Abstract
The Dot/Icm-translocated ankyrin B (AnkB) effector of Legionella pneumophila exhibits molecular mimicry of eukaryotic F-box proteins and is essential for intracellular replication in macrophages and protozoa. In addition to two eukaryotic-like ankyrin (ANK) domains, AnkB harbors a conserved eukaryotic F-box domain, which is involved in polyubiquitination of proteins throughout the eukaryotic kingdom. We have recently shown that the F-box domain of the AnkB effector is essential for decoration of the Legionella-containing vacuole (LCV) with polyubiquitinated proteins within macrophages and protozoan hosts. To decipher the role of the two ANK domains in the function of AnkB, we have constructed in-frame deletion of either or both of the ANK domain-encoding regions (ankBΔA1, ankBΔA2, and ankBΔA1A2) to trans-complement the ankB null mutant. Deletion of the ANK domains results in defects in intracellular proliferation and decoration of the LCV with polyubiquitinated proteins. Export of the truncated variants of AnkB was reduced, and this may account for the observed defects. However, while full-length AnkB ectopically expressed in mammalian cells trans-rescues the ankB null mutant for intracellular proliferation, ectopic expression of AnkBΔA1, AnkBΔA2, and AnkBΔA1A2 fails to trans-rescue the ankB null mutant. Importantly, ectopically expressed full-length AnkB is targeted to the host cell plasma membrane, where it recruits polyubiquitinated proteins. In contrast, AnkBΔA1, AnkBΔA2, and AnkBΔA1A2 are diffusely distributed throughout the cytosol and fail to recruit polyubiquitinated proteins. We conclude that the two eukaryotic-like ANK domains of AnkB are essential for intracellular proliferation, for targeting AnkB to the host membranes, and for decoration of the LCV with polyubiquitinated proteins.
Legionella pneumophila is ubiquitous in natural aquatic environments and man-made water systems, where it replicates within various amoebae and ciliated protozoan hosts (14, 17, 28). When inhaled in contaminated aerosols by humans, L. pneumophila can replicate within alveolar macrophages, causing pneumonia (14, 17, 28). Humans are thought to be the accidental host, while protozoan species are considered to be the natural hosts for L. pneumophila in the aquatic environment (14, 17, 28).
At the cellular and molecular levels, the intracellular life cycles of L. pneumophila within amoebae and human cells are very similar (14, 17, 28). Once inside alveolar macrophages or amoebae, L. pneumophila evades endocytic fusion and intercepts endoplasmic reticulum (ER)-to-Golgi vesicles to remodel its phagosome into a rough endoplasmic reticulum (RER)-derived vacuole (1, 18, 22, 41). The Dot/Icm type IV secretion system (40, 46) is required for L. pneumophila to modulate various mammalian and protozoan cellular processes through numerous effectors translocated into the host cell by the Dot/Icm type IV secretion apparatus (reviewed in reference 21). During late stages of proliferation within macrophages and amoebae, the Legionella-containing vacuole (LCV) becomes disrupted, followed by bacterial escape into the cytosol, where the last few rounds of proliferation occur (27). During this late transient residence of the bacteria in the cytosol, various virulence-related regulatory cascades triggered at the post-exponential phase are activated (30), resulting in phenotypic modulation and lysis of the host cell (29). Expression of all the ankyrin genes of L. pneumophila is temporally and spatially induced at the post-exponential phase (3).
Among >200 Dot/Icm-exported effectors, very few play a detectable role in intracellular proliferation of L. pneumophila, but the role of most of these effectors in the intracellular infection is not yet known. This may be due to the high level of redundancy among the effectors that likely allows L. pneumophila to exploit conserved cellular processes in different hosts in the environment (21).
The similarities in the intracellular life cycles of L. pneumophila within protozoan and mammalian cells at the molecular and cellular levels suggest that coevolution of this bacterium with protozoa has enabled it to acquire mechanisms to exploit conserved eukaryotic processes that facilitate its pathoadaptation to infect mammalian cells (14, 17, 28). Bioinformatic analyses of four L. pneumophila genomes have revealed the presence of many eukaryotic-like genes, such as ankyrin-encoding (ank) genes (5, 6, 10), which have been suggested to be acquired through horizontal gene transfer or by convergent evolution (5). Ankyrin proteins are eukaryotic proteins with a tandem 33-residue ankyrin (ANK) repeat involved in various eukaryotic protein-protein interactions, a repeat which is the most common protein domain in the eukaryotic kingdom (5). The eukaryotic ANK repeats have been recently found in various microbial pathogens, such as Anaplasma phagocytophilum, Coxiella burnetii, and L. pneumophila (4, 5, 15, 20, 33, 34).
Eleven ank genes have been recently characterized in L. pneumophila strain AA100/130b (3-5, 15, 16, 37). Among the 11 ank mutants, the ankB, ankJ, and ankH mutants exhibit defects in intracellular proliferation within amoebae, ciliated protozoa, and human macrophages, as well as a defect in vivo in intrapulmonary proliferation in the mouse model (4, 5, 15, 16, 37). All the ankyrin proteins of L. pneumophila are translocated into the host cell via the Dot/Icm type IV secretion system (4, 9, 15, 16, 21).
In addition to the two eukaryotic-like ANK domains, AnkB harbors an F-box eukaryotic domain (3-5, 15, 37), which is involved in ubiquitination of eukaryotic proteins (23, 36). Ubiquitination of proteins is an ancient and highly conserved eukaryotic posttranslational modification process that covalently links a 76-residue ubiquitin polypeptide to a specific protein to target it for proteasomal degradation or to modulate its function in a wide range of important cellular processes such as signaling, endocytosis, cell cycle regulation, and vesicular trafficking (23). A major group of ubiquitin ligases is the SCF1 (RBX1-CUL1-SKP1) trimolecular complex that binds the F-box domain (39), which is a highly conserved process throughout the eukaryotic kingdom, including yeasts and the social amoeba Dictyostelium discoideum (25, 38). The F-box family of human proteins also typically have leucine-rich repeat (LRR) or WD protein-protein interaction domains, which bind substrates and link them to the SCF1 ubiquitin ligase complex that is bound to the F-box domain (24, 42). However, the ANK domain has never been found in eukaryotic F-box proteins (23, 36), indicating a novel structure of the AnkB F-box protein compared to eukaryotic F-box proteins.
Most work examining ubiquitination and the SCF1 complex in amoebae has been performed in D. discoideum (12, 13, 26), which is also an established model system for infection of amoebae by L. pneumophila (43). Each of the major components of the yeast and mammalian SCF1 ubiquitin ligase complex has been identified in D. discoideum, appears to function in a manner identical to those in yeast and mammalian cells, and represents a key pathway for posttranslational modification of proteins (7, 12, 25, 26, 38, 44, 47). Protein ubiquitination has been also characterized in the ciliate Tetrahymena and Acanthamoeba (19, 32), and both are permissive protozoan hosts for L. pneumophila (14).
Many intracellular bacterial pathogens have been shown to exploit or intercept the host polyubiquitination machinery (45). The LCV is decorated with polyubiquitinated proteins (11), and the Dot/Icm-translocated AnkB effector is essential for this process (37). Importantly, the F-box domain and its conserved residues are essential for interaction of AnkB with the host Skp1 component of the SCF1 ubiquitin ligase and are essential for decoration of the LCV with polyubiquitinated proteins with macrophages, Acanthamoeba, and D. discoideum (37).
