Fig. 1. We used this strategy to produce three PR region fragments as recombinant fusion proteins in order to increase immunogenicity that could have otherwise been diminished due to their small size. In vector pET32a, the fusions are with mouse thioredoxin and a His tag, and in pET41-Ek/LIC, the fusions are with a 220-amino-acid glutathione S-transferase (GST) tag protein. Proteins with tag only were also purified for use as comparison immunization control proteins.
FIG. 1.
Regions of whole PspA (top) and portions of the full protein used in the constructs (bottom; see labels on the left) used in this paper. Amino acid sequences to the right of the construct diagrams are distinguished by region as follows: unbolded amino acids represent PspA sequence outside the proline-rich region, bolded amino acids represent proline-rich regions, underlined amino acids represent the NPB within a PR region, and boxed amino acids represent a hypothesized epitope for MAb PR-5C4.7. Diagrams in this figure are not drawn to scale.
Expression and purification of recombinant proteins.
All three vectors were transformed individually into BL21 Star (DE3) cells (Novagen) for protein production. After cultures reached an OD600 of 0.40 to 0.60, they were induced using 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) for 2 h and purified on either a Ni2+-nitrilotriacetic acid (NTA) resin (Novagen) for PR+NPB and PR-NPB or a Cobalt resin (ClonTech) for the N-terminal end of PR+NPB (PR+NPB_N), the C-terminal end of the PR+NPB (PR+NPB_C), and NPB by following the manufacturer's protocols for His tag purification. Purified protein was then dialyzed to remove imidazole. The first dialysis occurred at 4°C overnight in a buffer containing 2 mM Tris-HCl, 20 mM NaCl, 10 mM EDTA, and 0.1% glycine (pH 8.0). The second dialysis occurred at 4°C overnight in a buffer (pH 8.0) containing 50 mM Tris HCl, 20 mM NaCl, 0.1% glycine, and 0.1% Triton X-100.
Monoclonal antibodies.
MAb (PR-1A4.7, PR-5C4.6, and PR-6A5.12) from mice immunized with PR+NPB were generated at the University of Alabama at Birmingham Epitope Recognition and Immunoreagent Core Facility. MAb K67 was generated to a recombinant fragment of amino acids 288 to 588 of strain Rx1 PspA comprising the proline-rich region and the choline-binding domain of PspA generated as previously described (29). Rx1 PspA is identical to that of strain D39 (21). Production of MAb 2A4 to PspA's alpha-helical region and its epitope has been described previously (9). The approximate locations of epitopes for MAb were determined using Western blot analysis against purified recombinant proteins containing different PR region fragments (see Fig. 3). Two hybridoma cell lines that produced MAb that bound to the NPB only and one cell line that produced a MAb that bound elsewhere to the NPB were selected for analysis. The lines were subcloned, and high-titer antibody was generated in ascites fluid.
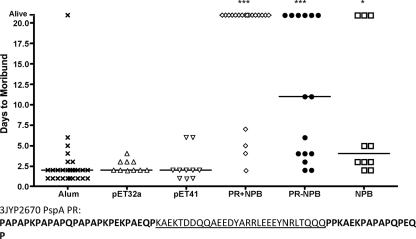
FIG. 3.
Active immunization with PR recombinant proteins protects against i.v. infection with pneumococcal type 3 strain 3JYP2670. CBA/N mice immunized with Imject alum control, vector tag protein, PR+NPB, PR-NPB, or NPB were infected i.v. with type 3 strain 3JYP2670 and monitored for the number of days to moribundity. Mice plotted as “Alive” were still not moribund at 21 days postinfection. None of the “Alive” mice showed ruffled fur, a hunched back, or any other signs of illness. All three proteins were found to protect significantly against infection compared to a pool of the alum and two vector controls (for Kruskal Wallis, P < 0.0001; P values were <0.001 [***] and <0.05 [*] compared to the pooled control using Dunn's multiple-comparison post hoc test). None of the control groups protected significantly more than any of the others in a separate post hoc analysis. The sequence of the PR region of infection strain 3JYP2670 PspA is distinguished by regions as in Fig. 1.
The concentrations of antibodies in ascites fluids were determined by separating the proteins in a sample of each ascites fluid on nitrocellulose using microzone electrophoresis, staining the proteins with Ponceau's stain, and scanning the gels with a densitometer. The percentage of the total serum proteins comprised by the MAb was determined using the density of the MAb band relative to the density of all the bands. The concentration of the MAb in μg/ml was then calculated from the total protein in the ascites fluid using a Bio-Rad protein assay (Bio-Rad).
Antibody titer.
The total antibody titer of mice to the PR region was determined by enzyme-linked immunosorbent assay (ELISA) using sera collected one day before infection. Briefly, plates were coated at a concentration of 2 μg/ml overnight with PR+NPB-6×, a PR+NPB construct linked only to a 6× His tag. Plates were then washed with phosphate-buffered saline-Tween at 0.1% (PBST) and incubated overnight with serially diluted serum samples. Plates were washed again and incubated first with goat anti-mouse BIOT antibody for an hour at room temperature (RT), washed, and then incubated with Strep-AP for an hour at RT before a final wash and development with SigmaFast 5-bromo-4-chloro-3-indolyl phosphate-nitroblue tetrazolium (Sigma Aldrich). An antibody index was calculated from the observed OD405 multiplied by the ELISA dilution at the OD reading. For this purpose, ODs were read at ELISA dilutions at which there was a linear relationship between the dilution and the OD. The average adjusted bindings were log transformed prior to statistical analysis. Analysis of variance (ANOVA) was performed for the groups, followed by Tukey's post hoc test to determine significant differences between average adjusted binding values.
