Abstract
Escherichia coli K-12 is able to grow under aerobic conditions on d-malate using DctA for d-malate uptake and the d-malate dehydrogenase DmlA (formerly YeaU) for converting d-malate to pyruvate. Induction of dmlA encoding DmlA required an intact dmlR (formerly yeaT) gene, which encodes DmlR, a LysR-type transcriptional regulator. Induction of dmlA by DmlR required the presence of d-malate or l- or meso-tartrate, but only d-malate supported aerobic growth. The regulator of general C4-dicarboxylate metabolism (DcuS-DcuR two-component system) had some effect on dmlA expression. The anaerobic l-tartrate regulator TtdR or the oxygen sensors ArcB-ArcA and FNR did not have a major effect on dmlA expression. DmlR has a high level of sequence identity (49%) with TtdR, the l- and meso-tartrate-specific regulator of l-tartrate fermentation in E. coli. dmlA was also expressed at high levels under anaerobic conditions, and the bacteria had d-malate dehydrogenase activity. These bacteria, however, were not able to grow on d-malate since the anaerobic pathway for d-malate degradation has a predicted yield of ≤0 ATP/mol d-malate. Slow anaerobic growth on d-malate was observed when glycerol was also provided as an electron donor, and d-malate was used in fumarate respiration. The expression of dmlR is subject to negative autoregulation. The network for regulation and coordination of the central and peripheral pathways for C4-dicarboxylate metabolism by the regulators DcuS-DcuR, DmlR, and TtdR is discussed.
Escherichia coli is able to use a large number of C4-dicarboxylates for growth under aerobic and anaerobic conditions. Under aerobic conditions the main C4-dicarboxylates used for growth are fumarate, succinate, and l-malate. The metabolism of these substrates involves uptake by the DctA transporter (encoded by the dctA gene), the citric acid cycle, and the malic enzyme/pyruvate dehydrogenase bypass to produce acetyl coenzyme A (acetyl-CoA) for feeding the citric acid cycle (9, 33, 35, 41). Anaerobically, fumarate is metabolized by fumarate respiration using the fumarate reductase FrdABCD (encoded by the frdABCD genes) and the fumarate/succinate antiporter DcuB (encoded by the dcuB gene) (for reviews, see references 22, 47, and 48). The C4-dicarboxylates l-malate and aspartate are transported to the cytoplasm by DcuB and then converted by fumarase B (fumB) and aspartase AspA to fumarate, which is used for fumarate respiration. In l-tartrate fermentation, the substrate is taken up by the l-tartrate/succinate antiporter TtdT (encoded by ttdT) and converted by the l-tartrate dehydratase TtdAB (encoded by ttdAB) to oxaloacetate (24, 38). Oxaloacetate is then metabolized by reactions of the central metabolism to fumarate and converted to succinate by fumarate respiration.
The pathways are regulated at the transcriptional level in response to O2, nitrate, and the C4-dicarboxylates. Many genes involved in aerobic catabolism, including dctA, are repressed under anaerobic conditions by the ArcB-ArcA two-component system (9, 20). The genes for fumarate respiration and related enzymes (dcuB, frdABCD, fumB, ttdAB, and ttdT) are subject to anaerobic induction by the oxygen sensor FNR and to nitrate repression by the NarX-NarL two-component system (15, 16, 37, 46). For regulation in response to C4-dicarboxylates two systems have been described. The DcuS-DcuR two-component system induces the dcuB, fumB, and frdABCD genes of central anaerobic C4-dicarboxylate (or fumarate respiration-linked) catabolism (13, 52). DcuS-DcuR also weakly induces expression of dctA (9). The expression of the ttdAB and ttdT genes involved in l-tartrate fermentation is transcriptionally activated by the cytoplasmic LysR-type regulator TtdR, which responds to l- and meso-tartrate (26, 36).
Recently, a metabolic route for aerobic degradation of d-malate was described for E. coli. d-Malate is carried to the cytoplasm by the transporter DctA and then oxidized by the (decarboxylating) d-malate dehydrogenase YeaU to pyruvate (39). An inducible enzyme of this type [d-malate + NAD(P) → pyruvate + CO2 + NAD(P)H + H+] was found previously in extracts of d-malate-grown E. coli (45). The pyruvate is then oxidized by pyruvate dehydrogenase and the citric acid cycle to CO2.
