Abstract
The pneumococcal histidine triad (Pht) proteins PhtA, PhtB, PhtD, and PhtE form a group of conserved pneumococcal surface proteins. Humans produce antibodies to Pht proteins upon exposure to pneumococcus, and immunization of mice has provided protective immunity against sepsis and pneumonia and reduced nasopharyngeal colonization. Pht proteins are candidates for inclusion in multicomponent pneumococcal protein vaccines. Their biological function in pneumococcal infections is not clear, but a role in complement inhibition has been suggested. We measured complement deposition on wild-type and Pht mutant strains in four genetic backgrounds: Streptococcus pneumoniae D39 (serotype 2) and R36A (unencapsulated derivative of D39) and strains of serotypes 3, 4, and 19F. PspA and PspC single and double mutants were compared to the wild-type and Pht-deficient D39 strains. Factor H binding was measured to bacterial cells, lysates, and protein antigens. Deletion of all four Pht proteins (Pht−) resulted in increased C3 deposition on the serotype 4 strain but not on the other strains. Pht antigens did not bind factor H, and deletion of Pht proteins did not affect factor H binding by bacterial lysates. The Pht− mutant serotype 4 strain bound slightly less factor H than the wild-type strain when binding was measured by flow cytometry. Pht proteins may play a role in immune evasion, but the mechanism of function is unlikely to be mediated by factor H binding. The relative contribution of Pht proteins to the inhibition of complement deposition is likely to be affected by the presence of other pneumococcal proteins and to depend on the genetic background.
Streptococcus pneumoniae heads the list of bacterial infections and deaths worldwide, causing bronchitis and ear and sinus infections as well as life-threatening pneumonia, meningitis, and septicemia. Pneumococcal virulence proteins play important roles in evading the components of immune defense and in the progression from nasopharyngeal colonization to infection of the lungs and bloodstream (17, 28). Immunization with a combination of proteins essential to the virulence of the bacterium could provide protection against pneumococci regardless of the capsular serotype (5, 36, 37).
The ability of pneumococci to evade complement attack is one of the key factors contributing to the pathogenicity of the bacterium (26, 51). Complement activation leads to opsonization of the pneumococcal surface with C3 degradation products C3b and iC3b, enabling intake of pneumococci by phagocytic cells through complement receptor-mediated phagocytosis (13, 47). Capsular polysaccharides mask the opsonins with the result that they are not recognized by phagocytic cells (6). Several pneumococcal proteins have been shown to inhibit complement deposition by interaction with complement components (27). Pneumococcal surface protein C (PspC) inhibits the activation of complement by binding factor H (9, 10), a serum protein that modulates the function of the complement (25, 26, 41). Binding of functionally active factor H promotes cleavage of C3b and decay of the alternative pathway C3 convertase on the bacterial surface, impairing opsonization and phagocytosis (26). Pneumococcal surface protein A (PspA) inhibits C3 deposition (51) by interfering with the C1q initiation step of the classical pathway (31), which is the dominant complement activation pathway in the innate host defense against pneumococci (7). Pneumolysin depletes complement by activating the classical pathway at a distance from the bacterium (56). In the absence of both PspA and PspC, C3 deposition on pneumococci increases as a result of complement activation through both the classical and alternative pathways (31). Nearly all clinical isolates of pneumococci have either a PspA family 1 or family 2 protein (19, 20). Members from PspA families 1 and 2 have the same inhibitory effect on deposition of human complement C3, suggesting similar roles for both in virulence (43). PspC is a highly polymorphic protein that is divided into 11 families based on variations in the outermost domain (23). Contribution of PspC to pneumococcal virulence varies between strains (29).
Based on indirect evidence, some pneumococcal molecules were reported to retain proteolytic activity against C3 and to degrade both chains of C3 in the absence of serum components (3, 22). The molecule was later found to belong to the open reading frame of phpA, named after the characteristic histidine triad motifs (58), presently known as the Pht protein family (2). PhtA, PhtB, PhtD, and PhtE form a group of conserved pneumococcal surface proteins (2). Humans produce antibodies to PhtB and PhtE upon natural exposure to pneumococci (16), and antibody concentrations to PhtA and PhtD increase during invasive infection (2). Concentration of serum antibodies to PhtB, PhtD, and PhtE rises in early childhood (21, 48) and declines in old age, with levels highest in young adults (21, 49). Immunization with PhtB has been shown to reduce nasopharyngeal colonization in mice after challenge with strain WU2 (serotype 3) and a serotype 14 strain (58), and PhtE and PhtB protected mice from pneumonia after challenge with a serotype 3 strain P4241 (16). The ability of antibodies to Pht proteins to protect from infection may depend on the characteristics of the infective strain; mice were protected against lethal sepsis induced by some but not all challenge strains when immunized with PhtA (53), PhtB, or PhtD (2). Immunization of mice with PhtB (53) or PhtE (16) conferred protection against sepsis from strain WU2 but neither PhtB nor PhtE protected mice from intravenous challenge with strain D39 or a serotype 6A strain (37).
The genetic background (1) and capsular serotype (33) of pneumococcal strains influence complement resistance, and the importance of certain virulence factors to the strain may vary depending on the genetic background. In this study we analyzed the effect of Pht proteins on complement deposition and factor H binding by comparing wild-type strains to Pht mutants generated in four different genetic backgrounds representing different capsular serotypes. The results suggest that Pht proteins may play a role in the inhibition of complement deposition, but the effect depends on the genetic background of the pneumococci as increased C3 deposition was seen only in one of the Pht mutant strains. The mechanism by which Pht proteins mediate their virulence functions remains unclear as factor H binding was not found to play a significant role in this study.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The pneumococcal strains used in this study are listed in Table 1. Pht mutants were prepared for this study. Previously prepared PspA− and PspC− mutant D39 strains, kindly provided by David Briles, were analyzed for comparison. Escherichia coli DH5α and JM109 (Gibco BRL Life Technology) strains were used for cloning.
TABLE 1.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Serotype | Descriptiona | Reference(s) or source |
|---|---|---|---|
| Strains | |||
| S. pneumoniae | |||
| D39 | 2 | Wild type | 4 |
| D39 PhtDE− | 2 | D39 derivative with phtD and phtE deletions (intermediate to quadruple mutant); Emr | This study |
| D39 ΔPht | 2 | D39 derivative with phtA, phtB, phtD, and phtE deletions; Emr Spr | This study |
| D39 ΔPspA | 2 | JY182; D39 derivative with pspA deletion; Emr | 55 |
| D39 ΔPspC | 2 | TRE108; D39 derivative with pspC deletion; Emr | 14 |
| D39 ΔPspA ΔPspC | 2 | TRE121; D39 derivative with pspA and pspC deletions; Emr Tcr | 14 |
| R36A | R | Wild-type (ATCC 27336), unencapsulated derivative of D39 | 4, 24, 30, 39 |
| R36A PhtAB− | R | R36A derivative with phtA and phtB deletions (intermediate to quadruple mutant); Spr | This study |
| R36A ΔPht | R | R36A derivative with phtA, phtB, phtD, and phtE deletions; Emr Spr | This study |
| 43 | 3 | Clinical isolate; naturally lacks phtA | Clinical isolate |
| 43 PhtDE− | 3 | 43 derivative with phtD and phtE deletions (intermediate to triple mutant); Emr | This study |
| 43 ΔPht | 3 | 43 derivative with phtB, phtD, and phtE deletions; Emr Spr | This study |
| 4-CDC | 4 | Wild-type (DS2382-94) | 45 |
| 4-CDC PhtDE− | 4 | 4-CDC derivative with phtD and phtE deletions (intermediate to quadruple mutant); Emr | This study |
| 4-CDC ΔPht | 4 | 4-CDC derivative with phtA, phtB, phtD, and phtE deletions; Emr Spr | This study |
| 2737 | 19F | Clinical isolate; naturally lacks phtA | Clinical isolate |
| 2737 PhtB− | 19F | 2737 derivative with phtB deletion (intermediate to triple mutant); Spr | This study |
| 2737 ΔPht | 19F | 2737 derivative with phtB, phtD, and phtE deletions; Emr Spr | This study |
| E. coli | |||
| DH5α | Cloning host strain | ||
| JM109 | Cloning host strain | 54 | |
| Plasmids | |||
| p-GEM-T | Promega | ||
| pJDC9 | Encoding ermB gene for erythromycin resistance | 8 | |
| pR350 | Encoding aad9 gene for spectinomycin resistance | 12 | |
| pGEM-T/eryR | Intermediate construct encoding ermB from pJDC9 | This study | |
| pGEM-T/ZR1 phtD | Intermediate construct encoding ∼400 bp before phtD gene | This study | |
| pGEM-T/ZR2 phtE | Intermediate construct encoding ∼200 bp after phtE gene | This study | |
| pGEM-T/ZR2 phtE-eryR | Intermediate construct encoding ermB and ∼200 bp after phtE gene | This study | |
| pRIT 15475 | Mutator vector encoding ∼400 bp before phtD gene, ermB, and ∼200 bp after phtE gene | This study | |
| pGEM-T/ZR1 phtA | Intermediate construct encoding ∼400 bp before phtA gene | This study | |
| pGEM-T/ZR2 phtA | Intermediate construct encoding ∼150 bp after phtA gene | This study | |
| pRIT 15472 | Mutator vector encoding ∼400 bp before phtA gene, aad9, and ∼150 bp after phtA gene | This study | |
| pGEM-T/ZR1 phtB | Intermediate construct encoding ∼640 bp before phtB gene | This study | |
| pGEM-T/ZR2 phtB | Intermediate construct encoding ∼640 bp after phtB gene | This study | |
| pGEM-T/ZR1 phtB-specR | Intermediate construct encoding ∼640 bp before phtB gene and aad9 | This study | |
| pRIT 15476 | Mutator vector encoding ∼640 bp before phtB gene, aad9, and ∼640 bp after phtB gene | This study | |
| pRIT 15473 | Mutator vector encoding ∼400-bp before phtA gene, aad9, and ∼640 bp after phtB gene | This study |
Emr, erythromycin resistance; Tcr, tetracycline resistance; Spr, spectinomycin resistance.