Protection by active immunization with PR proteins against i.v. challenge.
Dialyzed protein was diluted to a concentration of 50 μg/ml in a solution of Lactated Ringer's solution with 30% Imject alum (Pierce) and incubated for at least 8 h at 4°C. CBA/CaHNVrkxid/J (CBA/N) mice (Jackson Laboratories, Bar Harbor, ME) were immunized subcutaneously with 100 μl containing 5 μg protein on three occasions. There were rest periods of 2 and 3 weeks, respectively, between the first and second and second and third immunizations. Three weeks after the last immunization, 75 μl of blood was taken from the mice retro-orbitally for determining antibody titer to PR+NPB through ELISA and pneumococcal surface-binding antibodies through flow cytometry. Immunized CBA/N mice were infected intravenously (i.v.) the following day with 400 to 600 CFU of type 3 strain 3JYP2670 (Table 1) diluted from a frozen infection stock in Lactated Ringer's solution. Infected mice were observed for up to 21 days, and the number of days until the mice were moribund was recorded. A Kruskal-Wallis analysis was performed to determine any significant differences between the groups, followed by a Dunn's multiple-comparison post hoc test to determine significance compared to the control group.
Passive protection against i.v. infection.
To determine if the protection from active immunizations was antibody mediated, ascites fluid containing 20 μg of antibody to PR+NPB was injected into CBA/N mice intraperitoneally (i.p.). An hour later, mice were infected i.v. with either 600 CFU of WU2 or 10,000 CFU of BG12730 (Table 1). Mice were observed for 21 days to determine the number of days until they were moribund. A Kruskal-Wallis analysis was performed to determine significant differences between the groups, followed by a Dunn's multiple-comparison post hoc test to determine significant differences from the control group.
Definition of moribund.
Mice were scored as moribund if their surface temperature fell below 24°C or if they were unresponsive to touch. Surface temperature was determined with an MX2 Rayteck scanning thermometer (Santa Cruz, CA) (3) by examining the surface temperature across the lower back. A normal surface temperature of mice in our animal house is 30°C. All mice scored as moribund were euthanized by CO2 narcosis. Mice that developed paralysis and an inability to right themselves were also declared moribund and euthanized even though their temperature was >24°C.
Surface detection of antigen.
Flow cytometry analysis using Alexa Fluor 488-conjugated antibody to mouse IgG was utilized to detect antibody binding on the surface of live bacteria as done previously (12). All strains tested were encapsulated, and comparisons were made only between different antibodies for the same strain or between results for one antibody for strains with the same genetic background tested in parallel. Briefly, pneumococci were grown to an OD600 of 0.3, diluted to an OD600 of 0.1, and grown back to an OD600 of 0.3. Aliquots (250 μl) of bacteria were incubated with primary antibodies in 100-μl sample volumes. Primary antibodies were diluted in 1% bovine serum albumin (BSA)-PBS as follows: the tissue culture supernatant of MAb K67 was diluted 1:3; MAb PR1A4.7, PR5C4.6, and PR6A5.12 from ascites fluid were diluted 1:100; and the polyclonal sera from immunized mice were diluted 1:64. Fluorescence was determined as the number of times greater than control binding by dividing the geometric mean for samples treated with antibodies by the geometric mean for samples not treated with antibodies. ANOVA was performed for the groups to determine significant differences, followed by a Tukey's post hoc test to determine significant differences between groups.
Statistics.
Statistical evaluation has been described above in conjunction with each assay. Please note that in the figures, tables, and text, the error terms are always standard errors of the means.
RESULTS
PR region epitopes are surface exposed.
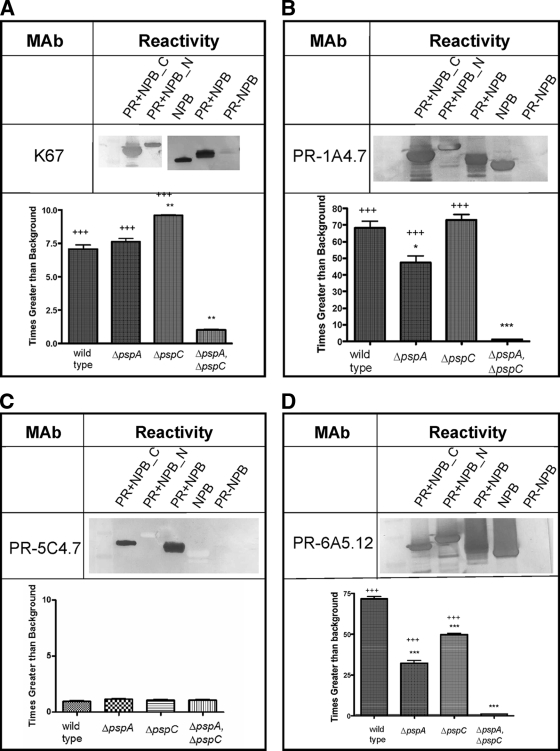
The recombinant PR proteins depicted in Fig. 1 are as follows: PR protein lacking the NPB (PR-NPB), PR protein containing the NPB (PR+NPB), the NPB alone (NPB), the N-terminal end of the PR+NPB (PR+NPB_N), and the C-terminal end of the PR+NPB (PR+NPB_C). These were analyzed for their reactivity with the MAb K67, generated to the PspA shared by strain D39 and previously shown to be cross-reactive with PspC (K. A. Benton, D. E. Briles, and S. K. Hollingshead, unpublished data). MAb K67 reacted to recombinant NPB in Western blot analysis and bound to native PspA and PspC of strain D39 (Fig. 2A). PspA produced by strain 3JYP2670 contains NPB sequences. Pooled sera from mice immunized with PR+NPB, which protects against infection with 3JYP2670 pneumococci, also contained an antibody reactive with live 3JYP2670 pneumococci as detected by flow cytometry analysis. Compared to preimmune pooled sera, the immune pooled sera provided a >10-fold increase in binding to live pneumococci, showing that antibodies elicited in mice to this region of PspA were capable of detecting the PR region of native protein on the pneumococcal surface (12.9 ± 0.2 fluorescence postimmunization compared to 1.2 ± 0.1 fluorescence preimmunization).