Bioinformatic and deletion studies have suggested that yeaU encoding d-malate dehydrogenase is transcriptionally regulated by the LysR-type regulator YeaT (39). YeaT is closely related to the TtdR regulator of l-tartrate metabolism (26, 36). In order to test the suggested role of YeaT and to differentiate this regulator from the DcuS-DcuR and TtdR regulators, the role of YeaT in the transcriptional regulation of yeaU and d-malate metabolism was studied. The substrate specificity of YeaT was compared to that of DcuS and TtdR, which respond to all C4-dicarboxylates (DcuS) or specifically to l- and meso-tartrate (TtdR) (26, 28). This study identified the compounds that stimulate d-malate metabolism and showed how transcriptional regulation of the various C4-dicarboxylate pathways is differentiated and coordinated. To this end, the expression of yeaU (YeaT dependent), dcuB (DcuS-DcuR dependent), and ttdT (TtdR dependent) in response to regulators and stimuli that are characteristic of the pathways was tested. It turned out that the TtdR and YeaT regulators respond to a narrow and specific spectrum of effectors, in contrast to DcuS-DcuR, which responds to all C4-dicarboxylates. Surprisingly, d-malate dehydrogenase was strongly induced under anaerobic conditions as well; therefore, d-malate degradation during anaerobic growth of E. coli was studied. Since yeaT and yeaU encode a regulator and an enzyme specifically involved in d-malate metabolism, the genes and proteins were designated dmlR and dmlA (d-malate degradation) and DmlR and DmlA, respectively.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
For genetic analyses the bacteria (Table 1) were grown aerobically in Luria-Bertani (LB) broth (40). For expression studies and growth experiments the bacteria were grown in eM9 medium (24, 31), which is M9 mineral medium (34) supplemented with acid-hydrolyzed casein (0.1%, wt/vol; Gibco BRL) and l-tryptophan (0.005%, wt/vol). For anaerobic growth the cultures were incubated at 37°C in degassed media in infusion bottles with rubber stoppers under N2. The medium was supplemented with glycerol (50 mM), dimethyl sulfoxide (DMSO) (20 mM), and sodium gluconate (50 mM). Fumarate, l-, d-, and meso-tartrate, succinate, d- and l-malate, and nitrate (sodium salts; 50 mM each) were added as indicated below. For aerobic growth cultures were incubated in shaken (170 rpm) Erlenmeyer flasks with baffles filled to at most 15% of the maximal volume with eM9 medium containing the substrates plus gluconate (50 mM), as indicated below.
TABLE 1.
Strains of E. coli and plasmids used
| Strain | Genotype and/or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| LJ1 | MG1655 fnr+ | 10 |
| MC4100 | F−araD139 (argF-lac)U169 rpsL150 (ΔlacZ) relA1 flbB530 deoC1 ptsF25 rbsR | 44 |
| IMW151a | MC4100 fnr::tet | This study |
| IMW262 | MC4100 dcuS::Camr | 52 |
| IMW523 | MC4100 ttdR::Camr | 26 |
| IMW530 | MC4100 λ(Φ[ttdR-lacZ]hyb) Ampr | 26 |
| IMW533 | MC4100 dmlR::Camr | This study |
| IMW540 | BW25113 yeaV::KanrdctA::Spcr | This study |
| IMW543 | MC4100 dmlR::Camr λ(Φ[ttdR-lacZ]hyb) Ampr | 26 |
| IMW544 | BW25113 dctA::Spcr | This study |
| IMW547 | MC4100 dmlR::Camr ΔttdR | 26 |
| IMW577 | MC4100 dmlR::CamrdcuR::Kanr | This study |
| BW25113 | BW24113 F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-4) lacIp-4000(lacIq) λ−rpoS396(Am) rph-1 Δ(rhaD-rhaB)568 rrnB-4 hsdR514 | 8 |
| JW5293 | BW25113 yeaV::Kanr | NBRP, Keio Collection |
| JW2240 | BW25113 yfaV::Kanr | NBRP, Keio Collection |
| JW1789 | BW25113 yeaU (dmlA)::Kanr | NBRP, Keio Collection |
| MI1443 | Δ(frdABCD ampC) Smr | 30 |
| RM313 | MC4100 arcA1zjj::Tn10 Tetr | 42 |
| Plasmids | ||
| pJL28 | (′lacZ bla+), protein fusion vector, Ampr | E. Bremer, Marburg,Germany |
| pJL29 | (′lacZ bla+), protein fusion vector, Ampr | E. Bremer, Marburg,Germany |
| pJL30 | (′lacZ bla+), protein fusion vector, Ampr | E. Bremer, Marburg,Germany |
| pMW99 | pJL29 with Φ(dcuB-lacZ)hyb, including dcuB promoter (560 bp), Ampr | 52 |
| pMW322 | pJL28 with Φ(ttdA-lacZ)hyb, including ttdA promoter (586 bp), Ampr | 26 |
| pMW323 | pJL30 with Φ(dmlA-lacZ)hyb, including dmlA promoter (1,066 bp), Ampr | This study |
| pET28a | Expression plasmid with T7 promoter and N-terminal His6 tag, Kanr | |
| pMW414 | pET28a with dmlR with its own promoter, Kanr | This study |
| pKD3 | oriRγ cat bla Δ(phoB-phoR)580 galU95 ΔuidA3::pir+ ΔendA::FRT | 8 |
| pKD46 | oriR101 repA101(Ts) araBp-gam-bet-exo bla | 8 |
| pCP20 | FLP+ λcI857 λ pRRepts cat bla | 8 |
| pMW806 | pJL28 with Φ(yeaT-lacZ)hyb, including yeaT promoter (190 bp), Ampr | This study |
Construction of dmlA- and dmlR-lacZ reporter gene fusions.
Gene fusions of dmlA and dmlR with lacZ were constructed essentially as described previously using the reporter gene fusion plasmids pJL28 and pJL30 (3, 52). To obtain the dmlA-lacZ and dmlR-lacZ fusions in a plasmid, the promoter regions of the dmlA and dmlR genes were amplified with primers yeaU_eco20 (5′-CGC GCA GAG AAT TCG GTA AT-3′) and yeaU_sal20 (5′-CCG ATT CCT GAA TGT CGA CT-3′) and primers yeaU_EcoRI_for (5′ CTT CTT TGC GAA TTC CGT CTC C) and yeaT_HindIII_rev (5′ GCG CAA AGC TTT CAG CAG C), respectively, from genomic DNA of E. coli LJ1. The corresponding fragments covered the complete upstream regulatory regions of dmlA and dmlR, respectively. The amplified promoter regions were cloned into the corresponding restriction sites of reporter gene fusion plasmids pJL28 and pJL30, yielding pMW323 and pMW806, which encode the DmlA-LacZ and DmlR-LacZ fusion proteins. The dmlR-lacZ gene fusion was transferred to the specialized transducing phage λRZ5, which was integrated into the chromosome (at the att site) of acceptor strains having the desired genotypes (4).
d-Malate dehydrogenase activity.
The d-malate dehydrogenase activity (decarboxylating) in cell-free homogenates was determined photometrically at 365 nm by determining the d-malate (4 mM)-dependent reduction of NAD+ (1 mM) in buffer containing triethanolamine (TRA) (90 mM, pH 7.6) and MgCl2 (4 mM). The bacteria used for preparation of the cell-free homogenates were grown in eM9 medium under aerobic conditions on d-malate and gluconate (50 mM each) to an optical density at 578 nm (OD578) of 0.5. Each cell-free homogenate was prepared from bacteria washed in TRA buffer (0.1 M, pH 7.6) at 4°C. The washed bacteria in TRA buffer with 2 mM dithiothreitol (DTT) were broken twice with a French press at 87 × 105 Pa. The homogenate was cleared by centrifugation at 10,000 × g for 15 min at 4°C and used for the enzyme assay at 37°C. Formation of 1 mol of NADH corresponded to degradation of 1 mol of d-malate. For controls the same assay was performed with anoxic buffers in anaerobic cuvettes with rubber stoppers under an N2 atmosphere.
β-Galactosidase activity assay.
Bacteria were grown in eM9 medium under aerobic or anaerobic conditions with the substrates indicated below for the individual experiments, as described previously (4, 26). The subcultures used for inoculation were grown under the same conditions and with the same media and substrates as the bacteria used for the main experiment. Using exponentially growing bacteria (OD578, 0.5 to 0.8), the β-galactosidase activity was determined as described by Miller (34). The activities were determined using three or more independent growth experiments, each performed at least in triplicate. Standard deviations were calculated.