For preparation of Pht mutants, pneumococci were cultured at 37°C with 5% CO2 on blood agar plates or in Todd-Hewitt broth supplemented with 0.5% yeast extract (THYE) and erythromycin (0.2 μg/ml) and/or spectinomycin (250 μg/ml) when appropriate. E. coli strains were cultured at 37°C in Luria-Bertani broth (LB; Difco-Becton Dickinson) with or without 1.5% (wt/vol) Bacto agar (Difco-Becton Dickinson), supplemented with erythromycin and/or spectinomycin (both at 100 μg/ml) when appropriate.
The S. pneumoniae strains were prepared for transformation by culturing in CAT medium (40) to an optical density at 600 nm (OD600) of ∼0.28 (Genesys 20; ThermoSpectronic). Bacteria were harvested by centrifugation, resuspended in the complete transformation medium (CTM) of Smith et al. (50) to an OD600 of ∼0.01, and cultured again to an OD600 of ∼0.1. The pellet from CTM culture was resuspended in CTM supplemented with 15% glycerol, and aliquots were frozen at −70°C. Frozen stocks were thawed and diluted in CTM medium with 2.5 to 5 μl of synthetic competence-stimulating peptide (CSP-1 or CSP-2; 100 ng/ml in CTM medium; Neosystem, Strasbourg, France). Competent pneumococci were produced by growing the cultures of recipient pneumococci at 37°C and removing the cells at 5, 10, 15, and 20 min.
Fresh pneumococcal cultures were used in flow cytometric assays measuring C3 deposition and factor H binding and in detection of Pht, PspA, and PspC on the bacterial surface as described previously (33), with a minor modification. Bacteria were cultured twice in THYE broth supplemented with 5% (wt/vol) fetal bovine serum (FBS) to logarithmic phase to a defined turbidity (OD600 of ∼0.26) (Spectronic Genesys spectrophotometer; Milton Roy). Bacterial concentrations of the samples were estimated from the OD620 versus viable count curve.
Construction of Pht mutants.
Genes for Pht proteins are arranged in tandem pairs in the chromosome, with phtA and phtB in the opposite orientation from phtD and phtE (2). Upstream and downstream regions of the different pht genes were amplified by PCR using the D39 genome as a template. Table 1 summarizes all the plasmids used in this study. Primers are listed in Table 2. Mutator vectors were constructed from plasmid pGEM-T (Promega Benelux, Leiden, Netherlands) that replicates in E. coli but not in S. pneumoniae. The mutator vectors contain recombinant zones that correspond to the upstream and downstream regions of the pht genes to be deleted surrounding an antibiotic resistance gene. To prepare quadruple mutant strains deficient of all pht genes, two different antibiotic resistance genes were used to combine deletions of the two different loci (phtD-phtE and phtA-phtB). An erythromycin resistance gene (ermB), amplified from a derivative of the pJDC9 (8), was selected for the phtD-phtE locus. For the phtA-phtB locus, a spectinomycin resistance gene (aad9), purified from plasmid pR350, kindly provided by James Paton (12), was used. Wild-type strains D39, 4-CDC, and 43 were first transformed using pRIT 15475. The resulting phtD phtE double mutants were then transformed with pRIT 15473, leading to the quadruple pht mutants. Wild-type strains 2737 and R36A were first transformed using pRIT 15473, and the resulting phtA phtB double mutants were then transformed with pRIT 15475, leading to the quadruple pht mutants.
TABLE 2.
Primers used in the study
| Primer name | Sequence | Amplification |
|---|---|---|
| EPDMI112 | 5′-CCAAGCTTGAAAAGAAAAACGAAATGATAC-3′ | ermB |
| EPDMI113 | 5′-ATAGAATTCCAAATTCCCCGTAGGCGCTAG-3′ | |
| EPDMI99 | 5′-AAAGGATCCCAAGCCTTTATCAAAAAAGCTCAG-3′ | ∼400 bp before phtD |
| EPDMI100 | 5′-GGGAAGCTTTCTTTCCTCACTTTAATTCTTCTG-3′ | |
| LT53 | 5′-CCGGAATTCGGAATAGCAGTAGAAAAAGTC-3′ | ∼200 bp after phtE |
| LT54 | 5′-GCTCTAGATTATTTATATGATGTCTTCATTATT-3′ | |
| EPDMI181 | 5′-AAGCTTCTTCTTGTCGGAATGGGCTTG-3′ | ∼400 bp before phtA |
| EPDMI182 | 5′-TCTAGAGGATCCTCATTTCTAAATTGCAGAAAC-3′ | |
| EPDMI183 | 5′-CCCGAATTCTGAAAAATGAAAGTCTCGATA-3′ | ∼150 bp after phtA |
| EPDMI184 | 5′-CTCGAGTCTAGACCTTTCCTCTTTTCTTAAGTA-3′ | |
| LT9 | 5′-GGAATTCATTACCATAATATTAAATTTGC-3′ | ∼640 bp before phtB |
| LT10 | 5′-CGGGATCCTCTTTCCTCTTTTCTTAAG-3′ | |
| LT11 | 5′-CCCAAGCTTAGGTAGCAGCATTTTCTAAC-3′ | ∼640 bp after phtB |
| LT12 | 5′-GCTCTAGATTAGCAAAACCTAGGTCAAAAAAG-3′ |
Characterization of the Pht mutants.
To assess the accuracy of the recombination, the genomic DNA of the mutant strains was purified, and the recombinant regions were sequenced. Phenotypes of the mutants and wild-type strains were confirmed by Western blot analysis of bacterial suspensions cultured to the logarithmic growth phase using a rabbit polyclonal antiserum raised against PhtD in detection. The rabbit anti-PhtD serum was tested by enzyme immunoassay (EIA) to be reactive with all four Pht proteins (data not shown). Binding of antibody to Pht on the membranes was detected with goat anti-rabbit IgG conjugated to alkaline phosphatase and visualized with an NBT/BCIP (nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate) substrate system (Bio-Rad Immun-Blot assay kit 170-6460).
Detection of Pht, PspA, and PspC on the bacterial surface.
Expression of Pht proteins on bacteria was detected using polyclonal rabbit antiserum to PhtD. Expression of specific Pht proteins was assessed using mouse monoclonal antibodies (MAbs) reactive in the EIA with PhtB and PhtD (H112-10A2) and with PhtE (HN1-10C12). The MAbs were prepared from fusions between spleen cells from BALB/c mice (Charles River Laboratories, Montreal, Canada) immunized with either PhtD (H112-10A2 MAb) or PhtE (HN1-10C12 MAb) proteins and mouse myeloma cells (SP2/0) according a protocol previously described (15).
Two pools of polyclonal antisera specific for either PspA family 1 or family 2 were used for detecting PspA on the bacterial surface. The antiserum for detection of PspA family 1 proteins came from rabbits immunized with two different family 1 PspA antigens, and the PspA family 2 antiserum was from rabbits immunized with three family 2 antigens, as described previously (34, 52).
A pool of polyclonal antisera from rabbits immunized with a PspC antigen (CbpA), received from GlaxoSmithKline Biologicals (Rixensart, Belgium), was used for detection of PspC on pneumococci. The CbpA antigen used in immunization was cloned from strain R6X, which is an unencapsulated derivative of D39 (30). D39 and R6X have been reported to represent PspC family 3.1 (23).
Bacteria (2 × 108 cells) were incubated with rabbit antiserum to PhtD, PspA family 1, PspA family 2, or PspC diluted in gelatin-Veronal (barbital) buffer ([GVB] 141 mM NaCl, 1.8 mM sodium barbital, 3.1 mM barbituric acid, pH 7.3 to 7.4, with 0.1% gelatin) for 45 min at 22°C in a horizontal shaker or with the monoclonal antibodies to Pht proteins for 1 h at 4°C. Unbound antibody was washed with Veronal-buffered saline ([VBS] 141 mM NaCl, 1.8 mM sodium barbital, 3.1 mM barbituric acid, pH 7.3 to 7.4). Binding of polyclonal anti-PhtD, anti-PspA, or anti-PspC and of monoclonal anti-Pht antibodies was detected by incubating the bacteria for 30 min at 22°C with Alexa Fluor 488-labeled goat anti-rabbit IgG or anti-mouse IgG (Molecular Probes, Invitrogen), respectively, diluted in GVB. Bacteria were washed and resuspended in 1 ml of VBS. Expression of proteins on the bacterial surface was analyzed by flow cytometry (FACSCalibur; Becton Dickinson), collecting data from 20,000 gated events. A sample with bacteria incubated in buffer instead of antiserum or MAbs was used as a negative control in setting the threshold. Each bacterial strain was analyzed in three independent experiments.
Serum samples used in the complement assays.
Normal human sera (NHS) from unimmunized subjects were used as a source of complement in the C3 deposition assay. The ethics committee of the National Public Health Institute, Helsinki, Finland, reviewed the protocol and approved the use of the sera. Pht-deficient strains and their wild-type counterparts were analyzed with 20 different NHS. In addition, wild-type and mutant strains in the D39 genetic background were analyzed with 6 of the 20 NHS. Sera were screened for antibodies to the corresponding capsular antigens (2, 3, 4, and 19F) and protein antigens PspA (family 1 and 2), PspC (CbpA), and PhtD as previously described (49). NHS contained very low concentrations of antibodies to capsular polysaccharides, whereas concentrations of antibodies to pneumococcal surface proteins, especially to PhtD and PspC, were much higher (Table 3). Geometric mean antibody concentrations in the six selected sera were similar to those in the 20 sera in general. Each strain was also analyzed twice with an agammaglobulinemic human serum (AGS) and once with a pooled serum from naïve mice (NMS) (657BL/6JRccHsd) with nondetectable antibody concentrations to the relevant protein and capsular antigens. All sera were divided into small volumes and stored at −70°C to preserve intact complement activity. Once thawed, a sample was used immediately as a source of complement for the C3 deposition assay. For the factor H binding assay, NHS from a single donor was inactivated by incubation at 56°C for 30 min, divided into small aliquots, and stored at −70°C before use as a source of factor H.
TABLE 3.
Antibody concentrations to pneumococcal antigens in NHS
| Antigen | Serum IgG concn (μg/ml)a |
|---|---|
| Capsular polysaccharide | |
| 2 | 0.25 (0.14-0.44) |
| 3 | 0.49 (0.27-0.87) |
| 4 | 0.11 (0.05-0.23) |
| 19F | 1.75 (0.85-3.60) |
| Protein | |
| PhtD | 20.1 (14.0-28.8) |
| PspC | 11.7 (6.87-19.9) |
| PspA family 1 | 2.03 (1.14-3.64) |
| PspA family 2 | 2.08 (1.40-3.08) |
Geometric mean antibody concentrations with 95% CI.
Complement C3 deposition assay.
Deposition of C3 on pneumococci was measured by flow cytometry as previously described (33), with minor modifications. Bacteria (2 × 108 cells) were incubated with 20% serum diluted in GVB2+ (GVB with 0.5 mM MgCl2 and 0.15 mM CaCl2) in a horizontal shaker for 30 min at 37°C. Bound C3 molecules (C3b and iC3b) were detected by incubating the bacteria with anti-C3c-fluorescein isothiocyanate (FITC)-conjugated rabbit polyclonal anti-human complement C3c antibodies (Dako Immunoglobulins, Denmark) or with FITC-conjugated goat anti-mouse complement C3 (Cappel, MP Biomedicals, Solon, OH) diluted in GVB for 30 min on ice. Binding of C3 on the bacteria was detected by fluorescence-activated cell sorting; data from 20,000 gated events were collected. A sample with bacteria incubated with serum diluted in GVB-EDTA (GVB with 10 mM EDTA), which blocks activation of both alternative and classical pathways of complement activation, was used as a negative control in each analysis.
Factor H binding to pneumococcal protein antigens.
Microtiter plates (catalog number 655061; Greiner Bio-One) were coated at concentration of 2 μg/ml with recombinant Pht proteins (PhtA, PhtB, PhtD, and PhtE) consisting of the full-length molecule as well as with truncated PspA family 1, PspA family 2, and PspC antigens consisting of the surface-exposed domain by incubation at 4°C overnight. Wells were blocked with 10% fetal bovine serum (FBS) diluted in phosphate-buffered saline (PBS) and incubated at 37°C for 1 h. Purified human factor H (Quidel) was added on the wells at two concentrations, 5 μg/ml and 15 μg/ml, diluted in 10% FBS-PBS, and the samples were incubated at 37°C for 1 h. Control wells coated with each of the protein antigens were incubated with 10% FBS-PBS instead. Binding of factor H was detected with murine anti-human factor H monoclonal antibody (Quidel) by incubation at 37°C for 1 h. Alkaline phosphatase-conjugated rabbit anti-mouse IgG (Jackson ImmunoResearch) incubated for 2 h at 37°C, followed with p-nitrophenyl phosphate substrate (Sigma Immunochemicals) incubated for 1 h at 37°C, was used for detection. Optical densities were measured at 405 nm with a photometer (Mithras; Berthold Technologies). The results were calculated from three separate analyses.
Factor H binding to bacterial cells.
Binding of factor H to pneumococci was measured by flow cytometry as previously described (33) with minor modifications. Pneumococci (2 × 108 cells) were incubated in 30% heat-inactivated serum diluted in VBS-1% bovine serum albumin (BSA) for 40 min at 37°C with shaking. Binding of factor H was detected by incubation for 30 min at 22°C with monoclonal murine anti-human factor H antibody (Quidel) diluted in VBS-BSA and secondary antibody Alexa Fluor 488-labeled goat anti-mouse IgG (Molecular Probes, Invitrogen). Binding of factor H to bacteria was detected by flow cytometry, collecting data from 20,000 gated events. Each bacterial strain was analyzed in three separate experiments with the same serum. Binding of factor H from serum was analyzed once in parallel with serum and purified human complement factor H (Quidel). Purified factor H was used at a concentration of 90 μg/ml, roughly corresponding to the factor H concentration in 30% human serum. Purified factor H and serum gave identical results (data not shown), as has been reported previously (10).
Factor H binding to cell lysates.
Binding of purified factor H to bacterial suspensions cultured to the logarithmic growth phase was measured by Western blot analysis. Samples were run in a nonreducing 10% SDS-PAGE gel and transferred to a nitrocellulose membrane. The membrane was blocked with 3% gelatin for 2 h and immersed in blotting buffer containing 10 μg/ml purified human factor H (Quidel) overnight at room temperature. Binding was detected with monoclonal murine anti-human factor H antibody (Quidel). Binding of the antibody on the membrane was detected with goat anti-mouse IgG conjugated to horseradish peroxidase (Bio-Rad Immun-Blot assay kit 170-6464).
Statistical methods.
Geometric means of serum antibody concentrations (GMC) and geometric mean intensities of fluorescence (GMF) for protein expression, C3 deposition, and factor H binding with 95% confidence intervals (CI) were analyzed. A Student's t test (two-tailed; paired) was applied on log-transformed data in comparisons of wild-type and mutant pneumococci. Arithmetic means and standard deviations were calculated from the ODs measured by EIA for factor H binding to pneumococcal protein antigens. Binding of factor H to the antigens was compared with the background ODs of wells not incubated with factor H using a Student's two-tailed t test for means. In all analyses P values of less than 0.05 were considered to indicate a statistically significant difference. The Pearson correlation was calculated from antibody concentrations measured by EIA and geometric mean intensities of fluorescence for C3 deposition of individual NHS.
RESULTS
Phenotypic characterization of the Pht mutant strains.
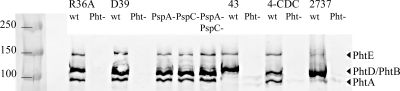
Wild-type and mutant strains were phenotypically characterized by immunoblotting. Lysates of pneumococci were blotted with a rabbit polyclonal anti-PhtD antibody, which recognized all four Pht isotypes (Fig. 1). PhtE bands were detectable in all strains but they were fainter in 2737. PhtA was detected in wild-type R36A, D39, and 4-CDC but not in strains 43 and 2737, which naturally lack PhtA. Deletion of PspA and/or PspC had no effect on the expression of Pht proteins on the D39 background. The quadruple mutants did not express any Pht proteins.
FIG. 1.
Western blot analysis of wild-type and mutant pneumococcal strains. Anti-PhtD antibody was used to probe extracts from the wild-type (wt) and PhtABDE− quadruple mutant (Pht−) strains (R36A, D39, 43, 4-CDC, and 2737) as well as PspA− and/or PspC− mutant derivatives of D39. The molecular sizes of the Pht proteins have previously been reported to range from 92 to 115 kDa (2). The positions of the different Pht bands are indicated on the right in the figure, and molecular mass markers are indicated on the left (kDa).
Expression of surface proteins.
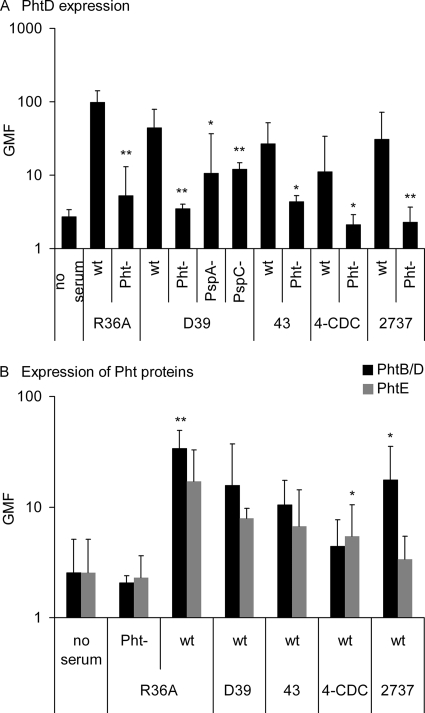
Surface expression of Pht proteins on wild-type and mutant pneumococci was compared using the rabbit polyclonal anti-PhtD antiserum. Expression of Pht proteins was readily detectable on the surface of all wild-type strains but not on the Pht-deficient mutants with deletions in all pht genes, as expected (Fig. 2A). Reactivity with the antiserum to PhtD was reduced in the D39 PspA and PspC mutants compared to the D39 wild type. To get an idea of the expression of specific Pht proteins on the surface of wild-type pneumococcal strains, binding of murine monoclonal antibodies was assessed. All wild-type strains bound PhtB/PhtD and PhtE (Fig. 2B). The monoclonal antibodies were not as reactive as the polyclonal serum, probably because the polyclonal anti-PhtD serum contains antibodies which detect a larger repertoire of epitopes on the Pht molecules. The unencapsulated R36A bound more of both PhtB/PhtD and PhtE antibodies than the encapsulated strains, indicating that the capsule masks some of the epitopes recognized by the monoclonal antibodies.
FIG. 2.
Expression of pneumococcal histidine triad proteins detected with polyclonal antiserum to PhtD (A) and monoclonal antibodies to PhtB/PhtD and PhtE (B) on the pneumococcal surface. GMF from three separate analyses with 95% CI are shown. Panel A shows mutants with the corresponding wild type, and panel B shows a comparison of PhtB/PhtD with PhtE expression by a Student's paired t test (two-tailed). *, P < 0.05; **, P < 0.01.
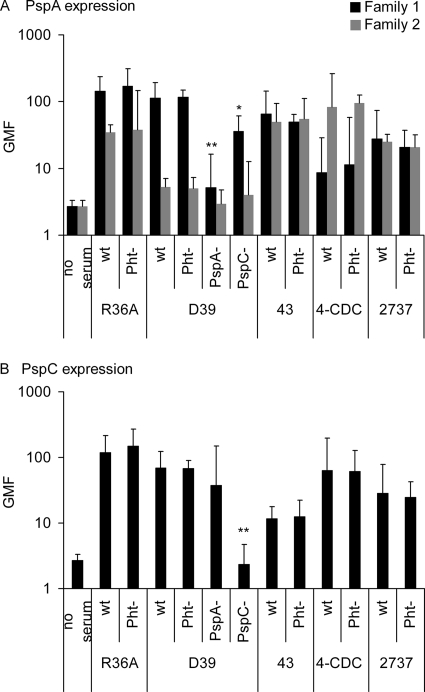
Pneumococcal surface proteins PspA and PspC were detected with rabbit polyclonal antiserum on the wild-type and Pht mutant strains using D39 PspA and PspC deletion mutants as negative controls. The rabbit antisera for PspA families 1 and 2 react more strongly with the homologous PspA family (34). Strains R36A and D39 reacted more with the PspA family 1 than with the family 2 antiserum (Fig. 3A), which is consistent with the D39 parent strain's having a family 1 PspA (1). Strain 4-CDC reacted more with family 2 antiserum, suggesting that it has a family 2 PspA. Strains 43 and 2737 reacted equally with both anti-PspA sera: their PspAs are highly cross-reactive or the strains have two PspAs. All strains were positive with the antiserum to PspC (Fig. 3B) although the PspC protein in serotype 3 diverges from the CbpA antigen used in immunization (23). No significant differences were observed in the reactivity of the wild-type and Pht mutant strains with PspA or PspC antiserum, suggesting that the deletion of Pht proteins had not affected the expression of PspA and PspC (Fig. 3). However, the expression of PspA was reduced in the D39 ΔpspC mutant compared to the wild-type D39 (Fig. 3A).
FIG. 3.
Expression of surface proteins PspA and PspC on the surface of pneumococcal strains. Expression of PspA family 1 and PspA family 2 proteins was detected with two different sera. GMF from three separate analyses with 95% CI are shown. *, P < 0.05; **, P < 0.01 (comparison of mutant with the corresponding wild type by a Student's two-tailed paired t test).
Deposition of C3 on pneumococci.
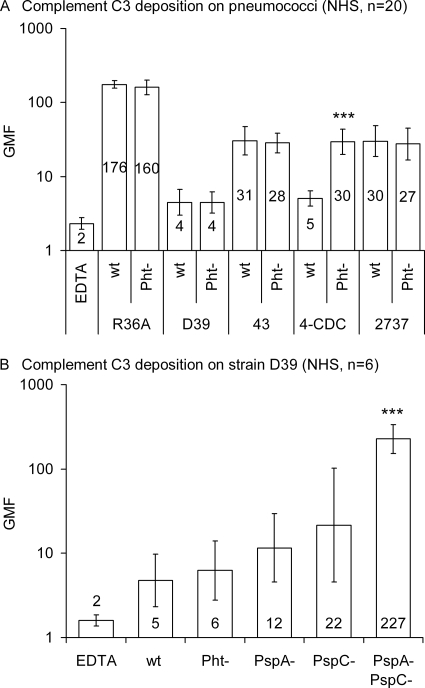
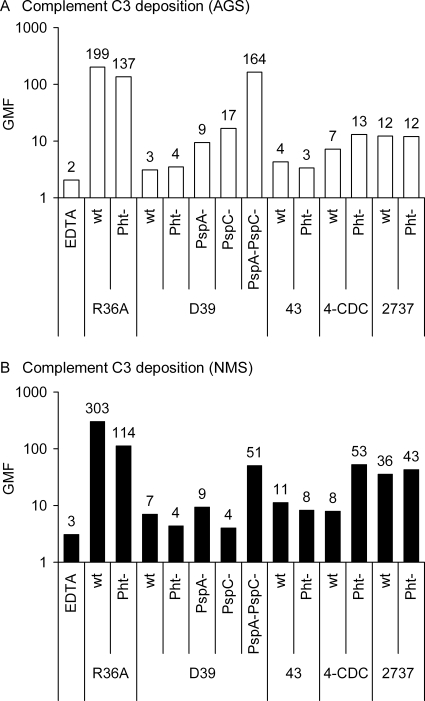
The effect of Pht proteins on the deposition of complement component C3 was assessed by flow cytometry by comparing C3 binding on the surface of wild-type and Pht-deficient mutant strains. The mutant 4-CDC strain lacking all Pht proteins bound significantly more C3 from NHS than the wild-type strain (Fig. 4A). No significant differences were found in C3 deposition between the wild-type and Pht mutant in any of the other genetic backgrounds. Individual NHS resulted in higher (or lower) C3 deposition on the Pht mutant, but on average similar amounts of C3 were deposited on wild-type and Pht mutant R36A, D39, 43, and 2737 strains. The relative influence of Pht deletion on C3 deposition compared to the deletion of PspA, PspC, or both was assessed by analyzing the D39 wild-type strain in parallel with the different mutants, each with six NHS, the AGS, and NMS. Deposition of C3 from NHS was significantly increased in the PspA− PspC− mutant compared to the wild-type D39, whereas only slightly (not significantly) more C3 was measured on the PspA− and PspC− single mutants (Fig. 4B). Deposition of C3 from AGS was 2-fold on the Pht− mutant compared to deposition on the wild-type 4-CDC, but no differences were observed between other Pht− mutants and their wild-type counterparts (Fig. 5A). C3 deposition from AGS was 3-fold on the PspA− mutant, 6-fold on the PspC− mutant, and 48-fold on the PspA− PspC− double mutant compared to deposition on the wild-type, whereas the amounts of C3 measured on the Pht− mutant D39 and on the wild-type were equally as small (Fig. 5A). C3 deposition from the NMS was 7-fold on the Pht-deficient 4-CDC and on the D39 PspA− PspC− double mutant compared to the corresponding wild-type strains, but deposition on the PspA− and PspC− single mutants was not increased (Fig. 5B). Encapsulated wild-type strains D39, 43, 4-CDC, and 2737 were clearly more resistant to complement deposition than the rough strain R36A when NHS (Fig. 4A), AGS (Fig. 5A), or NMS (Fig. 5B) was used as the source of complement.
FIG. 4.
Deposition of complement C3 on the pneumococcal surface of wild-type and mutant strains. (A) GMF with 95% CI from the analysis of wild-type and mutant pneumococci with 20 different NHS are shown. (B) GMF and 95% CI from the analysis of D39 wild type and mutants with six different NHS. Serum with 10 mM EDTA, which inhibits activation of complement via classical and alternative pathways, was used as a negative control. ***, P < 0.001 (comparison of mutant with the corresponding wild type by a Student's two-tailed paired t test).
FIG. 5.
Deposition of complement C3 on the pneumococcal surface from AGS and NMS. GMF from analysis of wild-type and mutant pneumococci are shown. The average fluorescence results of two separate analyses with AGS are shown in panel A, whereas analysis with the NMS was only performed once.
Correlation of C3 deposition with serum antibody concentration.
The concentrations of antibodies in the NHS to the capsular polysaccharides and protein antigens relevant for this study were measured by EIA (Table 3). The concentration of antibodies to pneumococcal protein antigens correlated positively with complement deposition on the encapsulated wild-type pneumococci. Serum concentration of antibody to PspA family 1 correlated with complement deposition on strains D39, 43, and 2737 (Table 4). Positive correlations were also found between the concentration of antibody to PspA family 2 and C3 deposition on these strains, but they were not statistically significant. 4-CDC was the only encapsulated strain with no correlation between anti-PspA concentration and C3 deposition. Instead, a weak positive correlation was found with this strain, unlike with the other strains, between the concentration of antibody to PhtD and C3 deposition (Table 4). No correlation between antibody concentration and C3 deposition was observed for R36A, probably because the unencapsulated strain is highly susceptible to complement even in the absence of antibodies (data not shown). The concentrations of polysaccharide-specific antibodies in the NHS were very low for serotypes other than 19F. Concentrations of antibody to serotype 19F correlated positively with C3 deposition on the serotype 19F strain (r = 0.56; P = 0.011, Pearson correlation).
TABLE 4.
Correlation of serum antibody concentration with C3 deposition
| Strain | Correlation with concn of antibody to:a |
|||
|---|---|---|---|---|
| PhtD | PspC | PspA1b | PspA2 | |
| D39 | −0.071 | 0.065 | 0.813** | 0.385 |
| 43 | 0.040 | 0.156 | 0.493* | 0.436 |
| 4-CDC | 0.164 | 0.042 | −0.007 | 0.005 |
| 2737 | −0.004 | 0.063 | 0.543* | 0.244 |
Pearson correlation coefficient.
*, P < 0.05; **, P < 0.001.
Factor H binding.
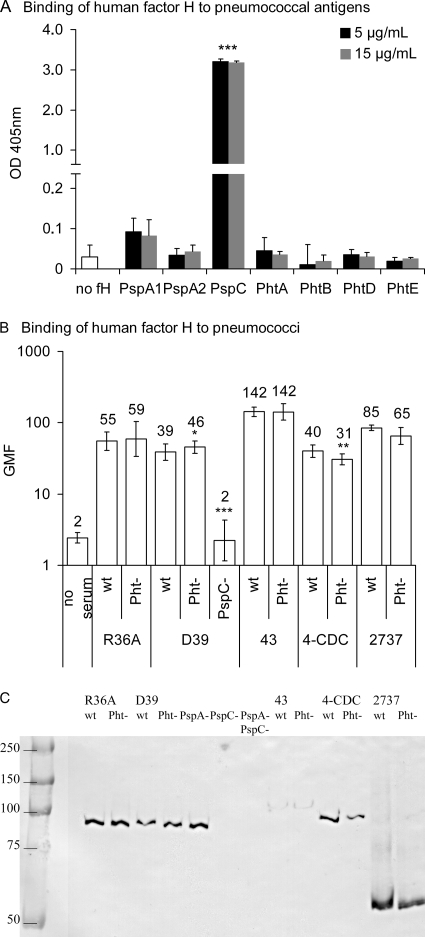
In our preliminary analyses direct binding of Pht antigens PhtB, PhtD, and PhtE to a number of human complement proteins (C3, C3b, C4BP, and factor H) was measured using Biacore surface plasmon resonance technology. PhtD showed weak binding to factor H, whereas no binding was observed to any of the other complement proteins tested (data not shown). Because data from a previous study suggested that Pht proteins could bind factor H with low affinity (38), binding of purified human factor H to pneumococcal protein antigens was measured by a similar EIA methodology using two different concentrations of factor H. Binding of factor H to the protein antigens was compared with the background ODs of wells coated with the same antigen not incubated with factor H. PspC bound factor H very strongly (P < 0.001, Student's t test for means), whereas no significant binding of factor H was observed to PhtA, PhtB, PhtD, or PhtE nor to the PspA antigens used as controls (Fig. 6A).
FIG. 6.
Binding of factor H to pneumococcal protein antigens and to pneumococcal wild-type (wt), PhtABDE− quadruple mutant (Pht−) and PspA− and PspC− mutant bacteria and cell lysates. (A) Binding of factor H to pneumococcal protein antigens measured by EIA. The average ODs of three separate experiments are given with standard deviations. Two different concentrations of purified human factor H were used. Average ODs of wells not incubated with factor H (no fH) were calculated from three separate experiments using each antigen; the different antigens had similar, low backgrounds. ***, P < 0.001 (comparison with the background ODs of the corresponding antigen by a Student's two-tailed t test). (B) Flow cytometric analysis of factor H binding to pneumococcal cells. GMF from three repeated analyses with 95% CI are shown. *, P < 0.05; **, P < 0.01 (comparison with the corresponding wild-type by a Student's two-tailed paired t test). (C) Western blot analysis of factor H binding to cell lysates. Mutants lacking Pht proteins exhibited binding patterns identical to those of the wild-type strains, whereas mutants lacking PspC bound no factor H. Molecular mass markers are indicated on the left (kDa). Binding was strongest on single bands corresponding to the molecular sizes of PspC proteins, which range from 59 to 105 kDa (46).
To assess the possible impact of Pht proteins expressed on the surface of bacterial cells, factor H binding was measured by flow cytometry on wild-type and Pht-deficient mutant strains R36A, D39, 3-43, 4-CDC, and 2737. Because PspC is to date the only pneumococcal protein shown to bind factor H, as a control factor H binding to the PspC− mutant D39 was analyzed. All wild-type strains bound factor H, and the lack of Pht proteins did not abolish this interaction (Fig. 6B). The PspC-deficient control strain bound no factor H at all. The Pht mutant 4-CDC strain bound factor H slightly less than the parent strain, whereas the D39 Pht mutant bound slightly more than the wild type.
Since it is possible that the polysaccharide capsule hinders factor H binding to pneumococcal surface proteins, binding of human factor H was measured by Western blotting using cell lysates of wild-type and Pht-deficient mutants. Lysates of PspA− and PspC− mutant strains were included as controls. Deletion of PspA or Pht proteins had no effect on factor H binding, whereas the PspC− strain and the PspA− PspC− double mutant strain bound no factor H (Fig. 6C). The samples were also blotted with rabbit antiserum to PspC, which reacted strongly with the bands of proteins that bound factor H (data not shown).
DISCUSSION
In this study we report increased C3 deposition on the Pht-deficient mutant 4-CDC strain compared to its wild-type counterpart. This suggests a role for Pht proteins in immune evasion. However, we did not observe any significant differences in complement deposition between wild-type and Pht mutant strains in genetic backgrounds of strains 43, 2737, or D39 or its unencapsulated derivative R36A. The influence of Pht on complement deposition may be affected by the presence of other pneumococcal proteins and could thus depend on the genetic background. The possible impact of Pht proteins on complement deposition is likely to be mediated by a mechanism other than what has been described for PspC since none of the Pht proteins bound factor H and since deletion of Pht proteins had no effect on factor H binding to cell lysates. Except for the slight reduction in factor H binding by the 4-CDC strain, deletion of Pht proteins had no effect on factor H binding to bacterial cells either. The major role of Pht proteins in infection may also be other than inhibition of complement at the C3 level.
Surface expression levels of PspA and PspC were similar in all Pht-deficient strains and their wild-type counterparts. This suggests that the increased C3 deposition on the mutant 4-CDC resulted from deletion of Pht proteins, not from reduced expression of PspA or PspC, which are known to play important roles in complement evasion. Increased deposition on the Pht-deficient 4-CDC strain was evident with NHS, AGS, and NMS. We did not observe differences between the wild type and the D39 Pht− mutant strain with any serum source. This is in contrast to a previous study, where significantly more C3 from NMS was deposited on the surface of D39 lacking all Pht proteins than on the wild-type strain (38).
Differences in methodology and the statistics used to describe the data are likely to explain why the influence of a single protein on complement deposition has been larger in some and smaller in other studies. Previous studies assessing the effects of loss of PspA, PspC, or pneumolysin in D39 have compared the arithmetic mean intensities and percentages of fluorescent bacteria, with inconsistent results (31, 42, 56). We compared the geometric mean intensities of fluorescence because the fluorescence axis is logarithmic, and the geometric mean describes the data better than the arithmetic mean. The percentage of fluorescently labeled bacteria correlated well with geometric mean intensity (r = 0.97), whereas the correlation with arithmetic mean fluorescence intensity was less perfect (r = 0.52, Pearson correlation) (data not shown).
Although the Pht genes are conserved (2, 16), it is likely that the genetic background of the strains partly explains why the lack of Pht proteins affected complement resistance of one but not all strains. Lack of PspA resulted in increased C3 deposition in the genetic backgrounds of D39 (51) and WU2 (43). Comparison of several pneumococcal strains with different genetic backgrounds indicated that the effects of loss of PspC on C3 deposition are strikingly varied; allelic variation in PspC structure may affect its interaction with the complement system (57). In previous studies PspC alone seemed to have little (42) or no effect (31) on complement deposition on D39, while deletion of both PspA and PspC resulted in stronger C3 deposition than when only one of the proteins was missing (31, 42). Comparison of PspA− and pneumolysin-deficient single and double mutants in D39 showed that the two proteins work in concert; the lack of both PspA and pneumolysin resulted in higher C3 deposition than either single mutation (56). Deposition of C3 was further increased when all three proteins were deleted; compared to the double and triple mutants of PspA, PspC, and Ply, deletion of a single protein had little effect on C3 deposition (42, 56). It is possible that the lack of Pht proteins combined with deletion of PspA, PspC, or pneumolysin would have a similar, synergistic effect on complement deposition.
In our previous studies we observed that high concentrations of polysaccharide-specific antibodies to 6B and 19F present in immune serum resulted in increased C3 deposition on these serotypes (33). The serum concentration of antibodies to PspA from immunized adults has been shown to correlate with C3 deposition on pneumococci (35). A similar correlation between the concentration of antibody to pneumococcal surface proteins and C3 deposition was observed in this study when NHS was the source of complement. The level of correlation reflected the genetic background of the strain; C3 deposition on D39, which possesses a family 1 PspA (1), correlated more strongly with family 1 than with family 2 antibodies, and the concentration of antibody to serotype 19F correlated positively with C3 deposition on the serotype 19F strain 2737. Antibody to either PspA family 1 or family 2 or both correlated positively with C3 deposition on all the encapsulated wild-type strains except 4-CDC, whereas antibody to PhtD correlated positively, although weakly, with only 4-CDC deposition. The efficacy of antibody-mediated protection has been shown to depend on the genetic background of the challenge strains (2, 18, 44). Interestingly, immunization with PspA from serotype 4 strain EF5668 protected mice better from WU2 expressing the same PspA than from EF5668 itself (18), whereas passive immunization of mice with rabbit anti-PhtA antiserum protected mice efficiently against EF5668, which could indicate that Pht proteins are more important than PspA to the EF5668 strain (2). Since immunization with Pht proteins failed to protect mice from sepsis caused by D39 (37) and since neither antibody to Pht proteins nor deletion of Pht proteins from D39 affected C3 deposition on D39, it is possible that the importance of Pht proteins in the genetic background of D39 is not as significant as it seems to be for 4-CDC.
Recombinant Pht proteins, and PhtD most importantly, were shown to bind human factor H in a previous study, although to a much lesser extent than PspC (38). The interaction of PspC with factor H has been shown to be species specific: clinical isolates bound only human factor H, not murine or any other factor H that was tested (32). Accordingly, we observed increased C3 deposition on the PspC mutant D39 when NHS or AGS was the sources of complement but not when NMS was used. If Pht proteins inhibited complement activation by a similar manner as PspC, increased complement activation would be expected with NHS but not with NMS, which was the source of complement in a previous study where Pht proteins were suggested to inhibit complement activation by recruitment of factor H (38). The results of a more recent study comparing the effects of PspC deletion in several different genetic backgrounds indicated that factor H binding is almost completely dependent on PspC as no detectable factor H binding was observed to any of six PspC-deficient strains and as only a low level of binding was detected to the seventh strain that was analyzed. The results suggest that Pht proteins could not contribute significantly to factor H binding (57). In this study the Pht-deficient 4-CDC strain bound less factor H than the wild type, but the difference was very small compared to the effect of deletion of PspC from strain D39, which completely abolished binding of factor H. Pht-deficient D39 bound slightly more factor H than the wild type. Because Pht protein antigens did not bind factor H and because binding of factor H to cell lysates was dependent on the presence of PspC but not Pht proteins, it is highly unlikely that Pht proteins would have any biological significance in factor H binding. It is possible, however, that the Pht proteins inhibit complement by interacting with other complement proteins. A recent study described the interaction of PspC of clinical isolates with complement protein C4bp, an inhibitor of the classical pathway; the interaction was allele specific and restricted to particular serotypes (11).
In conclusion, the results of this study suggest that Pht proteins may have a role in complement evasion, but the importance of Pht proteins most likely depends on the genetic background of the strain. In contrast with a previous report (38), deletion of Pht proteins did not result in increased C3 deposition on D39. Deletion of PspA or PspC in the D39 background resulted in only a small increase in C3 deposition, whereas C3 deposition increased significantly when both proteins were deleted, as reported in previous studies (31, 42). Other virulence proteins involved in inhibition of complement deposition may compensate the lack of a single protein. Because the genetic background of the pneumococcal strain can have a significant influence on its virulence, both in vivo and in vitro, it is important to study the effects of virulence factors using several different strains. Taken together with earlier studies, these studies strongly make the point that the protection of pneumococci against complement deposition involves the coordinated efforts of many different surface proteins. Combining several antigens which play a role in complement evasion could provide a vaccine formula effective against pneumococci of various genetic backgrounds.
Acknowledgments
This work was supported by GlaxoSmithKline Biologicals (GSK-Bio) and partly by the Infection Biology Research Program of the Medical Faculty of the University of Helsinki and by grants from the Academy of Finland and Sigrid Juselius Foundation.
We thank David Briles from the University of Alabama at Birmingham for critical reading of the manuscript and for generously supplying the D39 PspA and PspC mutant strains as well as the anti-PspA rabbit sera. Statistician Mika Lahdenkari is acknowledged for advice in statistical analysis of the data at the National Institute for Health and Welfare, Finland. We also acknowledge Laurence Turpin, Sylvie Vanderschrick, and Régine Masengo (GSK-Bio) for their support and their expertise in molecular bacteriology.
Editor: A. Camilli
Footnotes
Published ahead of print on 1 March 2010.
REFERENCES
- 1.Abeyta, M., G. G. Hardy, and J. Yother. 2003. Genetic alteration of capsule type but not PspA type affects accessibility of surface-bound complement and surface antigens of Streptococcus pneumoniae. Infect. Immun. 71:218-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamou, J. E., J. H. Heinrichs, A. L. Erwin, W. Walsh, T. Gayle, M. Dormitzer, R. Dagan, Y. A. Brewah, P. Barren, R. Lathigra, S. Langermann, S. Koenig, and S. Johnson. 2001. Identification and characterization of a novel family of pneumococcal proteins that are protective against sepsis. Infect. Immun. 69:949-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angel, C. S., M. Ruzek, and M. K. Hostetter. 1994. Degradation of C3 by Streptococcus pneumoniae. J. Infect. Dis. 170:600-608. [DOI] [PubMed] [Google Scholar]
- 4.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus type III. J. Exp. Med. 79:137-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briles, D. E., E. Ades, J. C. Paton, J. S. Sampson, G. M. Carlone, R. C. Huebner, A. Virolainen, E. Swiatlo, and S. K. Hollingshead. 2000. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect. Immun. 68:796-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, E. J., S. W. Hosea, C. H. Hammer, C. G. Burch, and M. M. Frank. 1982. A quantitative analysis of the interactions of antipneumococcal antibody and complement in experimental pneumococcal bacteremia. J. Clin. Invest. 69:85-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, J. S., T. Hussell, S. M. Gilliland, D. W. Holden, J. C. Paton, M. R. Ehrenstein, M. J. Walport, and M. Botto. 2002. The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. Proc. Natl. Acad. Sci. U. S. A. 99:16969-16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J. D., and D. A. Morrison. 1988. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene 64:155-164. [DOI] [PubMed] [Google Scholar]
- 9.Dave, S., A. Brooks-Walter, M. K. Pangburn, and L. S. McDaniel. 2001. PspC, a pneumococcal surface protein, binds human factor H. Infect. Immun. 69:3435-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dave, S., M. K. Pangburn, C. Pruitt, and L. S. McDaniel. 2004. Interaction of human factor H with PspC of Streptococcus pneumoniae. Indian J. Med. Res. 119 (Suppl.):66-73. [PubMed] [Google Scholar]
- 11.Dieudonne-Vatran, A., S. Krentz, A. M. Blom, S. Meri, B. Henriques-Normark, K. Riesbeck, and B. Albiger. 2009. Clinical isolates of Streptococcus pneumoniae bind the complement inhibitor C4b-binding protein in a PspC allele-dependent fashion. J. Immunol. 182:7865-7877. [DOI] [PubMed] [Google Scholar]
- 12.Dintilhac, A., G. Alloing, C. Granadel, and J. P. Claverys. 1997. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol. Microbiol. 25:727-739. [DOI] [PubMed] [Google Scholar]
- 13.Fearon, D. T., and W. W. Wong. 1983. Complement ligand-receptor interactions that mediate biological responses. Annu. Rev. Immunol. 1:243-271. [DOI] [PubMed] [Google Scholar]
- 14.Hakansson, A., H. Roche, S. Mirza, L. S. McDaniel, A. Brooks-Walter, and D. E. Briles. 2001. Characterization of binding of human lactoferrin to pneumococcal surface protein A. Infect. Immun. 69:3372-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamel, J., B. R. Brodeur, Y. Larose, P. S. Tsang, A. Belmaaza, and S. Montplaisir. 1987. A monoclonal antibody directed against a serotype-specific, outer-membrane protein of Haemophilus influenzae type b. J. Med. Microbiol. 23:163-170. [DOI] [PubMed] [Google Scholar]
- 16.Hamel, J., N. Charland, I. Pineau, C. Ouellet, S. Rioux, D. Martin, and B. R. Brodeur. 2004. Prevention of pneumococcal disease in mice immunized with conserved surface-accessible proteins. Infect. Immun. 72:2659-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hava, D. L., J. LeMieux, and A. Camilli. 2003. From nose to lung: the regulation behind Streptococcus pneumoniae virulence factors. Mol. Microbiol. 50:1103-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, X., and L. S. McDaniel. 2007. The genetic background of Streptococcus pneumoniae affects protection in mice immunized with PspA. FEMS Microbiol. Lett. 269:189-195. [DOI] [PubMed] [Google Scholar]
- 19.Hollingshead, S. K., L. Baril, S. Ferro, J. King, P. Coan, and D. E. Briles. 2006. Pneumococcal surface protein A (PspA) family distribution among clinical isolates from adults over 50 years of age collected in seven countries. J. Med. Microbiol. 55:215-221. [DOI] [PubMed] [Google Scholar]
- 20.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmlund, E., B. Simell, T. Jaakkola, M. Lahdenkari, J. Hamel, B. Brodeur, T. Kilpi, and H. Kayhty. 2007. Serum antibodies to the pneumococcal surface proteins PhtB and PhtE in Finnish infants and adults. Pediatr. Infect. Dis. J. 26:447-449. [DOI] [PubMed] [Google Scholar]
- 22.Hostetter, M. K. 1999. Opsonic and nonopsonic interactions of C3 with Streptococcus pneumoniae. Microb. Drug Resist. 5:85-89. [DOI] [PubMed] [Google Scholar]
- 23.Iannelli, F., M. R. Oggioni, and G. Pozzi. 2002. Allelic variation in the highly polymorphic locus pspC of Streptococcus pneumoniae. Gene 284:63-71. [DOI] [PubMed] [Google Scholar]
- 24.Iannelli, F., B. J. Pearce, and G. Pozzi. 1999. The type 2 capsule locus of Streptococcus pneumoniae. J. Bacteriol. 181:2652-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janulczyk, R., F. Iannelli, A. G. Sjoholm, G. Pozzi, and L. Bjorck. 2000. Hic, a novel surface protein of Streptococcus pneumoniae that interferes with complement function. J. Biol. Chem. 275:37257-37263. [DOI] [PubMed] [Google Scholar]
- 26.Jarva, H., R. Janulczyk, J. Hellwage, P. F. Zipfel, L. Bjorck, and S. Meri. 2002. Streptococcus pneumoniae evades complement attack and opsonophagocytosis by expressing the pspC locus-encoded Hic protein that binds to short consensus repeats 8-11 of factor H. J. Immunol. 168:1886-1894. [DOI] [PubMed] [Google Scholar]
- 27.Jarva, H., T. S. Jokiranta, R. Wurzner, and S. Meri. 2003. Complement resistance mechanisms of streptococci. Mol. Immunol. 40:95-107. [DOI] [PubMed] [Google Scholar]
- 28.Kadioglu, A., J. N. Weiser, J. C. Paton, and P. W. Andrew. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6:288-301. [DOI] [PubMed] [Google Scholar]
- 29.Kerr, A. R., G. K. Paterson, J. McCluskey, F. Iannelli, M. R. Oggioni, G. Pozzi, and T. J. Mitchell. 2006. The contribution of PspC to pneumococcal virulence varies between strains and is accomplished by both complement evasion and complement-independent mechanisms. Infect. Immun. 74:5319-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanie, J. A., W. L. Ng, K. M. Kazmierczak, T. M. Andrzejewski, T. M. Davidsen, K. J. Wayne, H. Tettelin, J. I. Glass, and M. E. Winkler. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189:38-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, J., D. T. Glover, A. J. Szalai, S. K. Hollingshead, and D. E. Briles. 2007. PspA and PspC minimize immune adherence and transfer of pneumococci from erythrocytes to macrophages through their effects on complement activation. Infect. Immun. 75:5877-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, L., Z. Ma, T. S. Jokiranta, A. R. Whitney, F. R. DeLeo, and J. R. Zhang. 2008. Species-specific interaction of Streptococcus pneumoniae with human complement factor H. J. Immunol. 181:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melin, M., H. Jarva, L. Siira, S. Meri, H. Kayhty, and M. Vakevainen. 2009. Streptococcus pneumoniae capsular serotype 19F is more resistant to C3 deposition and less sensitive to opsonophagocytosis than serotype 6B. Infect. Immun. 77:676-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melin, M. M., S. K. Hollingshead, D. E. Briles, W. P. Hanage, M. Lahdenkari, T. Kaijalainen, T. M. Kilpi, and H. M. Kayhty. 2008. Distribution of pneumococcal surface protein A families 1 and 2 among Streptococcus pneumoniae isolates from children in Finland who had acute otitis media or were nasopharyngeal carriers. Clin. Vaccine Immunol. 15:1555-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochs, M. M., W. Bartlett, D. E. Briles, B. Hicks, A. Jurkuvenas, P. Lau, B. Ren, and A. Millar. 2008. Vaccine-induced human antibodies to PspA augment complement C3 deposition on Streptococcus pneumoniae. Microb. Pathog. 44:204-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogunniyi, A. D., R. L. Folland, D. E. Briles, S. K. Hollingshead, and J. C. Paton. 2000. Immunization of mice with combinations of pneumococcal virulence proteins elicits enhanced protection against challenge with Streptococcus pneumoniae. Infect. Immun. 68:3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogunniyi, A. D., M. Grabowicz, D. E. Briles, J. Cook, and J. C. Paton. 2007. Development of a vaccine against invasive pneumococcal disease based on combinations of virulence proteins of Streptococcus pneumoniae. Infect. Immun. 75:350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogunniyi, A. D., M. Grabowicz, L. K. Mahdi, J. Cook, D. L. Gordon, T. A. Sadlon, and J. C. Paton. 2008. Pneumococcal histidine triad proteins are regulated by the Zn2+-dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J. [DOI] [PubMed]
- 39.Pearce, B. J., F. Iannelli, and G. Pozzi. 2002. Construction of new unencapsulated (rough) strains of Streptococcus pneumoniae. Res. Microbiol. 153:243-247. [DOI] [PubMed] [Google Scholar]
- 40.Porter, R. D., and W. R. Guild. 1976. Characterization of some pneumococcal bacteriophages. J. Virol. 19:659-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quin, L. R., S. Carmicle, S. Dave, M. K. Pangburn, J. P. Evenhuis, and L. S. McDaniel. 2005. In vivo binding of complement regulator factor H by Streptococcus pneumoniae. J. Infect. Dis. 192:1996-2003. [DOI] [PubMed] [Google Scholar]
- 42.Quin, L. R., Q. C. Moore III, and L. S. McDaniel. 2007. Pneumolysin, PspA, and PspC contribute to pneumococcal evasion of early innate immune responses during bacteremia in mice. Infect. Immun. 75:2067-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren, B., A. J. Szalai, O. Thomas, S. K. Hollingshead, and D. E. Briles. 2003. Both family 1 and family 2 PspA proteins can inhibit complement deposition and confer virulence to a capsular serotype 3 strain of Streptococcus pneumoniae. Infect. Immun. 71:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roche, H., B. Ren, L. S. McDaniel, A. Hakansson, and D. E. Briles. 2003. Relative roles of genetic background and variation in PspA in the ability of antibodies to PspA to protect against capsular type 3 and 4 strains of Streptococcus pneumoniae. Infect. Immun. 71:4498-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romero-Steiner, S., D. Libutti, L. B. Pais, J. Dykes, P. Anderson, J. C. Whitin, H. L. Keyserling, and G. M. Carlone. 1997. Standardization of an opsonophagocytic assay for the measurement of functional antibody activity against Streptococcus pneumoniae using differentiated HL-60 cells. Clin. Diagn. Lab Immunol. 4:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenow, C., P. Ryan, J. N. Weiser, S. Johnson, P. Fontan, A. Ortqvist, and H. R. Masure. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25:819-829. [DOI] [PubMed] [Google Scholar]
- 47.Rothlein, R., and T. A. Springer. 1985. Complement receptor type three-dependent degradation of opsonized erythrocytes by mouse macrophages. J. Immunol. 135:2668-2672. [PubMed] [Google Scholar]
- 48.Simell, B., P. Ahokas, M. Lahdenkari, J. Poolman, I. Henckaerts, T. M. Kilpi, and H. Kayhty. 2009. Pneumococcal carriage and acute otitis media induce serum antibodies to pneumococcal surface proteins CbpA and PhtD in children. Vaccine 27:4615-4621. [DOI] [PubMed] [Google Scholar]
- 49.Simell, B., M. Lahdenkari, A. Reunanen, H. Kayhty, and M. Vakevainen. 2008. Effects of ageing and gender on naturally acquired antibodies to pneumococcal capsular polysaccharides and virulence-associated proteins. Clin. Vaccine Immunol. 15:1391-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, M. D., N. B. Shoemaker, V. Burdett, and W. R. Guild. 1980. Transfer of plasmids by conjugation in Streptococcus pneumoniae. Plasmid 3:70-79. [DOI] [PubMed] [Google Scholar]
- 51.Tu, A. H., R. L. Fulgham, M. A. McCrory, D. E. Briles, and A. J. Szalai. 1999. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect. Immun. 67:4720-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vela Coral, M. C., N. Fonseca, E. Castaneda, J. L. Di Fabio, S. K. Hollingshead, and D. E. Briles. 2001. Pneumococcal surface protein A of invasive Streptococcus pneumoniae isolates from Colombian children. Emerg. Infect. Dis. 7:832-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wizemann, T. M., J. H. Heinrichs, J. E. Adamou, A. L. Erwin, C. Kunsch, G. H. Choi, S. C. Barash, C. A. Rosen, H. R. Masure, E. Tuomanen, A. Gayle, Y. A. Brewah, W. Walsh, P. Barren, R. Lathigra, M. Hanson, S. Langermann, S. Johnson, and S. Koenig. 2001. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 69:1593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 55.Yother, J., G. L. Handsome, and D. E. Briles. 1992. Truncated forms of PspA that are secreted from Streptococcus pneumoniae and their use in functional studies and cloning of the pspA gene. J. Bacteriol. 174:610-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuste, J., M. Botto, J. C. Paton, D. W. Holden, and J. S. Brown. 2005. Additive inhibition of complement deposition by pneumolysin and PspA facilitates Streptococcus pneumoniae septicemia. J. Immunol. 175:1813-1819. [DOI] [PubMed] [Google Scholar]
- 57.Yuste, J., S. Khandavilli, N. Ansari, K. Muttardi, L. Ismail, C. Hyams, J. Weiser, T. Mitchell, and J. S. Brown. 2010. The effects of PspC on complement-mediated immunity to Streptococcus pneumoniae vary with strain background and capsular serotype. Infect. Immun. 78:283-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, Y., A. W. Masi, V. Barniak, K. Mountzouros, M. K. Hostetter, and B. A. Green. 2001. Recombinant PhpA protein, a unique histidine motif-containing protein from Streptococcus pneumoniae, protects mice against intranasal pneumococcal challenge. Infect. Immun. 69:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]