While most eukaryotic F-box proteins harbor WD or LRR substrate binding domains in addition to the F-box domain, AnkB is a noncanonical F-box protein, as it harbors two ANK domains. It is not known whether the two ANK domains of AnkB of L. pneumophila are required for the function of AnkB. Here we show that the two eukaryotic-like ANK domains of AnkB are essential for intracellular proliferation, for targeting AnkB to the host membranes, and for decoration of the LCV with polyubiquitinated proteins.
MATERIALS AND METHODS
Bacterial strains, cell cultures, and infections.
L. pneumophila serogroup I parental strain AA100/130b (ATCC BAA-74) and the dotA, ankB, and complemented ankB mutants were grown as described previously (4). Escherichia coli strain DH5α was used for cloning purposes. Maintenance of the macrophage-like U937 cells was carried out as we previously described (15). HEK293 cell cultures were maintained as previously described (37).
For intracellular proliferation studies, infections of macrophages and Acanthamoeba polyphaga were performed as described previously (4, 15). Briefly, cells were infected at a multiplicity of infection (MOI) of 10 for 1 h followed by treatment with 50 μg/ml gentamicin for 1 h to kill extracellular bacteria, and this was considered the zero time point (T0). At each time point, the cells were lysed and plated on agar plates.
For single cell analysis studies, HEK293 cells were seeded onto glass coverslips in 24-well plates and transfected with plasmid DNA as described below. Transiently transfected HEK293 cells were then infected with stationary-phase wild-type (WT) and ankB mutant bacteria at an MOI of 10 for 1 h. Monolayers were then treated with 50 μg/ml gentamicin for 1 h to kill extracellular bacteria. At 2 h and 12 h postinfection, cells were fixed and permeabilized using methanol at −20°C and prepared for confocal microscopy as described below.
DNA manipulations.
DNA manipulations and restriction enzyme digestions were performed using standard procedures. Restriction enzymes and T4 DNA ligase were purchased from NEB (Madison, WI). To generate domain mutant alleles of ankB, an inverse PCR strategy was employed using pBCSK+ harboring the ankB gene as a template. Briefly, phosphorylated primers (Table 1) were designed to hybridize adjacent to DNA encoding the ANK domains and then the entire plasmid minus the domain of interest was PCR amplified using Phusion DNA polymerase (Finnzymes Oy, Finland). The resulting PCR product was then treated with DpnI restriction to remove residual template DNA from the reaction mixture and then allowed to religate using T4 DNA ligase. The ligation products were transformed into E. coli DH5α. DNA sequencing was used to confirm deletion of the ANK domains in ankB and to ensure the integrity of the ankB reading frame. Recombinant plasmids were then electroporated into the L. pneumophila ankB mutant.
TABLE 1.
Primers used in this study
| Primer name | Primer sequence (5′-3′) |
|---|---|
| AnkB A1 Fa | ACCAATAGCTACCACTTAAA |
| AnkB A1 Ra | TCGCTTTATATGCTGTTGTC |
| AnkB A2 Fa | AAAGATAACTATGGTGATTC |
| AnkB A2 Ra | GTGGTAGCTATTGGTCTGGG |
| AnkB A1A2 Ra | GTGGTAGCTATTGGTTCGC |
| AnkB cya F | GGATCCTTATGAAAAAGAATTTTTTTTCTG |
| AnkB cya R | CTGCAGTTAACAAACAAGGCACTTGCT |
Primer modified with 5′ phosphorylation.
The various pBCSK+ vectors harboring the mutant ankB alleles were used as templates to generate CYA fusions. Primers listed in Table 1 were used to PCR amplify the mutant ankB alleles, and the resulting PCR products were cloned into pCR2.1 via topoisomerization as described by the manufacturer (Invitrogen, Carlsbad, CA). The mutant ankB alleles were then subcloned into the BamHI-PstI sites of pCYA-ralF (31), resulting in replacement of the ralF gene with ankB alleles in frame with cya. Recombinant plasmids were electroporated into the wild-type strain AA100.
The various pBCSK+ vectors harboring the mutant ankB alleles were used as templates to generate 3×Flag fusions in plasmid p3X-flag CMV-10 as described previously (37). Primers used are listed in Table 1.
Production of rabbit anti-AnkB antiserum.
To generate anti-AnkB antiserum, we generated recombinant 6×His-AnkBΔF-box protein from E. coli and used this protein to immunize rabbits. Briefly, we cloned the ankBΔfbox gene into plasmid pET200 (Invitrogen, Carlsbad, CA). This plasmid was introduced into E. coli BL21 Star, and protein expression was induced by the addition of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to mid-log-phase cultures. Induction was performed overnight at 25°C. His-tagged protein was then purified from the E. coli cultures using Ni-NTA Superflow columns following the manufacturer's instructions (Qiagen, Valencia, CA). Removal of the F-box domain of AnkB resulted in high quantities of soluble protein in E. coli BL21 Star. The native protein antigen was used to immunize rabbits (Genscript, Piscataway, NJ). Final antiserum from the rabbits was used at a 1/60,000 dilution in Western blot assays.
Translocation of AnkB into macrophages.
For adenylate cyclase fusion assays, monolayers were infected with L. pneumophila at an MOI of 50 for 1 h at 37°C. Monolayers were washed extensively to remove extracellular bacteria and subsequently lysed in 200 μl of 0.1 M HCl plus 0.5% (vol/vol) Triton X-100. Cell lysates were processed following directions of the direct cyclic AMP enzyme immunoassay kit (Assay Designs), as we described previously (4). To confirm expression of CYA fusion proteins, bacterial lysates equivalent to 1 × 108 bacteria were electrophoresed on sodium dodecyl sulfate (SDS)-polyacrylamide gels and immunoblotted to nitrocellulose membranes by standard techniques. CYA fusions were detected using an anti-M45 antibody. To ensure equal loading among wells, the membranes were reprobed with anti-chloramphenicol acetyltransferase (anti-CAT) antibody as described previously (4).
Ectopic expression and distribution of AnkB in mammalian cells.
The human renal epithelial HEK293 cells were grown on circular glass coverslips (Fisher Scientific, Pittsburgh, PA) pretreated with 0.1 mg/ml of poly-l-lysine in 24-well culture plates at a concentration of 5 × 104 cells/ml overnight. The subconfluent cultures of HEK293 were transiently transfected with 2 μg of purified plasmid DNA for 24 h using Fugene HD reagent (Roche, Mannheim, Germany) as described previously (37). The transfected cells were used for infection as described above.
Confocal laser scanning microscopy.
Processing of infected cells for confocal microscopy was performed as described previously (15, 37). Polyclonal rabbit anti-L. pneumophila antiserum was detected with Alexa Fluor 488-conjugated donkey anti-rabbit IgG (1/1,000 dilution) (Invitrogen, Carlsbad, CA). Polyubiquitinated proteins were detected using anti-polyubiquitin FK1 antibody (1/50 dilution; Biomol International/Affiniti, Exeter, United Kingdom), followed by Alexa Fluor 555-conjugated goat anti-mouse IgM (Invitrogen, Carlsbad, CA). 3×Flag-tagged proteins were detected using mouse anti-Flag M2 antibody (1/200 dilution; Sigma, St. Louis, MO), followed by Alexa Fluor 488- or 555-conjugated anti-mouse IgG antibody (Invitrogen, Carlsbad, CA). The cells were examined using an Olympus FV1000 laser scanning confocal microscope as we described previously (37). On average, 8 to 15 0.2-μm serial Z sections of each image were captured and stored for further analyses, using Adobe Photoshop CS3.
RESULTS
Contribution of the ANK domains of AnkB to intracellular proliferation.