Identification of fermentation products by HPLC.
The products of d-malate fermentation in the supernatants from anaerobic growth experiments (late exponential growth phase) or from anaerobic cell suspensions in eM9 medium were determined after removal of bacteria by centrifugation. The substrates and products in each supernatant were analyzed by high-performance liquid chromatography (HPLC) with an Aminex HPX87H column (300 by 7.8 mm; Bio-Rad) at 65°C with buffer (6.5 mM H2SO4) at a flow rate of 550 μl/min and were quantified by UV (215 nm) and refractive index detection (25). The retention time was determined and quantitative calibration for d-malate was performed using a standard solution of d-malate. The amounts of CO2 and H2 formed were calculated by assuming that 1 mol of formate was formed per mol of acetate and mol of ethanol and by assuming that the difference between the theoretical and experimental amounts of formate was converted to CO2 and H2.
Inactivation of dmlR.
The dmlR gene was deleted using the method of Datsenko and Wanner (8) as described by Lehnen et al. (31) and Kim and Unden (24). For insertional inactivation, the PCR product of the chloramphenicol resistance (Cmr) cassette from plasmid pKD3 was used, which was flanked by FRT sequences. Primers yeaT_H1P1 (5′-GGA GAA AAT CAG GTT TTA ACC TGA TAT CAA CCC GAT AAT TGA ATC ATT AAG TGT AGG CTG GAG CTG CTT C-3′) and yeaT_H2P2 (5′-CAT CAC CAT CCT ATA ATT GAC TGA AAA GGA ATT AAT CCC CGT AAC TGC ATA TGA ATA TCC TCC TTA G-3′) contain parts of the regions adjacent to FRT and of the 5′ and 3′ regions of dmlR, respectively. The PCR products obtained with the primers were purified, concentrated, and used for transformation. Chloramphenicol-resistant colonies were tested to determine loss of the helper plasmid (pKD46) by examining ampicillin sensitivity. To delete the Cmr cassette, the dmlR::Cmr mutant was transformed with the FLP helper plasmid pCP20 and selected at 30°C (6). The dmlR::Cmr ΔdmlR mutant genotype was verified by performing PCR with test primers yeaT_test20 (5′-CCA GAT AGG TCC AGT AAT TC-3′), cat_frd (5′-GAG ATT ATG TTT TTC GTC TCA GCC AAT CC-3′), cat_rev (5′-CTA TCC CAT ATC ACC AGC TCA CCG TCT TTC-3′), and cat_mitte (5′-CTC TGG AGT GAA TAC CAC GAC-3′).
RESULTS
DmlR, a d-malate- and tartrate-responsive regulator of aerobic d-malate catabolism.
The dmlR dmlA (or yeaT yeaU) gene cluster of E. coli is thought to encode enzymes and proteins for the degradation of hydroxylated C4-decarboxylates (39). dmlR has an orientation different than that of the dmlA gene. DmlA, the product of dmlA is a (decarboxylating) d-malate dehydrogenase, and deletion of dmlA inhibited aerobic growth on d-malate (39). The LysR-type regulator DmlR was suggested to regulate expression of the dmlA gene (39). DmlR shows 49% sequence identity and 67% similarity to the l-tartrate regulator TtdR (26, 36). A large number of basic amino acid residues are conserved in the effector binding domains of DmlR, TtdR, and related LysR-type regulators.
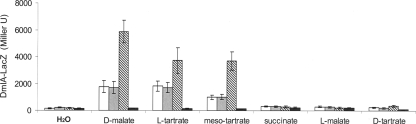
A plasmid-borne dmlA-lacZ reporter gene fusion was used to test transcriptional regulation of dmlA expression in response to the presence of C4-dicarboxylates during aerobic growth (Fig. 1A). In a wild-type background, d-malate and meso- and l-tartrate caused high levels of induction of dmlA-lacZ expression (up to 12.3-fold). With l-malate, succinate, and d-tartrate there was no or only weak induction. The inducers d-malate and l- and meso-tartrate are C4-dicarboxylates that have a hydroxyl group in the R configuration at the C-2 or C-3 position. l-Malate, which has a hydroxyl group in the S configuration at the C-2 position, does not function in dmlA-lacZ induction.
FIG. 1.
Transcriptional regulation of dmlA-lacZ gene fusion by C4-dicarboxylates and the regulators DmlR, TtdR, and DcuS during aerobic growth. The dmlA-lacZ gene fusion was present on plasmid pMW323, and its expression was measured in E. coli wild-type strain MC4100 and dmlR-, ttdR-, and dcuS-deficient derivatives. Bacteria were grown under aerobic conditions in eM9 medium containing gluconate (50 mM) and the effectors indicated (50 mM). The strains used were strains MC4100 (wild type) (open bars), IMW533 (dmlR) (black bars), IMW523 (ttdR) (gray bars), and IMW262 (dcuS) (striped bars).
There was almost no induction of dmlA expression by d-malate and l- and meso-tartrate in the strain lacking a functional dmlR gene (Fig. 1); induction was restored by introducing plasmid-borne dmlR (pMW414) into the mutant (890 Miller units). Inactivation of ttdR had only a small effect or no effect on dmlA-lacZ expression, whereas inactivation of dcuS resulted in a significant (2.0- to 3.7-fold) increase in dmlA-lacZ expression compared to the wild type. Inactivation of dcuR stimulated expression of dml-lacZ to the same extent as dcuS inactivation. In a dcuR dmlR double mutant the stimulating effect of the dcuR mutation was not observed, suggesting that the effect of DcuS-DcuR requires a functional DmlR regulator. Inspection of the dmlA promoter revealed a consensus site for DcuR (1) at positions −96 to −46 relative to the translational start site, which could be an indication that DcuS-DcuR directly represses dmlA.
DmlR and DmlA are required for aerobic growth on d-malate.
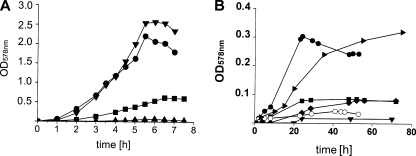
The induction of dmlA expression by d-malate and the tartrate isomers suggests that these compounds support aerobic growth. The bacteria were able to grow on d-malate under aerobic conditions at rates and with yields comparable to those obtained when the bacteria were grown on succinate (Fig. 2A), in agreement with the report of Reed et al. (39), whereas meso-tartrate (Fig. 2A) or d-tartrate (not shown) did not stimulate growth. l-Tartrate supported only slow growth in the enriched M9 medium (Fig. 2A). When M9 medium without amino acid supplementation was used, there was no growth on l-tartrate, whereas the growth on d-malate and succinate decreased only slightly (not shown). Aerobic growth on d-malate and succinate required the presence of an intact aerobic C4-dicarboxylate transporter, DctA (dctA gene) (not shown), as shown previously (39). Inactivation of the yeaV gene, which is located downstream of dmlA and codes for a secondary transporter belonging to the betaine-carnitine-choline (BCCT) family, had no effect on the growth with d-malate. Similarly, inactivation of the dcuA, dcuB, dcuC, ttdT, and yfaV genes, which encode other secondary transporters or putative transporters for C4-dicarboxylates, did not impair the growth on d-malate (not shown). We concluded that aerobic growth that depends on the dmlA dmlR genes occurs only on d-malate. The structurally related C4-dicarboxylates l- and meso-tartrate induce dmlA expression but do not support aerobic growth. DctA is the only or major transporter for d-malate uptake, as suggested previously (39).
FIG. 2.
Aerobic (A) and anaerobic (B) growth of E. coli wild-type and mutant strains on d-malate and related C4-dicarboxylates. (A) Wild-type strain MC4100 was grown in eM9 medium with 50 mM d-malate (•), l-tartrate (▪), meso-tartrate (▴), or succinate (▾). (B) E. coli wild-type strain MC4100 (•), frdABCD mutant MI1443 (▪), fumB mutant JW4083 (▾), dmlA mutant JW1789 (▸), and dcuS mutant IMW262 (⧫) were grown anaerobically using eM9 medium with d-malate plus glycerol (20 mM each. Wild-type strain MC4100 was also grown on d-malate without glycerol (○).
The effect of the C4-dicarboxylate regulators DcuS, TtdR, and DmlR on aerobic growth with d-malate and succinate was tested in the same way (not shown). The dmlR mutant did not grow on d-malate, as noted by Reed et al. (39). The growth of a mutant deficient in dcuS on d-malate and succinate was slightly impaired, whereas inactivation of ttdR had no significant effect (not shown). The significance of inactivation of dcuS for growth on d-malate and succinate can be explained by the stimulation of dctA expression by DcuS-DcuR (9).
Effect of electron acceptors on dmlA expression.
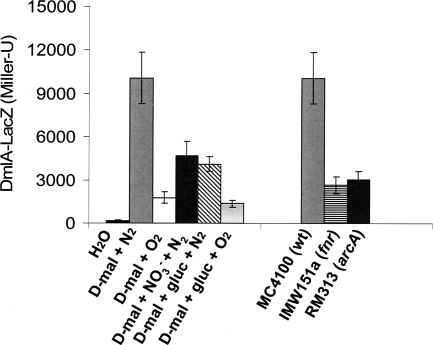
The expression of dmlA-lacZ at high levels was induced under anaerobic conditions in the presence of d-malate and was more than 5-fold greater than the expression under aerobic conditions (Fig. 3). Addition of nitrate during anaerobic growth repressed the expression about 2.2-fold, but the expression was still higher than the expression under aerobic conditions. The presence of glucose during anaerobic growth repressed dmlA expression to levels similar to those observed after nitrate addition, suggesting that there was some glucose repression (2.4-fold). No repression by glucose was observed during aerobic growth. The Prodoric virtual footprint database shows that there is a potential cyclic AMP receptor protein (CRP) binding site in the dmlR-dmlA promoter region (http://prodoric.tu-bs.de/vfp/vfp_promoter.php). A response to glucose repression only under anaerobic conditions is not expected for typical CRP-cyclic AMP-dependent regulation, and the significance of the CRP site has to be tested experimentally. d-Malate dehydrogenase requires divalent cations like Mn2+ or Mg2+ to function (18, 19, 45), and Easton et al. (10) observed increased expression of yeaU (dmlA) in the presence of Zn2+. During aerobic growth on d-malate plus gluconate the expression of dmlA increased 1.2- and 1.5-fold when the MgSO4 in the growth medium (eM9) was replaced by MnSO4 or ZnSO4, supporting the observation of Easton et al. (10).
FIG. 3.
Transcriptional regulation of the dmlA-lacZ fusion in response to electron acceptors and glucose (A) and mutations in fnr or arcB (B). (A) Wild-type strain MC4100 containing pMW323 (with the dmlA-lacZ gene fusion) was grown in eM9 medium containing gluconate (50 mM) for aerobic growth or in glycerol (50 mM) with DMSO (20 mM) for anaerobic growth. The following compounds were included in the individual experiments, as indicated on the x axis: H2O (no addition), d-malate (d-mal), d-malate plus sodium nitrate (NO3−), and d-malate plus glucose (gluc) (50 mM each). Growth was performed under aerobic (O2) or anaerobic (N2) conditions. (B) Strains MC4100 (wild type [wt]), IMW151a (fnr), and RM313 (arcA), each containing pMW323 (with the dmlA-lacZ gene fusion), were grown under anaerobic conditions in eM9 medium containing glycerol (50 mM), DMSO (20 mM), and d-malate (50 mM). The β-galactosidase activity of the bacteria was determined in the mid-exponential growth phase (OD578, 0.5 to 0.8).
The arcA and fnr genes code for regulators controlling the expression of genes in response to the presence of O2 (15, 20, 47). Inactivation of the fnr and arcA genes caused a decrease in dmlA expression (Fig. 3). When the effects of the mutations were tested with other substrates, like l-tartrate, d-malate plus nitrate, or d-malate plus glucose, only weak effects and repression of dmlA-lacZ were observed. This suggests that the effects of FNR and ArcA on dmlA-lacZ expression are not direct. Only the anaerobic d-malate-induced expression of dmlA-lacZ, which was very high for unknown reasons, was decreased by nitrate, glucose, and fnr and arcA inactivation. There are no clear FNR or ArcA sites in front of dmlA, supporting the view that the effects of FNR, ArcA, and oxygen are indirect. Overall, it appears that dmlA expression is induced under aerobic and anaerobic conditions by d-malate and l- and meso-tartrate.
Anaerobic growth with d-malate.
Wild-type E. coli was able to grow slowly on d-malate under anaerobic conditions, provided that glycerol was present as an additional electron donor (Fig. 2B). The growth rate on glycerol plus d-malate (about 0.02 to 0.04 h−1) was comparable to the anaerobic growth rate on d-tartrate plus glycerol (0.039 h−1) (25). Nearly equimolar amounts of d-malate and glycerol were consumed, and the major fermentation products were succinate, formate, acetate, and ethanol (Table 2). The overall fermentation balance was as follows: 1.0 d-malate + 1.0 glycerol → 0.9 succinate + 0.6 acetate + 0.6 ethanol + 0.7 formate + 0.5 CO2 + 0.5 H2.
TABLE 2.
Products of anaerobic d-malate fermentation in growing and resting cells of E. colia
| Substrate(s) | Substrate consumption (mol/mol d-malate) |
Product formation (mol/mol d-malate) |
||||||
|---|---|---|---|---|---|---|---|---|
| d-Malate | Glycerol | Succinate | Acetate | Ethanol | Formate | H2 | CO2 | |
| Glycerol + d-malate (growth) | 1.0 | 0.9 | 0.9 | 0.6 | 0.6 | 0.4 | 0.8 | 0.8 |
| Glycerol + d-malate (cell suspension) | 1.0 | 1.1 | 0.89 | 0.41 | 0.3 | 1.09 | 0 | 0 |
| d-Malate (cell suspension) | ND | ND | NP | NP | NP | NP | NP | NP |
Strain MC4100 (wild type) was grown in eM9 medium with glycerol and d-malate (20 mM each). The supernatants of early-stationary-phase cultures were analyzed to determine the decreases in the amounts of d-malate and glycerol and the amounts of succinate, acetate, ethanol, and formate produced. The bacteria used for the cell suspensions (OD578, 8) were grown on glycerol plus d-malate. The carbon yields for the fermentation balances of the growing bacteria and for the cell suspensions were 102% and 83%, respectively, based on the substrates and products shown. ND, not degraded; NP, no product.
The nearly 1:1 ratio of d-malate consumption to succinate production and the requirement for an electron donor suggest that there is conversion of d-malate to fumarate by fumarase, followed by reduction of fumarate to succinate by fumarate reductase. Oxidation of glycerol to formate, acetate, and ethanol supplies electrons for fumarate reduction. Strains of E. coli lacking fumarate reductase (frdABCD genes) or the anaerobic fumarase FumB were not able to grow anaerobically on d-malate (Fig. 2B), supporting the conclusion that anaerobic d-malate metabolism depends on fumarate respiration and expression of fumB and frdABCD. In addition, the anaerobic C4-dicarboxylate transporter DcuB could be important since expression of dctA is repressed under anaerobic conditions (9). Inactivation of dcuS inhibited anaerobic growth on d-malate (Fig. 2B), which can be explained by the requirement for DcuS-DcuR for fumB and dcuB induction. On the other hand, anaerobic growth on d-malate plus glycerol was not dependent on DmlA, since the growth of a dmlA mutant on d-malate was not impaired. However, a small portion of d-malate may be degraded via DmlA and pyruvate to ethanol, acetate, and formate. This possibility is supported by the substoichiometric succinate production compared to d-malate degradation and the excess acetate and ethanol production compared to glycerol degradation in some experiments (Table 2).
When a suspension of bacteria was incubated with d-malate and glycerol, the products were comparable to those obtained from growing bacteria (Table 2); only the yields and the relative amounts of the products showed some variation compared to the results obtained for the growing bacteria. When a cell suspension was incubated with d-malate under anaerobic conditions without glycerol, d-malate was not metabolized to a significant extent.
The low rates of growth and of d-malate metabolism under anaerobic conditions raised the question whether the anaerobically grown bacteria have significant d-malate dehydrogenase activity. To measure d-malate dehydrogenase activity, the bacteria were grown aerobically and anaerobically on d-malate plus cosubstrates to obtain induction of dmlA as described above (Fig. 1 and 3). The bacteria grown under aerobic conditions exhibited d-malate dehydrogenase activity (20 U/g protein), which was detected by the d-malate-dependent formation of NADH from NAD. NADH formation was abrogated when lactate dehydrogenase was included in the assay mixture, since the lactate dehydrogenase reoxidized the NADH at the expense of the pyruvate that was formed by d-malate dehydrogenase. The cell extracts were not able to use l-tartrate as a substrate for dehydrogenation instead of d-malate (<1 U/g protein in the cell extract). Therefore, d-malate is converted by the decarboxylating d-malate dehydrogenase and produces pyruvate, as shown previously for this type of enzyme (12, 39, 45), and the enzyme is specific for d-malate. The activity was 3.2-fold higher when the bacteria were grown under anaerobic conditions (64 U/g protein), which is in agreement with the results of the expression studies (Fig. 3). We concluded that anaerobically grown bacteria exhibit high d-malate dehydrogenase activities. However, this enzyme is not used in anaerobic metabolism irrespective of the presence of an additional electron donor (glycerol).
Transcriptional regulation of dmlR by C4-dicarboxylate-specific regulators.
The expression of LysR-type regulators is often subject to autoregulation or regulation by other regulators (43). The expression of dmlR in response to DmlR and other regulators (DcuS-DcuR and TtdR) of C4-dicarboxylate metabolism was tested by using a dmlR-lacZ gene fusion and the corresponding regulator mutants. A single copy of the dmlR-lacZ fusion was introduced into the genome to avoid distortion of expression data due to variations in the copy numbers of plasmids. Under aerobic conditions the level of expression of dmlR-lacZ was generally low (Fig. 4A). In the wild-type background d-malate or l-tartrate caused no or only weak induction. When the dmlR gene encoding DmlR was inactivated, the expression of dmlR-lacZ increased significantly in the absence of C4-dicarboxylates or when l-tartrate or fumarate was present (1.4- to 1.8-fold). Stimulation by dmlR inactivation was not observed, however, in the presence of d-malate. The derepression in the dmlR mutant suggests that there was negative autoregulation of dmlR by DmlR. The presence of fumarate also resulted in minor induction (or derepression) of dmlR expression in various backgrounds.
FIG. 4.
Effects of C4-dicarboxylates and the regulators TtdR, DcuS, and DmlR on the expression of dmlR-lacZ under aerobic (A) and anaerobic (B) growth conditions. (A) Strains IMW563 (wild type) (open bars), IMW564 (ttdR) (gray bars), IMW566 (dcuS) (striped bars), and IMW565 (dmlR) (black bars) carrying a chromosomal dmlR-lacZ gene fusion were grown in eM9 medium containing gluconate and the effectors indicated on the x axis (50 mM each) under aerobic conditions. (B) Anaerobic growth of the same strains in eM9 medium containing gluconate (50 mM), glycerol (50 mM), and DMSO (20 mM) with and without d-malate (50 mM).
During anaerobic growth the expression of dmlR-lacZ decreased further (Fig. 4B). Again, the expression increased in the dmlR mutant in the absence and in the presence of the inducers. Thus, the expression of dmlR appears to be negatively autoregulated under anaerobic conditions as well, but the level of expression was generally lower than that under aerobic conditions.
Role for DmlR in the transcriptional regulation of ttdA and dcuB.
To examine a potential regulatory effect of DmlR on the other pathways for C4-dicarboxylate degradation, the expression of DcuS-DcuR- and TtdR-regulated genes was measured in dmlR-positive and -negative backgrounds (Table 3). The dcuB and ttdA genes are major targets of DcuS-DcuR and TtdR, respectively, and were used due to their clear responses to the regulators. l-Tartrate, d-malate, and fumarate were tested as the key effectors for the sensors TtdR, DmlR, and DcuS-DcuR. As expected, expression of the TtdR-regulated ttdA-lacZ gene fusion was strongly induced in the wild-type strain by l-tartrate but was not induced or was only weakly induced by fumarate or d-malate. In the dmlR mutant the l-tartrate-induced expression was 26% lower, suggesting that DmlR has some (direct or indirect) effect on ttdA expression. Surprisingly, d-malate had a strong stimulatory effect on ttdA-lacZ expression in the dmlR mutant (53-fold), which was not observed for the wild type (2.3-fold). The increased expression required an intact ttdR gene, since in a ttdR dmlR double mutant there was not much stimulation.
TABLE 3.
Effect of dmlR mutation on the expression of ttdA-lacZ, ttdR-lacZ, and dcuB-lacZ gene fusionsa
| Inducer | