Abstract
Bacteria can detect, transmit, and react to signals from the outside world by using two-component systems (TCS) and serine-threonine kinases and phosphatases. Streptococcus mutans contains one serine-threonine kinase, encoded by pknB. A gene encoding a serine-threonine phosphatase, pppL, is located upstream of pknB. In this study, the phenotypes of pknB and pppL single mutants and a pknB pppL double mutant were characterized. All mutants exhibited a reduction in genetic transformability and biofilm formation, showed abnormal cell shapes, grew slower than the wild-type strain in several complex media, and exhibited reduced acid tolerance. The mutants had reduced cariogenic capacity but no significant defects in colonization in a rat caries model. Whole-genome transcriptome analysis revealed that a pknB mutant showed reduced expression of genes involved in bacteriocin production and genetic competence. Among the genes that were differentially regulated in the pknB mutant, several were likely to be involved in cell wall metabolism. One such gene, SMU.2146c, and two genes encoding bacteriocins were shown to also be downregulated in a vicK mutant, which encodes a sensor kinase involved in the response to oxidative stress. Collectively, the results lead us to speculate that PknB may modulate the activity of the two-component signal transduction systems VicKR and ComDE. Real-time reverse transcriptase PCR (RT-PCR) showed that genes downregulated in the pknB mutant were upregulated in the pppL mutant, indicating that PppL serves to counteract PknB.
Dental plaque is a complex biofilm on the tooth surface and principal habitat of more than 500 different bacterial species (33). Mutans streptococci, in particular Streptococcus mutans, that reside in this biofilm are the major cause of dental caries (26), due to their capacity to metabolize carbohydrates present in our diet to lactic acid. The resulting local decrease in pH causes demineralization of the tooth enamel and tooth dentin. Apart from production of acid from sugars, the ability to stick to the tooth surface and grow in biofilms, tolerance of low pH, and production of bacteriocins are important virulence factors of S. mutans.
All living organisms have constantly to adapt to changes in the environment. In bacteria, signals from the outside world are frequently detected and transmitted by two-component systems (TCS), which consist of a membrane-located sensor histidine kinase and response regulator, which is localized in the cytoplasm. In response to an environmental signal, the former protein is autophosphorylated on a histidine residue. The phosphate group is then transferred to an aspartate residue of the response regulator, which, in its active phosphorylated form, regulates the transcription of target genes. S. mutans contains 14 TCS (5, 23), some of which have been studied in detail. The ComDE TCS is essential for expression of bacteriocins (49) and is involved in the development of genetic competence (24). The VicKR TCS is required for the response to oxidative stress (10, 42).
Another type of signal transduction system comprises serine-threonine protein kinases (STPKs), which were first discovered in eukaryotes but later found to be widespread in bacteria as well. In streptococci, the presence of an STPK was first described for Streptococcus agalactiae (35). Most of the bacterial STPKs are predicted to consist of an N-terminal kinase domain in the cytoplasm, a central membrane-located domain, and a C-terminal sensory domain located extracellularly. Like histidine kinases, STPKs are autophosphorylated, but in this case on serine or threonine residues. In contrast to two-component response regulators, STPKs exert their effect by phosphorylation of target proteins. Relatively little information is available on these target proteins, but data obtained so far show little overlap between organisms (11, 17, 31, 37). Prokaryotic STPKs have been shown to regulate various cellular functions including virulence in streptococci (13, 17, 35). Dephosphorylation of STPKs is thought to be carried out by their cognate serine-threonine protein phosphatases (STPPs), which was confirmed in vitro (6, 17, 31, 35). Thus, STPPs might reverse the effect of STPKs.
S. mutans contains one STPK and one STPP, encoded by pknB and pppL, respectively (16). Mutation of pknB, which is located immediately downstream of pppL, results in a pleiotropic phenotype: defects in the development of genetic competence, in the ability to form biofilms, and in tolerance of low pH (16).
In the present study, we have carried out further phenotypic investigations of the pknB mutant, of a pppL mutant, and of a pknB pppL double mutant. Microarray analysis was used to analyze the transcriptome of a pknB mutant. It is shown that the pknB regulon overlaps with the vicRK regulon and comDE regulons, suggesting cross talk between PknB and these TCS.
MATERIALS AND METHODS
Organisms and growth conditions.
S. mutans strains (Table 1) were routinely grown at 37°C under anaerobic conditions or in an atmosphere of 5 to 10% CO2 and 90% air in brain heart infusion (BHI) medium (Oxoid) or in THY medium, which consists of Todd-Hewitt broth (Becton-Dickinson) with 0.3% yeast extract (Oxoid). To assay biofilm formation, the strains were grown in TSBY broth, which consists of tryptone soy broth (Oxoid) with 0.5% yeast extract (Oxoid). When required, antibiotics were added at the following concentrations: erythromycin, 10 μg/ml; kanamycin, 750 μg/ml; streptomycin, 700 μg/ml; spectinomycin, 700 to 1,000 μg/ml. For induction of the promoter in the complemented strains carrying pLR16T (see below), anhydrotetracycline (ATC; Sigma Aldrich) was used at a concentration of 0.5 μg/ml. Escherichia coli strain JM109, XL1-Blue, or DH5α was used as the host for propagation of plasmids and was grown in Luria-Bertani (LB) medium at 37°C with aeration. When required, antibiotics were added at the following concentrations: erythromycin, 200 to 400 μg/ml; kanamycin, 50 μg/ml; spectinomycin, 100 μg/ml.
TABLE 1.
S. mutans strains used in this study
| Strain designation | Genotype or characteristics | Source or reference |
|---|---|---|
| UA159 | Wild-type ATCC 700610 | ATCC |
| EA72 | UA159 ΔpknB::aphA3 Kmr | 16 |
| EA74 | UA159 ΔpppL::erm Ermr | This study |
| EA75 | UA159 ΔpknB::aphA3 ΔpppL::erm Kmr Emr | This study |
| OMZ 955 | UA159 Δhk11::ermAM | D. Cvitkovitch, University of Toronto |
| OMZ 953 | UA159 ΔvicK::ermAM | D. Cvitkovitch, University of Toronto |
| OMZ 1110 | UA159 ΔciaRH::erm | This study |
| OMZ 1111 | UA159 ΔcomDE::erm | This study |
| OMZ 1112 | UA159 ΔpknB::aphA3 ΔciaRH::erm | This study |
| EA82 | EA72 carrying pLR16T, with 0.7-kb fragment containing pknB plus RBS | This study |
| EA83 | EA74 carrying pLR16T with 1.8-kb fragment containing pppL with RBS | This study |
| EA84 | EA75 carrying pLR16T with 2.6-kb fragment containing pppL and pknB with RBS | This study |
| EA79 | EA72 carrying pDL278 containing Perm from pVA838 and 0.7-kb fragment containing pknB with RBS | This study |
| EA80 | EA74 carrying pDL278 containing Perm from pVA838 and 1.8-kb fragment containing pppL with RBS | This study |
| EA81 | EA75 carrying pDL278 containing Perm from pVA838 and 2.6-kb fragment containing pppL and pknB with RBS | This study |
| EA78 | EA72 carrying pVA838 containing Perm and pknB with RBS | 16 |
Construction of mutants and complemented strains.
An approach similar to that used for construction of the pknB mutant (16) was used to construct a pppL mutant (strain EA74): a 744-bp DNA fragment encompassing almost the entire pppL gene (SMU.483) was PCR amplified and cloned in pGEM-T Easy to produce plasmid pEA80. Outward-facing primers with 5′ restriction sites were designed to anneal to the insert in pEA80 such that inverse PCR resulted in deletion of a 97-bp fragment and introduction of a unique SmaI site. The erythromycin resistance gene and its promoter were amplified from pVA838 (27) and cloned into the unique SmaI site to generate plasmid pEA81. The insert was released from pEA81 and transformed into S. mutans UA159 as a linear fragment. Allelic replacement was confirmed by PCR using primers flanking pppL. To construct the pppL pknB double mutant (strain EA75), the insertionally inactivated pknB and pppL genes were liberated as linear fragments from pEA72 (16) and pEA81, respectively, and cotransformed into strain UA159. Allelic replacement was confirmed by PCR. Construction of ciaRH and comDE mutants was carried out as described previously (49).
For genetic complementation, two different plasmids were used, pDL278 (22) and pLR16T (37). The erythromycin resistance gene promoter from pVA838 (27) was cloned as an EcoRI/SalI fragment into the E. coli/Streptococcus shuttle vector pDL278 (22) to drive expression of the wild-type genes, which were cloned, together with a ribosome binding site (RBS), as SalI/SphI fragments. Complemented strains were also constructed using the shuttle vector pLR16T, which contains a tetracycline-inducible promoter (37). The wild-type genes plus RBS were cloned into pLR16T as SalI/SphI fragments.
Electron microscopy.
Strains UA159, EA72, EA74, and EA75 were grown in THY broth to the mid-exponential phase (optical density at 600 nm [OD600] = 0.3). Growth was arrested by fixation in 2.5% glutaraldehyde, followed by 1% osmium tetroxide in 0.185 M cacodylate buffer, and dehydration using a graded series of ethanol. Propylene oxide was then used to facilitate the infiltration with Epon. Ultrathin sectioning was done with a Reichert-Jung Ultracut E ultramicrotome, and the sections were mounted on uncoated 400 mesh copper grids. Samples were contrasted first with saturated, acidified uranyl acetate as described previously (47), omitting the addition of methanol, and then with lead citrate as described previously (50), both for 3 min each. The sections were viewed on a Philips EM400T transmission electron microscope (TEM) at 60 kV.
Biofilm assay.
Biofilm formation was quantified using a polystyrene microtiter plate assay. Overnight cultures in TSBY broth were diluted to an optical density of 0.1 into the wells of a 96-well microtiter tray (Sarstedt; no. 83.1835) containing 200 μl TSBY broth supplemented with sucrose (10 mg/ml) and incubated for 24 h without agitation. The medium was removed, and the wells were washed twice with sterile phosphate-buffered saline (PBS). The biofilms were first fixed for 15 min in 99% (vol/vol) methanol and then stained with 0.1% (vol/vol) safranin for 5 min. After staining, the wells were washed twice with PBS to remove excess dye and then dried at 50°C for 30 min. Biofilms were quantified by measuring the absorbance of the stained biofilms at 492 nm using a microplate reader (MRX II; Dynex Technologies). Statistical significance was determined using the Mann-Whitney test.
Genetic competence assays.
Overnight cultures in 10 ml of Todd-Hewitt broth supplemented with 10% horse serum were diluted 1 in 40 into 10 ml of fresh medium and grown for 3.5 h at 37°C. One microgram of genomic DNA from a spontaneously streptomycin-resistant variant of UA159 was added to 1 ml of each culture, and incubation was continued for a further 3 h before plating on agar containing streptomycin. A control without added DNA was included with every transformation to determine the rate of spontaneous mutation. Total viable counts were determined by plating dilutions on agar without antibiotic. Statistical significance was determined using the Mann-Whitney test. For assays using competence-stimulating peptide (CSP), overnight cultures were diluted 1 in 40 into Todd-Hewitt broth with 10% horse serum and incubated for 1 h. Each culture was then split into two 5-ml volumes, and CSP was added to one volume at a final concentration of 1 μg/ml. CSP with a purity of 99% was synthesized by Sigma-Genosys. The cultures were then incubated at 37°C for a further 3 h before plating on selective and nonselective agar.
Acid tolerance assay.
Overnight cultures in BHI broth were diluted 1:25 into fresh broth (containing anhydrotetracycline as appropriate) and grown until late exponential phase (OD ∼ 0.7). The cultures were then diluted, and 10-μl volumes were spotted onto THY agar (containing ATC at a final concentration of 0.5 μg/ml) at either pH 7.0 or pH 5.0 and incubated for 72 h.
RNA isolation, cDNA preparation and labeling, and microarray analysis.
Overnight cultures of S. mutans strains UA159 and EA72 grown in THY medium were brought to an OD550 of 1.5, diluted 100-fold, and cultured until the OD550 was around 0.3 for both strains. Total RNA was extracted from 5 ml of liquid culture using an RNeasy Mini kit (Qiagen) with on-column RNase-free DNase digestion according to the manufacturer's recommendation. All RNA samples were checked for integrity by denaturing formaldehyde agarose gel electrophoresis and by electrophoresis on a 2100 Bioanalyzer (Agilent). Total RNA (7 μg) was reverse transcribed and labeled using a CyScribe postlabeling kit (GE Healthcare). Approximately 500 ng of each Cy3- and Cy5-labeled cDNA was used for hybridization on an S. mutans whole-genome chip (46) using a fully automated hybridization station (Tecan). The arrays were scanned using a DNA microarray scanner (Agilent) at full laser power. GeneSpotter software (MicroDiscovery GmbH) was used for determining the average signal intensity and the local background for each spot. The data were then imported into the program Genespring (Agilent), normalized using Lowess normalization, and filtered, leaving out intensities below 100 in both channels. The statistical significance of ratios was determined by applying Student's t test. Genes were considered to be differentially expressed if the ratio was above 2 with a P value of ≤0.05. The experiment was done with 3 biological replicates, including one dye swap.
Quantitative RT-PCRs.
Quantitative reverse transcriptase PCRs (RT-PCRs) were performed with the same RNA samples that were used for microarray experiments. The gene-specific primers were designed using Primer Express software (Applied Biosystems) and are listed in Table 2. RNA samples were treated with DNase (Promega) before reverse transcription. A reverse transcription reaction mixture containing 500 ng total RNA, 200 ng random hexamers, and 10 mM deoxynucleoside triphosphates (dNTPs) was incubated at 65°C for 5 min. After the mixture was cooled on ice for at least 1 min, 4 μl of 5× first-strand buffer, 1 μl of 0.1 M dithiothreitol (DTT), 1 μl of RNaseOUT (Invitrogen), and 200 units of SuperScript III RT (Invitrogen) were added to the reaction mixture, which was incubated at 25°C for 5 min followed by 1 h at 50°C. Reactions were inactivated by incubation at 70°C for 15 min. The same samples but without reverse transcriptase enzyme were run in parallel to check for DNA contamination. The real-time PCR mixture (25 μl) contained 1× SYBR green PCR master mix (Applied Biosystems), 2.5 ng cDNA, and 50 nM appropriate forward and reverse PCR primers. Reactions were run on an ABI-Prism 7000 sequence detection system (Applied Biosystems) with the following parameters: initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. The amount of target gene mRNA was normalized to the amount of 16S rRNA. Each assay was performed with at least 2 independent RNAs in duplicate.
TABLE 2.
List of primers used for real-time PCR
| Target gene | Primer name | Sequence (5′-3′) |
|---|---|---|
| 16S rRNA | 16SrRNAf | CGCAGGCGGTCAGAAA |
| 16SrRNAr | TTCCAGAGCACACTATGGTTGAG | |
| SMU.20 (mreC) | SMU20f | TCCCTGATCTAACTGTTTTGTCATCT |
| SMU20r | TGTCACCCCATCATTTTTTCC | |
| SMU.503c | SMU503f | TGCAGCCTGGTTTCATCCTAA |
| SMU503r | TGGATAGGGATTGGGATGTGA | |
| SMU.609c | SMU609f | AGAAACACCCCAAGTTGAAGTTG |
| SMU609r | TTCTTTGGATTTTGCACCTTTGT | |
| SMU.625 (comEA) | SMU625f | GCATGAGGGCGTCTATACTTTACC |
| SMU625r | TTTGCGATCAGCCTTTTCAGA | |
| SMU.910 (gtfD) | SMU910f | CAGGCAGCCAACGCATTAA |
| SMU910r | AGCCCTCGCTCATCATAAGC | |
| SMU.984 | SMU984f | CAACGGGTAATCAGGCAAAAG |
| SMU984r | TCTACAAAAGCAAGATGACTAGAAGACTCT | |
| SMU.1882c | SMU1882f | ATTATTCAAAATGGCTGGGTGAA |
| SMU1882r | TTACTAAATCCTCTATTACCTTGAGCTGAGT | |
| SMU.1986c (bsmH) | SMU1896f | AGACATGTTAGCCGCTGTTGAAG |
| SMU1896r | AAGCGCCTGTTCCAATCGTA | |
| SMU.1910c | SMU1910f | TTTTCTGTCTACTGGCCGAATG |
| SMU1910r | CAAATGTATAAATGATTACCGTCTTCCT | |
| SMU.1914c (bsmA) | SMU1914f | GGTGCTGGGCAAGGTTATATG |
| SMU1914r | CCGATTCCTCCAGCAATAGC | |
| SMU.1984 (comYC) | SMU1984f | TCAGGCTGAGCTTTATGAGCTTAA |
| SMU1984r | CCTTGTATGAATTTGCCTGTTCTTG | |
| SMU.2146c | SMU2146f | TCAGGAGCTTCAGGTCTCTTTCA |
| SMU2146r | CCTTGGGCTTTATAAGCATTGATAG |
Bacteriocin assays.
Bacteriocin production was evaluated as described before (49). In brief, producer strains were grown overnight in THY broth, stab inoculated into THY agar, and grown in an environment of 10% CO2. After 24 h, 0.1 ml of an overnight culture of the indicator strain (Enterococcus faecalis CG110) was mixed with 4 ml of molten THY top agar and poured over the plate. The plates were then incubated for 24 h at 37°C. The diameter of the zone of inhibition around the producing strains was measured.
Rat caries model.
Specific-pathogen-free, caries-susceptible Osborne-Mendel rats (Institute of Oral Biology, University of Zurich) were used to investigate in vivo effects of the mutants on smooth-surface dental plaque and initial and advanced dentinal fissure caries, as well as on the establishment of S. mutans in the oral microbiota. Each experimental group consisted of 10 animals. Thirteen days after birth, the pups and their dams were transferred to stainless-steel screen bottom cages without bedding and fed a finely ground stock diet (diet no. 3433; Provimi Kliba AG) to prevent impaction of food and bedding particles in fissures. Tap water was given ad libitum. On day 20 after birth, the dams were removed, the oral cavities of 7 of the dams were swabbed to exclude the presence of S. mutans, and the littermates were distributed among the 5 different groups, 2 animals per cage. The cages and feeding equipment had been sterilized prior to being occupied by the animals. An uninfected control group was also included in the study as a control for background conditions. These animals were placed in a separate rack as far away as possible from the other racks in order to minimize the risk of cross-infection. On days 21 and 22, each rat was infected orally, twice daily, by inoculation of 200 μl of a heavy suspension (approximately 108 CFU) of the bacterial strains. The uninfected control group was given 200 μl of distilled water. To promote colonization of the bacteria, all rats, including the noninfected control group, received drinking water containing 2% sucrose and 2% glucose during days 20 to 22 as well as cariogenic diet 2000a (15), containing 40% sucrose. Following the period of colonization, on day 23, feeding with diet 2000a was continued but carbohydrates were omitted from the drinking water. Sterilization of the feeding and housing equipment was resumed. The trays containing urine and feces were sterilized daily, drinking bottles and food cups were sterilized every other day, and the cages for all the animals were sterilized weekly. Hand disinfection and the use of fresh disposable aprons were employed between handling the different groups. Shortly before the end of the study, swabs were taken from all 40 rats to obtain a final count of the microbial status of the animals. For each animal a sterile, moistened swab was rubbed twice against the lingual surfaces of molars situated in the upper and lower jaws on both sides of the mouth. Microbiological analysis of the swabs was as described previously (40). On day 52, at the end of the 28-day experimental period, the animals were sacrificed. The upper and lower jaws were dissected out and immersed in fixative (10% buffered formalin phosphate) for a minimum of 72 h. Maxillary molars were evaluated for plaque extent using the method previously described (38). Briefly, the removed jaws were dipped into 1% erythrosin for 15 s and destained by placing them under running tap water for 15 s. The extent of smooth-surface plaque on the first and second molars in the four quadrants was evaluated under a dissection microscope using a grading index from 0 to 4. Absence of smooth-surface plaque was scored 0, and complete covering of smooth surfaces with plaque was scored 4. Smooth-surface caries lesions were scored using Keyes area units E (18) for the first molar (6 units at risk) and the second molar (4 units at risk). By this method, the maximum score for both mandibles was 20. Mandibular molars were sectioned and scored for fissure caries (19). Here, the 12 fissures from the first and second mandibular molars were examined for the presence of initial and advanced dentinal lesions. Data were compared with a two-way analysis of variance, and least significant differences were calculated using the Anova statistics program.
Microarray data accession number.
Microarray data have been submitted to GEO under accession number GSE18355.
RESULTS
Defects in cell shape and cell division in the mutants.
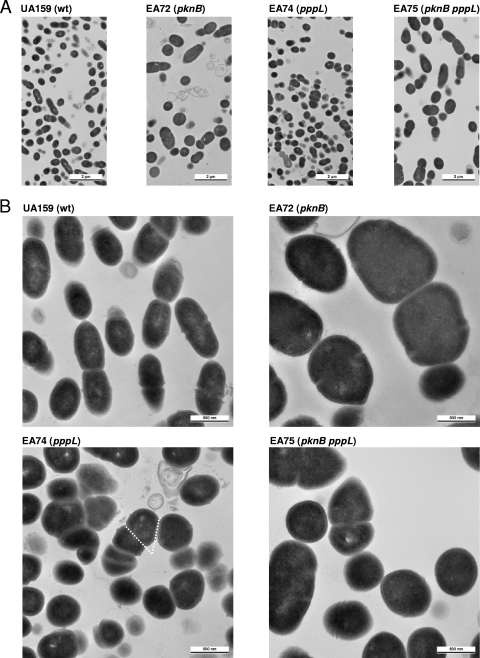
Transmission electron microscopy showed that cells of the pknB mutant had an abnormal shape (Fig. 1), appearing larger and more rounded than the wild-type strain. As opposed to the wild-type strain, a significant fraction (about 10%) of the cells of the pknB mutant appeared to have lysed, since we observed structures resembling ghost cells and debris. Cells of strain EA74 appeared the same size as the wild type but were also more rounded and showed irregular cell division. In several dividing cells of strain EA74, the division plane was at an angle or perpendicular to the plane of the previous division, in contrast to the wild-type strain, in which the division plane was parallel to the previous one. Strain EA75 showed morphology similar to that of strain EA72, except for the apparent absence of lysed cells. These results suggest a role for both pknB and pppL in the maintenance of cell shape and in cell division.
FIG. 1.
Transmission electron microscopy of S. mutans UA159 and the mutants EA72, EA74, and EA75 (see Table 1) at magnifications of ×2,500 (A) and ×12,000 (B). The dashed lines indicate planes of division which are in an angle. wt, wild type.
The mutants have growth defects and are sensitive to stress.
Growth of the mutants in BHI broth was compared with that of the wild-type strain. As all the mutants displayed an increased tendency to clump, the cultures were vortexed thoroughly before removing samples for measurement of optical density. Compared to the wild-type strain, strain EA72 and strain EA75 showed a reduction in growth rate in the early phases of the growth cycle, but by 24 h all three strains had reached similar cell densities. In contrast, growth of the pppL mutant (strain EA74) was significantly retarded throughout the growth cycle compared to that of the wild-type strain (data not shown). Similar results were obtained when the strains were grown in THY medium (data not shown). In contrast, TSBY supported improved growth of the mutant strains, with the kinase, phosphatase, and double mutants reaching mean ODs (in 3 experiments) of 1.0, 1.1, and 1.1, respectively, in 7 h, compared with a mean OD of 1.3 for the wild-type strain.
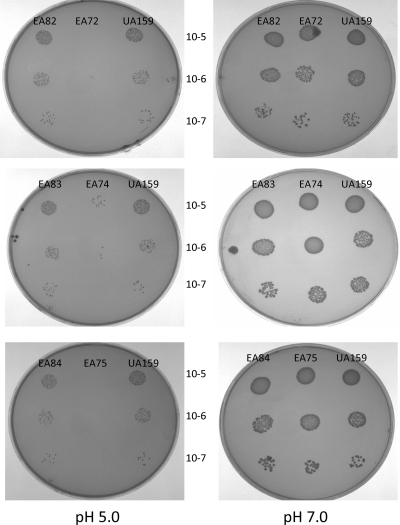
The ability of the mutants to grow on agar at pH 5.0 was examined. In three independent experiments, all the mutants showed a reduced ability to grow at pH 5.0 (Fig. 2). Interestingly, we did not see complementation with strains EA79, EA80, and EA81, which carry pDL278 derivatives in which the wild-type genes are expressed from an erythromycin resistance gene promoter (Perm), whereas we have previously reported complementation when pknB is expressed from Perm in pVA838 (16). Since pVA838 and pDL278 are derived from the same replicon (22), we assume that the expression levels of these genes are critical for acid tolerance and that the different manipulations each plasmid has undergone affected copy number. Therefore, complemented strains EA82, EA83, and EA84 were constructed using pLR16T, which was used previously for complementation of an STPK mutant strain of Streptococcus agalactiae (37) (pVA838 could not be used here as the pppL mutant was constructed using an erythromycin resistance gene). In all cases, delivery of the wild-type gene in pLR16T restored the ability to grow at pH 5.0 (Fig. 2).
FIG. 2.
Growth of mutants (see Table 1) at pH 7.0 and pH 5.0.
Addition of 25 mM paraquat (inducer for superoxide) to the growth medium (THY) resulted in an increase in the lag phase of the wild-type strain of about 7 h, whereas the pknB mutant did not grow at all (data not shown). Addition of 0.25 M NaCl to the growth medium also increased the lag phase of the pknB mutant but not that of the wild-type strain (data not shown). These results suggest that the pknB mutant is more sensitive to acid, oxidative, and osmotic stress than the wild-type strain.
The mutants have a biofilm defect.
We have previously demonstrated, using confocal laser scanning microscopy, that the pknB mutant (EA72) has a reduced ability to form biofilms (16). To determine the ability of the pppL mutant, the double mutant, and complemented strains to form biofilms, we used a polystyrene microtiter plate assay. As the mutants demonstrated planktonic growth comparable to that of the wild-type in TSBY broth (see above), this medium was used for the biofilm assay. Measurement of the optical density of safranin-stained biofilms showed that the thicknesses of biofilm produced by the kinase, phosphatase, and double mutants were 39%, 30%, and 54% of that of the wild-type strain, respectively (P < 0.00001; data not shown). Although it is possible that the requirements for growth in biofilms are different from those in broth, we conclude that the biofilm defect is not simply the result of a general growth defect. Genetic complementation (strains EA78, EA80, and EA81) resulted in partial restoration of the defect in all cases (P < 0.00001), demonstrating specifically the involvement of the pknB and pppL genes in the biofilm phenotype.
The mutants have reduced transformability.
A 3-fold decrease in transformation frequency was apparent in the pknB mutant (strain EA72) compared to the parent (P = 0.02) (Table 3). Introduction in trans of the wild-type gene under the control of the erythromycin resistance gene promoter (Perm) in pDL278 gave a 26-fold increase in transformation efficiency compared to that for the mutant strain (P = 0.005), resulting in a transformation efficiency higher than that of the wild type. The phosphatase mutant showed a 94-fold decrease in transformability compared to the wild type (P = 0.005), and again, transformability was restored to a level greater than that of the wild type by complementation. The double mutant showed a 70-fold reduction in transformation frequency compared to the wild type, and once again, competence was restored by complementation (Table 3).
TABLE 3.
Transformation efficiencies of wild type and mutantsa
| Strain | Count (CFU/ml) |
Transformation frequency | |
|---|---|---|---|
| Total | Transformants | ||
| UA159 | 5.6 × 109 (1.0 × 109-1.9 × 1010) | 4.4 × 103 (1.8 × 103-5.2 × 103) | 7.8 × 10−7 |
| EA72 | 4.6 × 108 (4.0 × 108-5.3 × 108) | 1.2 × 102 (3.0 × 101-1.6 × 102) | 2.6 × 10−7 |
| EA79 | 7.1 × 108 (6.5 × 108-8.0 × 108) | 4.9 × 103 (1.0 × 103-6.1 × 103) | 6.9 × 10−6 |
| EA74 | 6.0 × 108 (5.5 × 108-7.2 × 108) | 5 (0-20) | 8.3 × 10−9 |
| EA80 | 8.1 × 108 (6.2 × 108-9.0 × 108) | 2.2 × 103 (8.5 × 102-2.6 × 103) | 2.7 × 10−6 |
| EA75 | 4.2 × 108 (4.0 × 108-4.5 × 108) | 5 (0-10) | 1.1 × 10−8 |
| EA81 | 8.6 × 108 (8.0 × 108-9.0 × 108) | 1.5 × 103 (7.3 × 102-2.3 × 103) | 1.7 × 10−6 |
Results shown are the median counts (ranges) from three independent experiments, each performed in duplicate. All the mutant strains showed a significant reduction in transformability compared to the parent strain (P = 0.02 for EA72; P = 0.005 for EA74; P = 0.005 for EA75). The complemented strains showed a significant increase in transformability compared to the respective mutant strains (P = 0.005 for all three strains).
The addition of CSP at a final concentration of 1 μg/ml significantly increased the transformability of all strains, resulting in a 32-fold increase in transformation frequency in the parent strain (P < 0.01) and increases of approximately 5,000-fold, 130-fold, and 19,000-fold in the kinase, phosphatase, and double mutants, respectively (P < 0.01 for all mutants; data not shown).
The mutants have reduced cariogenicity in vivo.
In order to evaluate the effect of pknB and pppL mutations on plaque formation, cariogenic potential, and establishment in vivo, we used a rat model of dental caries (Table 4). The three mutant strains all exhibited increased plaque scores compared with the wild-type control (P < 0.001). The mutant strains in treatment groups 3 (pknB mutant) and 5 (pppL pknB mutant) displayed significantly less initial dentinal lesions than the wild type in treatment group 2 (P < 0.05), but the pppL mutant showed the same initial dentinal lesion incidence as the wild type. All three mutant strains showed significantly less advanced fissure lesions than the wild-type strain. Treatment groups 3 and 4 (pppL mutant) displayed reduced advanced dentinal lesion incidence at the borderline of significance (P < 0.05), whereas treatment group 5 showed a more significant difference (P < 0.01). The wild type exhibited significantly more enamel lesions than the mutants in treatment groups 3 and 5 (P < 0.01). No significant difference between the wild type and the pppL mutant was observed.
TABLE 4.
Influence of pknB and pppL deletion on smooth-surface plaque extent, initial and advanced dentinal fissure lesions, smooth-surface caries, and colonization properties
| Group (infecting strain) | Scorea for: |
Totala (log CFU) |
|||||
|---|---|---|---|---|---|---|---|
| Plaque extentb | Initial dentinal fissuresc | Advanced dentinal fissuresc | Smooth-surface cariesd | Bacteria | Streptococci | S. mutans bacteria | |
| 1 (none [water control]) | 2.8 ± 0.63*** | 9.5 ± 1.72*** | 6.6 ± 2.80*** | 0.5 ± 0.97*** | 7.6 ± 0.23 | 7.3 ± 0.33 | ND***e |
| 2 (wild type) | 1.5 ± 0.53 | 12.0 ± 0.00 | 11.2 ± 1.14 | 6.8 ± 4.34 | 7.5 ± 0.38 | 7.4 ± 0.36 | 7.2 ± 0.46 |
| 3 (EA72) | 2.6 ± 0.52*** | 10.9 ± 0.99* | 9.5 ± 0.71* | 1.4 ± 2.07** | 7.7 ± 0.26 | 7.6 ± 0.26 | 5.6 ± 2.97 |
| 4 (EA74) | 2.6 ± 0.84*** | 11.5 ± 0.71 | 9.6 ± 1.17* | 4.4 ± 3.89 | 7.3 ± 0.27* | 7.2 ± 0.34 | 6.4 ± 0.75 |
| 5 (EA75) | 2.7 ± 0.48*** | 10.9 ± 1.10* | 9.0 ± 1.33** | 1.6 ± 1.51** | 7.7 ± 0.18 | 7.5 ± 0.20 | 5.4 ± 2.92 |
***, P < 0.001; **, P < 0.01; *, P < 0.05. All groups were compared with group 2 (wild type).
Four units at risk.
Twelve fissures at risk.
Twenty units at risk.
ND, not detectable.
Compared with the group treated with the wild-type strain, treatment group 4 displayed a reduced number of total bacteria (P < 0.05); the other mutants were not significantly different from the wild type. None of the treatments with the three mutant strains were significantly different in the numbers of total streptococci from treatment with the wild-type strain. However, the pknB mutant strain in treatment group 3 displayed significantly more colonies than the pppL mutant strain in treatment group 4 (P < 0.01). With regard to the number of S. mutans CFU, there were no significant differences between any of the three mutant strains compared with the wild type in treatment group 2. Interestingly, for two animals from each of treatment groups 3 and 5, S. mutans could not be recovered after swabbing. Although this led to a reduction in the mean number of S. mutans CFU recovered, the differences were not statistically significant.
Transcriptome analysis of the pknB mutant.
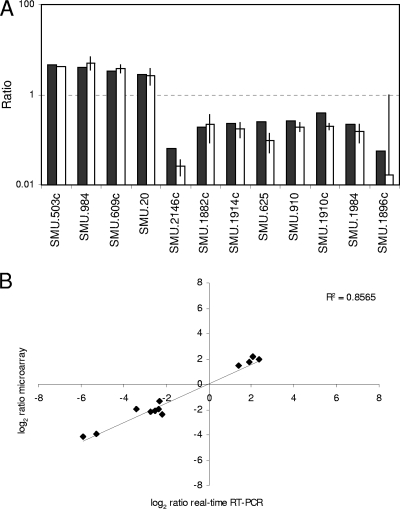
To identify genes whose expression is under direct or indirect control of PknB, the transcriptomes of strains UA159 and EA72 grown to the exponential growth phase (OD600 = 0.3) were compared. Data analysis revealed 26 upregulated genes and 41 downregulated genes in the pknB mutant (Table 5). Reverse transcriptase real-time PCR of 12 genes was performed to validate the results from microarray analysis. A good correlation between both methods was observed (Fig. 3) (r2 = 0.86).
TABLE 5.
Up- and downregulated genes in pknB mutant strain EA72
| ORF | Gene | Predicted protein or gene function | Change in expression (fold) |
|---|---|---|---|
| SMU.503c | Hypothetical protein | 4.61 | |
| SMU.984 | Hypothetical protein | 3.95 | |
| SMU.609 | Putative 40-kDa cell wall protein precursor | 3.31 | |
| SMU.1067c | Putative ABC transporter, permease protein | 2.84 | |
| SMU.334 | Argininosuccinate synthase (citrulline-aspartate ligase) | 2.81 | |
| SMU.21 | mreD | Putative cell shape-determining protein | 2.80 |
| SMU.753 | Conserved hypothetical protein | 2.79 | |
| SMU.20 | mreC | Putative cell shape-determining protein | 2.77 |
| SMU.614 | Hypothetical protein | 2.70 | |
| SMU.667 | nrdG | Putative ribonucleotide reductase, small subunit | 2.46 |
| SMU.985 | bglA | Putative β-glucosidase | 2.44 |
| SMU.1086 | kitH | Putative thymidine kinase | 2.44 |
| SMU.815 | Putative amino acid transporter, amino acid-binding protein | 2.43 | |
| SMU.510c | Hypothetical protein | 2.41 | |
| SMU.1422 | pdhB | Putative pyruvate dehydrogenase E1 component β-subunit | 2.37 |
| SMU.663 | argC | Putative N-acetyl-γ-glutamyl-phosphate reductase (N-acetyl-glutamate-γ-semialdehyde dehydrogenase) | 2.33 |
| SMU.1487 | Conserved hypothetical protein | 2.32 | |
| SMU.1424 | pdhD | Putative dihydrolipoamide dehydrogenase | 2.30 |
| SMU.1402c | Conserved hypothetical protein | 2.29 | |
| SMU.335 | Argininosuccinate lyase | 2.22 | |
| SMU.1070c | Conserved hypothetical protein | 2.18 | |
| SMU.877 | agaL | α-Galactosidase | 2.16 |
| SMU.529 | Hypothetical protein | 2.13 | |
| SMU.1404c | Conserved hypothetical protein | 2.07 | |
| SMU.879 | msmF | Multiple sugar-binding ABC transporter, permease protein | 2.05 |
| SMU.673 | Conserved hypothetical protein | 2.03 | |
| SMU.1896c | bmsH | Hypothetical protein | 0.06 |
| SMU.1895c | Hypothetical protein | 0.06 | |
| SMU.2146c | Hypothetical protein | 0.07 | |
| SMU.1882c | Hypothetical protein | 0.19 | |
| SMU.1063 | opuAa | Putative ABC transporter, ATP-binding protein, proline/glycine betaine transport system | 0.20 |
| SMU.1982c | Conserved hypothetical protein | 0.21 | |
| SMU.1981c | Conserved hypothetical protein | 0.21 | |
| SMU.1984 | comYC | Putative competence protein | 0.22 |
| SMU.1980c | Conserved hypothetical protein | 0.23 | |
| SMU.1987 | comYA | Putative ABC transporter, ATP-binding protein; late competence gene | 0.23 |
| SMU.285 | Hypothetical protein | 0.23 | |
| SMU.1914c | bsmA | Hypothetical protein | 0.23 |
| SMU.625 | comEA | Putative competence protein | 0.25 |
| SMU.277 | Hypothetical protein | 0.26 | |
| SMU.910 | gtfD | Glucosyltransferase-S | 0.26 |
| SMU.1912c | Hypothetical protein | 0.28 | |
| SMU.1908c | Hypothetical protein | 0.29 | |
| SMU.616 | Hypothetical protein | 0.30 | |
| SMU.278 | Hypothetical protein | 0.30 | |
| SMU.1062 | opuAb | Putative ABC transporter, proline/glycine betaine permease protein | 0.32 |
| SMU.453 | mraW | S-Adenosyl-methyltransferase | 0.33 |
| SMU.1906c | Hypothetical protein | 0.34 | |
| SMU.2089 | hexB | Putative mismatch repair protein | 0.34 |
| SMU.618 | Hypothetical protein | 0.34 | |
| SMU.1865 | mutY | Putative A/G-specific DNA glycosylase | 0.35 |
| SMU.281 | Hypothetical protein | 0.36 | |
| SMU.1444c | Conserved hypothetical protein | 0.36 | |
| SMU.1956c | Hypothetical protein | 0.37 | |
| SMU.1910c | Hypothetical protein | 0.39 | |
| SMU.1961c | Putative PTS, sugar-specific enzyme IIA component | 0.39 | |
| SMU.1909c | Hypothetical protein | 0.41 | |
| SMU.283 | Hypothetical protein | 0.42 | |
| SMU.2038 | pttB | Putative PTS, trehalose-specific IIABC component | 0.43 |
| SMU.992 | Hypothetical protein | 0.43 | |
| SMU.1979c | Conserved hypothetical protein | 0.44 | |
| SMU.2037 | treA | Putative trehalose-6-phosphate hydrolase | 0.45 |
| SMU.1957 | Putative PTS, mannose-specific IID component | 0.46 | |
| SMU.2116 | opuCa | Putative osmoprotectant amino acid ABC transporter, ATP-binding protein | 0.46 |
| SMU.279 | Hypothetical protein | 0.48 | |
| SMU.499 | Putative late competence protein | 0.48 | |
| SMU.1958c | Putative PTS, mannose-specific IIC component | 0.50 |
FIG. 3.
Inhibition of E. faecalis CG110 by S. mutans. wt, wild type.
Among the upregulated genes in the pknB mutant were several which are likely to be involved in synthesis, degradation, or remodeling of the cell wall. Expression of SMU.609, a putative murein hydrolase gene (7), was upregulated more than 3-fold in the pknB mutant. SMU.20 and SMU.21, which encode the putative cell shape-determining proteins MreC and MreD, respectively, were also upregulated. SMU.503c, encoding a probable lipoprotein of unknown function, was upregulated nearly 5-fold in the mutant. The two open reading frames (ORFs) adjacent to SMU.503c are transcribed in opposite directions, which implies that SMU.503 is a single-gene operon. Other upregulated genes included SMU.1067c, encoding a component of an ATP-binding cassette (ABC) transporter with unknown substrate, and SMU.334, as well as SMU.335, which is involved in biosynthesis of arginine.
SMU.1895c and SMU.1896c, probably encoding bacteriocins (49), were strongly downregulated in the pknB mutant. The expression of several other bacteriocins or bacteriocin-related genes was downregulated as well (SMU.1906, SMU.1908, SMU.1912, and SMU.1914). A cluster of genes encoding small proteins (SMU.277, SMU.278, SMU.279, SMU.281, SMU.283, and SMU.285) were all downregulated in the pknB mutant. The function of these genes is unknown, but of possible significance is that they are located immediately upstream of the comAB genes, which are required for export of CSP (49). Interestingly, use of BAGEL (9) resulted in the prediction that all six genes encoded bacteriocins.
SMU.2146c, encoding an amino acid sequence that shows similarity to those of transglycosylases involved in cell wall remodeling, was downregulated by approximately 5-fold in the pknB mutant. Six genes from the comY operon (28) were downregulated in the pknB mutant by 2- to 5-fold. These genes are directly involved in uptake of DNA during competence (28). Two other genes, SMU.625 and SMU.499, putatively encoding functions in competence development, were downregulated as well.
Other downregulated genes included SMU.1062 (opuAA), SMU.1063 (opuAB), and SMU.2116 (opuCA). These genes are thought to encode ABC transporters for uptake of compatible solutes and have been shown to be induced by osmotic stress (1). In addition, two phosphotransferase transport systems (PTSs), one for fructose/mannose (SMU.1956, SMU.1957, SMU.1958, SMU.1961, and SMU.1962) and one for trehalose (SMU.2037 and SMU.2038), were downregulated in the pknB mutant.
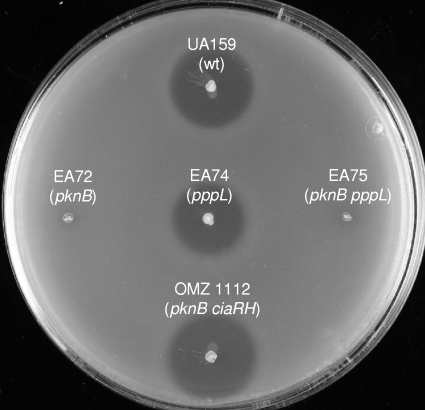
Bacteriocin production in mutants.
Production of bacteriocins was investigated in vitro by using deferred antagonism assays with Enterococcus faecalis CG110 (14) as the indicator strain. The pknB mutant and the double pknB pppL mutant, but not the pppL mutant, showed a strong reduction in inhibition (Fig. 4). The ciaRH two-component signal transduction system is known to regulate competence development and bacteriocin production in S. mutans (2, 34). A mutant in which pknB and both ciaR and ciaH had been inactivated inhibited growth of E. faecalis CG110 like the wild-type strain. Thus, deletion of ciaRH appears to counteract the pknB defect. Whereas the wild-type strain has immunity against its own bacteriocins, the pknB mutant was sensitive to the bacteriocins produced by the wild-type strain (data not shown).
FIG. 4.
(A) Comparison of microarray results and real-time RT-PCR results. Filled bars, results from microarray analysis; open bars, results from real-time RT-PCR analysis. (B) Correlation between gene expression data obtained from microarray analysis and real-time PCR analysis.
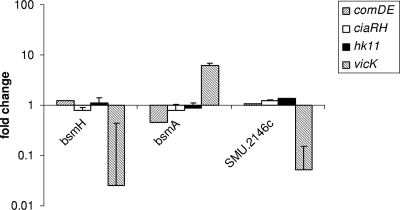
Expression of SMU.2146c and bsmH is downregulated in a vicK mutant.
In order to investigate whether TCS were involved in regulation of genes which were differentially expressed in the pknB mutant, real-time RT-PCR was employed to compare levels of expression of bsmA, bsmH (SMU.1896c), and SMU.2146c in the wild-type strain and in TCS mutants. RNA was extracted from cultures of S. mutans UA159 and derivatives thereof with mutations in comDE, HK11 (SMU.486), vicK, and ciaRH. The expression of SMU.2146c and bsmH was strongly downregulated in the vicK mutant but remained almost unchanged in the other mutants (Fig. 5). On the other hand, the expression of bsmA was upregulated in the vicK mutant but downregulated in the comDE mutant. The downregulation of bsmA in the comDE mutant corroborates previous results (49).
FIG. 5.
Expression of SMU.2146c, bsmH, and bsmA in hk11 (OMZ 955), vicK (OMZ 953), comDE (OMZ 1111), and ciaRH (OMZ 1110) mutants relative to that in the wild-type strain (UA159).
These results suggest that vicK exerts positive regulation on bsmH and SMU.2146c. The cognate response regulator of VicK, VicR, recognizes a consensus binding site (12). Inspection of the upstream sequences revealed the presence of a putative VicR binding site (TGTTATTTTCTTGCAATTATTTGACTT) upstream of bsmH but not of SMU.2146c.
Expression of PknB-regulated genes in a pppL mutant.
We hypothesized that the role of the putative PppL phosphatase is to counter the action of the PknB kinase. To validate this hypothesis, reverse transcriptase real-time PCR was used to analyze the expression levels of the PknB-regulated genes bsmA, bsmH, SMU.2146c, comYC, and mreC in strain EA74 (Fig. 6). Indeed, four genes that were downregulated in the pknB mutant were upregulated in the pppL mutant. However, the mreC gene, which was upregulated in the pknB mutant, was also slightly upregulated in the pppL strain.
FIG. 6.
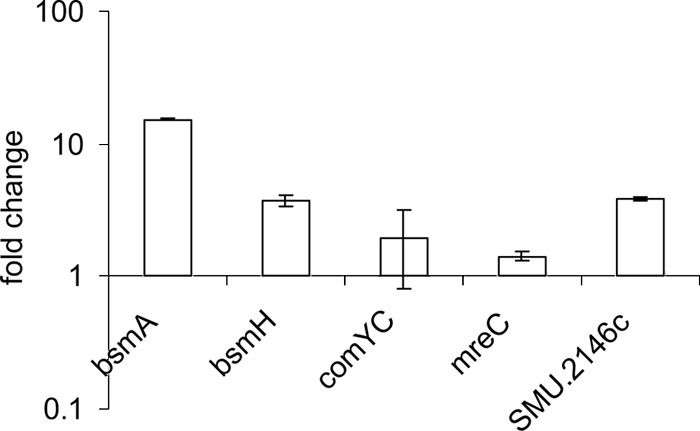
Real-time PCR analysis of expression of selected genes in the pppL mutant strain EA74 relative to that in the wild-type strain.
DISCUSSION
This study has shown that the pknB mutant of S. mutans is deficient in a variety of virulence properties: development of genetic competence, biofilm formation, acid tolerance, and production of bacteriocins. Electron microscopic analysis revealed aberrant cell sizes and shapes of the pknB mutant cells and of the double mutant, which is suggestive of cell wall changes that may render them more susceptible to different stress conditions, such as low pH, oxidative, and osmotic stress. Cells of the pppL mutant had a normal size but appeared affected in cell division. Analysis of the growth of the mutant strains in broths showed that they are retarded to various degrees, indicating the importance of the system for normal growth. All mutants showed a tendency to clump, which may be caused by changes in surface properties. In both BHI and THY media, whereas the kinase mutant and the double mutant showed significant defects only in the early phases of growth, the growth of the phosphatase mutant was severely retarded throughout the growth cycle. These growth defects were not seen in TSBY. The disparate growth of the mutants in the different complex media is intriguing, and a full investigation of the nutritional requirements of the mutants is under way. Although a role for the STPK in regulation of purine biosynthesis in group B streptococci (GBS) (37) and in Staphylococcus aureus (11) has been reported, we would not expect to see a deficiency in purine biosynthesis in complex medium.
The mutants showed defects in their ability to grow at low pH, an environmental stress frequently encountered by S. mutans within the oral cavity. In addition, the pknB mutant was unable to grow under conditions of oxidative stress. The Streptococcus pneumoniae stkP mutant also shows a reduced tolerance of acidic pH and other environmental stresses including oxidative stress, high temperature, and high osmolarity (39), suggesting that these signal transduction systems are concerned with transmitting information about environmental signals into the cell.
The phenotype of the pknB pppL double mutant was very similar to that of the single pknB mutant. It is interesting that the S. mutans kinase-phosphatase double mutant grew better in BHI and THY media than the phosphatase mutant; this suggests that inactivation of the kinase relieves some of the highly deleterious effects caused by mutation of pppL. These results are consistent with a model where the phosphatase PppL is required to return PknB to the unphosphorylated state and to keep a balance between phosphorylation and dephosphorylation. An excess of phosphorylated PknB, caused by the absence of phosphatase activity in the pppL mutant, is apparently harmful to the cell. In S. pneumoniae, Streptococcus agalactiae, and Streptococcus pyogenes, mutation of the orthologous STPP gene is apparently lethal (17, 32, 33), and as far as we are aware, the S. mutans pppL mutant is the first viable streptococcal STPP mutant reported. In future, it will be important to investigate whether the activity of STPPs is regulated and, if so, by which signals.
STPKs have been shown to be required for virulence in animal models of infection with other streptococci including S. pneumoniae (13) and S. agalactiae (35). In S. pyogenes, an STPK isogenic mutant is defective in expression of virulence factors, in its ability to adhere to human pharyngeal cells, and in its ability to resist phagocytosis (17), suggesting that it also has an important role in the virulence of this Streptococcus. Here, the colonizing properties of the mutants in the rat model were not affected significantly, which suggests that the defects in biofilm formation and acid tolerance response are not necessarily critical for establishment in vivo. However, the mutants exhibited a significant decrease in advanced dentinal fissure lesions and smooth-surface caries. This reduction in cariogenicity may be caused by a decrease in production of lactic acid, although this hypothesis requires further investigation. Interestingly, in these experiments the presence of the wild-type strain resulted in a reduced plaque score compared to the water control, suggesting that S. mutans produces a factor that reduces polysaccharide production by the normal microbiota. The fact that colonization with the mutants did not produce this effect suggests that these strains may be defective in production of this unknown factor.
It has been shown before that the ComDE TCS is essential for the expression of a number of bacteriocin-encoding genes (20, 49). ComDE is also involved in development of competence, but the mechanism by which competence is induced has not yet been elucidated. Transcriptomic analysis showed that genes required for genetic competence and genes encoding bacteriocins were downregulated in the pknB mutant. This was reflected by a decrease in transformation efficiency and by a decrease of inhibition of Enterococcus faecalis, respectively. Despite this, the mutants showed an increase in transformability in response to CSP, indicating that interaction with the receptor, ComD, was unaffected. We previously reported loss of CSP responsiveness in a pknB mutant (16). Further investigation revealed that this strain had been subcultured, which we assume resulted in secondary mutations that would explain the loss of responsiveness to CSP. Although we have not observed an increased rate of spontaneous mutation to streptomycin resistance in the experiments reported here, an increased mutation rate in an S. pneumoniae STPK mutant has recently been reported and presumably accounts for the same discrepancy in reports of the competence phenotype of this mutant (32). In the work reported here, the mutants were taken directly from freezer stocks for all experiments to minimize the accumulation of secondary mutations.
We also observed that the expression of bsmH (SMU.1896c) and SMU.2146c was downregulated in both pknB and vicK mutants. These results suggest that the PknB/PppL regulatory circuit interacts with the ComDE and VicKR TCS. Two different models can be envisaged for this interaction. Phosphorylation by PknB of as yet unknown target proteins might cause changes in the signals perceived by ComDE or VicKR. Alternatively, PknB might act directly by phosphorylation of the sensor kinase or the response regulator of the TCS. Phosphoproteomic studies have been used to determine the substrates for STPKs (31, 44, 45), but no indication was found for phosphorylation of a TCS. However, it has recently been shown that the S. agalactiae STPK, Stk1, can phosphorylate the CovR response regulator in vitro at a threonine residue located in close vicinity to the aspartate residue phosphorylated by the CovS histidine kinase (36). Phosphorylation by Stk1 prevented CovR from binding to the promoter (25).
Previous studies have shown that the VicRK TCS is involved in the response to oxidative stress (5, 10, 41). VicRK was recently shown to be also involved in acid survival, since a vicK mutant produced less lactic acid and was more resistant to acid stress than the wild-type strain (40). Interestingly, transcriptome analysis revealed that SMU.1895c, SMU.1896c, and SMU.2146c were among the most highly downregulated genes in the vicK mutant grown at pH 5.5 (40). Our RT-PCR data, although collected at neutral pH, confirm these results. Differential regulation of several other common genes in vicK and pknB mutants was observed (not shown), which suggests that the vicKR regulon overlaps at least in part with the pknB regulon. It is thus not surprising that there are similarities between the phenotypes of pknB and vicK mutants.
In S. pneumoniae, overlap of genes regulated by VicRK and pknB has also been observed (39). Notably, PknB and VicRK exert positive regulation on PcsB, an essential murein hydrolase involved in cell wall synthesis (29). For several streptococcal species, including S. mutans and S. pneumoniae, it has not been possible to isolate vicR mutants, suggesting that vicR is essential for normal growth (4, 41, 51). In S. pneumoniae, the requirement for VicR could be traced back to its function as the transcriptional activator of pcsB, since vicR mutants which constitutively expressed pcsB were found to be viable (30). In both S. mutans and S. pneumoniae, vicK mutants are viable, which suggests that phosphorylation of VicR is not solely dependent on the presence of VicK but that VicR may be phosphorylated by other histidine kinases or by other phosphate donors such as PknB. In agreement with the latter hypothesis, we were unable to obtain a vicK pknB double mutant of S. mutans (data not shown).
The defects of pknB and pppL mutants in cell shape and size and the increased sensitivity to stress allude to an important role of the system in sustaining cell wall integrity. This was also corroborated by the transcriptome analysis of the pknB mutant, since several of the differentially regulated genes are likely involved in cell wall biosynthesis. Among these genes were mreC and mreD. Depletion of MreC or MreD in Bacillus subtilis resulted in altered cell shape (21), whereas an mreD mutant of Streptococcus thermophilus formed very long chains and was sensitive to oxidative stress (48). SMU.984, one of the most strongly upregulated genes, encodes a protein that shows similarity to several autolysins and N-acetylmuramoyl-l-alanine amidases. Although its function is unknown, the SMU.984 protein contains a CHAP domain (3), suggesting that it might have cell wall-degrading activity. However, a signal sequence for translocation across the membrane could not be detected (data not shown). The genomic location of SMU.984 is somewhat peculiar in the context of its possible function, since it is located within a cluster of genes required for β-glucoside metabolism (8). SMU.984 is preceded by a canonical E. coli σ70-dependent promoter and followed by a transcriptional terminator. Nevertheless, the gene downstream of SMU.984, bglA (SMU.985), was also upregulated in the pknB mutant.
Most STPKs, including S. mutans PknB, contain multiple so-called PASTA (penicillin and serine threonine kinase-associated) domains, which are thought to bind unlinked peptidoglycan and as a result could activate expression of cell wall biosynthesis proteins (52). This hypothesis was recently confirmed for an STPK involved in exit from spore dormancy in Bacillus subtilis (43). It will be of interest to determine whether the same molecule also functions as a signal for PknB in S. mutans.
Acknowledgments
We thank Verena Osterwalder, Steve Reese, Hilary Holmes, Martin Gander, Catharine Aquino, and Ilse Seyfarth for excellent technical assistance. We also thank Howard Jenkinson for providing pVA838.
This study was supported in part by grant 226 from the research foundation of the Swiss Dental Association SSO.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 15 March 2010.
REFERENCES
- 1.Abranches, J., J. A. Lemos, and R. A. Burne. 2006. Osmotic stress responses of Streptococcus mutans UA159. FEMS Microbiol. Lett. 255:240-246. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, S.-J., Z. T. Wen, and R. A. Burne. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 74:1631-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., and N. D. Rawlings. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234-237. [DOI] [PubMed] [Google Scholar]
- 4.Bhagwat, S. P., J. Nary, and R. A. Burne. 2001. Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans UA159. FEMS Microbiol. Lett. 205:225-230. [DOI] [PubMed] [Google Scholar]
- 5.Biswas, I., L. Drake, D. Erkina, and S. Biswas. 2008. Involvement of sensor kinases in the stress tolerance response of Streptococcus mutans. J. Bacteriol. 190:68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boitel, B., M. Ortiz-Lombardia, R. Durán, F. Pompeo, S. T. Cole, C. Cerveñansky, and P. M. Alzari. 2003. PknB kinase activity is regulated by phosphorylation in two Thr residues and dephosphorylation by PstP, the cognate phospho-Ser/Thr phosphatase, in Mycobacterium tuberculosis. Mol. Microbiol. 49:1493-1508. [DOI] [PubMed] [Google Scholar]
- 7.Catt, D. M., and R. L. Gregory. 2005. Streptococcus mutans murein hydrolase. J. Bacteriol. 187:7863-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cote, C. K., D. Cvitkovitch, A. S. Bleiweis, and A. L. Honeyman. 2000. A novel β-glucoside-specific PTS locus from Streptococcus mutans that is not inhibited by glucose. Microbiology 146:1555-1563. [DOI] [PubMed] [Google Scholar]
- 9.de Jong, A., S. A. F. T. van Hijum, J. J. E. Bijlsma, J. Kok, and O. P. Kuipers. 2006. BAGEL: a web-based bacteriocin genome mining tool. Nucleic Acids Res. 34:W273-W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, D. M., M. J. Liu, J. M. ten Cate, and W. Crielaard. 2007. The VicRK system of Streptococcus mutans responds to oxidative stress. J. Dent. Res. 86:606-610. [DOI] [PubMed] [Google Scholar]
- 11.Donat, S., K. Streker, T. Schirmeister, S. Rakette, T. Stehle, M. Liebeke, M. Lalk, and K. Ohlsen. 2009. Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus. J. Bacteriol. 191:4056-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubrac, S., P. Bisicchia, K. M. Devine, and T. Msadek. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70:1307-1322. [DOI] [PubMed] [Google Scholar]
- 13.Echenique, J., A. Kadioglu, S. Romao, P. W. Andrew, and M.-C. Trombe. 2004. Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 72:2434-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gawron-Burke, C., and D. B. Clewell. 1984. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J. Bacteriol. 159:214-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guggenheim, B., K. G. König, E. Herzog, and H. R. Mühlemann. 1966. The cariogenicity of different dietary carbohydrates tested on rats in relative gnotobiosis with a Streptococcus producing extracellular polysaccharide. Helv. Odontol. Acta 10:101-113. [PubMed] [Google Scholar]
- 16.Hussain, H., P. Branny, and E. Allan. 2006. A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans. J. Bacteriol. 188:1628-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin, H., and V. Pancholi. 2006. Identification and biochemical characterization of a eukaryotic-type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: their biological functions and substrate identification. J. Mol. Biol. 357:1351-1372. [DOI] [PubMed] [Google Scholar]
- 18.Keyes, P. H. 1958. Dental caries in the molar teeth of rats. II. A method for diagnosing and scoring several types of lesions simultaneously. J. Dent. Res. 37:1088-1099. [DOI] [PubMed] [Google Scholar]
- 19.König, K. G., T. M. Marthaler, and H. R. Mühlemann. 1958. Methodik der kurzfristig erzeugten Rattenkaries. Dtsch. Zahn Mund Kieferheilkd. 29:99-127. [Google Scholar]
- 20.Kreth, J., D. C. I. Hung, J. Merritt, J. Perry, L. Zhu, S. D. Goodman, D. G. Cvitkovitch, W. Shi, and F. Qi. 2007. The response regulator ComE in Streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis. Microbiology 153:1799-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leaver, M., and J. Errington. 2005. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol. Microbiol. 57:1196-1209. [DOI] [PubMed] [Google Scholar]
- 22.LeBlanc, D. J., L. N. Lee, and A. Abu-al-Jaibat. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130-145. [DOI] [PubMed] [Google Scholar]
- 23.Lévesque, C. M., R. W. Mair, J. A. Perry, P. C. Y. Lau, Y. H. Li, and D. G. Cvitkovitch. 2007. Systemic inactivation and phenotypic characterization of two-component systems in expression of Streptococcus mutans virulence properties. Lett. Appl. Microbiol. 45:398-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Y. H., P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, W.-J., D. Walthers, J. E. Connelly, K. Burnside, K. A. Jewell, L. J. Kenney, and L. Rajagopal. 2009. Threonine phosphorylation prevents promoter DNA binding of the group B Streptococcus response regulator CovR. Mol. Microbiol. 71:1477-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macrina, F. L., J. A. Tobian, K. R. Jones, R. P. Evans, and D. B. Clewell. 1982. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene 19:345-353. [DOI] [PubMed] [Google Scholar]
- 28.Merritt, J., F. Qi, and W. Shi. 2005. A unique nine-gene comY operon in Streptococcus mutans. Microbiology 151:157-166. [DOI] [PubMed] [Google Scholar]
- 29.Ng, W.-L., K. M. Kazmierczak, and M. E. Winkler. 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol. Microbiol. 53:1161-1175. [DOI] [PubMed] [Google Scholar]
- 30.Ng, W.-L., G. Robertson, T., K. M. Kazmierczak, J. Zhao, R. Gilmour, and M. E. Winkler. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae. Mol. Microbiol. 50:1647-1663. [DOI] [PubMed] [Google Scholar]
- 31.Nováková, L., L. Sasková, P. Pallová, J. Janeček, J. Novotná, A. Ulrych, J. Echenique, M.-C. Trombe, and P. Branny. 2005. Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. FEBS J. 272:1243-1254. [DOI] [PubMed] [Google Scholar]
- 32.Osaki, M., T. Arcondéguy, A. Bastide, C. Touriol, H. Prats, and M.-C. Trombe. 2009. The StkP/PhpP signaling couple in Streptococcus pneumoniae: cellular organization and physiological characterization. J. Bacteriol. 191:4943-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi, F., J. Merritt, R. Lux, and W. Shi. 2004. Inactivation of the ciaH gene in Streptococcus mutans diminishes mutacin production and competence development, alters sucrose-dependent biofilm formation, and reduces stress tolerance. Infect. Immun. 72:4895-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajagopal, L., A. Clancy, and C. E. Rubens. 2003. A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J. Biol. Chem. 278:14429-14441. [DOI] [PubMed] [Google Scholar]
- 36.Rajagopal, L., A. Silvestroni, A. Vo, and C. E. Rubens. 2006. Regulation of cytotoxin expression by converging eukaryotic-type and two-component signalling mechanisms in Streptococcus agalactiae. Mol. Microbiol. 62:941-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajagopal, L., A. Vo, A. Silvestroni, and C. E. Rubens. 2005. Regulation of purine biosynthesis by a eukaryotic-type kinase in Streptococcus agalactiae. Mol. Microbiol. 56:1329-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Regolati, B., and P. Hotz. 1972. Cariostatic effect of glycerophosphate. Helv. Odontol. Acta 16:13-18. [PubMed] [Google Scholar]
- 39.Sasková, L., L. Nováková, M. Basler, and P. Branny. 2007. Eukaryotic-type serine/threonine protein kinase StkP is a global regulator of gene expression in Streptococcus pneumoniae. J. Bacteriol. 189:4168-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senadheera, D., K. Krastel, R. Mair, A. Persadmehr, J. Abranches, R. Burne, and D. G. Cvitkovitch. 2009. Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J. Bacteriol. 191:6415-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senadheera, M. D., B. Guggenheim, G. A. Spatafora, Y.-C. C. Huang, J. Choi, D. C. I. Hung, J. S. Treglown, S. D. Goodman, R. P. Ellen, and D. G. Cvitkovitch. 2005. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J. Bacteriol. 187:4064-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senadheera, M. D., A. W. C. Lee, D. C. I. Hung, G. A. Spatafora, S. D. Goodman, and D. G. Cvitkovitch. 2007. The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J. Bacteriol. 189:1451-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah, I. M., M.-H. Laaberki, D. L. Popham, and J. Dworkin. 2008. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135:486-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silvestroni, A., K. Jewell, W.-J. Lin, J. Connelly, M. Ivancic, W. Tao, and L. Rajagopal. 2009. Identification of serine/threonine kinase substrates in the human pathogen group B Streptococcus. J. Proteome Res. 8:2563-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soufi, B., F. Gnad, P. R. Jensen, D. Petranovic, M. Mann, I. Mijakovic, and B. Macek. 2008. The Ser/Thr/Tyr phosphoproteome of Lactococcus lactis IL1403 reveals multiply phosphorylated proteins. Proteomics 8:3486-3493. [DOI] [PubMed] [Google Scholar]
- 46.Sztajer, H., A. Lemme, R. Vilchez, S. Schulz, R. Geffers, C. Y. Y. Yip, C. M. Lévesque, D. G. Cvitkovitch, and I. Wagner-Döbler. 2008. Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation. J. Bacteriol. 190:401-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tandler, B. 1990. Improved uranyl acetate staining for electron microscopy. J. Electron Microsc. Tech. 16:81-82. [DOI] [PubMed] [Google Scholar]
- 48.Thibessard, A., F. Borges, A. Fernandez, B. Gintz, B. Decaris, and N. Leblond-Bourget. 2004. Identification of Streptococcus thermophilus CNRZ368 genes involved in defense against superoxide stress. Appl. Environ. Microbiol. 70:2220-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Ploeg, J. R. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 187:3980-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venable, J. H., and R. Coggeshall. 1965. A simplified lead citrate stain for use in electron microscopy. J. Cell Biol. 25:407-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner, C., A. de Saizieu, H. J. Schonfeld, M. Kamber, R. Lange, C. J. Thompson, and M. G. Page. 2002. Genetic analysis and functional characterization of the Streptococcus pneumoniae vic operon. Infect. Immun. 70:6121-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeats, C., R. D. Finn, and A. Bateman. 2002. The PASTA domain: a β-lactam-binding domain. Trends Biochem. Sci. 27:438-440. [DOI] [PubMed] [Google Scholar]