Abstract
Previous work suggested that the underlying mechanisms by which the Streptococcus mutans ClpXP protease affects virulence traits are associated with accumulation of two orthologues of the Spx regulator, named SpxA and SpxB. Here, a thorough characterization of strains lacking the spx genes (ΔspxA, ΔspxB, and ΔspxA ΔspxB) revealed that Spx, indeed, participates in the regulation of processes associated with S. mutans pathogenesis. The ΔspxA strain displayed impaired ability to grow under acidic and oxidative stress conditions and had diminished long-term viability at low pH. Although the ΔspxB strain did not show any inherent stress-sensitive phenotype, the phenotypes observed in ΔspxA were more pronounced in the ΔspxA ΔspxB double mutant. By using two in vivo models, we demonstrate for the first time that Spx is required for virulence in a Gram-positive pathogen. Microarrays confirmed the global regulatory role of SpxA and SpxB. In particular, SpxA was shown to positively regulate genes associated with oxidative stress, a finding supported by enzymatic assays. SpxB had a secondary role in regulation of oxidative stress genes but appeared to play a larger role in controlling processes associated with cell wall homeostasis. Given the high degree of conservation between Spx proteins of low-GC Gram-positive bacteria, these results are likely to have broad implications.
Streptococcus mutans is an important oral pathogen, directly associated with the initiation and progression of dental caries. The ability to survive the periodic shifts in pH, oxygen tension, and nutrient availability encountered in dental biofilms is a key factor in the virulence of this organism (29). Therefore, the dissection of the mechanisms controlling stress tolerance could facilitate the development of novel therapeutic agents against diseases caused by S. mutans and related pathogens.
Clp proteolytic complexes play an essential role in cell homeostasis by assisting in refolding or degrading damaged proteins that accumulate in the cytoplasm following exposure to stress-inducing conditions (20). In addition to playing a role in protein quality control, Clp proteases also contribute to cellular homeostasis by controlling the stability of regulatory proteins (16, 22). Due to their multiple roles in cell physiology, it is not surprising that Clp proteases have been consistently implicated in bacterial pathogenesis (15, 18, 21). In addition, studies demonstrated that purified ClpP could elicit serotype-independent protection against pneumococcal infections in mice (8, 26, 34), and two new classes of antibiotics that target ClpP have recently been discovered (5, 7).
In S. mutans, the Clp complex is composed of multiple subunits of the ClpP peptidase in combination with subunits of Clp ATPases (ClpC, ClpE, or ClpX) (30). We along with others have demonstrated that strains bearing deletions in the clpP or clpX genes display important phenotypes associated with S. mutans virulence, including slow growth under stress conditions, reduced autolysis, increased sensitivity to antibiotics, altered biofilm formation, and increased acid tolerance (11, 13, 24, 30). The enhanced acid tolerance was unexpected as mutations of clpP or clpX genes of low-GC Gram-positive bacteria have normally resulted in stress-sensitive phenotypes (13, 14, 19, 49), with the exception of a Staphylococcus aureus clpX mutant that showed enhanced heat tolerance (15). Because a clear picture of the biological role of Clp proteolysis in bacterial pathogenesis has yet to emerge, an important observation that arose from our previous study was that inactivation of either one of two S. mutans orthologues of the Spx (suppressor of clpP and clpX) transcriptional regulator (spxA or spxB) was sufficient to alleviate phenotypes associated with the clpP and clpX mutations (24). More specifically, we demonstrated that Spx accumulated in the ΔclpP and ΔclpX strains and that the enhanced ability of the clpP and clpX mutants to survive lethal pH treatment, as well as the increased long-term persistence at nonlethal low pH, was abolished by inactivation of either spxA or spxB (24).
The Spx global regulator is highly conserved among low-GC Gram-positive bacteria and has been studied in detail in the soil organism Bacillus subtilis (32, 36-39, 41). Elegant studies from the laboratory of Nakano et al. demonstrated that Spx is a substrate of ClpXP proteolysis (41) and that accumulation of Spx was responsible for the pleiotropic phenotypes associated with clpXP mutations (36). Similar observations have been made in Lactococcus lactis (17), suggesting that the interactions between ClpXP and Spx are relatively conserved within Gram-positive bacteria.
The B. subtilis Spx has been shown to participate in the disulfide stress response by interacting with the C-terminal domain of the RNA polymerase α-subunit (α-CTD) to repress a variety of cellular processes while activating transcription of genes involved in oxidative stresses (38, 40). The Spx protein is activated via a CXXC redox disulfide motif, which senses intracellular disulfide generated by toxic oxidants (38). Transcriptional repression is a result of Spx's interfering with the interactions between the RNA polymerase and specific activator proteins (54), while activation of transcription requires contact between the Spx/RNA polymerase complex and upstream promoter DNA, thereby allowing Spx-induced engagement of RNA polymerase subunits with the core promoter (48).
Aside from the extensive characterization in B. subtilis, the genetic and physiologic significance of Spx in pathogenic bacteria has not been adequately explored. To date, a very small number of studies have associated Spx regulation with the expression of virulence attributes in Gram-positive bacteria. In Listeria monocytogenes, a putative spx gene was found highly upregulated during intracellular growth, suggesting that Spx might play a role in intracellular invasion, survival, or both (10). More recently, Spx was shown to affect growth, general stress tolerance, and biofilm formation in S. aureus (44), as well as induction of the X-state (competence) in Streptococcus pneumoniae (51).
Because we have shown an intimate association between the accumulation of Spx and the underlying mechanisms by which ClpXP proteolysis affects virulence traits in S. mutans, we sought to conduct here a detailed characterization of the spx genes in S. mutans. The results presented below clearly indicate that global regulation by Spx proteins modulates stress tolerance, survival, and virulence in S. mutans.
MATERIALS AND METHODS
Growth conditions of bacterial strains.
The S. mutans strains used in this study are listed in Table 1. S. mutans UA159 and its derivatives were routinely grown in brain heart infusion (BHI) medium at 37°C in a 5% CO2 atmosphere. When appropriate, spectinomycin (1.5 mg ml−1), erythromycin (10 μg ml−1), or kanamycin (1 mg ml−1) was added to the medium. An automated growth reader (Bioscreen C; Oy Growth Curves Ab, Ltd.) was used to monitor the ability of bacterial strains to grow in the presence of various stresses. To maintain an anaerobic environment, an overlay of 50 μl of sterile mineral oil was included in each well. For the Bioscreen C growth experiments, overnight cultures were diluted 1:20 into BHI medium and grown in a 5% CO2 atmosphere to an optical density at 600 nm (OD600) of 0.3, at which point 5 μl of the culture was used to inoculate wells of a 100-well plate containing 300 μl of the appropriate medium.
TABLE 1.
S. mutans strains used in this study
| Strain | Relevant genotype or description | Source or reference |
|---|---|---|
| UA159 | Wild type | Laboratory stock |
| UA159StR | Spontaneous Str mutanta | 12 |
| JL12 (ΔspxA) | spxA::Spec | 24 |
| JL13 (ΔspxB) | spxB::Erm | 24 |
| JL21 (ΔspxA ΔspxB) | spxA::Spec spxB::Erm | 24 |
| JL22 (Δsmu1651) | smu1651::Kmb | This study |
| JL23 (ΔclpP Δsmu1651) | clpP::Erm smu1651::Kmb | This study |
| JL24 (ΔspxA complement) | JL12 with pMC340B harboring spxA | This study |
| JL25 (ΔspxB complement) | JL13 with pMC340B harboring spxB | This study |
Str, streptomycin resistance.
Insertion of a nonpolar Km cassette.
Construction of mutant strains.
Creation of strains bearing mutations in spxA, spxB, or both was described previously (24). Similarly, a PCR ligation mutagenesis approach was used to create the Δsmu1651 mutant strain. Briefly, PCR fragments flanking the smu1651 gene were obtained and ligated to a nonpolar kanamycin (Kan) resistance marker, and the ligation mix was used to transform S. mutans UA159. A double Δsmu1651 ΔclpP mutant was constructed by further transformation of an erythromycin-resistant ΔclpP strain (24) using chromosomal DNA isolated from the Δsmu1651 mutant. Mutant strains were isolated on BHI plates supplemented with the appropriate antibiotic(s). The deletions were confirmed as correct by PCR sequencing of the insertion site and flanking sequences. For complementation studies with the spx single mutants, the full-length spxA or spxB gene was cloned into the kanamycin resistance integration vector pMC340B (23), which was then integrated into the appropriate mutant strain at the mtlA1 locus.
Long-term survival.
The ability of the S. mutans strains to survive a period of several days was assessed via a long-term survival assay, in which an overnight culture of cells was diluted 1:20 in TY (tryptone and yeast extract) medium containing 50 mM glucose. Growth of the cultures was monitored until stationary phase was reached, at which point an aliquot was removed for serial dilution into 0.1 M glycine, pH 7.0, and plating on BHI agar. Cultures continued to be incubated at 37°C in 5% CO2 for several days, with serial dilutions of the cultures plated daily until growth was no longer detected. Colonies were counted after 48 h of incubation.
Acid killing.
For killing experiments, exponentially grown cultures (OD600 of ≈0.5) were washed once with one culture volume of 0.1 M glycine buffer (pH 7) and resuspended in one-fifth of the original volume in 0.1 M glycine buffer (pH 2.85) for up to 90 min. Every 30 min, aliquots were serially diluted, plated on BHI agar, and incubated for 48 h before colonies were counted.
Galleria mellonella infection.
For the G. mellonella killing assays, insects in the final instar larval stage were purchased from Vanderhorst, Inc. (St. Marys, OH), stored at 4°C in the dark, and used within 7 days of shipment. Groups of 20 larvae, ranging from 200 to 300 mg in weight and with no signs of melanization, were randomly chosen and used for subsequent infection. A 10-μl Hamilton syringe was used to inject 5-μl aliquots of bacterial inoculum into the hemocoel of each larva via the last left proleg. Bacterial colony counts on BHI plates were used to confirm initial inocula. Groups injected with saline solution or with heat-inactivated (10 min at 75°C) S. mutans UA159 were used as controls in each experiment. After injection, larvae were incubated at 37°C, and appearance (signs of melanization) and survival were recorded at selected intervals. Larvae were scored as dead when they displayed no movement in response to touch. Kaplan-Meier killing curves were plotted, and estimations of differences in survival were compared using a log rank test. A P value of ≤0.05 was considered significant. All data were analyzed with GraphPad Prism, version 4.0, software. Experiments were repeated three independent times with similar results. To determine the number of bacteria per larva during the infectious process, two larvae were randomly chosen at 3-h intervals and homogenized using a bead beater (Biospec Products) for 30 s in the presence of 1 ml of 0.9% saline. The homogenate was serially diluted in 0.9% saline solution and plated on mitis salivarius and BHI agar for colony counts.
Rat colonization.
To test the infectivity of the spx mutants in vivo, we conducted a study to evaluate the ability of the strains to colonize the teeth of animals using an established rodent model of dental caries (6). Briefly, pathogen-free Sprague-Dawley rat pups 19 days of age were infected separately for three consecutive days by means of cotton swab with actively growing ΔspxA, ΔspxB, and ΔspxAΔspxB strains and UA159StR, a spontaneous streptomycin-resistant strain (12). Animals were fed a cariogenic diet (Diet-2000 containing 56% sucrose) and 5% (wt/vol) sucrose-water ad libitum. The experiment proceeded for 15 days, at the end of which the animals were euthanized by CO2 asphyxiation, and the lower jaws were removed for microbiological assessment (25). The number of mutans streptococci recovered from the animals was expressed as the number of CFU ml−1 of jaw sonicate. The data were subjected to analysis of variance for significance and Grubb's test to detect outliers (none were detected). This study was reviewed and approved by the University of Rochester Committee on Animal Resources.
RNA extraction.
RNA from S. mutans cells was isolated as described previously (1). Briefly, RNA was isolated from homogenized S. mutans cells grown in BHI medium, buffered to pH 7.0, to an OD600 of 0.4 by repeated hot acid-phenol-chloroform extractions, and the nucleic acid was precipitated with 1 volume of ice-cold isopropanol and 1/10 volume 3 M sodium acetate (pH 5) at −20°C overnight. RNA pellets were resuspended in 80 μl of nuclease-free H2O and treated with DNase I (Ambion) at 37°C for 30 min. RNA concentrations were determined using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and 1-μl samples were run on an agarose gel to verify RNA integrity. The RNA was purified again using an RNeasy mini-kit (Qiagen), including a second on-column DNase treatment that was performed as recommended by the supplier. Final RNA concentrations were determined in triplicate.
Microarray experiments.
S. mutans UA159 version 1 microarray slides were provided by the J. Craig Venter Institute Pathogen Functional Genomics Resource Center (PFGRC) (http://pfgrc.jcvi.org/index.php/microarray). The microarray protocols have been previously published in detail (1, 31), so only a brief description is included. A reference RNA prepared from S. mutans UA159 cells grown in BHI medium to an OD600 of 0.5 was used in every experiment, as described previously (1). Four cDNA samples generated from 2 μg of RNA originating from four independent cultures of S. mutans UA159 and its derivatives were hybridized to the arrays along with the reference cDNA. Test cDNA was coupled with Cy3-dUTP, while reference cDNA was coupled with Cy5-dUTP (Amersham Biosciences). The slides were hybridized to a mixture containing equal amounts of test and reference cDNA for 16 h at 42°C in a MAUI (MicroArray User Interface) hybridization chamber (BioMicro Systems). Hybridized slides were washed and scanned using a GenePix scanner (Axon Instruments). Data were analyzed using software available at the J. Craig Venter Institute PFGRC website. Single-channel images of the slides were loaded into Spotfinder and overlaid. MIDAS software was used to normalize the spot intensity data using LOWESS and iterative log mean centering with the default setting, followed by in-slide replicate analysis. Statistical analysis was carried out with BRB Array Tools (http://linus.nci.nih.gov/BRB-ArrayTools.html) with a cutoff P value of 0.001. Additional details regarding array protocols are available at http://pfgrc.jcvi.org/index.php/microarray/protocols.html.
Real-time quantitative PCR.
A subset of genes was selected to validate the microarray analysis by real-time quantitative reverse transcription-PCR (qRT-PCR). Gene-specific primers (see Table S1 in the supplemental material) were designed using Beacon Designer, version 2.0, software (Premier Biosoft International). Reverse transcription and real-time reverse transcriptase PCR were carried out according to protocols described elsewhere (1, 31). A Student's t test was performed to verify significance of the real-time PCR quantifications.
Enzymatic assays.
For enzymatic assays, mid-log-phase cultures of S. mutans UA159 and its derivatives grown in BHI medium were harvested by centrifugation and washed with sterile water, and the cell pellets were stored at −80°C until use. Cells were resuspended in 1 ml of lysis buffer (10 mM Tris, pH 7.5, 50 mM NaCl, 1 mM EDTA). An equal volume of 0.1-mm-diameter, acid-washed glass beads was added, and the mixture was homogenized in a bead beater for two 2-min intervals. Homogenized cells were then centrifuged at 6,600 × g for 10 min at 4°C, and the cleared lysates were used for the enzymatic assays. All reactions were carried out at room temperature. Protein concentration of the cell lysates was determined by the method of Bradford. All assay methods have been previously published, so only a brief description was included below (9, 43, 46). The mixture used to determine NADH oxidase activity (46) contained 50 μg of protein lysate in 100 mM potassium phosphate buffer, pH 7, with 0.3 mM EDTA. NADH was added to a final concentration of 0.16 mM to initiate the reaction. The decrease in absorbance at 340 nm was recorded over 4-min intervals using a spectrophotometer (DU640; Beckman Coulter). One unit of NADH activity is defined as the amount of enzyme needed to catalyze the oxidation of 1 mmol of NADH min−1. A glutathione oxidoreductase (GOR) assay (9) was performed by adding 1 mM GSSG (glutathione disulfide) (Sigma) to a mix containing 50 μg of protein lysate, 100 mM potassium phosphate buffer, 1 mM EDTA, and 0.1 mM NADPH to catalyze the reaction. The decrease in absorbance was measured at 340 nm over 4-min intervals. One unit of GOR activity is defined as the amount of enzyme needed to catalyze the oxidation of 1 μmol of NADPH per minute. Superoxide dismutase (SOD) activity was measured by a method using cytochrome c, xanthine, and xanthine oxidase (43). Briefly, 1-ml reaction mixtures containing 50 mM potassium phosphate buffer, pH 7.8, 10 μM cytochrome c, and 50 μM xanthine were mixed with various amounts of cell extract. The concentration of xanthine oxidase (Sigma) used in the assay was empirically determined by the amount necessary to produce a rate of reduction of cytochrome c of 0.025 U min−1 at an absorbance of 550 nm. One unit of SOD activity is defined as the amount of enzyme needed to inhibit the reduction of cytochrome c by 50%.
Microarray data accession number.
Microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under GEO Series accession number GSE19656.
RESULTS
Identification of Spx homologues in S. mutans.
A BLAST search of the S. mutans UA159 genome (2) performed using the B. subtilis Spx sequence revealed three putative homologues: Smu1142, Smu1651, and Smu2084. Pfam searches placed all three proteins as members of the arsenate reductase family, which include true arsenate reductases (ArsC) and Spx proteins. Previously, we demonstrated that inactivation of smu1142 and smu2084 alleviated phenotypes of the ΔclpP and ΔclpX strains; hence, the genes were named spxA and spxB, respectively (24). However, in that study we did not investigate whether smu1651 encoded a third Spx protein. Of note, it has been suggested that the number of Spx paralogues may vary among different species, with as many as seven spx-like genes identified in L. lactis (17, 52).
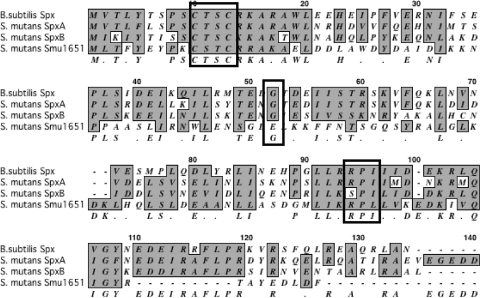
Because Spx is highly conserved among Gram-positive bacteria, sequence similarity and the presence of certain motifs can be used to identify its homologues. The CXXC motif in the amino terminus, involved in sensing the redox state by means of disulfide bond formation (38, 42), is present in SpxA, SpxB, and Smu1651 (Fig. 1). In addition to the CXXC motif, an RPI motif in the carboxyl terminus of Spx has been implicated in the modulation of the CXXC motif in the presence of sulfate (47). This exact motif is present in SpxA but is found as SPI in SpxB and RPL in Smu1651. Finally, the conserved Gly52 responsible for the interaction of the B. subtilis Spx with the RNA polymerase α-CTD is found only in SpxA and SpxB and not in Smu1651.
FIG. 1.
Alignment of B. subtilis Spx and S. mutans SpxA, SpxB, and Smu1651 amino acid sequences. Similar residues are shaded, and a consensus is shown below the sequences. Conserved motifs discussed in the text are outlined in a box. The GenBank accession numbers are as follows: B. subtilis Spx, NP_389032.1; S. mutans SpxA, SMU.1142c; S. mutans SpxB, SMU.2084c; S. mutans Smu1651, SMU.1651.
To evaluate whether accumulation of Smu1651 could be associated with phenotypes observed in the clpP and clpX mutants, the smu1651 gene was replaced by a nonpolar antibiotic resistance marker and used to isolate a Δsmu1651 ΔclpP double mutant. The inactivation of smu1651 failed to suppress the slow growth or tendency to clump in broth that was common to the ΔclpP strain (data not shown). Given this result and the fact that Smu1651 lacks the conserved Gly52 residue necessary for interactions with the RNA polymerase, this protein was not considered to be a true Spx protein, and further characterizations of the Δsmu1651 mutants were not continued. Thus, we conclude that S. mutans harbors only two Spx proteins.
Growth characteristics of Δspx strains.
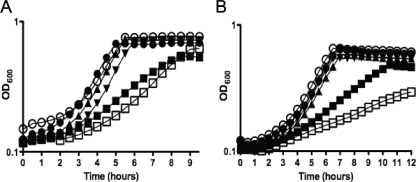
Whereas our previous study examined the effects of mutations in spxA or spxB in combination with ΔclpP or ΔclpX strains, here we seek to characterize S. mutans strains bearing mutations in only the spx genes. The growth characteristics of the ΔspxA, ΔspxB, and ΔspxA ΔspxB strains, as well as mutant strains complemented with a copy of the spxA or spxB gene, were assessed in an automated growth reader (Fig. 2). When grown in BHI medium at 37°C with a mineral oil overlay to maintain an anaerobic environment, the ΔspxB strain did not appear to have any inherent growth deficiency, while the ΔspxA and ΔspxA ΔspxB strains demonstrated moderate reductions in growth rates (Fig. 2A). We also tested the ability of the mutant strains to grow under aerobic (in BHI medium with shaking) conditions (Fig. 2B). Slow-growth phenotypes were again observed for the ΔspxA and the double mutant ΔspxA ΔspxB strains though this trend was more pronounced in the double mutant ΔspxA ΔspxB strain, which also failed to reach the same final OD600 as the other strains. As seen under anaerobic growth conditions, the ΔspxB strain grew as well as the parent strain under aerobic conditions. Of note, growth of all strains in glass flasks in a 5% CO2 atmosphere was similar to the growth patterns of the strains in the Bioscreen growth monitor with a mineral oil overlay (data not shown).
FIG. 2.
Growth characteristics of the Δspx strains using the Bioscreen C growth reader. Growth curves of S. mutans UA159 (•), ΔspxA (▪), ΔspxB (○), ΔspxAB (□), complemented ΔspxA (▴), and complemented ΔspxB (▾) strains performed in a Bioscreen C growth reader in BHI medium at 37°C under anaerobic (A) and aerobic (B) conditions. The curves shown are representative of those of at least five experiments.
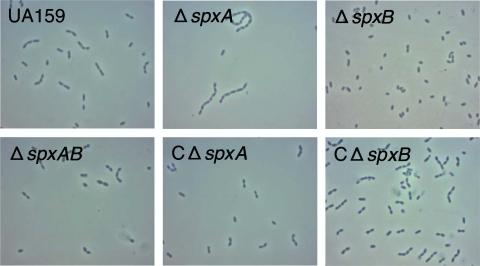
When observed under the microscope during all stages of growth, the ΔspxB strain was completely unable to form cell chains. On the other hand, the ΔspxA strain showed a tendency to form chains longer than those formed by the parent UA159 strain upon entry into late log and stationary growth phases. Interestingly, the double mutant ΔspxA ΔspxB strain was capable of forming chains of lengths typical of those of the parent strain during all growth stages (Fig. 3).
FIG. 3.
Phase-contrast microphotographs of S. mutans UA159, ΔspxA, ΔspxB, ΔspxAB, complemented ΔspxA (cΔspxA), and complemented ΔspxB (cΔspxB) strains.
Complementation of the ΔspxA and ΔspxB mutants restored wild-type phenotypes.
To confirm that the lack of spxA and spxB was responsible for the phenotypes of the ΔspxA and ΔspxB mutants, respectively, each gene was integrated in the mtlA1 locus of its respective mutant strain using the integration vector pMC340B. Complementation of each spx single mutant completely reverted the slow-growth (ΔspxA) (Fig. 2) and altered-chain length (ΔspxA and ΔspxB) (Fig. 3) phenotypes of the spx single mutants.
Inactivation of the spx genes, in particular spxA, affects long-term survival at low pH.
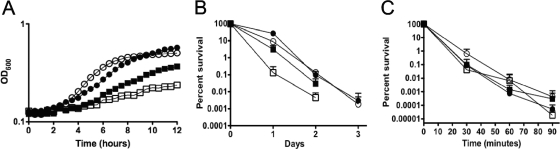
Previously, we reported an enhanced ability of the ΔclpP and ΔclpX strains to survive short-term and long-term acid challenges (24). Inactivation of either spxA or spxB reverted the increased acid tolerance of the ΔclpP and ΔclpX mutants, suggesting that accumulation of Spx was directly involved with this phenotype. Here, we first assessed the growth characteristics of the Δspx strains at an acidic pH (BHI medium, pH 5.5 or pH 5, with mineral oil overlay). Similar to the trends observed under aerobic and anaerobic conditions, the ΔspxA and ΔspxA ΔspxB strains grew at significantly lower rates than the parent at pH 5.5 (Fig. 4A), and the ΔspxA ΔspxB strain was completely unable to grow at pH 5 (data not shown). On the other hand, the ΔspxB strain displayed growth rates that were comparable to those for the wild-type strain (Fig. 4A and data not shown).
FIG. 4.
Acid stress tolerance of S. mutans UA159 (•), ΔspxA (▪), ΔspxB (○), and ΔspxAB (□) strains. (A) Growth in BHI medium, pH 5.5, at 37°C determined using the Bioscreen C growth reader monitor. (B) Long-term survival assay. Aliquots of cultures were first plated on BHI agar when the stationary growth phase was reached (day 0). (C) Acid killing. Aliquots were plated on BHI agar upon suspension in 0.1 M glycine (pH 2.85) and each 30 min thereafter. The growth curve in panel A at pH 5.5 is a representative of at least five independent experiments, and the curves shown in panels B and C are the averages with standard deviations of the results from three independent experiments.
After observing that growth of strains lacking spxA was significantly impaired at low pH, we further characterized the acid tolerance of the Δspx strains by performing long-term survival and acid killing experiments. In the long-term survival assay, survival of the cells in TY medium containing 50 mM glucose, in which a final pH of approximately 4.2 was attained by all strains upon entry into stationary phase, was monitored daily. Compared to the parent strain, long-term viability of the ΔspxA and ΔspxA ΔspxB strains was substantially reduced, with no viable cells detected after 2 days. However, survival of the ΔspxB strain was identical to that of UA159 (Fig. 4B). Interestingly, there were no significant differences among the survival profiles of the Δspx mutant strains and the parent strain in acid killing experiments (Fig. 4C).
The ΔspxA and ΔspxA ΔspxB strains, but not ΔspxB, showed reduced tolerance to oxidative stress agents.
The B. subtilis Spx is recognized as a global regulator of oxidative stress responses by activating the transcription of genes involved in thiol homeostasis (38, 39). Growth curves of the S. mutans spx mutants under aerobic conditions suggested that SpxA and, to a lesser extent, SpxB play a role in oxygen tolerance. To further evaluate this possibility, we assessed the capacity of the Δspx mutants to grow in the presence of 0.25 mM diamide (a specific oxidant of thiols) or 0.5 mM H2O2 (Fig. 5). In the presence of 0.5 mM H2O2, both the ΔspxA and ΔspxA ΔspxB strains showed a slow-growth pattern compared to the parental strain. In the presence of 0.25 mM diamide, the ΔspxA strain showed a prolonged lag phase that was followed by a slightly lower growth rate than that of the parent. Interestingly, the ΔspxA ΔspxB double mutant was highly sensitive to diamide and was completely unable to grow in the presence of 0.25 mM diamide.
FIG. 5.
Growth curves of S. mutans UA159 (•), ΔspxA (▪), ΔspxB (○), and ΔspxAB (□) strains under oxidative stress conditions. Exponentially grown cultures were inoculated in BHI medium containing 0.5 mM H2O2 (A) or 0.25 mM diamide (B), and growth at 37°C was monitored using the Bioscreen C growth reader. The curves shown are representative of a typical experiment performed at least five times.
Inactivation of the spx genes reduces virulence of S. mutans in the greater wax worm model.
The intrahemocoelic infection of the larvae of the greater wax worm G. mellonella has been used to study the virulence of various bacteria and fungi, including Gram-positive pathogens such as Bacillus cereus, Enterococcus faecalis, and S. aureus (27, 45, 50). In G. mellonella, the production of superoxide via a respiratory burst by insect-specialized phagocytic cells, known as hemocytes, is an important line of defense against microbial infections (4). Given that Spx is an important regulator of oxidative stress responses (39) and considering that the ΔspxA and ΔspxA ΔspxB strains showed increased sensitivity to oxidative stress agents, we asked whether the greater wax worm model would be appropriate for studying the virulence and fitness of S. mutans. Upon demonstration that intrahemocoelic injection with S. mutans UA159 cultures can effectively kill the larvae of G. mellonella in a dose-dependent manner (Fig. 6A), we infected three groups of 20 larvae each with 1 × 107 CFU of S. mutans UA159, ΔspxA, ΔspxB, and ΔspxA ΔspxB cultures. As shown in Fig. 6B, the rates of killing were significantly lower in larvae infected with any of the three Δspx strains than in those infected with the parent UA159 strain (P ≤ 0.001). After 18 h of infection, approximately 90% of the larvae infected with UA159 were dead, whereas 50, 40, and 25% of the larvae infected with the ΔspxA, ΔspxB, and ΔspxA ΔspxB strains, respectively, were dead (Fig. 6B). Both of the complemented spx strains were more virulent than the respective ΔspxA or ΔspxB single mutant strains and showed virulence similar to that of parent strain (data not shown). To further confirm that killing was due to an infectious process and not to toxic effects of bacterial components such as the cell wall lipoteichoic acid (LTA), we assessed survival of larvae injected with heat-killed S. mutans and confirmed that no significant killing was observed after several days.
FIG. 6.
Killing of G. mellonella larvae infected with S. mutans UA159 and its derivatives at 37°C. (A) Dose-dependent killing by S. mutans UA159. Survival (Kaplan-Meier) plots of larvae injected with S. mutans cells at 1 × 105 (solid black line), 1 × 106 (broken black line), 1 × 107 (solid gray line), or 1 × 108 (broken gray line) CFU/larva. (B) Kaplan-Meier plots of S. mutans UA159 (solid black line), ΔspxA (broken black line), ΔspxB (solid gray line), and ΔspxAB (broken gray line) strains injected at 1 × 107 CFU/larva. There was no killing of larvae injected with saline (data not shown) and minimum killing of larva injected with heat-killed S. mutans UA159 cells (dotted line). The experiments were repeated three times, and the results are representative of a typical experiment. Compared to the wild-type UA159 strains, all Δspx strains showed attenuated virulence (P ≤ 0.05).
The ΔspxA and ΔspxA ΔspxB mutants, but not ΔspxB, showed reduced ability to colonize the teeth of rats.
To further demonstrate the involvement of Spx with host infection, we investigated the capacity of the Δspx mutants to colonize the teeth of pathogen-free rat pups. Following a 15-day infection period, colonies of S. mutans UA159 and all three of the Δspx strains were recovered from the jaws of infected pups, demonstrating that the Δspx strains were capable of establishing in the rats (Fig. 7). While the average number of recovered CFU of the ΔspxB strain was similar to that of the parent strain, colonies of both the ΔspxA and ΔspxA ΔspxB mutant strains were recovered in significantly fewer numbers (P ≤ 0.05).
FIG. 7.
Colonization of S. mutans UA159 and derivative strains on the teeth of rats after 15 days. The symbols shown represent the recovered bacterial colonies from each individual rat while the horizontal line represents the mean recovery per bacterial strain. Grubb's test did not indicate the presence of outliers among each strain. *, P ≤ 0.02.
Microarray analysis further revealed the global regulatory role of SpxA and SpxB.
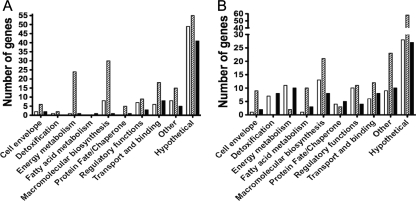
In an effort to disclose the scope of the Spx regulon in S. mutans and to explore the differences in the regulatory roles of each Spx protein, the transcriptome of mid-exponential-phase cultures of S. mutans UA159, ΔspxA, ΔspxB, and ΔspxA ΔspxB strains were analyzed by microarrays. Confirming the global role of Spx proteins in gene regulation, a large number of genes were differentially regulated in the double mutant and in each individual Δspx strain compared to the parent (172 genes in ΔspxA, 313 genes in ΔspxB, and 148 genes in ΔspxA ΔspxB; P ≤ 0.001). There was minimal overlap between genes that appeared differentially expressed in ΔspxA and ΔspxB arrays (only 16 genes with the same trends in expression appeared in both arrays), suggesting distinct regulatory duties for each Spx protein. The genes that appeared on these microarray analyses have been grouped into functional categories (Fig. 8), and the complete list of altered genes is presented in Table S2 in the supplemental material. A subset of 26 genes was selected and used for qRT-PCR analysis for validation of the microarray data (Table 2 and data not shown).
FIG. 8.
Number of genes separated by functional categories that were differentially expressed by S. mutans ΔspxA (open bars), ΔspxB (hatched bars), and ΔspxAB (dark bars) compared to the parent UA159 strain in microarray analysis. (A) Upregulated genes. (B) Downregulated genes.
TABLE 2.
Expression ratios of genes involved in detoxification of oxidative stresses identified in S. mutans microarray analysis
| Gene name | Function | ΔspxA strain |
ΔspxB strain |
Δspx ΔspxB strain |
|||
|---|---|---|---|---|---|---|---|
| Arraya | RT-PCR | Array | RT-PCR | Array | RT-PCR | ||
| trxA | Thioredoxin | ND | 1.967 | 0.639 | 1.574 | 0.227 | 0.134** |
| trxB | Thioredoxin reductase | 0.355 | 0.800 | ND | 1.900 | ND | 0.233** |
| dpr | Peroxide resistance protein | 0.133 | 0.263* | ND | 0.289* | 0.198 | 0.021** |
| sodA | Superoxide dismutase | 0.054 | 0.058 | ND | 0.129 | 0.068 | 0.004 |
| ahpC | Alkyl hydroperoxide reductase | 0.136 | 0.236*** | ND | 0.491** | 0.072 | 0.055*** |
| ahpF | Alkyl hydroperoxide reductase | 0.167 | 0.236*** | ND | 0.542** | 0.079 | 0.060*** |
| gor | Glutathione reductase | 0.258 | 0.462* | ND | 0.962 | 0.298 | 0.104** |
| tpx | Thiol peroxidase | 0.173 | 0.088*** | ND | 0.500* | 0.069 | 0.003*** |
| nox | H2O-forming NADH oxidase | 0.183 | 0.284*** | ND | 0.261*** | 0.046 | 0.010*** |
Array data are relative levels of expression compared to expression in S. mutans UA159. RT-PCR data are gene copy numbers relative to those of S. mutans UA159. ND, no significant differences in expression were determined by microarray analysis. *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.0005.
Genes involved in oxidative stress responses are positively regulated by SpxA and, to a lesser extent, by SpxB.
The expression of several genes involved in defense against oxidative stress (trxB, thioredoxin reductase; dpr, peroxide resistance protein; sod, superoxide dismutase; ahpC and ahpF, alkyl hydroperoxide reductase C and F subunits; gor, glutathione reductase; tpx, thiol peroxidase; and nox-2, water-forming NADH oxidase) was downregulated in ΔspxA and ΔspxA ΔspxB strain arrays (Table 2; see also Table S2 in the supplemental material). This trend of decreased expression of genes involved in oxidative stress tolerance was not observed for the ΔspxB strain (with the exception of trxA, thioredoxin, which was downregulated). However, the fold change in expression was frequently greater in the ΔspxA ΔspxB double mutant than in the ΔspxA strain, suggesting that SpxB may also play a role in the regulation of these genes. In fact, qRT-PCR validations of the microarray data, a technique much more sensitive than microarrays, revealed that expression levels of ahpC, ahpF, dpr, nox, and sodA were also downregulated in the ΔspxB strain (Table 2).
Inactivation of spxB affects expression of genes involved in cell envelope structure and composition.
Analysis of the ΔspxB microarrays revealed a change in expression of a number of genes involved in cell division, cell envelope, or fatty acid metabolism (4, 11, and 10 genes, respectively). The change in expression of these genes may affect the normal cell division cycle and chain formation and may help explain the lack of chains observed from planktonic cultures of the ΔspxB strain. Interestingly, only five of the genes in these categories also appeared in the microarray analysis of the ΔspxA ΔspxB strain, which, in turn, was capable of forming chains of normal length.
Spx regulation affects expression of genes associated with key virulence properties of S. mutans.
In addition to sharing common functions with the well-characterized B. subtilis Spx (for example, positive regulation of oxidative stress genes), the transcriptomes of the Δspx mutants indicate that Spx might also control traits that are particularly important in S. mutans. Among other genes/pathways whose expression was altered in the Δspx mutants were genes involved in DNA repair (mutY and smxA downregulated in both the ΔspxA and ΔspxA ΔspxB strains), malolactic fermentation (mleS and mleP downregulated in ΔspxA ΔspxB), proton-translocating F-ATPase activity (atpC, atpD, and atpG upregulated in ΔspxB), and monounsaturated fatty acid biosynthesis (fabM downregulated in ΔspxA ΔspxB). Notably, these genes/pathways are all associated with the acid tolerance of S. mutans (29) and help to explain the phenotypes of the Δspx strains in relation to acid stress. Interestingly, genes essential for biofilm formation namely, gtfB, gtfD, and gpbA, were found to be upregulated in the ΔspxB strain. The gtfB and gtfD genes encode glucosyltransferase enzymes (GTFs) required for the production of glucans from sucrose, whereas gbpA encodes a glucan binding protein that plays a role in adhesion and accumulation of dental biofilms (3, 35, 53).
The microarray analyses also revealed downregulation of clpX mRNA in both ΔspxB and ΔspxA ΔspxB strains. Whereas some clp genes (clpC, clpE, and clpP) have been shown to fall under the transcriptional control of the CtsR repressor (30), the regulatory mechanisms controlling clpX expression are largely unknown. It is possible that Spx proteins could serve as activators of clpX, forming a regulatory loop, as the ClpXP protease modulates cellular levels of Spx. Finally, a large number of genes that appeared in each of the microarrays represented uncharacterized hypothetical genes (corresponding to 45, 36, and 46% for ΔspxA, ΔspxB, and ΔspxA ΔspxB, respectively), indicating that the identity of a significant portion of the genetic network controlled by Spx remains unknown.
Activities of selected oxidative stress enzymes show a dramatic reduction in the absence of SpxA.
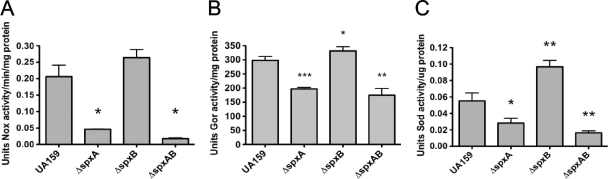
The microarray analysis revealed that a number of genes involved in detoxification were downregulated in the ΔspxA and ΔspxA ΔspxB strains compared to the parent strain. Genes encoding two S. mutans proteins with NADH oxidase activity, nox-2 (NADH oxidase) and ahpF (alkyl hydroperoxide reductase), showed more than 5-fold decrease in expression levels in the ΔspxA strain compared to the parent; this difference was elevated to more than 10-fold (nox-2) or 20-fold (ahpF) in the ΔspxA ΔspxB double mutant. To verify that these changes in gene expression could be detected at the enzyme activity level, NADH oxidase activity assays were performed with lysates from cultures of each of the spx mutant strains. Consistent with the gene expression profile, NADH oxidase activity was reduced in the ΔspxA strain and was even lower in the ΔspxA ΔspxB strain (Fig. 9A). Surprisingly, the NADH oxidase activity of the ΔspxB mutant was slightly enhanced compared to that of the parent strain. In addition to ahpCF and nox-2, microarrays revealed that expression levels of gor and sod were decreased by approximately 3- and 14-fold, respectively, in the ΔspxA and ΔspxA ΔspxB strains. The corresponding enzymatic assays were also performed to determine activities of Gor and Sod in the S. mutans Δspx strains. Similar to the NADH oxidase assay, Gor and Sod activities were proportionally reduced in the ΔspxA and ΔspxA ΔspxB strains though enhanced in the ΔspxB strain (Fig. 9B and C).
FIG. 9.
Enzymatic activities of NADH oxidase (A), glutathione reductase (B), and superoxide dismutase (C) determined from lysates of S. mutans cultures. Values shown are averages with standard deviations of the results from three independent experiments. *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.005.
DISCUSSION
Despite the strong linkage of ClpXP proteolysis with bacterial pathogenesis (5, 7, 15, 18, 21, 26) and the close association of Spx accumulation with phenotypes resulting from clpXP mutations (17, 24, 36), the effects of global regulation by Spx have not been characterized in pathogenic bacteria. In our previous work, we showed that ClpXP proteolytic control of two Spx orthologues is involved in processes underlying S. mutans pathogenesis (24). Here, we demonstrate that global regulation by Spx modulates stress tolerance, survival, and virulence in S. mutans and that SpxA and SpxB fulfill common, as well as distinct, roles within the cells. Phenotypic characterization of ΔspxA, ΔspxB, and ΔspxA ΔspxB mutants suggest that SpxA plays a major role in cell survival, likely by regulating the expression of pathways that are associated with oxygen tolerance and metabolism. Although the ΔspxB strain did not show impaired growth or survival under the circumstances tested, virulence of ΔspxB was attenuated in the wax worm model, and the phenotypes of the ΔspxA strain were often more pronounced in the ΔspxA ΔspxB double mutant.
Evidence is now accumulating that multiple Spx proteins with similar regulatory functions may be found in the genomes of low-GC Gram-positive bacteria (24, 47, 51, 52). However, a direct linkage of multiple Spx proteins with attenuation of phenotypes observed in clpP or clpX mutants has been demonstrated only in S. mutans (24). In B. subtilis, an Spx paralogue (MgsR, for modulator of the general stress response) was shown to control a subset of genes belonging to the general stress regulon, including products linked to oxidative stress resistance (47). It is not known whether there is any type of interaction between MgsR and Spx. In L. lactis, another Spx paralogue (named SpxB) was shown to positively regulate O-acetylation of the cell wall peptidoglycan, a process that confers increased resistance to lysozyme (52). Interestingly, it was demonstrated that inactivation of TrmA in L. lactis, a proven Spx protein shown to be involved in the heat-sensitive phenotype of a clpP mutant (17), resulted in a marked increase in lysozyme resistance (52). Since both TrmA and SpxB were shown to interact with the RNA polymerase, the authors proposed that these proteins might compete for binding targets or may have a synergistic (or antagonistic) effect such that different binding combinations direct the RNA polymerase complex to different promoters (52). Finally, two Spx homologues (named SpxA1 and SpxA2) were recently identified in S. pneumoniae (51). Although single mutations in either of the S. pneumoniae spx genes could be stably isolated, a double mutation was apparently lethal, supporting the idea of potential overlap in the roles of the Spx proteins (51). Nevertheless, it was demonstrated that transcriptional repression by SpxA1 has a negative effect in the development of the X-state (competence) (51).
The impaired growth of strains bearing an spxA deletion (ΔspxA and ΔspxA ΔspxB), coupled with our microarray analysis, indicates that SpxA is the primary regulatory protein responsible for transcriptional activation of genes involved in oxidative stress and thiol homeostasis in S. mutans. In addition to the phenotypic and transcriptomic characterization of the Δspx mutants, amino acid sequence analysis of SpxA and SpxB provided another indication that SpxA might, indeed, respond more efficiently to oxidative stress than SpxB. ClustalW alignment revealed that the S. mutans SpxA shares higher levels of similarity with the well-characterized B. subtilis Spx protein (80% similarity versus 73% for SpxB). Moreover, it has been shown that the N-terminal CXXC motif, the C-terminal RPI motif, and the conserved Gly52 responsible for Spx-RNA polymerase interactions are essential residues for Spx activity (55). While the CXXC motif and Gly52 were conserved in the S. mutans SpxA and SpxB proteins, the RPI motif, implicated in modulation of the CXXC motif in the presence of sulfate, was present in SpxA but found as SPI in the SpxB protein. Thus, it is possible that a single amino acid substitution in the RPI C-terminal region of the S. mutans SpxB may affect the ability of this protein to sense the intracellular redox potential of the cell. Work is under way to test this hypothesis.
Despite the fact that an oxidation-sensitive phenotype was not observed in ΔspxB, the increased sensitivity of the ΔspxA ΔspxB strain to diamide suggested that SpxB might play a secondary (and additive) role in controlling thiol homeostasis. The role of Spx in regulation of pathways that cope with products from oxidation was further confirmed by reduced expression of NADH oxidase, Gor, and Sod activities in the ΔspxA and ΔspxA ΔspxB strains. Unexpectedly, enzymatic activities were actually enhanced in the ΔspxB mutant compared to the parent strain. These findings indicate a potential cross talk between SpxA and SpxB that could also involve other regulators in a complex system to fine-tune oxidative stress pathways. Based on our current knowledge, we hypothesize that SpxA-SpxB interactions could affect oxidative stress pathways in ways that may not be mutually exclusive. First, the two Spx proteins could compete for binding to the RNA polymerase. In this case, assuming that these proteins have similar capacity to bind to the RNA polymerase α-CTD domain, the absence of one regulator would facilitate binding of the other. This appears to be the case in L. lactis, where two Spx homologs (TrmA and SpxB) were shown to physically interact and compete for binding with the RNA polymerase in an ex vivo (yeast two-hybrid) assay (52). Second, SpxA (or SpxB) could affect ClpXP-dependent degradation of Spx proteins by modulating clpP or clpX transcriptional levels. In fact, clpX mRNA levels were significantly downregulated in the ΔspxB and ΔspxA ΔspxB strains, suggesting that SpxB may be a positive regulator of clpX transcription. In this scenario, Spx levels would be kept in check by increased ClpXP proteolysis. It is also possible that the Spx-RNA polymerase interactions are even more complex and involve different binding combinations of the two Spx proteins and, possibly, other regulators.
It is widely accepted that there is a substantial overlap in different stress pathways, including overlaps between the acid and oxidative stress pathways (28, 33). Along these lines, the ΔspxA and ΔspxA ΔspxB strains that displayed increased sensitivity to oxidative agents also showed impaired ability to grow at an acidic pH, as well as reduced low-pH long-term survival capacities. In addition to genes involved in oxidative stress and thiol homeostasis, several other genes that might help explain the acid-sensitive phenotypes of ΔspxA and ΔspxA ΔspxB were identified in the microarrays. In particular, the expression of genes involved in pathways linked to S. mutans acid tolerance (28, 29), including DNA repair (mutY and smxA), monounsaturated fatty acid biosynthesis (fabM), and malolactic fermentation (mleS and mleP), were found downregulated in either one or both of the ΔspxA and ΔspxA ΔspxB strains. In addition, a large number of the differentially transcribed genes encoded hypothetical proteins, suggesting that the identity of a significant portion of the genetic network influenced by Spx is still unknown. Conceivably, the identification of Spx-regulated genes encoding hypothetical proteins might lead to the identification of novel factors involved in S. mutans stress tolerance.
Upon characterization of the spx mutants in vitro, we used two in vivo models to assess the role of Spx in S. mutans pathogenesis: killing of the larva of the greater wax moth G. mellonella and the ability to colonize the teeth of pathogen-free rats. In G. mellonella, the rates and overall levels of killing were significantly lower in all Δspx mutants (ΔspxA, ΔspxB, and ΔspxA ΔspxB strains), revealing that Spx contributes significantly to virulence in this nonmammalian model. To test the correlation of the G. mellonella model with an established mammalian model of infection, we tested the ability of the Δspx strains to colonize the teeth of rats fed a high-sucrose diet. The results of the rat infection experiments fit well with the overall trends that we have observed with the in vitro experiments. In other words, the recovery of the ΔspxB strain was similar to that of the parent, but recovery of the ΔspxA strain was significantly reduced, and that of the ΔspxA ΔspxB strain was even further diminished. While these results show a positive correlation between the ability of the ΔspxA and ΔspxA ΔspxB strains to kill G. mellonella and to implant on the teeth of rats, they did not correlate with the reduced virulence of ΔspxB in the insect model. This difference points out an important limitation of G. mellonella for studies involving S. mutans pathogenesis. S. mutans is primarily a dental pathogen that lives in multispecies biofilms in dental plaque, where the ability to produce glucan polymers from sucrose via GTFs plays a key role in adhesion and accumulation of biofilms and dramatically affects the biophysical properties of dental plaque, including changes in the pH values of the microenvironment (3, 35, 53). In fact, an enhanced ability to synthesize glucans might offer a possible explanation for the successful colonization of the ΔspxB strain on the teeth of rats. The microarray analysis revealed that expression of gtfB and gtfD, responsible for the production of water-insoluble and water-soluble glucans, respectively, and of the glucan-binding protein gbpA was significantly enhanced in the ΔspxB strain. The upregulation of these genes suggests that the ΔspxB strain might have an enhanced capacity to form biofilms in sucrose and that the high-sucrose diets provided to the rats may have given ΔspxB an important advantage toward establishing in the rat model. Nevertheless, the in vivo studies strongly support the association of Spx with the virulence of S. mutans and demonstrate that killing of G. mellonella should be considered, with associated caveats, a useful addition to studies involving S. mutans pathogenesis.
In summary, the present work clearly demonstrates that Spx regulates physiological traits that are important for S. mutans virulence and indicates that multiple Spx proteins may have evolved to control common as well as unique cellular processes in bacteria. The S. mutans SpxA resembles the B. subtilis Spx and is specialized in controlling the expression of genes involved in oxidative stress. SpxB appears to play a secondary (and additive) role in controlling oxidative stress responses but seems to play a larger role in processes associated with cell wall homeostasis. Most importantly, by using two in vivo models of infection, this study demonstrates, for the first time, that Spx is required for full virulence in a Gram-positive pathogen.
Supplementary Material
Acknowledgments
This study was supported by NIH-NIDCR awards DE019783 to J.A.L. and DE017425 to R.G.Q. J.K.K., I.R.R., J.A., and A.M.D. were supported by the NIDCR training program in oral science grant T32DE007202. P.L.R. was supported by CAPES/MEC (BEX 2827/07-7) and CNPq/MCT (302222/2008-1) from Brazil.
Footnotes
Published ahead of print on 16 March 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abranches, J., M. M. Candella, Z. T. Wen, H. V. Baker, and R. A. Burne. 2006. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 188:3748-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banas, J. A., and M. M. Vickerman. 2003. Glucan-binding proteins of the oral streptococci. Crit. Rev. Oral Biol. Med. 14:89-99. [DOI] [PubMed] [Google Scholar]
- 4.Bergin, D., E. P. Reeves, J. Renwick, F. B. Wientjes, and K. Kavanagh. 2005. Superoxide production in Galleria mellonella hemocytes: identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect. Immun. 73:4161-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottcher, T., and S. A. Sieber. 2008. beta-Lactones as specific inhibitors of ClpP attenuate the production of extracellular virulence factors of Staphylococcus aureus. J. Am. Chem. Soc. 130:14400-14401. [DOI] [PubMed] [Google Scholar]
- 6.Bowen, W. H., S. K. Pearson, and D. A. Young. 1988. The effect of desalivation on coronal and root surface caries in rats. J. Dent. Res. 67:21-23. [DOI] [PubMed] [Google Scholar]
- 7.Brotz-Oesterhelt, H., D. Beyer, H. P. Kroll, R. Endermann, C. Ladel, W. Schroeder, B. Hinzen, S. Raddatz, H. Paulsen, K. Henninger, J. E. Bandow, H. G. Sahl, and H. Labischinski. 2005. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 11:1082-1087. [DOI] [PubMed] [Google Scholar]
- 8.Cao, J., D. Li, Y. Gong, N. Yin, T. Chen, C. K. Wong, W. Xu, J. Luo, X. Zhang, C. W. Lam, and Y. Yin. 2009. Caseinolytic protease: a protein vaccine which could elicit serotype-independent protection against invasive pneumococcal infection. Clin. Exp. Immunol. 156:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlberg, I., and B. Mannervik. 1985. Glutathione reductase. Methods Enzymol. 113:484-490. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chattoraj, P., A. Banerjee, S. Biswas, and I. Biswas. 2010. ClpP of Streptococcus mutans differentially regulates expression of genomic islands, mutacin production and antibiotics tolerance. J. Bacteriol. 192:1312-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clancy, K. A., S. Pearson, W. H. Bowen, and R. A. Burne. 2000. Characterization of recombinant, ureolytic Streptococcus mutans demonstrates an inverse relationship between dental plaque ureolytic capacity and cariogenicity. Infect. Immun. 68:2621-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng, D. M., J. M. ten Cate, and W. Crielaard. 2007. The adaptive response of Streptococcus mutans towards oral care products: involvement of the ClpP serine protease. Eur. J. Oral Sci. 115:363-370. [DOI] [PubMed] [Google Scholar]
- 14.Frees, D., A. Chastanet, S. Qazi, K. Sorensen, P. Hill, T. Msadek, and H. Ingmer. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54:1445-1462. [DOI] [PubMed] [Google Scholar]
- 15.Frees, D., S. N. Qazi, P. J. Hill, and H. Ingmer. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48:1565-1578. [DOI] [PubMed] [Google Scholar]
- 16.Frees, D., K. Savijoki, P. Varmanen, and H. Ingmer. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63:1285-1295. [DOI] [PubMed] [Google Scholar]
- 17.Frees, D., P. Varmanen, and H. Ingmer. 2001. Inactivation of a gene that is highly conserved in Gram-positive bacteria stimulates degradation of non-native proteins and concomitantly increases stress tolerance in Lactococcus lactis. Mol. Microbiol. 41:93-103. [DOI] [PubMed] [Google Scholar]
- 18.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286-1294. [DOI] [PubMed] [Google Scholar]
- 19.Gerth, U., E. Kruger, I. Derre, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28:787-802. [DOI] [PubMed] [Google Scholar]
- 20.Gottesman, S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19:565-587. [DOI] [PubMed] [Google Scholar]
- 21.Ibrahim, Y. M., A. R. Kerr, N. A. Silva, and T. J. Mitchell. 2005. Contribution of the ATP-dependent protease ClpCP to the autolysis and virulence of Streptococcus pneumoniae. Infect. Immun. 73:730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenal, U., and R. Hengge-Aronis. 2003. Regulation by proteolysis in bacterial cells. Curr. Opin. Microbiol. 6:163-172. [DOI] [PubMed] [Google Scholar]
- 23.Jung, C. J., Q. H. Zheng, Y. H. Shieh, C. S. Lin, and J. S. Chia. 2009. Streptococcus mutans autolysin AtlA is a fibronectin-binding protein and contributes to bacterial survival in the bloodstream and virulence for infective endocarditis. Mol. Microbiol. 74:888-902. [DOI] [PubMed] [Google Scholar]
- 24.Kajfasz, J. K., A. R. Martinez, I. Rivera-Ramos, J. Abranches, H. Koo, R. G. Quivey, Jr., and J. A. Lemos. 2009. Role of Clp proteins in expression of virulence properties of Streptococcus mutans. J. Bacteriol. 191:2060-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koo, H., S. K. Pearson, K. Scott-Anne, J. Abranches, J. A. Cury, P. L. Rosalen, Y. K. Park, R. E. Marquis, and W. H. Bowen. 2002. Effects of apigenin and tt-farnesol on glucosyltransferase activity, biofilm viability and caries development in rats. Oral Microbiol. Immunol. 17:337-343. [DOI] [PubMed] [Google Scholar]
- 26.Kwon, H. Y., A. D. Ogunniyi, M. H. Choi, S. N. Pyo, D. K. Rhee, and J. C. Paton. 2004. The ClpP protease of Streptococcus pneumoniae modulates virulence gene expression and protects against fatal pneumococcal challenge. Infect. Immun. 72:5646-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebreton, F., E. Riboulet-Bisson, P. Serror, M. Sanguinetti, B. Posteraro, R. Torelli, A. Hartke, Y. Auffray, and J. C. Giard. 2009. ace, Which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect. Immun. 77:2832-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemos, J. A., J. Abranches, and R. A. Burne. 2005. Responses of cariogenic streptococci to environmental stresses. Curr. Issues Mol. Biol. 7:95-107. [PubMed] [Google Scholar]
- 29.Lemos, J. A., and R. A. Burne. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154:3247-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemos, J. A., and R. A. Burne. 2002. Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 184:6357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemos, J. A., M. M. Nascimento, V. K. Lin, J. Abranches, and R. A. Burne. 2008. Global regulation by (p) ppGpp and CodY in Streptococcus mutans. J. Bacteriol. 190:5291-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, J., and P. Zuber. 2000. The ClpX protein of Bacillus subtilis indirectly influences RNA polymerase holoenzyme composition and directly stimulates sigma-dependent transcription. Mol. Microbiol. 37:885-897. [DOI] [PubMed] [Google Scholar]
- 33.Marquis, R. E. 2004. Applied and ecological aspects of oxidative-stress damage to bacterial spores and to oral microbes. Sci. Prog. 87:153-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morsczeck, C., T. Prokhorova, J. Sigh, M. Pfeiffer, M. Bille-Nielsen, J. Petersen, A. Boysen, T. Kofoed, N. Frimodt-Moller, P. Nyborg-Nielsen, and P. Schrotz-King. 2008. Streptococcus pneumoniae: proteomics of surface proteins for vaccine development. Clin. Microbiol. Infect. 14:74-81. [DOI] [PubMed] [Google Scholar]
- 35.Munro, C., S. M. Michalek, and F. L. Macrina. 1991. Cariogenicity of Streptococcus mutans V403 glucosyltransferase and fructosyltransferase mutants constructed by allelic exchange. Infect. Immun. 59:2316-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakano, M. M., F. Hajarizadeh, Y. Zhu, and P. Zuber. 2001. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 42:383-394. [DOI] [PubMed] [Google Scholar]
- 37.Nakano, M. M., S. Nakano, and P. Zuber. 2002. Spx (YjbD), a negative effector of competence in Bacillus subtilis, enhances ClpC-MecA-ComK interaction. Mol. Microbiol. 44:1341-1349. [DOI] [PubMed] [Google Scholar]
- 38.Nakano, S., K. N. Erwin, M. Ralle, and P. Zuber. 2005. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol. Microbiol. 55:498-510. [DOI] [PubMed] [Google Scholar]
- 39.Nakano, S., E. Kuster-Schock, A. D. Grossman, and P. Zuber. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 100:13603-13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakano, S., M. M. Nakano, Y. Zhang, M. Leelakriangsak, and P. Zuber. 2003. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. U. S. A. 100:4233-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakano, S., G. Zheng, M. M. Nakano, and P. Zuber. 2002. Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J. Bacteriol. 184:3664-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newberry, K. J., S. Nakano, P. Zuber, and R. G. Brennan. 2005. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 102:15839-15844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen, P. T., J. Abranches, T. N. Phan, and R. E. Marquis. 2002. Repressed respiration of oral streptococci grown in biofilms. Curr. Microbiol. 44:262-266. [DOI] [PubMed] [Google Scholar]
- 44.Pamp, S. J., D. Frees, S. Engelmann, M. Hecker, and H. Ingmer. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 188:4861-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peleg, A. Y., D. Monga, S. Pillai, E. Mylonakis, R. C. Moellering, Jr., and G. M. Eliopoulos. 2009. Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J. Infect. Dis. 199:532-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phan, T. N., P. T. Nguyen, J. Abranches, and R. E. Marquis. 2002. Fluoride and organic weak acids as respiration inhibitors for oral streptococci in acidified environments. Oral Microbiol. Immunol. 17:119-124. [DOI] [PubMed] [Google Scholar]
- 47.Reder, A., D. Hoper, C. Weinberg, U. Gerth, M. Fraunholz, and M. Hecker. 2008. The Spx paralogue MgsR (YqgZ) controls a subregulon within the general stress response of Bacillus subtilis. Mol. Microbiol. 69:1104-1120. [DOI] [PubMed] [Google Scholar]
- 48.Reyes, D. Y., and P. Zuber. 2008. Activation of transcription initiation by Spx: formation of transcription complex and identification of a cis-acting element required for transcriptional activation. Mol. Microbiol. 69:765-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robertson, G. T., W. L. Ng, R. Gilmour, and M. E. Winkler. 2003. Essentiality of clpX, but not clpP, clpL, clpC, or clpE, in Streptococcus pneumoniae R6. J. Bacteriol. 185:2961-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salamitou, S., F. Ramisse, M. Brehelin, D. Bourguet, N. Gilois, M. Gominet, E. Hernandez, and D. Lereclus. 2000. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology 146:2825-2832. [DOI] [PubMed] [Google Scholar]
- 51.Turlan, C., M. Prudhomme, G. Fichant, B. Martin, and C. Gutierrez. 2009. SpxA1, a novel transcriptional regulator involved in X-state (competence) development in Streptococcus pneumoniae. Mol. Microbiol. 73:492-506. [DOI] [PubMed] [Google Scholar]
- 52.Veiga, P., C. Bulbarela-Sampieri, S. Furlan, A. Maisons, M. P. Chapot-Chartier, M. Erkelenz, P. Mervelet, P. Noirot, D. Frees, O. P. Kuipers, J. Kok, A. Gruss, G. Buist, and S. Kulakauskas. 2007. SpxB regulates O-acetylation-dependent resistance of Lactococcus lactis peptidoglycan to hydrolysis. J. Biol. Chem. 282:19342-19354. [DOI] [PubMed] [Google Scholar]
- 53.Yamashita, Y., W. H. Bowen, R. A. Burne, and H. K. Kuramitsu. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect. Immun. 61:3811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, X., Y. Hu, X. Guo, E. Lescop, Y. Li, B. Xia, and C. Jin. 2006. The Bacillus subtilis YkuV is a thiol:disulfide oxidoreductase revealed by its redox structures and activity. J. Biol. Chem. 281:8296-8304. [DOI] [PubMed] [Google Scholar]
- 55.Zuber, P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 186:1911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.