Abstract
A gene cluster was identified which contains genes involved in the biosynthesis of actinomycin encompassing 50 kb of contiguous DNA on the chromosome of Streptomyces chrysomallus. It contains 28 genes with biosynthetic functions and is bordered on both sides by IS elements. Unprecedentedly, the cluster consists of two large inverted repeats of 11 and 13 genes, respectively, with four nonribosomal peptide synthetase genes in the middle. Nine genes in each repeat have counterparts in the other, in the same arrangement but in the opposite orientation, suggesting an inverse duplication of one of the arms during the evolution of the gene cluster. All of the genes appear to be organized into operons, each corresponding to a functional section of actinomycin biosynthesis, such as peptide assembly, regulation, resistance, and biosynthesis of the precursor of the actinomycin chromophore 4-methyl-3-hydroxyanthranilic acid (4-MHA). For 4-MHA synthesis, functional analysis revealed genes that encode pathway-specific isoforms of tryptophan dioxygenase, kynurenine formamidase, and hydroxykynureninase, which are distinct from the corresponding enzyme activities of cellular tryptophan catabolism in their regulation and in part in their substrate specificity. Phylogenetic analysis indicates that the pathway-specific tryptophan metabolism in Streptomyces most probably evolved divergently from the normal pathway of tryptophan catabolism to provide an extra or independent supply of building blocks for the synthesis of tryptophan-derived secondary metabolites.
The actinomycins are chromopeptide lactones produced by various Streptomyces strains (34). They consist of a phenoxazinone chromophore (actinocin) with two pentapeptide lactone rings attached in amide linkage (Fig. 1). The various naturally occurring actinomycins differ by amino acid substitutions in specific positions of their peptide chains, while the actinocin moiety remains unchanged (34, 49). Due to the planar structure of actinocin, actinomycins can intercalate into GC-rich DNA. This leads to inhibition of transcription in both prokaryotes and eukaryotes (67). Actinomycins have strong antineoplastic activities, which have attracted much interest in their chemistry and biosynthesis (47, 49). The high toxicity of the compounds, however, has restricted their therapeutic use to only a few tumor types (18). Structural redesign of this compound class to minimize toxicity is a main goal of research on actinomycins (3).
FIG. 1.
Structure of actinomycin C1. Actinomycin C1 (I) is the main component of the actinomycin C mixture (C1, C2, and C3) elaborated by S. chrysomallus. Actinomycins C2 and C3 differ from C1 by the replacement of one or both d-valine residues with d-allo-isoleucine. (II) Structure of 4-MHA pentapeptide lactone (half molecule of actinomycin C1). (III) Structure of 4-MHA. Sar, sarcosine (N-methylglycine); MeVal, N-methyl-l-valine.
Studies on the biosynthesis of actinomycins have shown that—as in their chemical synthesis—they are formed by the oxidative condensation of two aroyl pentapeptide lactone molecules (actinomycin halves, Fig. 1, structure II) (38). Their formal amino terminus is 4-methyl-3-hydroxyanthranilic acid (4-MHA) (Fig. 1, structure III), which, owing to its o-aminophenol structure, can be converted to a phenoxazinone by oxidative condensation with a second 4-MHA pentapeptide lactone molecule. This can occur spontaneously by exposure of the aminophenol to air or to chemicals such as ferricyanide (7). However, many enzymes can oxidize aminophenols to phenoxazinones, such as phenoxazinone synthases (66), peroxidases (13), catalases (52), laccases (16), tyrosinases (68, 71), or even cytochromes (50).
4-MHA pentapeptide lactone formation in actinomycin C-producing Streptomyces chrysomallus is catalyzed by four nonribosomal peptide synthetase (NRPS) enzyme subunits (actinomycin synthetases [ACMSs]) (58). Two stand-alone NRPS domains, ACMSI (4-MHA adenylating domain, 51 kDa) and AcmACP (4-MHA carrier domain, 10 kDa), serve as the initiation module. Two large multifunctional NRPS subunits, ACMSII and ACMSIII (280 and 480 kDa, respectively), contain the five elongation modules, with a thioesterase domain at the carboxy-terminal end of ACMS III catalyzing the assembly and release of the peptide lactone (lower part of Fig. 2). The acmA, -B, -C, and -D genes that encode the ACMS enzymes were cloned, sequenced, and shown to be arranged as two invertedly oriented gene pairs (62) (Fig. 2). In contrast, little is known about the genes that encode 4-MHA biosynthesis and the genes involved in regulation and self-resistance. It was shown previously that 4-MHA is derived from tryptophan (34), and enzymatic studies revealed pathway-specific isoforms of tryptophan oxygenase, kynurenine formamidase, and kynureninase activities in the actinomycin producers Streptomyces parvulus and S. chrysomallus (8, 9, 17, 22, 24, 35, 72). Moreover, an enzyme which methylates 3-hydroxyanthranilic acid (3-HA) at its 4 position was purified from S. antibioticus and characterized (32). Despite these achievements, however, the exact roles of these enzymes in actinomycin synthesis remained unknown.
FIG. 2.
Actinomycin biosynthetic gene cluster. The functions of the 28 genes that make up the gene cluster, including the 4 already known NRPS genes acmA to -D (66), are represented by arrows as follows: black, regulation; dark gray, 4-MHA biosynthesis; white, unknown role or function; gray, NRPS genes. In the lower part is shown the assembly line of the four NRPS subunits which catalyze the formation of 4-MHA pentapeptide lactone.
Here we present an analysis of the DNA regions flanking the NRPS genes in S. chrysomallus. The regions are highly similar to each other, each containing genes involved in 4-MHA synthesis and genes responsible for regulation and self-resistance. Nine genes in either arm have paralogous counterparts in the other, all arranged in the same order but in the opposite orientation, resulting in a large inverted repeat (IR) separated by the NRPS genes. This unique biosynthetic gene cluster most probably arose by duplication of one of the arms.
MATERIALS AND METHODS
Chemicals.
l-Kynurenine and 3-hydroxy-dl-kynurenine were from Sigma (Deisenhofen, Germany). Formyl-l-kynurenine was from Calbiochem-Behring (La Jolla, CA). Anthranilic acid, 3-hydroxyanthranilic acid (3-HA), and silica gel 60-coated alumina sheets were from Merck (Darmstadt, Germany). 4-Methyl-3-hydroxy-anthranilic acid (4-MHA) was synthesized as described previously (39). Authentic actinomycin D (C1) was from Applichem (Darmstadt, Germany). All other chemicals were of the highest purity commercially available.
Analytical methods.
Solvent systems for thin-layer chromatography (TLC) were solvent system I (ethyl acetate-methanol-water, 20:1:1, by volume), solvent system II (ethyl acetate-hexane-acetic acid, 8:2.5:0.5, by volume), and solvent system III (n-butanol-acetic acid-water, 4.1:1, by volume). High-performance liquid chromatography (HPLC) separation of formyl-l-kynurenine, l-kynurenine, anthranilic acid, 3-HA, and 4-MHA was done on a Eurosphere100-C18 column (Knauer AG, Berlin, Germany) with a water-acetonitrile gradient (containing 0.1% trifluoroacetic acid) of 0 to 60% acetonitrile in 15 min at a flow rate of 1 ml min−1. The detection wavelengths were 280 nm, 320 nm (formylkynurenine), and 365 nm (kynurenine). The actinomycin contents of cultures were determined by extraction of culture aliquots with ethyl acetate and subsequent spectrophotometric determination at 452 nm using authentic actinomycin D as the standard. In some cases, prior to quantitation, actinomycin samples were separated by TLC using solvent system I and yellow bands were scraped off the plates and extracted with ethanol. Protein determination was according to reference 5.
Enzyme assays.
Kynurenine formamidase activity was measured by incubating enzyme (50 to 150 μg of protein) in the presence of 2 mM formyl-l-kynurenine-25 mM potassium phosphate (pH 7.2) in a total volume of 700 μl at 32°C. The reaction was monitored spectrophotometrically by the increase in absorbance at 365 nm (22) in a Perkin-Elmer model 551S spectrometer. Alternatively, 100-μl aliquots were taken from the reaction mixture at different times and subjected to HPLC separation. The kynurenine formed was quantitated by using a calibration curve made with known amounts of kynurenine. Kynureninase and hydroxykynureninase were assayed by incubation of 25 μl enzyme (5 to 15 μg of protein in 25 mM potassium phosphate (pH 7.2) with 75 μl mix containing 2 mM kynurenine, 12 μM pyridoxal phosphate (PLP), and 25 mM potassium phosphate (pH 6.8) or 2 mM 3-hydroxy-dl-kynurenine, 12 μM PLP, and 25 mM potassium phosphate (pH 7.2), respectively, at 25°C in wells of a black well microtiter plate (Greiner Bio-One, Frickenhausen, Germany) at 25°C. The progress of the reaction was monitored by measuring the fluorescence of the anthranilic acid or 3-HA formed at an emission wavelength of 410 nm (excitation wavelength, 340 nm) in an Infinite M200 spectrophotometer (Tecan, Crailsheim, Germany) at 1-min intervals for up to 1 h. Calibration curves were recorded with samples containing various amounts of anthranilic acid or 3-HA in the same buffer.
Strains and cultures.
Strain Sc1 of S. chrysomallus was isolated as a single colony from a streaked culture of S. chrysomallus ATCC 11523 (wild type) on CM agar (22). Streptomyces coelicolor M145 was from the John Innes strain collection. Maintenance of strains was on CM agar (S. chrysomallus) or on R2YE (S. coelicolor) (26). Growth in liquid culture for the preparation of protein extracts from both strains was in either liquid CM using 300-ml Erlenmeyer flasks containing 100 ml medium and steel springs as baffles (22) or a glutamate-mineral salts medium (36) supplemented with 1% maltose using 500-ml Erlenmeyer flasks containing 200 ml medium. Incubation in the two media was done for up to 44 h or 72 h, respectively, at 28°C and 220 rpm in a New Brunswick G25 shaker. Induction of cultures with tryptophan was done by adding 2.5 mM tryptophan 8 h prior to harvest. Mycelium for enzyme preparation was harvested at different times of cultivation by suction filtration on Buechner funnels and, after washing with distilled water, frozen at −80°C. Escherichia coli strains DH5α (23), JM 105, and JM 109 (73) were grown on agar plates containing LB agar or in liquid cultures with 2× YT (61).
DNA manipulation.
DNA manipulations were done according to reference 61. The cosmid gene bank of S. chrysomallus DNA in E. coli was constructed using cosmid pV34. pV34 is a derivative of cosmid pHC79 (25). Chromosomal DNA of S. chrysomallus strain Sc1 was partially digested with Sau3A and ligated to BamHI-cleaved pV34. The average insert size was 30 to 40 kb. For infection with phage lysate, strain DH5α was used. Cosmids cosA1 and cosP1 were obtained by Southern colony hybridization screening of the cosmid library with two 32P-labeled synthetic oligonucleotide probes derived from internal peptide sequences of ACMSII and ACMSI (62). Cosmid 5a1 was obtained by Southern colony hybridization screening of the cosmid library with a 2.3-kb BamHI fragment covering most of the acmG gene (nucleotide positions 35698 to 38017) obtained from cosmid A1, which had been 32P labeled by nick translation (performed at Hartmann Analytics, Braunschweig, Germany). Subcloning of restriction fragments was done with vector pTZ18U (48). The insert of cosmid cosP1 ranged from 0 to 32 kb, that of cosmid cosA1 ranged from 4 to 41 kb, and that of cosmid cos5a1 ranged from 34 to 66 kb. Sequencing of cosmids cosA1 and cosP1 was performed at Eurogenetech (Brussels, Belgium) and ActinoDrug GmbH (Hennigsdorf, Germany). Cosmid 5a1 was sequenced in its entirety by Agowa AG (Berlin, Germany). Restriction digests of all three cosmids with HindIII and EcoRI, along with digests of chromosomal DNA of S. chrysomallus using the labeled cosmids as probes, revealed no detectable rearrangements of chromosomal DNA in the cosmids that might have arisen during the cloning procedure (not shown). PCR cloning of the catabolic kynureninase gene of S. chrysomallus, kynU, was performed with two primers, Form_F (5′-CCCAGTTCAACTCCTACCG-3′) and TrpO_R (5′-GTAGAAGTCGACGACCTGGT-3′), each derived from a conserved protein motif of the kynurenine formamidase gene in Streptomyces griseus subsp. griseus NBRC 13350 (SGR_3412) and from a conserved motif of the tryptophan dioxygenase gene of the same organism (SGR_3414), respectively. These two genes flank the kynureninase gene in the S. griseus chromosome, which is supposed to be highly similar to that of S. chrysomallus. The template DNA was chromosomal DNA of strain Sc-white, which lacks acm pathway-specific kynureninase sequences (see below). PCR was performed under standard conditions. The resulting 1,991-bp fragment was cloned into plasmid pJet1.2 using the CloneJet PCR cloning kit (Fermentas, St. Leon-Rot, Germany), subcloned into E. coli plasmid pSP72 (Promega, Mannheim, Germany) via KpnI and XbaI, and subsequently sequenced with universal and reverse primers (at Agowa AG, Berlin).
Enzyme preparation.
All operations were carried out at 2°C. Mycelium (5 g) of S. chrysomallus or S. coelicolor M145 was suspended in 10 ml of 0.1 M Tris-HCl (pH 8.3)-1 mM EDTA-2 mM benzamidine-4 mM dithioerythritol-1 mM phenylmethylsulfonyl fluoride-15% glycerol and passed through a French press (Aminco) at 15,000 lb/in2. To the homogenate were added MgCl2 (10 mM final concentration) and 50 μg/ml DNase I (grade II; Sigma) with gentle stirring until the viscosity had drastically decreased. The mixture was centrifuged at 20,000 × g at 2°C for 15 min. The supernatant was desalted in 1-ml portions on PD10 columns (Amersham Biosciences, Freiburg, Germany) previously equilibrated with 0.1 M Tris-HCl (pH 8.3)-1 mM EDTA-2 mM benzamidine-1 mM dithioerythritol-1 mM phenylmethylsulfonyl fluoride-10% glycerol. For enzymatic analysis, the solution was serially diluted with 25 mM potassium phosphate (pH 7.2) to 100 to 200 μg ml−1 protein.
Gene disruption of the actinomycin biosynthetic gene cluster.
Two fragments, A and B, were generated as “arms” of a hygromycin resistance cassette for replacement of part of the actinomycin biosynthetic gene cluster. To generate fragment A, a 5,986-kb KpnI fragment from nucleotide positions 5797 to 11783 of the gene cluster was cloned into KpnI-cleaved E. coli plasmid pTZ18U. From the resultant plasmid with the desired insert orientation, a 5,304-bp PstI fragment (right-end PstI site located in acmK) was obtained, which was cloned into PstI-cleaved E. coli plasmid pIJ2925 (31). The correct orientation of fragment A was assessed by restriction analysis of the resultant plasmid by digestion with PstI and SphI. Fragment B was generated by cloning a 4,068-bp PstI-KpnI fragment from nucleotide positions 38700 (PstI) to 42768 (KpnI) into PstI-KpnI-cleaved E. coli plasmid pTZ18U. From the resultant plasmid, a 4,076-bp HindIII-KpnI fragment was obtained, which was cloned into a HindIII-KpnI-cleaved fragment A plasmid, yielding a fragment A-B plasmid. A hygromycin resistance cassette, including transcriptional terminators, was obtained from a pBR322 derivative (4) as a 2.3-kb HindIII fragment, which was cloned into the HindIII site between fragments A and B in the fragment A-B plasmid. Five micrograms of the resultant fragment A-hyg-fragment B plasmid was denatured (0.2 M NaOH, 10 min, 37°C), chilled on ice, and neutralized by rapid addition of HCl. Transformation of protoplasts of S. chrysomallus Sc1 was performed by addition of neutralized plasmid and 20% polyethylene glycol 1000. After washing with medium P, the protoplast suspension was plated in a soft agar overlay containing 2 volumes medium P and 1 volume R2 (total volume, 4 ml) on R2YE plates (26), except that these media contained 0.22 M sucrose instead of 0.3 M sucrose. After incubation at 30°C for 1 day, the plate was overlaid with 4 ml soft agar containing hygromycin at a final concentration of 100 μg ml−1. Six colonies which appeared after 3 to 4 days were streaked after 7 days of growth on media with and without hygromycin. Two hygromycin-resistant transformants attracted attention by their good growth and whitish (instead of yellow) appearance. The other four grew poorly and, upon restreaking, lost their ability to grow on hygromycin-containing medium. They were not further characterized. The two well-growing transformants were repeatedly streaked onto solid media until their phenotypic stability was confirmed, and they were characterized as actinomycin nonproducers by inhibition tests and mass spectrometry. One of them, designated Sc-white, was used for further biochemical analysis. PCR analysis of Sc-white chromosomal DNA revealed the correct position of the hygromycin cassette between acmK and acmH as in the plasmid construct. Moreover, PCR analysis of Sc-white chromosomal DNA showed that both acmH and acmK were not present as full-length copies. Similarly, no amplification products were obtained from either acmA or acmB when using various primer pairs derived from their sequences. These results confirmed that the region in the actinomycin biosynthetic gene cluster between acmK and acmH was deleted by substitution with the hygromycin resistance cassette.
Sequence analysis and data handling.
Sequence analysis was done with PC/Gene (Intelligenetics, Mountain View, CA) and BlastP (National Center for Biotechnology Information). Multiple alignments were performed with ClustalW2 (70) and edited with GeneDoc (51); tree analysis was done with ClustalW and edited with Drawtree (http://www.phylodiversity.net) or by using the phylogeny.fr web service (15; http://www.phylogeny.fr/). Open reading frames (ORFs), operons, transcriptional start points, promoters, and terminators were identified using various computer programs such as FGENES-B (Softberry Inc.), SAK (21), BPROM (Softberry Inc.), and FindTerm (Softberry Inc.).
Nucleotide sequence accession numbers.
The gene sequences of the actinomycin biosynthetic gene cluster and the kynureninase kynU sequence were deposited in GenBank under accession no. HM038106 and HM038107, respectively.
RESULTS AND DISCUSSION
Organization of the actinomycin biosynthetic gene cluster.
Sequences of regions flanking the actinomycin synthetase genes were obtained from the inserts of three overlapping cosmids. These encompass 66,068 bp of contiguous DNA of which a 50-kb stretch contains 28 actinomycin biosynthetic genes, including the four already known, acmA, acmB, acmC, and acmD, which encode the NRPS subunits (58, 62) (Fig. 2). The left end of the gene cluster is bordered by an IS element containing transposase and integrase gene fragments. At the right end, another IS element with a transposase gene comes before the rightmost biosynthetic gene (see below). Downstream of the latter (i.e., beyond the right end) are several IS elements containing transposase gene fragments and a separate integrase gene (the latter not shown). All of these IS elements apparently have undergone rearrangements, indicating a hot-spot region of integration and recombination at this locus. The presence of IS elements is a frequently observed feature of antibiotic biosynthetic gene clusters perhaps reflecting transposition or horizontal gene transfer events in their origins (29, 45). All biosynthetic ORFs, their nucleotide coordinates, and their attributed functions are shown in Table 1.
TABLE 1.
Deduced functions of ORFs located within the actinomycin biosynthetic gene cluster of S. chrysomallus ATCC 11523
| ORF | No. of amino acid residues | Nucleotide position of 5′ end | Proposed ORF product | Source of most similar protein (% identity) | Accession no. |
|---|---|---|---|---|---|
| int_1 | 89 | 679 (−) | Integrase fragment | Streptomyces sp. C (84) | ZP_05509891 |
| orf41 | 41 | 727 (+) | IS493-like transposase fragment | Streptomyces sp. SPB78 (62) | ZP_05485094 |
| acmrC | 756 | 3206 (−) | UvrA-like protein | Streptosporangium roseum DSM 43021 (64) | ZP_04478111 |
| acmrB | 255 | 4060 (−) | ABC 2-type transporter | Streptomyces sp. AA4 (63) | ZP_05483228 |
| acmrA | 327 | 5033 (−) | ABC transporter ATPase subunit | Streptomyces sp. AA4 (66) | ZP_05483227.1 |
| acmQ | 294 | 5997 (−) | Siderophore interacting proteins | S. erythraea NRRL 2338 (58) | YP_001105422 |
| acmP | 280 | 6112 (+) | TetR family transcriptional regulator | S. erythraea NRRL 2338 (59) | YP_001105423 |
| acmO | 215 | 7181 (+) | LmbU-like protein | Streptomyces lincolnensis (52) | ABX00623 |
| acmN | 127 | 7856 (+) | Ferredoxin (nonfunctional) | Streptomyces tubercidicus (56) | AAT34970 |
| acmM | 414 | 9381 (−) | Cytochrome P450 monooxygenase (nonfunctional) | Streptomyces sp. I-1525 (67) | AAT45261 |
| acmL | 346 | 10495 (−) | Methyltransferase | Streptomyces lavendulae (54) | ABI22137 |
| acmK | 299 | 11447 (−) | Kynureninase | Micromonospora sp. ATCC 39149 (63) | ZP_04603982 |
| acmT | 211 | 12233 (−) | Hypothetical protein | Actinosynnema mirum DSM 43827 (36) | YP_003099816 |
| acmS | 186 | 12804 (+) | Hypothetical protein | Catenulispora acidiphila DSM 44928 (25) | YP_003111727 |
| acmR | 66 | 13017 (−) | MbtH-like proetin | Streptomyces roseosporus NRRL 15998 (74) | ZP_04691533 |
| acmD | 78 | 13320 (−) | 4-MHA carrier protein | S. chrysomallus (100) | AAD30112 |
| acmA | 472 | 14735 (−) | Peptide synthetase | S. chrysomallus (100) | AAD30111 |
| acmB | 2,611 | 15166 (+) | Peptide synthetase | S. chrysomallus (100) | AAC38442 |
| acmC | 4,247 | 22998 (+) | Peptide synthetase | S. chrysomallus (100) | AAF42473 |
| acmE | 235 | 35738 (+) | Hypothetical protein | A. mirum DSM 43827 (40) | YP_003099816 |
| acmF | 304 | 36480 (+) | Aryl formamidase | S. viridochromogenes DSM 40736 (49) | ZP_05534764 |
| acmG | 280 | 37391 (+) | Tryptophan 2,3-dioxygenase | S. viridochromogenes DSM 40736 (49) | ZP_05534761 |
| acmH | 420 | 38294 (+) | Kynureninase | Micromonospora sp. ATCC 39149 (69) | ZP_04603982 |
| acmI | 346 | 39628 (+) | Methyltransferase | S. lavendulae (55) | ABI22137 |
| acmJ | 211 | 41439 (−) | LmbU-like protein | Frankia alni ACN14a (54) | YP_714812 |
| acmU | 296 | 42450 (−) | TetR family transcriptional regulator | Saccharopolyspora erythraea NRRL 2338 (59) | YP_001105423 |
| acmV | 298 | 42565 (+) | Siderophore-interacting proteins | S. erythraea NRRL 2338 (60) | YP_001105422 |
| acmW | 327 | 43561 (+) | ABC transporter ATPase subunit | Streptomyces sp. AA4 (67) | ZP_05483227 |
| acmX | 255 | 44534 (+) | ABC 2-type transporter | Streptomyces sp. AA4 (63) | ZP_05483228 |
| acmY | 687 | 45372 (+) | UvrA-like protein fragment | S. roseum DSM 43021 (63) | ZP_04478111 |
| orf120 | 120 | 47818 (−) | Transposase IS4 protein | Streptomyces sp. Mg1 (90) | ZP_04996912.1 |
| orf157 | 157 | 48435 (−) | IS1647-like transposase | Streptomyces sp. Mg1 (88) | ZP_04999232.1 |
| acmY' | 70 | 48508 (+) | UvrA-like protein fragment | S. roseum DSM 43021 (66) | ZP_04478111 |
| orf42 | 42 | 49093 (−) | Transposase fragment | Streptomyces coelicolor A3(2) (90) | NP_624691 |
| orf238 | 238 | 49372 (+) | Transposase | Streptomyces sp. SPB74 (78) | ZP_04990820 |
The four peptide synthetase genes, acmA to -D, lie in the center of the cluster and are flanked by 13 genes to the left and 11 to the right (not counting IS elements). Nine genes on the right have paralogues on the left in the same order but in the opposite orientation. This results in two large IRs on either side of the central NRPS gene set. The symmetry of these repeats is interrupted in the left arm by the lack of paralogues of acmF and acmG from the right arm. Instead, the left arm contains four extra genes, acmR and acmS at the inner border and acmM and acmN in the middle (Fig. 2; Table 1). Sequence identities between the various paralogous proteins range between 57 and 84%, and the nearly identical arrangement of their genes suggests that they originated by gene duplication. Dot plot matrix analysis of the complete sequence reveals this symmetry also at the DNA level. There are nearly identical nucleotide sequence blocks indicating synteny between the arms (not shown).
BLASTP analysis of translated gene sequences allowed us to assign plausible functions to most of the genes. Besides the four already known NRPS genes, there are six genes (two paralogous pairs and two solitary genes) most probably involved in the biosynthesis of 4-MHA. Two pairs of paralogous genes encode putative regulatory proteins. There are four pairs of paralogous genes that encode three types of putative self-resistance mechanisms (discussed below). Four genes (one pair of paralogues and two solitary genes) encode proteins with unknown functions and another two genes, which encode a putative cytochrome P450 and a ferredoxin, respectively, with unknown roles in actinomycin C formation are discussed in the context of their physical linkage with the other genes.
Actinomycin peptide chain assembly genes.
The actinomycin synthetase enzymes have been described previously (58, 62). Their genes, acmA, acmB, acmC, and acmD, in the middle of the cluster, are organized as two transcription units pointing away from each other (Fig. 2). Computer analysis using programs such as FGENES-B and FindTerm suggest not only that acmB is cotranscribed with acmC but also that this operon includes acmE, acmF, acmG, acmH, and acmI, after which a large IR is present. Similarly, in the opposite direction, acmA and the subsequent genes, acmD, acmR, acmS, and acmT, are most probably transcribed as one transcript terminated by an IR between acmT and acmK. BLASTP analysis of the translated sequence of acmR, at a 66-bp distance from the 3′ end of the 4-MHA carrier protein gene acmD, indicated that it encodes a protein of 66 amino acids resembling MbtH-like proteins. MbtH-like proteins are conserved proteins of 60 to 75 amino acid residues. Genes for MbtH-like proteins are found in many, but not all, bacterial NRPS gene clusters, such as mbtH in that for siderophore biosynthesis in Mycobacterium tuberculosis (60) or as cdaX or cchK in those for the biosynthesis of calcium-dependent antibiotic or coelichelin, respectively, in S. coelicolor A(3)2 (1). The MbtH-like protein gene orf1 in the phosphinothricin tripeptide biosynthesis gene cluster of Streptomyces viridochromogenes is cotranscribed with two of its NRPS genes (63). Although MbtH-like protein genes cdaX and cchK in S. coelicolor were claimed to complement each other, it could not be determined whether MbtH-like proteins are essential for nonribosomal peptide synthesis (41). Therefore, their function remains unknown. Downstream of acmR comes acmS at a distance of 12 bp. It encodes a hypothetical protein of 186 residues. AcmS has relevant similarity to only one protein in the database (YP_003111727, 25% identity) from Catenulispora acidiphila DSM 44928. Its function is unknown. The putative RBS of acmT, which is 10 bp downstream of acmS, overlaps the stop codon of acmS, suggesting the cotranscription of at least acmS and acmT. acmT encodes a protein of 211 amino acids which is a paralogue (57% identity) of that encoded by the above-mentioned acmE gene in the right arm of the gene cluster. acmE encodes a protein of 212 amino acid residues. Both AcmT and AcmE show the highest similarities to several hypothetical proteins such as those from Actinosynnema mirum (EEH74708, 40% identity) or S. griseus subsp. griseus NBRC 13350 (SGR_898, 36% identity). As for AcmT and AcmE, the function of these proteins is unknown.
4-MHA biosynthesis genes: acmH and acmK, which encode kynureninase-like enzymes.
Situated 3′ to acmE in the right arm of the cluster are four genes in the order acmF, acmG, acmH, and acmI, all in the same orientation (Fig. 2). They encode proteins resembling kynurenine formamidases (acmF), tryptophan dioxygenases (acmG), kynureninases (acmH), and methyltransferases (acmI), respectively. Their presence agrees with the previous detection of pathway-specific isoforms of tryptophan oxygenase, kynurenine formamidase, and hydroxykynureninase in partially purified protein fractions from actinomycin-producing S. antibioticus, S. parvulus, and S. chrysomallus (8, 9, 17, 22, 72). These enzymes were considered to be involved in the conversion of tryptophan to 4-MHA (Fig. 3). Remarkably, the left arm of the gene cluster contains the genes acmK and acmL, which also encode a kynureninase and a methyltransferase, respectively. AcmH and AcmK are similar to each other (59% identity over 299 amino acid residues). However, AcmK is smaller than AcmH (299 versus 420 amino acid residues) because of the deletion of its first 121 amino-terminal amino acid residues with respect to the AcmH sequence, but AcmK still possesses the complete PLP binding domain.
FIG. 3.
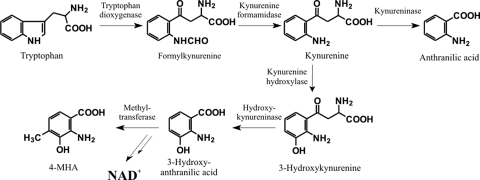
Pathways of tryptophan metabolism. The catabolic pathway leading from tryptophan to anthranilic acid, found in a number of bacteria, is inducible by tryptophan (56). The pathway leading from kynurenine to 3-HA is constitutive and found in all eukaryotes, including S. cerevisiae (64) and mammals (69). In the eukaryotic cell, 3-HA serves as a precursor of NAD+ or can be metabolized further. In actinomycin synthesis, 3-HA is converted to 4-MHA. NAD+, nicotine adenine dinucleotide.
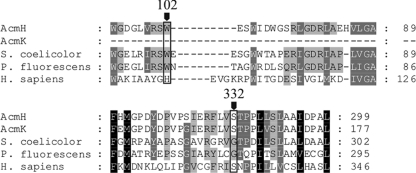
There are two different types of kynureninase in nature with different substrate specificities. The inducible enzyme, found in a number of bacteria, reacts with l-kynurenine and is involved in bacterial tryptophan catabolism leading to anthranilic acid (56). The constitutively expressed kynureninase (hydroxykynureninase), found in all eukaryotes, including Saccharomyces cerevisiae (64) and mammals (69), preferentially reacts with 3-hydroxy-l-kynurenine, yielding 3-HA. Besides being degraded as a step in tryptophan catabolism, 3-HA serves as a precursor of NAD+ in the eukaryotic cell (Fig. 3). BLASTP analysis revealed that AcmH and AcmK have the highest similarity to kynureninases such as that from Micromonospora sp. ATCC 39149 (ZP_04603982, 69% identity) or Geodermatophilus obscurus DSM 43160 (YP_003408639, 61% identity) and several others, the role of which in their hosts is not known. Previous analysis of the kynureninase mixture in actinomycin-producing S. parvulus or S. chrysomallus revealed two enzyme isoforms with Mrs of ∼82,000 and ∼56,000, respectively (22, 72). Since kynureninase-type enzymes are homodimers (30), their sizes would fit with the calculated molecular masses of AcmK (32.2 kDa) and of AcmH (45.5 kDa) and principally also the putative inducible kynureninase of S. chrysomallus. From comparison with published sequences of kynureninase genes of other streptomycetes, the latter enzyme could have a size similar to that of AcmH. In fact, the analysis of the two enzyme fractions from S. parvulus or S. chrysomallus revealed that the kynureninase isoform with the higher Mr preferred kynurenine as a substrate. In contrast, the smaller form preferred hydroxykynurenine and was named a hydroxykynureninase (72). However, both enzyme fractions could process the alternate substrate more efficiently than typical kynureninases and hydroxykynureninases, which each have a 100-fold difference in their Kms for either kynurenine or hydroxykynurenine, respectively (44, 65). Functional and structural studies of kynureninase-type enzymes have ascribed the substrate discrimination between kynurenine and hydroxykynurenine to specific signatures in the substrate-binding pockets of kynureninases and hydroxykynureninases. These are residue positions 102 (H) and 332 (S) in the human hydroxykynureninase sequence (44). In contrast, kynureninases show W102 and G332 signatures. Alignments of AcmH and AcmK sequences with various kynureninases of prokaryotic and eukaryotic origins revealed that AcmH has a kynureninase signature (W) at position 102, whereas at position 332 it has a hydroxykynureninase signature (S) (Fig. 4). AcmK lacks the first 121 amino acids relative to AcmH and thus lacks any signature at position 102 but, like AcmH, has a hydroxykynureninase signature at position 332 (S). This may explain—at least in part—the relaxed substrate specificity seen in the two enzyme fractions. In contrast, the translated sequences of kynureninase genes of S. griseus SGR_3413 (53), Streptomyces avermitilis SAV_4527 (54), and S. coelicolor SCO364 (1) all show the bacterial W102 and G332 consensus, suggesting that these enzymes may be true kynureninases that specifically convert kynurenine to anthranilic acid (Fig. 3).
FIG. 4.
Alignments of substrate-binding pocket sequences of eukaryotic and prokaryotic kynureninase-like enzymes. The alignments show the locations of the kynureninase and hydroxykynureninase active-site signatures based on the structural and functional analysis of the human (hydroxy)kynureninase sequence (48). Accession numbers: NP_003928.1, kynureninase isoform A (Homo sapiens); YP_257899.1, Pseudomonas fluorescens Pf-5 kynureninase; NP_627839.1, S. coelicolor A3(2) hydrolase.
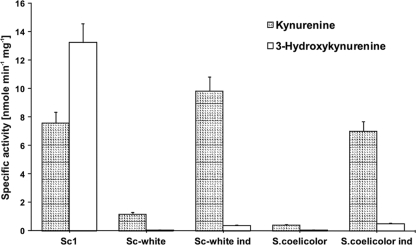
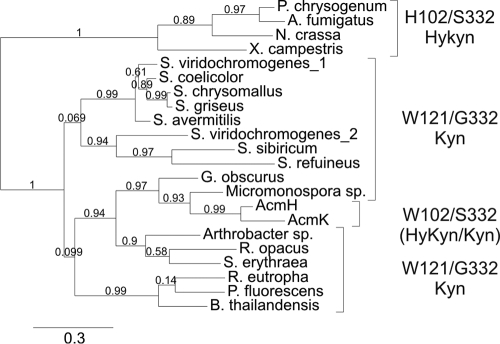
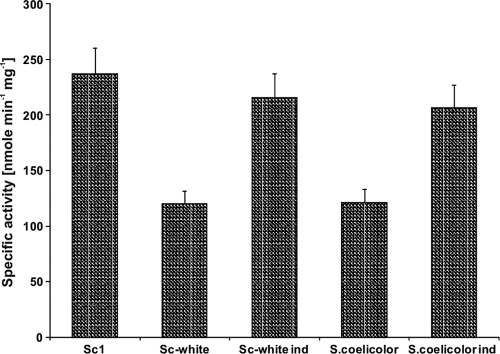
A hygromycin resistance cassette was inserted by recombination into the actinomycin biosynthetic gene cluster of S. chrysomallus strain Sc1 between nucleotide positions 11068 (corresponding to nucleotide [nt] 379 of acmK) and 38700 (corresponding to nt 407 of acmH) as described in Materials and Methods (Fig. 5). The cassette replaces ca. 57% of acmK and 67% of acmH, including acmE, acmF, and acmG on the right, acmA, -B, -C, and -D and acmR in the middle, and acmS and acmT on the left of the cluster. This results in the loss not only of large portions of acmK and acmH from their 5′ ends but also of their promoters and all other identified genes. The resultant mutant, named Sc-white, had lost the ability to produce actinomycin, as expected (see Materials and Methods). Enzymatic analysis of protein extracts of mutant Sc-white revealed that it completely lacked hydroxykynureninase activity compared with extract from actinomycin-producing wild-type strain Sc1, but it still had a significant level of kynureninase activity (Fig. 6). Strain Sc-white thus resembles a streptomycete which possesses only catabolic kynureninase activity such as S. coelicolor A3(2), which was tested as a control (Fig. 6). When 2.5 mM tryptophan was added to cultures of strain Sc-white or S. coelicolor, the kynureninase activity in their cell extracts dramatically increased more than 10-fold, with only a small concomitant increase in hydroxykynureninase activity, which apparently stems from the low natural cross-reactivity of the kynureninase with the alternate substrate hydroxykynurenine. These findings clearly indicate that S. chrysomallus, besides the pathway-specific hydroxykynureninase activity represented by AcmH and AcmK, has a kynureninase gene for normal tryptophan catabolism, like other streptomycetes, e.g., S. coelicolor. The kynureninase in S. coelicolor is most likely encoded by SCO3645, the only kynureninase-like sequence in its genome (1). To address the presence of a catabolic kynureninase in S. chrysomallus, we PCR cloned its gene by using primers derived from genes flanking the kynureninase gene SGR_3413 in the genome of S. griseus and by using Sc-white chromosomal DNA as the template. The translated sequence of that gene indeed encoded a kynureninase, the gene for which we designated kynU (see Materials and Methods). KynU is very similar to its S. griseus orthologue SGR_3413 (93% identity) or to SCO3645 of S. coelicolor (81% identity), which strongly suggests that kynU encodes the tryptophan-inducible kynureninase of S. chrysomallus. A phylogenetic analysis of AcmH, AcmK, and KynU, along with different kynureninase sequences from actinomycetes, bacteria, and fungi, is shown in Fig. 7. It can be seen that the fungal kynureninase sequences all assemble in a clade together with one bacterial sequence which is from Xanthomonas campestris pv. campestris B100 (YP_001904112). All of these kynureninase sequences, including the one from Xanthomonas, show a typical eukaryotic hydroxykynureninase signature in their substrate binding pocket. This agrees well with the fact that Xanthomonas species, in contrast to many bacteria, can synthesize NAD+ from 3-HA (10). The second main clade containing exclusively bacterial kynureninase sequences contains several subclades. In one of them lie AcmH and AcmK together with their most similar BLASTP hits from other bacteria (Fig. 7). They are distant from the clade formed by the kynureninases from streptomycetes. The clade of the latter branches into two subclades, one of which comprises catabolic kynureninase sequences such as the gene products of S. griseus SGR_3413 and S. coelicolor SCO3645 or KynU of S. chrysomallus. In contrast, the second subclade contains kynureninases apparently involved in secondary metabolism such as the gene products encoded by the genes orf16 and sibQ in the biosynthetic gene clusters for anthramycin in Streptomyces refuineus (27) and for sibiromycin in Streptosporangium sibiricum (43). These two antibiotics, like actinomycin, also contain 4-MHA as a building block in their structures, and their gene clusters should principally encode the same enzymatic makeup for the conversion of tryptophan to 4-MHA as the actinomycin biosynthetic gene cluster. However, both Orf16 and SibQ and their companions in that clade are significantly more distant from AcmH and AcmK than from the catabolic kynureninases. In addition, they both have a 102W and 332G kynureninase signature, like all other bacterial kynureninases, which underscores the uniqueness of AcmH and AcmK. Whether Orf16 and SibQ have a preference for kynurenine or hydroxykynurenine is not known.
FIG. 5.
Disruption of 4-MHA biosynthetic genes in the actinomycin gene cluster. Disruption was performed with a hygromycin cassette using an E. coli pUC18 derivative as a suicide vector as described in Materials and Methods.
FIG. 6.
Kynureninase and hydroxykynureninase activities in S. chrysomallus strains Sc1 and Sc-white. Gray bars, kynureninase activity; white bars, hydroxykynureninase activity. Specific activities were measured in crude extracts from 60-h-old mycelia grown in glutamate-mineral salts medium supplemented with maltose as described in Materials and Methods. The ratio of the specific activities of hydroxykynureninase and kynureninase in a culture of strain Sc1 reproducibly increased concomitantly with actinomycin production in culture up to an approximate value of 2 to 2.5:1 after 55 to 65 h of growth (not shown). Crude extracts from strain Sc-white and S. coelicolor strain M145, which does not produce actinomycins, were obtained from mycelium grown under the same conditions as strain Sc1 of S. chrysomallus. Ind, extract prepared from each strain after induction with 2.5 mM tryptophan. For details, see Materials and Methods.
FIG. 7.
Phylogenetic tree of kynureninase-like enzyme sequences from various bacteria and fungi, including AcmH and AcmK. Accession numbers: S. coelicolor A3(2), NP_627839.1; S. avermitilis, NP_825704.1; S. griseus subsp. griseus, YP_001824925.1; S. refuineus, ABW71847.1; S. viridochromogenes_1, ZP_05533056.1; S. viridochromogenes_2, ZP_05534763.1; Streptosporangium sibiricum, ACN39740.1; Arthrobacter sp., YP_830926.1; Saccharopolyspora erythraea, YP_001106253.1; Rhodococcus opacus, YP_002778666.1; Penicillium chrysogenum, XP_002565957.1)], Neurospora crassa, XP_956782.2; Aspergillus fumigatus, XP_755369.1; Pseudomonas fluorescens, YP_257899.1; Micromonospora sp. ATCC 39149, EEP69912.1; Geodermatophilus obscurus DSM 43160, YP_003408639.1; Burkholderia thailandensis, YP_441266.1; Ralstonia eutropha, CAJ93893.1; X. campestris pv. campestris strain B100, YP_001904112.1; S. chrysomallus, HM038107. The various clades are denoted with their kynureninase/hydroxykynureninase signatures.
4-MHA biosynthesis genes: acmF, which encodes kynurenine formamidase.
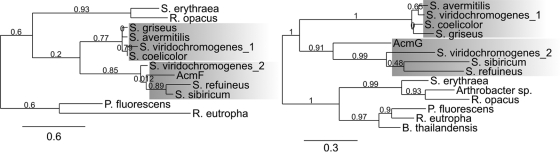
Based on the gene disruption, strain Sc-white should also lack actinomycin biosynthetic genes acmF (kynurenine formamidase) and acmG (tryptophan dioxygenase). AcmF has the highest similarity to the gene product of the gene ZP_05534764 from S. viridochromogenes DSM 40736, which encodes a putative esterase. Testing of enzyme extracts from strains Sc-white and Sc1 for kynurenine formamidase activity revealed that strain Sc-white contained about 50% less kynurenine formamidase activity than actinomycin-producing strain Sc1 (Fig. 8). The reduced activity is most likely due to loss of acmF, and the persisting activity in Sc-white must be encoded by the catabolic kynurenine formamidase gene. In other streptomycetes, kynurenine formamidase genes are represented, e.g., by S. coelicolor A3(2) (SCO3644), S. griseus (SGR_3412), and S. avermitilis (SAV_4531), which are clustered together with the catabolic kynureninase and tryptophan dioxygenase genes (1, 53, 54). This is also the case in S. chrysomallus, where the catabolic kynureninase gene kynU is flanked by genes that encode a tryptophan dioxygenase and a kynurenine formamidase (I. Crnovcic and U. Keller, unpublished data). Katz et al. (35) reported two kynurenine formamidase enzymes in actinomycin-producing S. parvulus, one of which was formed at the onset of actinomycin production whereas the other was constitutive. Apparently, strain Sc-white possesses only the constitutive form. Induction with tryptophan of cultures of strain Sc-white led to a ca. 2-fold increase in kynurenine formamidase activity, which was also seen in the case of S. coelicolor, which was used as a control (Fig. 8). These findings support the assumption that the kynurenine formamidase observed in strain Sc-white is the catabolic enzyme because induction by tryptophan is a known feature of this class of enzymes in bacteria (56). Phylogenetic analysis of various bacterial kynurenine formamidase sequences along with AcmF (Fig. 9) revealed two sister clades of streptomycete enzymes, one containing catabolic kynurenine formamidases and the second containing AcmF together with kynurenine formamidases encoded by the genes orf20 and sibK from the above-mentioned biosynthetic gene clusters for anthramycin in S. refuineus (27) and sibiromycin in S. sibiricum (43). AcmF thus shows phylogenetic linkage to these kynurenine formamidases involved in the formation of secondary metabolites, which suggests that they all have coevolved from a common ancestor which they share with the catabolic enzymes. Interestingly, it can be seen from this tree that S. viridochromogenes possesses not only a catabolic kynurenine formamidase (ZP_05533057.1) but also a kynurenine formamidase (ZP_05534764.1) phylogenetically related to AcmF and its homologues, which suggests that S. viridochromogenes may also produce a tryptophan-derived secondary metabolite.
FIG. 8.
Kynurenine formamidase activities in S. chrysomallus strains Sc1 and Sc-white. Specific activities were measured in crude extracts as described in Materials and Methods. Crude extracts from S. coelicolor strain M145, which does not produce actinomycins, were also tested for comparison. Ind, extract prepared from each strain after induction with 2.5 mM tryptophan. For details, see Materials and Methods.
FIG. 9.
Phylogenetic trees of kynurenine formamidases and tryptophan dioxygenases from different streptomycetes and bacteria. (Left) Kynurenine formamidases from S. coelicolor A3(2) (CAB42031.1), S. avermitilis (NP_825708.1), S. griseus subsp. griseus (YP_001824924.1), S. refuineus (ABW71851.1), S. viridochromogenes_1 (ZP_05533057.1), S. viridochromogenes_2 (ZP_05534764.1), Streptosporangium sibiricum (ACN39734.1), Saccharopolyspora erythraea (YP_001104168.1), Rhodococcus opacus (YP_002779035.1), Pseudomonas fluorescens (YP_257889.1), and Ralstonia eutropha (CAJ95273.1). There are two sister clades (shaded) containing streptomycete kynurenine formamidase, one containing catabolic sequences and the other containing sequences involved in secondary metabolism, such as AcmF in 4-MHA biosynthesis. (Right) Tryptophan dioxygenases from S. coelicolor A3(2) (NP_627840.1), S. avermitilis (NP_825703.1), S. griseus subsp. griseus (YP_001824926.1), S. refuineus (ABW71848.1), S. viridochromogenes_1 (ZP_05533055.1), S. viridochromogenes_2 (ZP_05534761.1), Streptosporangium sibiricum (ACN39739.1), Arthrobacter sp. (A0JUV5.1|T23O_ARTS2), Saccharopolyspora erythraea (A4FH01.1|T23O_SACEN), Rhodococcus opacus (YP_002778667.1), Pseudomonas fluorescens (AAY96155.1), Burkholderia thailandensis (YP_441265.1), and Ralstonia eutropha (YP_727262.1). There are two sister clades (shaded) containing streptomycete tryptophan dioxygenases, one containing catabolic sequences and the other containing sequences apparently involved in secondary metabolism, such as AcmG in 4-MHA biosynthesis.
MHA biosynthesis genes: acmG, which encodes tryptophan dioxygenase.
Foster and Katz (17) analyzed the regulation of tryptophan dioxygenase activity in S. parvulus and showed that its activity in protein extracts becomes detectable at the onset of actinomycin synthesis and remains at detectable levels throughout the antibiotic production phase. In contrast to pathway-specific kynureninase and kynurenine formamidase, however, there was no increase in tryptophan dioxygenase activity after induction with tryptophan (24). Measuring tryptophan dioxygenase activity in protein extracts of strain Sc1 actively synthesizing actinomycin revealed low but significant levels (not shown). In contrast, extracts of mutant strain Sc-white lacked detectable tryptophan dioxygenase activity, in agreement with loss of acmG. The low and unstable activity of tryptophan dioxygenase prevented the study of this enzyme in more detail. acmG encodes a protein of 280 amino acid residues which is similar in size and sequence to various tryptophan dioxygenases from bacteria, including the catabolic enzyme species from S. coelicolor A3(2) (SCO3646) S. avermitilis (SAV_4526), or S. griseus (SGR_3414). Phylogenetic analysis of a number of different bacterial tryptophan dioxygenases revealed that the tryptophan dioxygenases from streptomycetes formed a clade with two subclades, one containing apparently catabolic sequences exemplified by the corresponding sequences from the genomes of S. coelicolor, S. avermitilis, and S. griseus. As expected from its location in the actinomycin biosynthetic gene cluster, AcmG is located in the sister clade also containing the tryptophan dioxygenases encoded by the genes orf17 and sibP from the anthramycin and sibiromycin biosynthetic gene clusters, respectively (27, 43). This suggests specialized roles for AcmG, Orf17, and SibC in their hosts, which all can make 4-MHA. In the same sister clade is also a tryptophan dioxygenase (ZP_05534761.1) from S. viridochromogenes which, like its corresponding kynureninase (Fig. 7) and kynurenine formamidase (Fig. 9A), may also be involved in the biosynthesis of a tryptophan-derived secondary metabolite. This streptomycete—like S. chrysomallus—also possesses the catabolic versions of all of these enzymes (Fig. 7 and 9). In summary, the data presented here indicate a common lineage not only of the various tryptophan dioxygenases but also of the kynurenine formamidases and kynureninases of the 4-MHA biosynthetic pathway. Moreover, the branching of catabolic and 4-MHA-related enzyme sequences in phylogenetic trees (Fig. 7 and 9) suggests that these enzymes have coevolved after a gene duplication of a common ancestor gene cluster. This view is also supported by the fact that in most bacteria, the catabolic genes are clustered, which is mirrored by the tandem arrangement of acmF, acmG, and acmH from the actinomycin biosynthetic gene cluster with one permutational change in their sequential order (Fig. 10). A less conserved arrangement of the corresponding genes is observed in the sibiromycin and anthramycin biosynthetic gene clusters, where kynureninase and tryptophan dioxygenase are still linked whereas the kynurenine formamidase is disconnected from them by the insertion of other biosynthetic genes (Fig. 10).
FIG. 10.
Chromosomal clustering of genes related to the anthranilate pathway and the 4-MHA biosynthetic pathway in S. chrysomallus and other streptomycetes. The upper five gene clusters are catabolic sequences. They are pinned around the orthologues of SCO3645 (kynureninase like). The lower clusters represent the three 4-MHA-related clusters of tryptophan degradation. Orthologues and homologues of the various pathways are labeled with matching patterns as follows: dark gray, kynurenine formamidase-like enzyme; gray, tryptophan dioxygenase-like enzyme; white, kynureninase-like enzyme. Interspersed genes are indicated by small arrows.
Inspection of both anthramycin and sibiromycin biosynthetic gene clusters also revealed that they both contain genes that encode a putative kynurenine hydroxylase, orf23 and sibC, respectively (27, 43). Kynurenine hydroxylase is normally present in eukaryotes and those bacteria which can synthesize NAD+ from 3-HA but is apparently absent from bacteria which exclusively degrade tryptophan via anthranilate (65). Whether the kynurenine hydroxylase gene has been acquired by bacteria through horizontal gene transfer is still unclear. A kynurenine hydroxylase gene is missing in the actinomycin biosynthetic gene cluster presented here. However, previous genetic analysis of S. chrysomallus has shown that actinomycin synthetic genes lie in two different but closely linked loci represented by two complementation groups identified in crosses between various actinomycin-negative mutants (22). Attempts by our group are currently under way to identify that gene locus and to see whether the kynurenine hydroxylase or hypothetical anthranilate-hydroxylating enzyme genes are located there. Moreover, one could expect to identify in this locus also a gene that probably encodes an enzyme that catalyzes phenoxazinone formation during actinomycin synthesis. This would be the final gene in the actinomycin biosynthetic chain.
4-MHA biosynthesis genes: 3-HA methyltransferases.
acmL and acmI both encode methyltransferases of 346 amino acids. Their protein sequences are nearly identical (82%). BLASTP analysis of their translation products showed that they have the highest similarity (54% identity) to a methyltransferase (ABI22137) encoded by sfmM2 in the saframycin gene cluster of Streptomyces lavendulae (42), which was suggested to be implicated in methylation of the aromatic ring of tyrosine during saframycin synthesis. There is also high similarity to a methyltransferase encoded by sibL (54% identity) of the sibiromycin gene cluster of S. sibiricum (43) (FJ768674) or by orf19 of the anthramycin biosynthetic gene cluster in S. refuineus with 50% identity (27), which, like acmI and acmL, may be involved in 4-MHA synthesis. On the other hand, BLAST searches also indicated that AcmI and AcmL resemble O-methyltransferases of bacteria and plants. Structure modeling using the Phyre server (40) revealed the highest structural homology with the family of O-methyltransferases (type 1) from plants (74). These form the largest group of methyltransferases, with a great diversity of substrates. A prototype of these enzymes is caffeic acid 3-O-methyltransferase (COMT) from alfalfa (Medicago sativa), with which AcmI and AcmL share all significant AdoMet binding motifs at equivalent positions of their equally long protein sequences. The substrate of COMT is caffeic acid/5′-hydroxyferulic acid (74). Curiously, AcmI and AcmL have low similarity to the C-methyltransferases NovO and CouO, both from streptomycetes, which C-methylate aromatic phenolic intermediates in the biosynthesis of coumermycin and novobiocin (55). Therefore, it remains unclear whether AcmI and AcmL are orthologues of the 3-HA-methylating enzyme previously isolated from actinomycin-producing S. antibioticus (32).
Next, to the left of acmL in the left arm of the cluster, is a pair of genes, acmM and acmN, inversely oriented to each other. acmM encodes a protein of 415 amino acids resembling cytochrome P450 monooxygenases, whereas acmN encodes a ferredoxin protein. acmN has a frameshift mutation in its 3′ region, resulting in elongation of the normally 65-amino-acid-residue ferredoxin to an elongated protein of 124 amino acids without precedent in the database. The potential function of the P450 monooxygenase is unclear because there is no known reaction in actinomycin C formation that requires a P450 monooxygenase. Moreover, the nucleotide sequence of acmM has a point mutation in the middle that converts a coding codon to a stop codon. After this, the sequence continues normally to its 3′ end, thus encoding—if there is no stop codon—a standard-size cytochrome P450 protein of ca. 430 amino acids. Taken together, all of these facts leave little possibility that AcmM and AcmN have a functional role in actinomycin biosynthesis in S. chrysomallus. Importantly, acmN contains a large IR in its 3′ region (actually noncoding) that may serve as a terminator in the transcription of the upstream genes acmO and acmP.
Genes involved in regulation.
The orientation of acmP and acmO in the left arm and acmU and acmJ in the right arm suggests that they may be transcribed as polycistronic transcripts from promoters upstream of acmP or acmU. According to computer analysis by FGENES-B, these transcripts could comprise acmP, acmO, and acmN in the left arm and acmU and acmJ in the right arm, as deduced from the presence of large IRs in the 3′ regions of acmN and acmJ. acmP and acmU encode highly similar proteins of 280 and 296 amino acids (68% identity). BLASTP analysis revealed that they resemble the TetR_N repressor family of proteins, such as TetR-like regulators from actinomycetes and myxobacteria, e.g., from Saccharopolyspora erythraea (YP_001105423, 58% identity), Myxococcus xanthus (MXAN 2794, 36% identity), and S. coelicolor (SCO4358, 35% identity). In the 5′ regions of acmP and acmU is a block of four genes that apparently encode antibiotic resistance determinants. These genes are all in the orientation opposite to that of acmP and acmU, and all four possibly form an operon, as predicted by computer analysis. The 114-bp intergenic regions upstream of acmP and acmU are highly similar to each other (67.6% identity), and analysis of the upstream regions of acmP and acmU revealed similarity to a promoter region directing bldN transcription in S. coelicolor (2) with tentatively assigned transcription start points at nucleotide positions 6099 and 42464, respectively. The bldN promoter belongs to the family of sigE-type promoters which have been shown to play a role in the expression of actinomycin synthetase genes in S. antibioticus. Disruption of sigE in that strain resulted in loss of acmA expression and actinomycin synthesis (33).
Coupled 3′ to acmP and acmU follow acmO and acmJ, respectively. They consist of 215 and 211 amino acid residues, respectively, and resemble each other (70% identity). Homology searches indicated high similarity to LmbU (50% identity) and NovE (44% identity) from lincomycin-producing Streptomyces lincolnensis (57) and novobiocin-producing Streptomyces spheroides, respectively. NovE is a transcriptional regulator of novobiocin biosynthesis in S. spheroides, which acts in conjunction with an unknown protein (14). Disruption of novE results in a drastic reduction of novobiocin production. A TTA codon in the homologue lmbU was postulated as a possible regulatory element, as in the regulatory proteins of other secondary metabolic gene clusters (57). TTA codons are absent from acmO and acmJ and from any part of the actinomycin biosynthetic gene cluster. How the pairs AcmP-AcmU and AcmJ-AcmO regulate actinomycin synthesis is not known yet.
Genes involved in resistance.
At the ends of the two arms of the cluster lie two highly similar gene blocks that encode resistance proteins, i.e., acmrA, acmrB, acmrC, and acmQ and acmY, -X, -W and acmV, respectively. These genes resemble in their deduced functions the set of daunorubicin resistance genes in Streptomyces peucetius (29), except that they are all in tandem and therefore could possibly be transcribed as one transcription unit. From their similarities to known proteins, the four resistance genes appear to encode three self-resistance mechanisms: export, modification, and binding of actinomycin. The translated sequences of acmQ and its paralogue acmV consist of 294 and 298 amino acid residues, respectively. They strongly resemble each other (81% identity) and siderophore-interacting proteins such as to MxcB (38% identity) from the myxochelin biosynthetic gene cluster of Stigmatella aurantiaca (20) and ViuB (31% identity), an intracellular protein from Vibrio cholerae (12). ViuB affects the release of iron from the vibriobactin-iron complex, like the Fes protein of the Fep assembly of iron sequestration proteins from E. coli (6). ViuB can replace Fes in its role of iron release from enterobactin, but Fes cannot substitute for ViuB in the release of iron from vibriobactin, suggesting a different mechanism of ViuB compared to that of Fes (12). Like ViuB and MxcB, AcmQ and AcmV have a flavin cofactor binding site and are NADH-dependent oxidoreductases. Both mxcB and viuB are iron inducible and possess FUR binding sites in their promoter regions, which is characteristic of iron regulation (12, 20). acmQ and acmV have no FUR binding consensus in their upstream regions, indicating that they merely share the mechanism of interaction with the siderophore rather than the mode of gene regulation. From this, it can be inferred that both the acmQ and acmV gene products most likely modify actinomycin or its peptide precursor in an as-yet-unknown fashion. Interestingly, acmQ and acmV resemble DrrD, a protein involved in self-resistance of the daunorubicin producer S. peucetius (29). Daunorubicin, like actinomycin, is a DNA-intercalating molecule. DrrD, which is larger than acmQ and acmV (485 amino acid residues versus 294 and 298 amino acids), is also a flavin-containing, NADH-dependent oxidoreductase which has been suspected of reoxidizing reduced daunorubicin, preventing cross-linking of the reduced compound with DNA (29). However, its role in self-resistance in daunorubicin-producing S. peucetius is speculative, as for AcmQ and AcmV.
acmQ and acmV are followed by the gene pairs acmrA-acmrB and acmW-acmX, respectively. The translated gene products, AcmrA and AcmW, encode highly similar proteins of 327 amino acids (83% identity) and strongly resemble ATP-binding cassettes of ABC transporters such as those from S. erythraea (YP_001105422, 60% identity). The membrane-binding subunits of the two ABC transporters are encoded by acmrB and acmX, which are highly similar to each other (82% identity), both with 255 amino acid residues. The ABC transporter pairs AcmrA-AcmrB and AcmW-AcmX strongly resemble the corresponding transporter pair DrrA-DrrB (39 and 21% identity, respectively) from the daunorubicin gene cluster in S. peucetius (29, 37) or the ABC transporter for mithramycin resistance MtrA and MtrB from Streptomyces argillaceus (45) and most probably are export pumps for actinomycin.
The genes at the end of each arm are acmrC and acmY. acmrC encodes a protein of 756 amino acids that strongly resembles UvrA (28) from E. coli (39% identity) and DrrC (58% identity), a protein that confers resistance to daunorubicin in S. peucetius. DrrC binds to DNA in an ATP- and daunorubicin-dependent manner (19). It has DNA-binding characteristics similar to those of UvrA from E. coli and in fact can, when introduced into E. coli or Streptomyces lividans, confer resistance to daunorubicin (46). However, it cannot protect E. coli against UV radiation like UvrA does. Compared to UvrA, both DrrC and AcmrC lack a segment of ca. 100 amino acids that may play a role in the different mechanisms of protection. The apparent similarity of AcmrC to DrrC (and UvrA) indicates that AcmrC might be a DNA-binding protein that either protects DNA from binding by actinomycin or can repair DNA with its exonuclease function when actinomycin is present. The sequence of acmrC is highly similar to that of its paralogous counterpart acmY (75% identity) on the right of the actinomycin biosynthetic gene cluster. Remarkably, the sequence of the acmY gene is interrupted at nucleotide position 47435 by orf157, resembling a putative IS1647-like transposase (58% identity to the SGR_118t putative IS1647-like transposase from S. griseus subsp. griseus NBRC 13350). Beyond orf157, the missing remainder of acmY which encodes the carboxy-terminal 79 amino acids of the protein, was found to encode the region between nucleotide positions 48511 and 48720. The translated sequence of the reconstructed acmY sequence reveals a protein of 758 amino acids, which perfectly aligns with AcmrC and other exonucleases in the database. Whether the truncated acmY gene is still functional is not known, but if it is not, possibly the intact acmrC gene may take on its function. Similarly to acmY and acmrC, this may also apply to the other paralogous gene pairs in the gene cluster, not all of which are known to be truly functional; if some of them were mutated, the presence of an intact paralogous partner could complement the mutation in the other, thus conferring a selection for the maintenance of the palindromic structure of the actinomycin biosynthetic gene cluster.
How the palindromic structure of the actinomycin gene cluster evolved cannot be answered yet. Current mechanisms postulated for the generation of large palindromes in eukaryotic and prokaryotic genomes imply short IR sequences near a double-strand break. Such IRs can guide the formation of a hairpin at the broken end which is converted by bidirectional DNA replication to a palindrome with the hairpin exactly in the middle (11, 59). Alternative models favor the possibility that two identical molecules with the same IR can undergo recombination between the IRs, resulting in a head-to-tail arrangement of the two ligated molecules (11). In the case of the actinomycin biosynthetic gene cluster, similar mechanisms of duplication are difficult to envision since the palindromic arms are separated by more than 25 kb represented by the NRPS genes. Investigations of the structural arrangement of actinomycin biosynthetic genes in other streptomycetes such as S. parvulus, S. antibioticus, and others may shed light on how far their sequential order is conserved between different actinomycetes. This could provide additional insights into the evolution of the actinomycin biosynthesis gene cluster.
Acknowledgments
We thank David Hopwood for useful comments on the manuscript and Hildgund Schrempf for helpful advice.
This work was supported previously by the Deutsche Forschungsgemeinschaft (Ke 452/8-5) and more recently by the Unicat Research Center of Excellence at the Technical University of Berlin.
Footnotes
Published ahead of print on 19 March 2010.
REFERENCES
- 1.Bentley, S. D., K. F. Chater, A. M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 2.Bibb, M. J., V. Molle, and M. J. Buttner. 2000. Sigma (BldN), an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2). J. Bacteriol. 182:4606-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bitzer, J., M. Streibel, H. J. Langer, and S. Grond. 2009. First Y-type actinomycins from Streptomyces with divergent structure-activity relationships for antibacterial and cytotoxic properties. Org. Biomol. Chem. 7:444-450. [DOI] [PubMed] [Google Scholar]
- 4.Blondelet-Rouault, M. H., J. Weiser, A. Lebrihi, P. Branny, and J. L. Pernodet. 1997. Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene 190:315-317. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microorganism quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248. [DOI] [PubMed] [Google Scholar]
- 6.Brickman, T. J., and M. A. McIntosh. 1992. Overexpression and purification of ferric enterobactin esterase from Escherichia coli. Demonstration of enzymatic hydrolysis of enterobactin and its iron complex. J. Biol. Chem. 267:12350-12355. [PubMed] [Google Scholar]
- 7.Brockmann, H., and H. Muxfeldt. 1958. Konstitution und Synthese des Actinomycin-Chromophors. Chem. Ber. 91:1242-1265. [Google Scholar]
- 8.Brown, D. D., M. J. Hitchcock, and E. Katz. 1980. Evidence for a constitutive and inducible form of kynurenine formamidase in an actinomycin-producing strain of Streptomyces parvulus. Arch. Biochem. Biophys. 202:18-22. [DOI] [PubMed] [Google Scholar]
- 9.Brown, D. D., M. J. Hitchcock, and E. Katz. 1986. Purification and characterization of kynurenine formamidase activities from Streptomyces parvulus. Can. J. Microbiol. 32:465-472. [DOI] [PubMed] [Google Scholar]
- 10.Brown, A. T., and C. Wagner. 1970. Regulation of enzymes involved in the conversion of tryptophan to nicotinamide adenine dinucleotide in a colorless strain of Xanthomonas pruni. J. Bacteriol. 101:456-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler, D. K., D. Gillespie, and B. Steele. 2002. Formation of large palindromic DNA by homologous recombination of short inverted repeat sequences in Saccharomyces cerevisiae. Genetics 161:1065-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butterton, J. R., and S. B. Calderwood. 1994. Identification, cloning, and sequencing of a gene required for ferric vibriobactin utilization by Vibrio cholerae. J. Bacteriol. 176:5631-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christen, S., P. T. Southwell-Keely, and R. Stocker. 1992. Oxidation of 3-hydroxyanthranilic acid to the phenoxazinone cinnabarinic acid by peroxyl radicals and by compound I of peroxidases or catalase. Biochemistry 31:8090-8097. [DOI] [PubMed] [Google Scholar]
- 14.Dangel, V., A. S. Eustáquio, B. Gust, and L. Heide. 2008. novE and novG act as positive regulators of novobiocin biosynthesis. Arch. Microbiol. 190:509-519. [DOI] [PubMed] [Google Scholar]
- 15.Dereeper, A., V. Guignon, G. Blanc, S. Audic, S. Buffet, F. Chevenet, J. F. Dufayard, S. Guindon, V. Lefort, M. Lescot, J. M. Claverie, and O. Gascuel. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server issue):W465-W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eggert, C., U. Temp, J. F. Dean, and K. E. Eriksson. 1995. Laccase-mediated formation of the phenoxazinone derivative, cinnabarinic acid. FEBS Lett. 376:202-206. [DOI] [PubMed] [Google Scholar]
- 17.Foster, J. W., and E. Katz. 1981. Control of actinomycin D biosynthesis in Streptomyces parvulus: regulation of tryptophan oxygenase activity. J. Bacteriol. 148:670-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frei, E., III. 1974. The clinical use of actinomycin. Cancer Chemother. Rep. 58:49-54. [PubMed] [Google Scholar]
- 19.Furuya, K., and C. R. Hutchinson. 1998. The DrrC protein of Streptomyces peucetius, a UvrA-like protein, is a DNA-binding protein whose gene is induced by daunorubicin. FEMS Microbiol. Lett. 168:243-249. [DOI] [PubMed] [Google Scholar]
- 20.Gaitatzis, N., B. Kunze, and R. Müller. 2005. Novel insights into siderophore formation in myxobacteria. ChemBioChem 6:365-374. [DOI] [PubMed] [Google Scholar]
- 21.Gordon, L., A. Y. Chervonenkis, A. J. Gammerman, I. A. Shahmuradov, and V. Solovyev. 2003. Sequence alignment kernel for recognition of promoter regions. Bioinformatics 19:1964-1971. [DOI] [PubMed] [Google Scholar]
- 22.Haese, A., and U. Keller. 1988. Genetics of actinomycin C production in Streptomyces chrysomallus. J. Bacteriol. 170:1360-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning: a practical approach, vol. 1. IRL Press, McLean, VA.
- 24.Hitchcock, M. J., and E. Katz. 1988. Purification and characterization of tryptophan dioxygenase from Streptomyces parvulus. Arch. Biochem. Biophys. 261:148-160. [DOI] [PubMed] [Google Scholar]
- 25.Hohn, B., and J. Collins. 1980. A small cosmid for efficient cloning of large DNA fragments. Gene 11:291-298. [DOI] [PubMed] [Google Scholar]
- 26.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation in Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 27.Hu, Y., V. Phelan, I. Ntai, C. M. Farnet, E. Zazapoulos, and B. O. Bachmann. 2007. Benzodiazepine biosynthesis in Streptomyces refuineus. Chem. Biol. 14:691-701. [DOI] [PubMed] [Google Scholar]
- 28.Husain, I., B. Van Houten, D. C. Thomas, and A. Sancar. 1986. Sequences of Escherichia coli uvrA gene and protein reveal two potential ATP binding sites. J. Biol. Chem. 261:4895-4901. [PubMed] [Google Scholar]
- 29.Hutchinson, C. R., and A. L. Colombo. 1999. Genetic engineering of doxorubicin production in Streptomyces peucetius: a review. J. Ind. Microbiol. Biotechnol. 23:647-652. [DOI] [PubMed] [Google Scholar]
- 30.Jansonius, J. N. 1998. Structure, evolution and action of vitamin B6-dependent enzymes. Curr. Opin. Struct. Biol. 8:759-769. [DOI] [PubMed] [Google Scholar]
- 31.Janssen, G. R., and M. J. Bibb. 1993. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene 124:133-134. [DOI] [PubMed] [Google Scholar]
- 32.Jones, G. H. 1987. Actinomycin synthesis in Streptomyces antibioticus: enzymatic conversion of 3-hydroxyanthranilic acid to 4-methyl-3-hydroxyanthranilic acid. J. Bacteriol. 169:5575-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, G. H., M. S. Paget, L. Chamberlin, and M. J. Buttner. 1997. Sigma-E is required for the production of the antibiotic actinomycin in Streptomyces antibioticus. Mol. Microbiol. 23:169-178. [DOI] [PubMed] [Google Scholar]
- 34.Katz, E. 1967. Actinomycin, p. 276-341. In D. Gottlieb and P. D. Shaw (ed.), Antibiotics, vol II. Springer Verlag, New York, NY.
- 35.Katz, E., D. Brown, and M. J. Hitchcock. 1987. Arylformamidase from Streptomyces parvulus. Methods Enzymol. 142:225-234. [DOI] [PubMed] [Google Scholar]
- 36.Katz, E., P. Pienta, and A. Sivak. 1958. The role of nutrition in the synthesis of actinomycin. Appl. Microbiol. 6:236-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaur, P., and J. Russell. 1998. Biochemical coupling between the DrrA and DrrB proteins of the doxorubicin efflux pump of Streptomyces peucetius. J. Biol. Chem. 273:17933-17939. [DOI] [PubMed] [Google Scholar]
- 38.Keller, U., and F. Schauwecker. 2001. Nonribosomal biosynthesis of microbial chromopeptides. Prog. Nucleic Acid Res. Mol. Biol. 70:233-289. [DOI] [PubMed] [Google Scholar]
- 39.Keller, U., H. Kleinkauf, and R. Zocher. 1984. 4-Methyl-3-hydroxyanthranilic acid (4-MHA) activating enzyme from actinomycin-producing Streptomyces chrysomallus. Biochemistry 23:1479-1484. [DOI] [PubMed] [Google Scholar]
- 40.Kelley, L. A., and M. J. E. Sternberg. 2009. Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 4:363-371. [DOI] [PubMed] [Google Scholar]
- 41.Lautru, S., D. Oves-Costales, J. L. Pernodet, and G. L. Challis. 2007. MbtH-like protein-mediated cross-talk between non-ribosomal peptide antibiotic and siderophore biosynthetic pathways in Streptomyces coelicolor M145. Microbiology 153:1405-1412. [DOI] [PubMed] [Google Scholar]
- 42.Li, L., W. Deng, J. Song, W. Ding, Q. F. Zhao, C. Peng, W. W. Song, G. L. Tang, and W. Liu. 2008. Characterization of the saframycin A gene cluster from Streptomyces lavendulae NRRL 11002 revealing a nonribosomal peptide synthetase system for assembling the unusual tetrapeptidyl skeleton in an iterative manner. J. Bacteriol. 190:251-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, W., A. Khullar, S. Chou, A. Sacramo, and B. Gerratana. 2009. Biosynthesis of sibiromycin, a potent antitumor antibiotic. Appl. Environ. Microbiol. 75:2869-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lima, S., S. Kumar, V. Gawandi, C. Momany, and R. S. Phillips. 2009. Crystal structure of the Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex: insights into the molecular basis of kynureninase substrate specificity. J. Med. Chem. 52:389-396. [DOI] [PubMed] [Google Scholar]
- 45.Lombó, F., A. F. Braña, C. Méndez, and J. A. Salas. 1999. The mithramycin gene cluster of Streptomyces argillaceus contains a positive regulatory gene and two repeated DNA sequences that are located at both ends of the cluster. J. Bacteriol. 181:642-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lomovskaya, N., S. K. Hong, S. U. Kim, L. Fonstein, K. Furuya, and R. C. Hutchinson. 1996. The Streptomyces peucetius drrC gene encodes a UvrA-like protein involved in daunorubicin resistance and production. J. Bacteriol. 178:3238-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mauger, A. B., and H. Lackner. 2005. The actinomycins, p. 281-298. In D. G. I. Kingston, G. M. Cragg, and D. J. Newman (ed.), Anticancer agents from natural products. CRC Press, Inc., Boca Raton, FL.
- 48.Mead, D. A., A. Szczesna-Skoruep, and B. Kemper. 1986. Single stranded DNA ‘blue’ T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1:67-74. [DOI] [PubMed] [Google Scholar]
- 49.Meienhofer, J., and E. Atherton. 1973. Structure-activity relationships in the actinomycins. Adv. Appl. Microbiol. 16:203-300. [PubMed] [Google Scholar]
- 50.Nagasawa, H. T., H. R. Gutmann, and M. A. Morgan. 1959. The oxidation of o-aminophenols by cytochrome c and cytochrome oxidase. J. Biol. Chem. 234:1600-1604. [PubMed] [Google Scholar]
- 51.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW.NEWS 4:14. http://www.embnet.org/files/shared/EMBnetNews/embnet_news_4_2.pdf. [Google Scholar]
- 52.Ogawa, H., Y. Nagamura, and I. Ishiguro. 1983. Cinnabarinate formation in malpighian tubules of the silkworm, Bombyx mori: reaction mechanism of cinnabarinate formation in the presence of catalase and manganese ions. Hoppe Seylers Z. Physiol. Chem. 364:1507-1518. [DOI] [PubMed] [Google Scholar]
- 53.Ohnishi, Y., J. Ishikawa, H. Hara, H. Suzuki, M. Ikenoya, H. Ikeda, A. Yamashita, M. Hattori, and S. Horinouchi. 2008. The genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190:4050-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. U. S. A. 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pacholec, M., J. Tao, and C. T. Walsh. 2005. CouO and NovO: C-methyltransferases for tailoring the aminocoumarin scaffold in coumermycin and novobiocin antibiotic biosynthesis. Biochemistry 44:14969-14976. [DOI] [PubMed] [Google Scholar]
- 56.Palleroni, N. J., and R. Y. Stanier. 1964. Regulatory mechanisms governing synthesis of the enzymes for tryptophan oxidation by Pseudomonas fluorescens. J. Gen. Microbiol. 35:319-334. [DOI] [PubMed] [Google Scholar]
- 57.Peschke, U., H. Schmidt, H. Z. Zhang, and W. Piepersberg. 1995. Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78-11. Mol. Microbiol. 16:1137-1156. [DOI] [PubMed] [Google Scholar]
- 58.Pfennig, F., F. Schauwecker, and U. Keller. 1999. Molecular characterization of the genes of actinomycin synthetase I and of a 4-methyl-3-hydroxyanthranilic acid carrier protein involved in the assembly of the acylpeptide chain of actinomycin in Streptomyces. J. Biol. Chem. 274:12508-12516. [DOI] [PubMed] [Google Scholar]
- 59.Qin, Z., and S. N. Cohen. 2000. Long palindromes formed in Streptomyces by nonrecombinational intra-strand annealing. Genes Dev. 14:1789-1796. [PMC free article] [PubMed] [Google Scholar]
- 60.Quadri, L. E., J. Sello, T. A. Keating, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 62.Schauwecker, F., F. Pfennig, N. Grammel, and U. Keller. 2000. Construction and in vitro analysis of a new bi-modular polypeptide synthetase for synthesis of N-methylated acyl peptides. Chem. Biol. 7:287-297. [DOI] [PubMed] [Google Scholar]
- 63.Schwartz, D., N. Grammel, E. Heinzelmann, U. Keller, and W. Wohlleben. 2005. Phosphinothricin tripeptide synthetases in Streptomyces viridochromogenes Tü494. Antimicrob. Agents Chemother. 49:4598-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shetty, A. S., and F. H. Gaertner. 1973. Distinct kynureninase and hydroxykynureninase activities in microorganisms: occurrence and properties of a single physiologically discrete enzyme in yeast. J. Bacteriol. 113:1127-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shetty, A. S., and F. H. Gaertner. 1975. Kynureninase-type enzymes of Penicillium roqueforti, Aspergillus niger, Rhizopus stolonifer, and Pseudomonas fluorescens: further evidence for distinct kynureninase and hydroxykynureninase activities. J. Bacteriol. 122:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith, A. W., A. Camara-Artigas, M. Wang, J. P. Allen, and W. A. Francisco. 2006. Structure of phenoxazinone synthase from Streptomyces antibioticus reveals a new type 2 copper center. Biochemistry 45:4378-4387. [DOI] [PubMed] [Google Scholar]
- 67.Sobell, H. M. 1985. Actinomycin and DNA transcription. Proc. Natl. Acad. Sci. U. S. A. 82:5328-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki, H., Y. Furusho, T. Higashi, Y. Ohnishi, and S. Horinouchi. 2006. A novel o-aminophenol oxidase responsible for formation of the phenoxazinone chromophore of grixazone. J. Biol. Chem. 281:824-833. [DOI] [PubMed] [Google Scholar]
- 69.Tanizawa, K., and K. Soda. 1979. Purification and properties of pig liver kynureninase. J. Biochem. 85:901-906. [DOI] [PubMed] [Google Scholar]
- 70.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toussaint, O., and K. Lerch. 1987. Catalytic oxidation of 2-aminophenols and ortho hydroxylation of aromatic amines by tyrosinase. Biochemistry 26:8567-8571. [DOI] [PubMed] [Google Scholar]
- 72.Troost, T., M. J. Hitchcock, and E. Katz. 1980. Distinct kynureninase and hydroxykynureninase enzymes in an actinomycin-producing strain of Streptomyces parvulus. Biochim. Biophys. Acta 612:97-106. [DOI] [PubMed] [Google Scholar]
- 73.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved Ml3 phage cloning vectors and host strains: nucleotide sequence of the M13mpl8 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 74.Zubieta, C., X. Z. He, R. A. Dixon, and J. P. Noel. 2001. Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Biol. 8:271-279. [DOI] [PubMed] [Google Scholar]