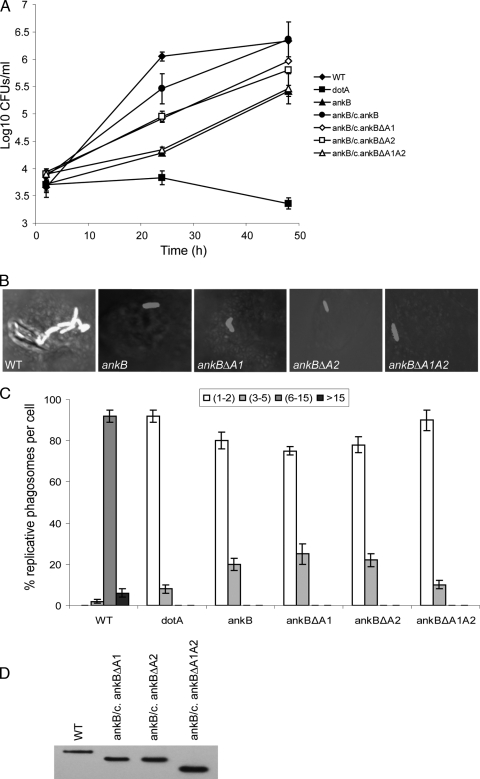
The ankB null mutant is impaired for intracellular replication in human macrophages and protozoa and for intrapulmonary proliferation in the mouse model (4, 5, 37). To determine if the two ANK eukaryotic-like domains of AnkB are crucial for the function of this effector in intracellular proliferation, we engineered mutant alleles of ankB with in-frame deletions of either or both of the two ANK domains (ankBΔA1, ankBΔA2, and ankBΔA1A2) (Fig. 1). Immunoblot analyses showed no detectable differences in the expression of the AnkB variants in L. pneumophila (Fig. 1D). We performed infections of human U937 macrophages with the wild-type strain AA100, the dotA mutant, the ankB mutant, or the ankB mutant harboring the WT ankB allele or one of the mutant alleles (ankBΔA1, ankBΔA2, or ankBΔA1A2). The infection was performed for 1 h using an MOI of 10, followed by 1 h of treatment with gentamicin to kill extracellular bacteria. At several time points after infection, the CFU were determined following growth on agar plates. Compared to the wild-type strain, the ankB mutant had a significant defect in intracellular growth (Fig. 1A). Complementation of the ankB mutant strain with a full-length ankB gene restored the intracellular growth defect. In contrast, complementation of the ankB mutant with the ankBΔA1 or ankBΔA2 allele resulted in a partial restoration of robust intracellular growth (Fig. 1A). However, complementation of the ankB mutant with the ankBΔA1A2 allele completely failed to restore intracellular growth, with CFU recovered at 24 and 48 h being similar to those of the ankB null mutant (Fig. 1A). As expected, a dotA mutant control failed to replicate intracellularly (Fig. 1A).
FIG. 1.
The two ANK domains of AnkB are essential for intracellular growth of L. pneumophila in macrophages. (A) Monolayers of U937 macrophages were infected with the WT strain and the isogenic dotA or ankB mutants or the ankB mutant complemented with either WT ankB (c/ankB) or one of the ankB mutant alleles. The infection was carried out in triplicate with an MOI of 10 for 1 h followed by 1 h of gentamicin treatment to kill extracellular bacteria. The infected monolayers were lysed at different time points and plated onto agar plates for colony enumeration. The results are representative of three independent experiments performed in triplicate. Error bars represent standard deviations. (B and C) Single cell analyses of L. pneumophila replicative phagosomes. At 10 h postinfection, 100 infected cells were analyzed by laser scanning confocal microscopy for formation of replicative phagosomes, and representative images are shown in panel B. L. pneumophila was stained by a polyclonal anti-L. pneumophila antibody and Alexa Fluor 555-conjugated anti-rabbit IgG (red). Quantification of the number of bacteria/cell at 10 h is shown in panel C. The dotA mutant was used as a negative control. Infected cells from multiple coverslips were examined in each experiment. The results are representative of three independent experiments performed in triplicate. Error bars represent standard deviations. (D) Immunoblot analysis shows no detectable differences in the expression levels of the AnkB variants in L. pneumophila. Total bacterial proteins equivalent to 1 × 108 bacteria were loaded onto SDS-polyacrylamide gels and immunoblotted using an anti-AnkB rabbit antiserum (1:60,000 dilution).
To confirm the above results, we performed single cell analysis by confocal microscopy to score the formation of replicative vacuoles (Fig. 1). At 2 h after infection, approximately 95% of the cells infected with the different strains harbored a single organism (data not shown). After 10 h of infection, approximately 90% of the cells harboring the wild-type strain contained 6 to 15 bacteria (Fig. 1B and C). In contrast, the ankB mutant showed no detectable replication at 10 h, similarly to the dotA or htrA mutant (2, 35) controls (Fig. 1B and C and data not shown). In cells harboring the ankB mutant complemented with the ankBΔA1, ankBΔA2, or ankBΔA1A2 allele, only 1 or 2 bacteria/cell were detected at 10 h in 80 to 90% of the infected cells (Fig. 1B and C). These results confirmed the growth kinetics data for the essential role of the two ANK domains in intracellular proliferation.
trans-rescue of the ankB mutant within cells harboring the wild-type strain.
The dot/icm mutants are rescued for their intracellular defect within cells harboring the wild-type strain of L. pneumophila (8). However, L. pneumophila mutants defective in intracellular replication due to a defect in stress response genes (such as htrA or rpoS) are not rescued within cells harboring the wild-type strain (2, 35). To examine the role of the ANK domains in rescue of the ankB mutant, we utilized the ankB mutant harboring the ankBΔA1, ankBΔA2, or ankBΔA1A2 alleles in a series of coinfection experiments. Monolayers were coinfected with the wild-type (WT) strain and the ankB mutant strains to determine whether the mutant replicated in cells coinfected with the wild-type strain. Coinfections with wild-type L. pneumophila and the isogenic dotA or htrA mutant were used as positive and negative controls, respectively. In all coinfections, only ∼5% of the cells were coinfected with the wild-type strain. Single cell analyses at 10 h postinfection showed that when the ankB mutant resided in cells harboring the wild-type strain, the ankB mutant replicated robustly (Fig. 2 and data not shown). The ankB mutant harboring any of the in-frame ANK domain deletion alleles was also rescued for intracellular proliferation when it coinhabited cells with the wild-type strain. Infection by any of the mutants alone showed that there was no detectable replication at 12 h (data not shown). In control coinfections of L. pneumophila and the isogenic dotA or htrA mutant, replication of the dotA but not the htrA mutant was rescued in cells containing the wild-type strain (Fig. 2). Our data show that the wild-type strain can rescue the in-frame ANK domain deletion mutants of AnkB for robust intracellular growth. We conclude that the loss of the two ANK domains of the Dot/Icm-translocated AnkB effector can be complemented in trans by the WT strain expressing the full-length AnkB.
FIG. 2.
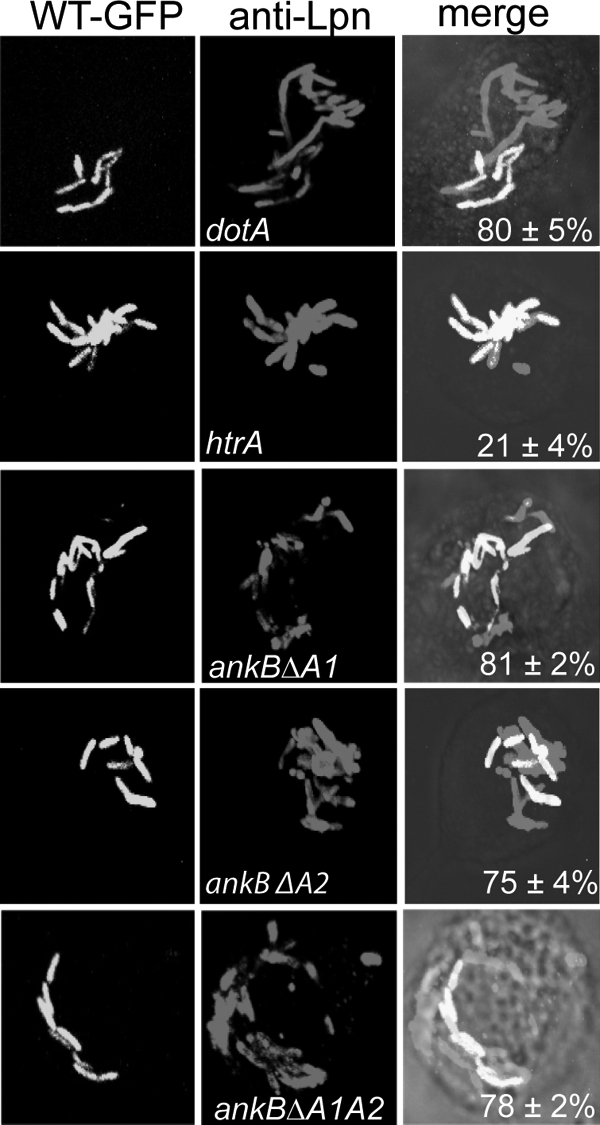
L. pneumophila bacteria expressing mutant alleles of AnkB are rescued within cells harboring the wild-type bacteria. Coinfections were performed using the wild-type L. pneumophila strain AA100 and the ankB mutant or using the ankB mutant harboring one of the mutant alleles ankBΔA1, ankBΔA2, and ankBΔA1A2. Infected U937 cells were gentamicin treated, washed, and fixed at 10 h after infection. Bacterial phagosomes were scored for all infected cells in randomly selected fields by confocal microscopy. Cells harboring phagosomes containing both strains (WT-GFP [green fluorescent protein] and a mutant) were scored. The htrA mutant was used as a negative control while the dotA mutant was used as a positive control. Representative confocal images are shown in panel A, while quantitative analyses are shown in panel B. Rescue was calculated by quantifying the percentage of the cells harboring the ankB mutant replicating within cells harboring the WT strain, and the numbers are shown in the merged images. The results are representative of three independent experiments performed in triplicate by analyses of 100 infected cells. Error bars represent standard deviations.
The role of the ANK domains of AnkB in acquisition of polyubiquitinated proteins within human macrophages and A. polyphaga.
The F-box domain of AnkB is essential for decoration of the LCV with polyubiquitinated proteins with macrophages, Acanthamoeba, and D. discoideum (37). While most eukaryotic F-box proteins harbor WD or LRR substrate binding domains, in addition to the F-box domain, AnkB is considered a noncanonical F-box protein, as it harbors two ANK domains. It is not known whether the two ANK domains of AnkB are functional domains required for decoration of the LCV with polyubiquitinated proteins.
Therefore, we analyzed the role of the two eukaryotic-like ANK domains of AnkB in acquisition of polyubiquitinated proteins by the LCV. Macrophages were infected as described above by the wild-type strain, the dotA or ankB mutant, or the ankB mutant harboring plasmids containing the WT ankB or one of the mutant ankB alleles and analyzed by confocal microscopy for acquisition of polyubiquitinated proteins by the LCV at 2 h postinfection. The data showed that within macrophages, ∼85% of the LCVs harboring the wild-type strain or the ankB mutant complemented with the WT ankB acquired polyubiquitinated proteins (Fig. 3 and data not shown). In contrast, LCVs harboring the ankB mutant complemented with the ankBΔA1, ankBΔA2, or ankBΔA1A2 allele failed to be decorated with polyubiquitinated proteins, similarly to the ankB null mutant or the dotA mutant control (22% ± 3% colocalization) (Fig. 3 and data not shown).
FIG. 3.
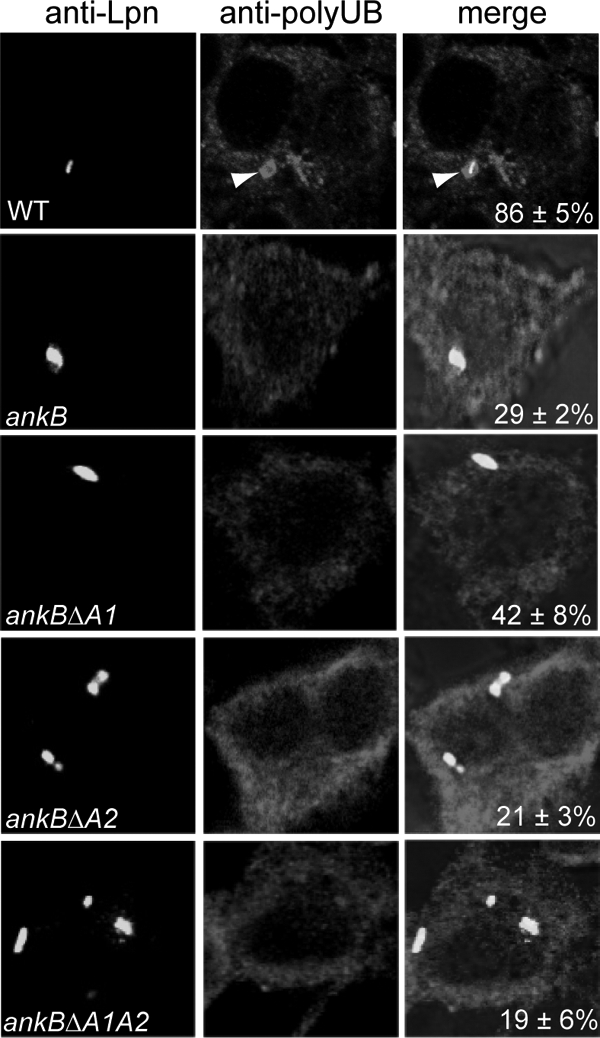
The two ANK domains of AnkB are essential for acquisition of polyubiquitinated proteins by the LCV within macrophages. Representative confocal images of infected macrophages for colocalization of phagosomes with polyubiquitinated proteins at 2 h post-infection of U937 cells by the wild-type strain of L. pneumophila, the isogenic ankB mutant, or the ankB mutant harboring one of the mutant ankB alleles on a plasmid. The cells were labeled with anti-Lpn antibody (green) and antipolyubiquitin (red). The arrowheads indicate heavy colocalization of polyubiquitin with the bacterium. Quantification of colocalization of the LCVs with polyubiquitin at 2 h postinfection has been determined by analysis of 100 infected cells and is shown in the merged images. The data are representative of three independent experiments.
Since the F-box domain of AnkB is essential for decorating the LCV with polyubiquitinated proteins within A. polyphaga, we utilized the eukaryotic domain deletion alleles (ankBΔA1, ankBΔA2, and ankBΔA1A2) to determine whether the two ANK domains also played a role in acquisition of polyubiquitinated proteins by the LCV within A. polyphaga. The data showed that within A. polyphaga, most of the LCVs (∼88%) harboring the wild-type strain or the ankB mutant complemented with the wild-type ankB allele were decorated with polyubiquitinated proteins (Fig. 4 and data not shown). In contrast, the LCVs harboring the ankB mutant complemented with the ankBΔA1, ankBΔA2, or ankBΔA1A2 allele were defective in acquisition of polyubiquitinated proteins, similarly to the ankB mutant (Fig. 4). Therefore, deletion of either of the two ANK domains of AnkB results in failure to decorate the LCV by polyubiquitinated proteins within mammalian and protozoan cells.
FIG. 4.
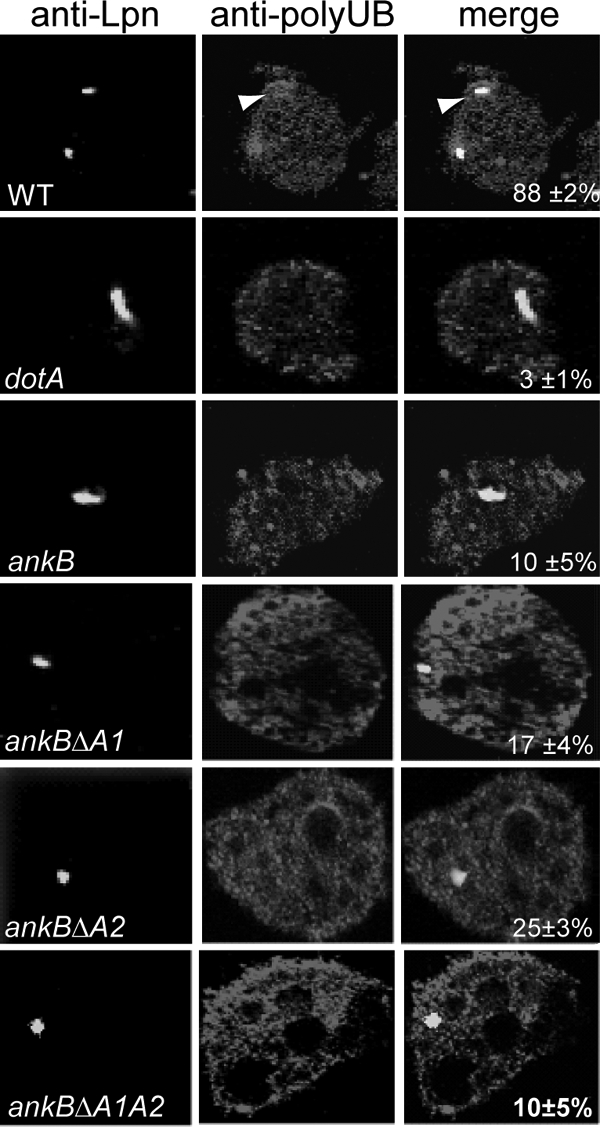
The two ANK domains of AnkB are essential for acquisition of polyubiquitinated proteins by the LCV within A. polyphaga. Representative confocal images of infected A. polyphaga for colocalization of phagosomes with polyubiquitinated proteins at 2 h post-infection of A. polyphaga by the wild-type strain of L. pneumophila, the ankB isogenic mutant, or the ankB mutant harboring one of the mutant ankB alleles on a plasmid. The cells were labeled with anti-Lpn antibody (green) and antipolyubiquitin (red). The arrowheads indicate heavy colocalization of polyubiquitin with the bacterium. Quantification of colocalization of the LCVs with polyubiquitin at 2 h postinfection is shown in the merged images and has been determined by analysis of 100 infected cells. The data are representative of three independent experiments.
Translocation of the AnkB variants into the host cell.
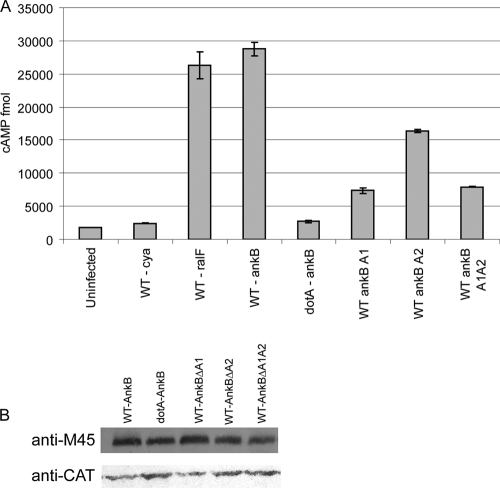
Dot/Icm-mediated translocation of AnkB into the host cell is essential for its crucial role in intracellular proliferation and for decoration of the LCV with polyubiquitinated proteins (4, 37). We examined whether inability of the AnkB deletion mutants (ankBΔA1, ankBΔA2, or ankBΔA1A2) to proliferate intracellularly and to recruit polyubiquitinated proteins to the LCV was due to failure of the Dot/Icm system to translocate these AnkB variants. We utilized the calmodulin-dependent adenylate cyclase-AnkB (CyaA-AnkB) fusion assay as a reporter system to assess translocation of the truncated AnkB variants. The data showed that there was less than a 2-fold reduction in translocation of the truncated AnkBΔA2 variant compared to the full-length AnkB while a ∼3-fold reduction in translocation was exhibited by the AnkBΔA1 and AnkBΔA1A2 truncated variants (Fig. 5A and data not shown). To ensure that this was not due to reduced expression or stability of the truncated proteins, Western blots of bacterial cell lysates were analyzed and showed equivalent expression levels of the Cya-AnkB fusions among all the tested strains (Fig. 5A). Thus, the defect in polyubiquitination and intracellular proliferation exhibited due to the loss of the ANK domains is not due to reduced expression or stability of the AnkB variant proteins. However, we cannot exclude the possibility that the observed reduction in translocation of the truncated variants of AnkB, compared to the WT AnkB, may account for the defect in recruitment of polyubiquitinated proteins to the LCV and for the defect in intracellular proliferation (Fig. 5A and data not shown).
FIG. 5.
Translocation of the AnkB variants into host cells. (A) Translocation of AnkB encoded by mutant alleles into U937 cells was determined at 1 h postinfection using adenylate cyclase assays and cyclic AMP (cAMP) levels as readout. Data points are the average cyclic AMP concentrations per well for a representative experiment performed three times in triplicate. Error bars represent standard deviations. (B) Equivalent expression of the AnkB-Cya fusion alleles in L. pneumophila was determined by immunoblot assays of Cya-AnkB fusions expressed in L. pneumophila. Proteins derived from equivalent numbers of bacteria (1 × 108) were loaded onto an SDS-polyacrylamide gel, and Cya fusion proteins were detected on Western blots probed with an anti-M45 antibody, recognizing the N-terminal M45 epitope on all Cya fusions. The blots were reprobed with anti-CAT antibodies, which showed equivalent expression of another protein encoded on the same reporter plasmid.
The role of the ANK domains in distribution of AnkB in mammalian cells.
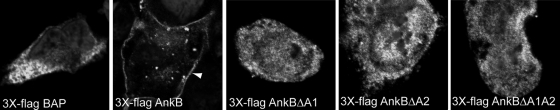
Although AnkB is clearly translocated into the host cell by the Dot/Icm system, we have been unable to detect AnkB during infections using AnkB-specific antibodies (4, 37). When ectopically expressed in HEK293 cells, the 3×Flag AnkB exhibits a striking localization to the host cell plasma membrane (37). Interestingly, similarly to the direct role of AnkB in decorating the LCV with polyubiquitinated proteins, ectopically expressed 3×Flag AnkB is localized to the host cell plasma membrane, where it recruits high levels of polyubiquitinated proteins. Interestingly, targeting of AnkB to the host cell membrane is essential for its function in recruitment of polyubiquitinated proteins (37). To examine whether the ANK domains were essential for targeting of AnkB to the host cell membrane, we determined localization of the 3×Flag AnkBΔA1, AnkBΔA2, and AnkBΔA1A2 truncated variants during ectopic expression in HEK293 cells. 3×Flag-tagged bacterial acid phosphatase (BAP) was used as a control. Transient transfection of HEK293 cells with a plasmid encoding 3×Flag AnkB revealed localization of AnkB to the plasma membrane (Fig. 6), as expected (37). In contrast, the AnkBΔA1, AnkBΔA2, and AnkBΔA1A2 truncated variants exhibited uniform distribution throughout the cytosol, similarly to the BAP control (Fig. 6). These data show that each of the two ANK domains of AnkB plays key roles in the peripheral localization of 3×Flag AnkB to the host cell plasma membrane.
FIG. 6.
Ectopic expression of 3×Flag AnkBΔA1, AnkBΔA2, or AnkBΔA1A2 in HEK293 cells shows altered cellular distribution compared with WT 3×Flag AnkB. Representative confocal images of HEK293 cells transiently transfected with plasmid DNA encoding 3×Flag AnkB, AnkBΔA1, AnkBΔA2, or AnkBΔA1A2 or the BAP control. At 24 h posttransfection, fixed and permeabilized HEK293 cells were labeled with mouse anti-Flag M2 antibody and an Alexa Fluor 488 secondary antibody (green). Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) (blue). The white arrowhead indicates localization of 3×Flag AnkB to the cell plasma membrane.
Next, we examined whether failure of the AnkBΔA1, AnkBΔA2, and AnkBΔA1A2 truncated variants to be targeted to the plasma membrane was associated with a defect in recruitment of polyubiquitinated proteins to the plasma membrane. Transfected cells were labeled with antipolyubiquitin and examined by confocal microscopy. Our data showed that while the WT AnkB recruited high levels of polyubiquitinated proteins to the plasma membrane, the AnkBΔA1, AnkBΔA2, and AnkBΔA1A2 truncated variants were defective in this process, similar to the BAP control (Fig. 7). This is consistent with failure to target the truncated AnkB variants to the plasma membrane (Fig. 6). We conclude that the two ANK domains are involved in targeting AnkB to host membranes and for the recruitment of polyubiquitinated proteins.
FIG. 7.
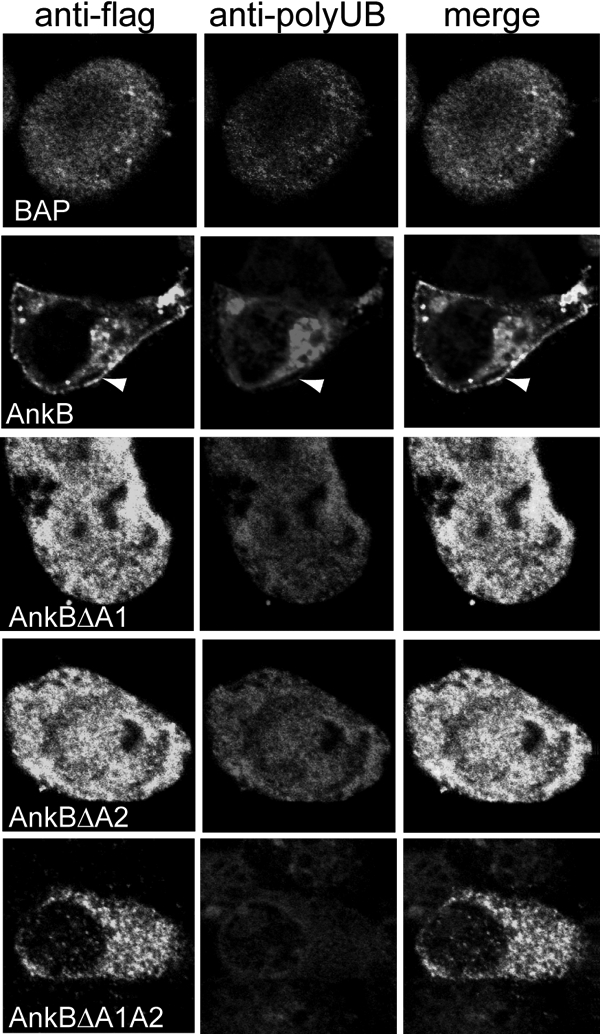
Ectopically expressed 3×Flag AnkBΔA1, AnkBΔA2, and AnkBΔA1A2 do not recruit polyubiquitinated proteins to the cell plasma membrane. At 24 h following transfection, fixed and permeabilized cells were labeled using a rabbit anti-Flag antibody (green) and a mouse antipolyubiquitin antibody (red). Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) (blue). The white arrowheads indicate colocalization of 3×Flag AnkB with polyubiquitinated proteins at the cell periphery.
Ectopic expression of AnkBΔA1, AnkBΔA2, and AnkBΔA1A2 in mammalian cells fails to trans-rescue the growth defect of the ankB mutant.
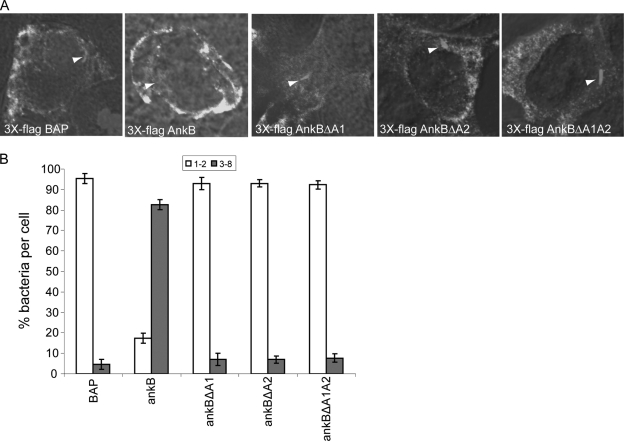
Our data above showed that the ankBΔA1, ankBΔA2, or ankBΔA1A2 alleles failed to complement the ankB mutant for intracellular replication and recruitment of polyubiquitinated proteins to the LCV. However, translocation of the AnkBΔA1, AnkBΔA2, and AnkBΔA1A2 truncated variants of AnkB was reduced compared to the WT AnkB. The reduced translocation of the AnkB variants might account for the reduced intracellular growth and recruitment of polyubiquitinated proteins observed for the ankB mutants complemented with the ankBΔA1, ankBΔA2, or ankBΔA1A2 alleles. We have shown that ectopic expression of 3×Flag AnkB in HEK293 cells is sufficient to restore intracellular growth to the ankB mutant bacteria, similarly to the WT strain (37). To overcome the caveat of reduced translocation of the AnkBΔA1, AnkBΔA2 and AnkBΔA1A2 variants in determining the functional role of the ANK domains, we examined the ability of the truncated proteins to trans-rescue the growth defect of the ankB mutant when they are ectopically expressed in HEK293 cells. Therefore, we examined whether transient ectopic expression of the AnkBΔA1, AnkBΔA2, and AnkBΔA1A2 variants trans-rescued the ankB mutant for its intracellular growth defect. Intracellular replication of the ankB mutant was examined in infected cells transiently transfected with plasmids encoding the 3×Flag AnkBΔA1, AnkBΔA2, or AnkBΔA1A2. The transiently transfected HEK293 cells were infected with the WT strain or the ankB mutant bacteria at an MOI of 10 for 1 h, followed by 1 h of treatment with gentamicin, and the number of intracellular bacteria was determined at 2 and 12 h postinfection by single cell analyses using microscopy. At 2 h postinfection, most cells harbored a single organism (data not shown). The ankB mutant failed to replicate in nontransfected cells or cells transfected with the 3×Flag BAP control (Fig. 8A and B). In contrast, ectopically expressed 3×Flag AnkB restored intracellular growth to the ankB mutant at 12 h postinfection, similarly to the wild-type bacteria in transfected or nontransfected cells. Importantly, the ankB mutant failed to replicate in cells expressing 3×Flag AnkBΔA1, AnkBΔA2, or AnkBΔA1A2 (Fig. 8A and B). This indicated that even when the AnkBΔA1, AnkBΔA2, or AnkBΔA1A2 proteins were supplied in trans, to overcome their reduced translocation, they did not restore intracellular replication to the ankB mutant. We conclude that the two ANK domains of AnkB are essential for intracellular proliferation of L. pneumophila in the host cell.