FIG. 2.
Reactivity of MAb to PR regions. Western blot of PR recombinant proteins reacted with MAb to PR regions (upper panels). Flow cytometry analysis of binding of MAb to pneumococcal strain D39 and its ΔpspA, ΔpspC, and ΔpspA ΔpspC mutants (lower panels). The ΔpspA ΔpspC mutant bound significantly less antibody than the wild type for all three MAb that bound the wild type (P < 0.0001 by ANOVA). Results for the Dunnett's multiple-comparison test against the wild-type control are as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Comparisons to the ΔpspA ΔpspC double mutant are as follows: +, P < 0.05; ++, P < 0.01; and +++, P < 0.001.
Capacity of rPR region peptides to elicit protection.
To determine if immunizations with regions of the PR region could elicit protective immune responses, groups of mice were immunized subcutaneously with one of the following: adjuvant alone, a pET32 purification tag without a pneumococcal insert, a pET4Ek/LIC purification tag without a pneumococcal insert, PR+NPB, PR-NPB, or NPB. All groups of mice were challenged intravenously with the capsular type 3 challenge strain 3JYP2670, whose family 2, clade 4 PspA differs from the family 1 PspAs of AC094 and BG9739 from which the recombinant PR (rPR) regions were derived. Strain 3JYP2670 has PR and NPB epitopes in its PspA but lacks either of them in its PspC (Table 1).
Immunization with neither of the control proteins, consisting of the purification tags, nor alum alone elicited protection in the mice against morbidity. Thus, for statistical comparisons to the groups immunized with PR constructs, the results for the control-protein-immunized mice and the alum-immunized mice were pooled to form a single control group. The mice immunized with each of the different PR proteins took significantly longer to become moribund than did the control mice (Fig. 3). Immunization with PR+NPB resulted in the greatest increase in the median number of days to morbidity, and 80% of the mice lived until day 21 postinfection, when the experiment was terminated (Fig. 3).
Measurement of antibody elicited by recombinant PR region peptides.
The immune sera collected from the immunized mice one day before pneumococcal challenge were incubated with the live 3JYP2670 challenge strain, and antibody bound to the bacterial surface was detected using flow cytometry. The PR region of the challenge strain 3JYP2670 contained a PR region with many sequence similarities to the recombinant PR+NPB used for immunization. Of the three vaccine constructs, PR+NPB elicited severalfold more antibody able to bind the 3JYP2670 surface than either of the other two constructs. The mean immunofluorescence following immunization with PR-NPB, PR-NPB, or NPB was 8.9 ± 1.7, 1.7 ± 0.4, or 2.9 ± 0.07, respectively. The greater immunofluorescence with antisera to PR+NPB was <0.01 compared to either PR-NPB or NPB by Tukey's post hoc test.
We also examined the ELISA titers of antibody to the three recombinant peptides. Antibody titers were determined to the PR+NPB peptide (PR+NPB-6×) that contains all of the epitopic regions of PR recombinants but lacks their pET32a tag. Based on its sequence, PR+NPB-6× should react with antibodies elicited by the PR epitopes of PR+NPB and PR-NPB and by NPB epitopes. Since it lacks the pET32a tag, it should not detect any antibodies that would have been elicited by the tag portions of the constructs. As in the case of the surface-binding assay, we found that of the three PR peptides used for immunization, the PR+NPB peptide elicited the highest antibody levels (Table 2).
TABLE 2.
Binding of polyclonal sera from immunized mice to PR+NPB-6× measured by ELISA
| Antigen | Avg binding (±SE)b |
P valuea |
|
|---|---|---|---|
| Alum | PR+NPB | ||
| PR+NPB | 2.72 (0.050) | <0.0001 | |
| PR-NPB | 1.97 (0.065) | <0.0001 | <0.0001 |
| NPB | 1.72 (0.080) | <0.0001 | <0.0001 |
| pET41 | 0.851 (0.084) | >0.05 | <0.0001 |
| Alum | 0.620 (0.025) | <0.0001 | |
Binding of polyclonal sera to PR+NPB-6× was significantly different by ANOVA (P < 0.0001). P values in the table are Tukey's post hoc comparisons of immunization antigen groups to either an alum-immunized group or a PR+NPB-immunized group. PR-NPB and NPB groups did not have significantly different binding.
Measured by antibody index.
We also compared the antibody levels in the sera of the immunized mice that subsequently survived the challenge with 3JYP2670 with the antibody levels of the immunized mice that became moribund. Although the mice that survived challenge had higher antibody levels than those that became moribund (antibody index of 257 multiplied or divided by 1.7 and antibody index of 100 multiplied or divided by 1.3) the difference was not statistically significant (P = 0.11, Student's t test). In contrast, when the amounts of antibody that could be detected binding the surface of the challenge strain were examined, the mean fluorescence level of binding of antibody in the sera of mice that survived was 7.550 ± 2.310 compared to 2.814 ± 0.6264, the fluorescence level of antibody in the sera of mice that became moribund. This difference was statistically significant at a P value of 0.02 (Student's t test). Thus, the ability of the elicited antibody to be detected binding the surface of the challenge strain was found to be a better indication of protective potential than the antibody levels detected by ELISA.