FIG. 8.
Ectopic expression of 3×Flag AnkBΔA1, AnkBΔA2, or AnkBΔA1A2 fails to rescue intravacuolar replication of the ankB mutant. HEK293 cells were transiently transfected with plasmids encoding 3×Flag BAP, 3×Flag AnkB, 3×Flag AnkBΔA1, 3×Flag AnkBΔA2, or 3×Flag AnkBΔA1A2 for 24 h prior to infection. Transfected HEK293 cells were then infected with the WT strain or the ankB or dotA mutant strain, and after 2 and 12 h, 100 infected cells were analyzed by confocal microscopy for formation of replicative phagosomes. (A) Representative confocal microscopy images of transfected HEK293 cells at 12 h postinfection. Ectopically expressed 3×Flag proteins were detected using antibody (green) while bacteria were detected using a rabbit anti-L. pneumophila antibody (red). Nuclei were stained with DAPI (blue). White arrowheads indicate bacteria inside cells. (B) Quantitation of single cell analysis of ankB mutant replicative phagosomes in transfected HEK293 cells 12 h postinfection. At 2 h and 12 h post-infection of HEK293 cells, 100 infected cells were analyzed by confocal microscopy for formation of replicative phagosomes. The WT strain and the dotA mutant bacteria were used as positive and negative controls, respectively. Infected cells from multiple coverslips were examined in each experiment. All the results are representative of three independent experiments performed in triplicate. Error bars represent standard deviations.
DISCUSSION
Among ∼200 Dot/Icm-exported effectors of L. pneumophila, very few have been shown to play a major role in intracellular proliferation within both protozoan and mammalian cells (4, 15, 21, 37). In contrast, AnkB plays a major role in the intracellular infection of both protozoan hosts and mammalian cells and is essential in vivo for intrapulmonary proliferation in the mouse model of Legionnaires' disease (4, 37). We have shown that the F-box domain of AnkB is essential for intracellular proliferation and for decoration of the LCV with polyubiquitinated proteins within mammalian and protozoan cells (37), which indicates that the host targets of AnkB are relatively conserved in mammalian and protozoan cells. Since the polyubiquitination machinery is highly conserved through evolution (12, 13, 26), it may not be surprising that the F-box domain exploits the polyubiquitination machinery within evolutionarily distant hosts. Exploitation of the polyubiquitination machinery is the first example of manipulation of conserved eukaryotic processes by L. pneumophila within mammalian and protozoan cells.
Deletion of either of the two ANK domains abolishes recruitment of polyubiquitinated proteins to the LCV within macrophages and amoebae. In addition, deletion of either of the two ANK domains fails to complement the ankB null mutant for its defect in intracellular replication. However, deletion of either or both of the ANK domains results in a decrease in export of the truncated proteins by the Dot/Icm system. The decrease in translocation may be due to improper folding of the protein to be presented for passage through the Dot/Icm channel, or the ANK domains may have specific translocation signals that aid in export of AnkB through the Dot/Icm system. It is difficult at this stage to determine the biological and biochemical threshold of export of AnkB essential for recruitment of polyubiquitinated proteins to the LCV. Therefore, it is not known whether the reduced translocation might have a drastic functional effect. Our data exclude the possibility that the reduced translocation of the truncated variants of AnkB is responsible for the functional defect, and this is based on several lines of evidence. First, ectopic expression of the AnkBΔA1, AnkBΔA2, or AnkBΔA1A2 truncated variants in mammalian cells results in failure to target the truncated variant proteins to the host plasma membrane, while the full-length AnkB is mainly targeted to the plasma membrane. Second, while ectopically expressed full-length AnkB recruits polyubiquitinated proteins to the plasma membrane, the AnkBΔA1, AnkBΔA2, or AnkBΔA1A2 truncated variants fail to exhibit the same process. Third, ectopic expression of the WT AnkB trans-rescues the ankB mutant for its defect in intracellular proliferation while the AnkBΔA1, AnkBΔA2 or AnkBΔA1A2 truncated variants fail to trans-rescue the ankB null mutant for its defect in intracellular proliferation. Taken together, we conclude that the two ANK domains of AnkB are essential for its function in decoration of the LCV with polyubiquitinated proteins and for intracellular proliferation.
Although most F-box proteins typically have an LRR or WD protein-protein interaction domain, the ANK protein-protein interaction domains have never been found in eukaryotic F-box proteins (23, 24, 36, 42). This indicates a noncanonical structure for the AnkB F-box protein compared to eukaryotic F-box proteins. While the F-box domain of AnkB is essential for interaction with the host Skp1 component of the SCF1 ubiquitin ligase, the two ANK domains are likely involved in binding to substrates targeted for polyubiquitination and linking them to the SCF1 ubiquitin ligase complex bound to the F-box domain to trigger their polyubiquitination. Since AnkB is targeted to host membranes, it is more likely that some of the targets of AnkB are localized to or associated with host cell membranes. It is likely that some of the substrates targeted by AnkB are highly conserved in mammalian and protozoan cells, since the two ANK domains are essential for the function of AnkB in both hosts. The identity of the substrates that bind the two ANK domains in mammalian and protozoan cells is one of our major future directions.
In summary, we have characterized an essential role for the two ANK domains of the F-box effector AnkB of L. pneumophila. Both ANK domains are essential for the function of AnkB to be targeted to membranes, to recruit polyubiquitinated proteins, and to enable intracellular bacterial proliferation. The two ANK domains are most likely involved in binding the substrates and linking them to the SCF complex bound to the F-box domain to trigger their polyubiquitination. Since the two ANK domains of AnkB are essential for decoration of the LCV by polyubiquitinated proteins within mammalian and protozoan cells, we predict that the two ANK domains bind evolutionarily conserved eukaryotic substrates. Deciphering the mammalian and protozoan substrates targeted by AnkB will facilitate our understanding of pathogenic evolution of L. pneumophila from a protozoan-interacting bacterium into a mammalian pathogen.
Acknowledgments
Y.A.K. is supported by Public Health Service awards R01AI43965 and R01AI069321 from NIAID and by the Commonwealth of Kentucky Research Challenge Trust Fund.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 1 March 2010.
REFERENCES
- 1.Abu Kwaik, Y. 1996. The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl. Environ. Microbiol. 62:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu-Zant, A., R. Asare, J. E. Graham, and Y. Abu Kwaik. 2006. Role for RpoS but not RelA of Legionella pneumophila in modulation of phagosome biogenesis and adaptation to the phagosomal microenvironment. Infect. Immun. 74:3021-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Khodor, S., T. Al-Quadan, and Y. Abu Kwaik. Temporal and differential regulation of expression of the eukaryotic-like ankyrin effectors of L. pneumophila. Environ. Microbiol., in press. [DOI] [PubMed]
- 4.Al-Khodor, S., C. T. Price, F. Habyarimana, A. Kalia, and Y. Abu Kwaik. 2008. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol. Microbiol. 70:908-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Khodor, S., C. T. D. Price, A. Kalia, and Y. Abu Kwaik. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol. 18:132-139. [DOI] [PMC free article] [PubMed]
- 6.Cazalet, C., C. Rusniok, H. Bruggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, and C. Buchrieser. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165-1173. [DOI] [PubMed] [Google Scholar]
- 7.Chung, C. Y., T. B. Reddy, K. Zhou, and R. A. Firtel. 1998. A novel, putative MEK kinase controls developmental timing and spatial patterning in Dictyostelium and is regulated by ubiquitin-mediated protein degradation. Genes Dev. 12:3564-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coers, J., C. Monahan, and C. R. Roy. 1999. Modulation of phagosome biogenesis by Legionella pneumophila creates an organelle permissive for intracellular growth. Nat. Cell Biol. 1:451-453. [DOI] [PubMed] [Google Scholar]
- 9.de Felipe, K. S., R. T. Glover, X. Charpentier, O. R. Anderson, M. Reyes, C. D. Pericone, and H. A. Shuman. 2008. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 4:e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Felipe, K. S., S. Pampou, O. S. Jovanovic, C. D. Pericone, S. F. Ye, S. Kalachikov, and H. A. Shuman. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 187:7716-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorer, M. S., D. Kirton, J. S. Bader, and R. R. Isberg. 2006. RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog. 2:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ennis, H. L., D. N. Dao, S. U. Pukatzki, and R. H. Kessin. 2000. Dictyostelium amoebae lacking an F-box protein form spores rather than stalk in chimeras with wild type. Proc. Natl. Acad. Sci. U. S. A. 97:3292-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ennis, H. L., D. N. Dao, M. Y. Wu, and R. H. Kessin. 2003. Mutation of the Dictyostelium fbxA gene affects cell-fate decisions and spatial patterning. Protist 154:419-429. [DOI] [PubMed] [Google Scholar]
- 14.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 15.Habyarimana, F., S. Al-Khodor, A. Kalia, J. E. Graham, C. T. Price, M. T. Garcia, and Y. A. Kwaik. 2008. Role for the ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ. Microbiol. 10:1460-1474. [DOI] [PubMed] [Google Scholar]
- 16.Habyarimana, F., C. T. Price, M. Santic, S. Al-Khodor, and Y. A. Kwaik. 2010. Molecular characterization of the Dot/Icm-translocated AnkH and AnkJ eukaryotic-like effectors of Legionella pneumophila. Infect. Immun. 78:1123-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harb, O. S., L. Y. Gao, and Y. Abu Kwaik. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2:251-265. [DOI] [PubMed] [Google Scholar]
- 18.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158:1319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, Q., and H. R. Henney, Jr. 1997. An Acanthamoeba polyubiquitin gene and application of its promoter to the establishment of a transient transfection system. Biochim. Biophys. Acta 1351:126-136. [DOI] [PubMed] [Google Scholar]
- 20.IJdo, J. W., A. C. Carlson, and E. L. Kennedy. 2007. Anaplasma phagocytophilum AnkA is tyrosine-phosphorylated at EPIYA motifs and recruits SHP-1 during early infection. Cell. Microbiol. 9:1284-1296. [DOI] [PubMed] [Google Scholar]
- 21.Isberg, R. R., T. J. O'Connor, and M. Heidtman. 2009. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7:13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kagan, J. C., and C. R. Roy. 2002. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 4:945-954. [DOI] [PubMed] [Google Scholar]
- 23.Kerscher, O., R. Felberbaum, and M. Hochstrasser. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22:159-180. [DOI] [PubMed] [Google Scholar]
- 24.Kobe, B., and A. V. Kajava. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11:725-732. [DOI] [PubMed] [Google Scholar]
- 25.Kondo-Okamoto, N., K. Ohkuni, K. Kitagawa, J. M. McCaffery, J. M. Shaw, and K. Okamoto. 2006. The novel F-box protein Mfb1p regulates mitochondrial connectivity and exhibits asymmetric localization in yeast. Mol. Biol. Cell 17:3756-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohanty, S., S. Lee, N. Yadava, M. J. Dealy, R. S. Johnson, and R. A. Firtel. 2001. Regulated protein degradation controls PKA function and cell-type differentiation in Dictyostelium. Genes Dev. 15:1435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molmeret, M., D. M. Bitar, L. Han, and Y. A. Kwaik. 2004. Disruption of the phagosomal membrane and egress of Legionella pneumophila into the cytoplasm during the last stages of intracellular infection of macrophages and Acanthamoeba polyphaga. Infect. Immun. 72:4040-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molmeret, M., M. Horn, M. Wagner, M. Santic, and Y. Abu Kwaik. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71:20-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molmeret, M., S. Jones, M. Santic, F. Habyarimana, M. T. Esteban, and Y. Abu Kwaik. 2010. Temporal and spatial trigger of post-exponential virulence-associated regulatory cascades by Legionella pneumophila after bacterial escape into the host cell cytosol. Environ. Microbiol. 12:704-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molofsky, A. B., and M. S. Swanson. 2004. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 53:29-40. [DOI] [PubMed] [Google Scholar]
- 31.Nagai, H., E. D. Cambronne, J. C. Kagan, J. C. Amor, R. A. Kahn, and C. R. Roy. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. U. S. A. 102:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neves, A. M., P. Guerreiro, and C. Rodrigues-Pousada. 1991. The macronuclear polyubiquitin gene of the ciliate Tetrahymena pyriformis. DNA Seq. 2:173-180. [DOI] [PubMed] [Google Scholar]
- 33.Pan, X., A. Luhrmann, A. Satoh, M. A. Laskowski-Arce, and C. R. Roy. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, J., K. J. Kim, K. S. Choi, D. J. Grab, and J. S. Dumler. 2004. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell. Microbiol. 6:743-751. [DOI] [PubMed] [Google Scholar]
- 35.Pedersen, L. L., M. Radulic, M. Doric, and Y. Abu Kwaik. 2001. HtrA homologue of Legionella pneumophila: an indispensable element for intracellular infection of mammalian but not protozoan cells. Infect. Immun. 69:2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6:9-20. [DOI] [PubMed] [Google Scholar]
- 37.Price, C. T., S. Al-Khodor, T. Al-Quadan, M. Santic, F. Habyarimana, A. Kalia, and Y. Abu Kwaik. 2009. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 5:e1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell, I. D., A. S. Grancell, and P. K. Sorger. 1999. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145:933-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schulman, B. A., A. C. Carrano, P. D. Jeffrey, Z. Bowen, E. R. Kinnucan, M. S. Finnin, S. J. Elledge, J. W. Harper, M. Pagano, and N. P. Pavletich. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408:381-386. [DOI] [PubMed] [Google Scholar]
- 40.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U. S. A. 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin, S., and C. R. Roy. 2008. Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell. Microbiol. 10:1209-1220. [DOI] [PubMed] [Google Scholar]
- 42.Smith, T. F., C. Gaitatzes, K. Saxena, and E. J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24:181-185. [DOI] [PubMed] [Google Scholar]
- 43.Solomon, J. M., A. Rupper, J. A. Cardelli, and R. R. Isberg. 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 68:2939-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teng-umnuay, P., H. R. Morris, A. Dell, M. Panico, T. Paxton, and C. M. West. 1998. The cytoplasmic F-box binding protein SKP1 contains a novel pentasaccharide linked to hydroxyproline in Dictyostelium. J. Biol. Chem. 273:18242-18249. [DOI] [PubMed] [Google Scholar]
- 45.Veiga, E., and P. Cossart. 2005. Ubiquitination of intracellular bacteria: a new bacteria-sensing system? Trends Cell Biol. 15:2-5. [DOI] [PubMed] [Google Scholar]
- 46.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 47.West, C. M., E. Kozarov, and P. Teng-umnuay. 1997. The cytosolic glycoprotein FP21 of Dictyostelium discoideum is encoded by two genes resulting in a polymorphism at a single amino acid position. Gene 200:1-10. [DOI] [PubMed] [Google Scholar]