Monoclonal antibodies to PR+NPB.
To examine the protective epitopes of antibodies elicited by PR+NPB, we prepared a panel of hybridoma antibodies using spleen cells from mice immunized with PR+NPB. The MAb were tested for reactivity with each of our PR constructs to determine their reactivity with epitopic regions within the PR region (Fig. 2A to D).
Epitopes of MAb PR-1A4.7 and PR-6A5.12 (as well as MAb K67, described above) were assumed to be within NPB because they bound only peptides containing NPB (Fig. 2B and D). PR-1A4.7 and PR-6A5.12 were the IgG 1 isotype. MAb PR-C54.7 (IgG 2a) was found to have a different specificity. PR-C54.7 bound to the proline-rich sequence containing a central NPB (PR+NBP) and to a fragment containing NPB and the C-terminal proline-rich flanking sequence (PR+NBP_C). It did not bind the peptide containing the N-terminal flanking sequence (PR+NBP_N) or NPB by itself (Fig. 2C). MAb PR-C54.7 may bind to the sequence PKPEQ, because it is the only sequence in the two reactive peptides that is not in any of the other peptides tested. However, at this stage of analysis, it was also possible that the epitope of MAb PR-C54.7 was a conformational epitope that requires the presence of both the NPB and the C-terminal proline-rich sequence.
To determine if the epitopes identified in Western blot analysis were surface accessible, mutant strains of capsular type 2 strain D39 in which either pspA or pspC or both genes had been inactivated were stained with MAb and analyzed using flow cytometry. Staining of mutant strains using the MAb K67, PR-1A4.7, and PR-6A5.12 in flow cytometry showed that the NPB epitopic site in both PspA and PspC is antibody accessible on the surface of live pneumococci (Fig. 2A, B, and D). Moreover, a mutant lacking both PspA and PspC (Table 1), as expected, showed no surface binding of NPB-specific antibodies (Fig. 2A, B, and D). The PR regions of D39's PspA and PspC do not contain the unique PR region sequence PKPEQ, which we have hypothesized is recognized by MAb PR-5C4.7. Consistent with this hypothesis, neither D39 nor any of its mutants were bound by MAb PR-5C4.7 (Fig. 2C). However, MAb PR-5C4.7 did exhibit surface binding to S. pneumoniae strain TIGR4 (data not shown), which expresses the PKPEQ epitope in its PspA sequence (41). These additional data with D39 and TIGR4 provide confirmation that MAb PR-5C4.7 indeed binds PKPEQ and show that when it was present, the PKPEQ epitope was surface accessible.
Passive protection with MAb.
To further understand the role of antibodies to the NBP and PKPEQ epitopes in the protection elicited by PR region immunization, we assessed the ability of each of the three MAb elicited to PR+NPB to passively protect against pneumococcal infection. Mice were injected i.p. with 20 μg of antibody from ascites fluid and infected an hour later with the type 3 capsular serotype strain WU2, which contains NPB only in its PspA. The PspC of WU2 lacks a PspA-like PR region (Table 1). WU2 was chosen as the challenge strain for these studies because it has been used extensively in passive-protection studies with MAb to epitopes in the alpha-helical domain of PspA (29, 30). To determine if the relative ability of MAb to bind to the surface of the infection strain was associated with any differences in passive protection observed, the surface binding of each MAb to the live WU2 strain was also determined using flow cytometry.
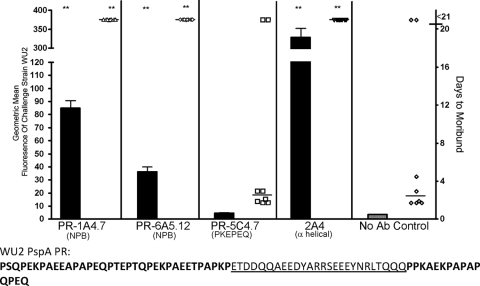
The two MAb that bound the NPB in Western blot analysis, PR1A4.7 and PR6A5.12 (Fig. 2A and D), each bound strongly to the surface of WU2 pneumococci and significantly protected against morbidity compared to that of the control mice injected with only Lactated Ringer's solution (Fig. 4). MAb K67, which was also shown to bind NBP, was not tested because we could not obtain the MAb in sufficient quantities. WU2's PspA sequence does not contain the PKPEQ sequence, and as expected, MAb PR5C4.7 neither bound WU2 nor protected against morbidity caused by WU2 (Fig. 4). As a positive control for this assay, we also included MAb 2A4, which binds to the alpha-helical domain of PspA of WU2 (9). MAb 2A4 bound the surface of WU2 even more strongly than did PR-1A4.7 or PR-6A5.12, and like those two MAb, it protected against pneumococcal infection in all eight of the challenge mice (Fig. 4).
FIG. 4.
Surface binding of MAb (bars) and passive protection against infection (symbols) for pneumococcal challenge strain WU2. Monoclonal antibodies for each panel are listed on the x axis, and the sequence of the single PR region in the PspA of strain WU2 is given below. For each antibody, the log binding to the infection strain determined by flow cytometry is indicated as a bar graph along the left y axis, and the number of days to reach moribundity is indicated as a dot plot along the right y axis. During challenge, mice were injected with antibody and infected i.v. 1 h later. Two MAb, PR-1A4.7 and PR-6A5.12, protected significantly compared to the control group that received no antibody (Kruskal Wallis P value, 0.0002; **, P < 0.01 using Dunn's multiple-comparison test to the control). MAb 2A4, which interacts with the alpha-helical region of family 1 PspA, was used as a positive control. The protective MAb (PR-1A4.7, PR-6A5.12, and 2A4) bound significantly more strongly to the infection strain (WU2) than did the nonprotective MAb (PR-5C4.7) or the saline (no-antibody) control (ANOVA P value, <0.001. ***, P < 0.01 compared to nonprotective strains using Tukey's post hoc test).
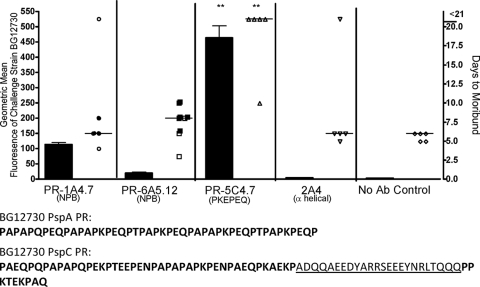
The same four MAb were also tested for binding and protection against capsular type 6A strain BG12730. Sequence analysis of pspA and pspC of strain BG12730 revealed no NPB in PspA and one copy of NPB in the PspC protein. The NPB in its PspC was located only a few amino acids upstream of the choline-binding repeats that anchor the protein to the cell surface. Strain BG12730's PspA, but not PspC, had multiple repeats of the PR region sequence PKPEQ, which we believe is the epitope for the MAb PR-5C4.7. MAb PR-5C4.7 also bound BG12730 and provided significant protection against morbidity from challenge with BG12730 compared to the control (Fig. 5). The two NPB-specific MAb did not bind BG12730 as well as PR-5C4.7 did and, unlike PR-5C4.7, did not protect against infection with BG12730. MAb 2A4 did not bind BG12730 and did not protect against infection with BG12730 (Fig. 5).
FIG. 5.
Surface binding of MAb (bars) and passive protection against infection (symbols) for pneumococcal challenge strain BG12730. Monoclonal antibodies for each panel are listed on the x axis, and the sequence of the PR regions in the PspA and the PspC of strain BG12730 is given below the x axis. Mice indicated with solid symbols were scored as moribund and euthanized due to neurologic symptoms. Mice indicated with open symbols were scored as moribund and euthanized due to a surface temperature of <24°C. For each antibody, the binding to the infection strain determined by flow cytometry is indicated as a bar graph along the left y axis, and the number of days to reach moribundity is indicated as a dot plot along the right y axis. During challenge, mice were injected with antibody and infected i.v. 1 h later. In this challenge, MAb PR-5C4.7 protected significantly well compared to the control group that received no antibody (Kruskal Wallis P value, 0.01; **, P < 0.01 using Dunn's multiple-comparison test to the control). Monoclonal antibody 2A4, which does not bind the PspA of this strain, was included as a negative control.
Strains WU2 and BG12730, used in the passive protection studies, displayed significant differences in pathogenesis. WU2 caused a rapid septic morbidity, and for all mice that became moribund, the median time to moribundity was 2.5 days. In the case of mice infected with WU2, the declaration of moribund status was always based on a marked drop in body temperature, to below 24°C. For BG12730, all 5 of the control mice and 4 of the 5 mice given passive MAb 2A4 were scored as moribund based on a drop in surface body temperature to below 24°C; however, the median time to moribundity for these 9 mice was 6 days. Seven mice given PR-1A4.7 or PR-6A5.12 were scored as moribund because they exhibited symptoms of neurologic pathology—partial paralysis or spinning when held by the tail (median time to moribundity was 7 days). These seven mice are indicated by solid symbols in Fig. 5. All mice scored as moribund because of neurologic symptoms maintained surface temperatures above 24°C, so if the temperature drop had been the cutoff, they would not have been considered moribund until a later time. To determine if the bacteria had crossed from the blood into the brain in BG12730-infected mice, numbers of CFU per brain were determined from moribund mice exhibiting each of the two types of behavior on day 6. The mice displaying neurologic problems were found to have >3,000 CFU per brain. A mouse that was euthanized due to lower body temperature had only 110 CFU of BG12730 in its brain.
DISCUSSION
The studies described herein show that the PR regions of both PspA and PspC are antibody accessible on the surface of live pneumococci. Moreover, recombinant PR fragments that lack any other PspA or PspC sequence are capable of eliciting protective immunity.
Although the ability of antibodies to PR regions to protect is dependent on both the expression of the relevant epitope by the challenge strain and on the surface exposure of the epitope, it was not related to the alpha-helix-based family or clade serotype of the PspA. Both PR+NPB and PR-NPB were from family 1, clade 1 PspAs and protected against infection with strain containing family 2, clade 4 PspAs. Moreover, antibodies to both PR and NPB epitopes were found to always be protective when their respective epitopes were present on surface-expressed PspA of pneumococci. In addition, MAb elicited by a single PR region of a family 1, clade 1 PspA could protect against challenge strains containing serologically different PspAs—either a family 1, clade 2 PspA or a family 2, clade 3 PspA. These findings are important, since cross-protection elicited by alpha-helical PspA sequences decreases as sequence differences between and among families and clades increase (14, 37). The correlation between the protective effect of our individual MAb and their ability to bind surface-exposed epitopes was reminiscent of the finding by Gor et al. (17) that “surface” proteins that elicited the best protection against pneumococci were those most accessible to antibody on wild-type pneumococci.
Both PR and NPB epitopes were protective against strains with surface-exposed epitopes. Two of the MAb (PR-1A4.7 and PR-6A5.12) recognized the NPB and protected mice infected with a capsule type 3 strain with a family 1, clade 2 PspA from becoming moribund. The third MAb (PR-5C4.7) bound PspA fragments containing the proline-rich sequence PKPEQ and protected against strain BG12370, which express PKPEQ epitopes in PspA. Additionally, PR5C4.7 did not bind or protect against sepsis with strain WU2, which lacks the PKPEQ epitope. Our expectation that PR-5C4.7 binds the PKPEQ epitope is based on a statistical association. Of the total of 9 strains and PR region fragments examined, 4 expressed the epitope and 5 did not. The binding of PR-5C4.7 gave a perfect association with epitope expression (P = 0.008 by Fisher's exact test).
Strain BR12370 expresses the NPB in its PspC but not its PspA. Both of our MAb reactive with the NPB bound this strain, but neither provided statistically significant protection. This may be because in this particular strain, the NPB is just 9 amino acids from the choline-binding repeats at the C-terminal end of PspC. In WU2, which these MAb protected against, the NPB was 15 amino acids from the choline-binding repeats. A bigger problem may be that in blood infections, PspC (also called CbpA) is reported to be more poorly expressed than is PspA (26, 27). However, until additional experiments are performed with additional strains, it should not be concluded that PspC cannot serve as an effective target of protective antibody for blood infections. It is also possible that at mucosal sites, where PspC is more strongly expressed than in the blood, it will be a better target of protective immunity. Even with the concerns about whether PspC can be an effective target of immunity to the PR region, our present immunization and passive protection data make it clear that epitopes in both the NPB and portions of the PR domain lacking the NPB are able to elicit protection against pneumococcal infection when at least PspA can be a target. About 56% of strains express NPB, and 46% of strains express an exact copy of PKEPEQ. Approximately 75% of strains would be expected to express one or the other. Moreover, about 87% of strains express a one-amino-acid variant of PKEPEQ, and it is likely that there are other proline-containing epitopes in the proline-rich region that may be protection eliciting (21).
The observation that partially protected mice infected with the capsular type 6A strain BG12730 appeared to have developed meningitis and/or ear infections was not surprising, since capsular group 6 strains have long been known to exhibit a longer time to morbidity/mortality in mice than type 4 or type 3 strains (4, 5), and during these lengthy infections, neurologic symptoms have been observed (Randall Harris and D. E. Briles, unpublished data). In our passive-protection studies, we observed that PR-5C4.7 reduced the occurrence of neurologic symptoms and significantly increased the time to moribundity. The fact that PR-5C4.7 could prevent deaths as well as the neurologic symptoms indicates that immunity to the PR region can prevent both sepsis and the associated neurologic disease.
These results indicate that the PR regions of PspA and PspC may be important immunogens for use in broadly cross-protective potential vaccines for use against S. pneumoniae of diverse capsular serotypes. The robustness of the results is supported by that fact that immunization with PR epitopes or passive protection with anti-PR MAb elicited protection against each of the three challenge strains examined. Giefing et al. arrived at a similar conclusion with regard to titers to PR epitopes containing prolines (16). This study, however, provides the first evidence that immunization with either the highly conserved PR or NPB region can be an important means of eliciting protection against pneumococci that should be effective across capsular types and that antibodies generated to this region alone are capable of protecting against pneumococcal infection.
Supplementary Material
Acknowledgments
We acknowledge Larry McDaniel for materials used in making MAb K67, Flora Gathof for administrative assistance, and Shaper Mirza for technical advice and revision of the manuscript.
The findings and conclusions in this article have not been formally disseminated by the FDA and should not be construed to represent the determination or policy of any federal agency. Kimberly Benton's work for this study was performed prior to her employment at the Food and Drug Administration.
In the 1980s, Barry M. Gray (presently at the University of Illinois in Peoria) generously gave us access to his large collection of pneumococci, including strain BG12730.
These studies were supported by NIH grants R01-AI021458 and R01-AI053749 and support from PATH, Seattle, WA. Calvin C. Daniels was supported in part by NIH grant T32 HL07553.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 1 March 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Balachandran, P., A. Brooks-Walter, A. Virolainen-Julkunen, S. K. Hollingshead, and D. E. Briles. 2002. Role of pneumococcal surface protein C in nasopharyngeal carriage and pneumonia and its ability to elicit protection against carriage of Streptococcus pneumoniae. Infect. Immun. 70:2526-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barocchi, M. A., S. Censini, and R. Rappuoli. 2007. Vaccines in the era of genomics: the pneumococcal challenge. Vaccine 25:2963-2973. [DOI] [PubMed] [Google Scholar]
- 3.Bast, D. J., M. Yue, X. Chen, D. Bell, L. Dresser, R. Saskin, L. A. Mandell, D. E. Low, and J. C. de Azavedo. 2004. Novel murine model of pneumococcal pneumonia: use of temperature as a measure of disease severity to compare the efficacies of moxifloxacin and levofloxacin. Antimicrob. Agents Chemother. 48:3343-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benton, K. A., J. C. Paton, and D. E. Briles. 1997. Differences in virulence for mice among Streptococcus pneumoniae strains of capsular types 2, 3, 4, 5, and 6 are not attributable to differences in pneumolysin production. Infect. Immun. 65:1237-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briles, D. E., M. J. Crain, B. M. Gray, C. Forman, and J. Yother. 1992. Strong association between capsular type and virulence for mice among human isolates of Streptococcus pneumoniae. Infect. Immun. 60:111-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briles, D. E., S. Hollingshead, A. Brooks-Walter, G. S. Nabors, L. Ferguson, M. Schilling, S. Gravenstein, P. Braun, J. King, and A. Swift. 2000. The potential to use PspA and other pneumococcal proteins to elicit protection against pneumococcal infection. Vaccine 18:1707-1711. [DOI] [PubMed] [Google Scholar]
- 7.Briles, D. E., S. K. Hollingshead, J. King, A. Swift, P. A. Braun, M. K. Park, L. M. Ferguson, M. H. Nahm, and G. S. Nabors. 2000. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J. Infect. Dis. 182:1694-1701. [DOI] [PubMed] [Google Scholar]
- 8.Brooks-Walter, A., D. E. Briles, and S. K. Hollingshead. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect. Immun. 67:6533-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crain, M. J., W. D. Waltman II, J. S. Turner, J. Yother, D. F. Talkington, L. S. McDaniel, B. M. Gray, and D. E. Briles. 1990. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect. Immun. 58:3293-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniels, C., P. Coan, D. E. Briles, and S. K. Hollingshead. 2006. Immunogenicity of mosaic pneumococcal surface proteins PspA and PspC. Natl. Conf. Gram-Pos. Pathog., p. 68. University of Nebraska, Omaha, NE.
- 11.Daniels, C., P. Coan, M. Darrieux, J. King, D. L. Dupraw, D. E. Briles, and S. K. Hollingshead. 2007. Non-coiled coil regions of Streptococccus pneumoniae surface proteins PspA and PspC can elicit protection from fatal sepsis, abstr. E-069, p. 140. Abstr. 107th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 12.Daniels, C. C., T. C. Briles, S. Mirza, A. P. Hakansson, and D. E. Briles. 2006. Capsule does not block antibody binding to PspA, a surface virulence protein of Streptococcus pneumoniae. Microb. Pathog. 40:228-233. [DOI] [PubMed] [Google Scholar]
- 13.Darrieux, M., E. N. Miyaji, D. M. Ferreira, L. M. Lopes, A. P. Lopes, B. Ren, D. E. Briles, S. K. Hollingshead, and L. C. Leite. 2007. Fusion proteins containing family 1 and family 2 PspA fragments elicit protection against Streptococcus pneumoniae that correlates with antibody-mediated enhancement of complement deposition. Infect. Immun. 75:5930-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darrieux, M., A. T. Moreno, D. M. Ferreira, F. C. Pimenta, A. L. de Andrade, A. P. Lopes, L. C. Leite, and E. N. Miyaji. 2008. Recognition of pneumococcal isolates by antisera raised against PspA fragments from different clades. J. Med. Microbiol. 57:273-278. [DOI] [PubMed] [Google Scholar]
- 15.Ferreira, D. M., M. Darrieux, D. A. Silva, L. C. Leite, J. M. Ferreira, Jr., P. L. Ho, E. N. Miyaji, and M. L. Oliveira. 2009. Characterization of protective mucosal and systemic immune responses elicited by pneumococcal surface protein PspA and PspC nasal vaccines against a respiratory pneumococcal challenge in mice. Clin. Vaccine Immunol. 16:636-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giefing, C., A. L. Meinke, M. Hanner, T. Henics, M. D. Bui, D. Gelbmann, U. Lundberg, B. M. Senn, M. Schunn, A. Habel, B. Henriques-Normark, A. Ortqvist, M. Kalin, A. von Gabain, and E. Nagy. 2008. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J. Exp. Med. 205:117-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gor, D. O., X. Ding, D. E. Briles, M. R. Jacobs, and N. S. Greenspan. 2005. Relationship between surface accessibility for PpmA, PsaA, and PspA and antibody-mediated immunity to systemic infection by Streptococcus pneumoniae. Infect. Immun. 73:1304-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haber, M., A. Barskey, W. Baughman, L. Barker, C. G. Whitney, K. M. Shaw, W. Orenstein, and D. S. Stephens. 2007. Herd immunity and pneumococcal conjugate vaccine: a quantitative model. Vaccine 25:5390-5398. [DOI] [PubMed] [Google Scholar]
- 19.Hakansson, A., H. Roche, S. Mirza, L. S. McDaniel, A. Brooks-Walter, and D. E. Briles. 2001. Characterization of binding of human lactoferrin to pneumococcal surface protein A. Infect. Immun. 69:3372-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hicks, L. A., L. H. Harrison, B. Flannery, J. L. Hadler, W. Schaffner, A. S. Craig, D. Jackson, A. Thomas, B. Beall, R. Lynfield, A. Reingold, M. M. Farley, and C. G. Whitney. 2007. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998-2004. J. Infect. Dis. 196:1346-1354. [DOI] [PubMed] [Google Scholar]
- 21.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu, H. E., K. A. Shutt, M. R. Moore, B. W. Beall, N. M. Bennett, A. S. Craig, M. M. Farley, J. H. Jorgensen, C. A. Lexau, S. Petit, A. Reingold, W. Schaffner, A. Thomas, C. G. Whitney, and L. H. Harrison. 2009. Effect of pneumococcal conjugate vaccine on pneumococcal meningitis. N. Engl. J. Med. 360:244-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iannelli, F., M. R. Oggioni, and G. Pozzi. 2002. Allelic variation in the highly polymorphic locus pspC of Streptococcus pneumoniae. Gene 284:63-71. [DOI] [PubMed] [Google Scholar]
- 24.King, Q. O., B. Lei, and A. G. Harmsen. 2009. Pneumococcal surface protein A contributes to secondary Streptococcus pneumoniae infection after influenza virus infection. J. Infect. Dis. 200:537-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyaw, M. H., R. Lynfield, W. Schaffner, A. S. Craig, J. Hadler, A. Reingold, A. R. Thomas, L. H. Harrison, N. M. Bennett, M. M. Farley, R. R. Facklam, J. H. Jorgensen, J. Besser, E. R. Zell, A. Schuchat, and C. G. Whitney. 2006. Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N. Engl. J. Med. 354:1455-1463. [DOI] [PubMed] [Google Scholar]
- 26.LeMessurier, K. S., A. D. Ogunniyi, and J. C. Paton. 2006. Differential expression of key pneumococcal virulence genes in vivo. Microbiology 152:305-311. [DOI] [PubMed] [Google Scholar]
- 27.Mahdi, L. K., A. D. Ogunniyi, K. S. LeMessurier, and J. C. Paton. 2008. Pneumococcal virulence gene expression and host cytokine profiles during pathogenesis of invasive disease. Infect. Immun. 76:646-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDaniel, L. S., D. O. McDaniel, S. K. Hollingshead, and D. E. Briles. 1998. Comparison of the PspA sequence from Streptococcus pneumoniae EF5668 to the previously identified PspA sequence from strain Rx1 and ability of PspA from EF5668 to elicit protection against pneumococci of different capsular types. Infect. Immun. 66:4748-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDaniel, L. S., B. A. Ralph, D. O. McDaniel, and D. E. Briles. 1994. Localization of protection-eliciting epitopes on PspA of Streptococcus pneumoniae between amino acid residues 192 and 260. Microb. Pathog. 17:323-337. [DOI] [PubMed] [Google Scholar]
- 30.McDaniel, L. S., G. Scott, J. F. Kearney, and D. E. Briles. 1984. Monoclonal antibodies against protease-sensitive pneumococcal antigens can protect mice from fatal infection with Streptococcus pneumoniae. J. Exp. Med. 160:386-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDaniel, L. S., J. S. Sheffield, P. Delucchi, and D. E. Briles. 1991. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect. Immun. 59:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDaniel, L. S., J. S. Sheffield, E. Swiatlo, J. Yother, M. J. Crain, and D. E. Briles. 1992. Molecular localization of variable and conserved regions of pspA and identification of additional pspA homologous sequences in Streptococcus pneumoniae. Microb. Pathog. 13:261-269. [DOI] [PubMed] [Google Scholar]
- 33.Meinke, A., T. Henics, M. Hanner, D. B. Minh, and E. Nagy. 2005. Antigenome technology: a novel approach for the selection of bacterial vaccine candidate antigens. Vaccine 23:2035-2041. [DOI] [PubMed] [Google Scholar]
- 34.Nabors, G. S., P. A. Braun, D. J. Herrmann, M. L. Heise, D. J. Pyle, S. Gravenstein, M. Schilling, L. M. Ferguson, S. K. Hollingshead, D. E. Briles, and R. S. Becker. 2000. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies to heterologous PspA molecules. Vaccine 18:1743-1754. [DOI] [PubMed] [Google Scholar]
- 35.Ogunniyi, A. D., M. C. Woodrow, J. T. Poolman, and J. C. Paton. 2001. Protection against Streptococcus pneumoniae elicited by immunization with pneumolysin and CbpA. Infect. Immun. 69:5997-6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park, I. H., D. G. Pritchard, R. Cartee, A. Brandao, M. C. Brandileone, and M. H. Nahm. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 45:1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roche, H., A. Hakansson, S. K. Hollingshead, and D. E. Briles. 2003. Regions of PspA/EF3296 best able to elicit protection against Streptococcus pneumoniae in a murine infection model. Infect. Immun. 71:1033-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roche, H., B. Ren, L. S. McDaniel, A. Hakansson, and D. E. Briles. 2003. Relative roles of genetic background and variation in PspA in the ability of antibodies to PspA to protect against capsular type 3 and 4 strains of Streptococcus pneumoniae. Infect. Immun. 71:4498-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schenkein, J. G., M. H. Nahm, and M. T. Dransfield. 2008. Pneumococcal vaccination for patients with COPD: current practice and future directions. Chest 133:767-774. [DOI] [PubMed] [Google Scholar]
- 40.Singleton, R. J., T. W. Hennessy, L. R. Bulkow, L. L. Hammitt, T. Zulz, D. A. Hurlburt, J. C. Butler, K. Rudolph, and A. Parkinson. 2007. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA 297:1784-1792. [DOI] [PubMed] [Google Scholar]
- 41.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 42.Wardlaw, T., E. White Johansson, and M. Hodge. 2006. Pneumonia: the forgotten killer of children. The United Nations Children's Fund (UNICEF)/World Health Organization (WHO), Geneva, Switzerland.
- 43.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, N. M. Bennett, R. Lynfield, A. Reingold, P. R. Cieslak, T. Pilishvili, D. Jackson, R. R. Facklam, J. H. Jorgensen, and A. Schuchat. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737-1746. [DOI] [PubMed] [Google Scholar]
- 44.Xin, W., S. Y. Wanda, Y. Li, S. Wang, H. Mo, and R. Curtiss III. 2008. Analysis of type II secretion of recombinant pneumococcal PspA and PspC in a Salmonella enterica serovar Typhimurium vaccine with regulated delayed antigen synthesis. Infect. Immun. 76:3241-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yother, J., and D. E. Briles. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J. Bacteriol. 174:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yother, J., G. L. Handsome, and D. E. Briles. 1992. Truncated forms of PspA that are secreted from Streptococcus pneumoniae and their use in functional studies and cloning of the pspA gene. J. Bacteriol. 174:610-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yother, J., and J. M. White. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 176:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.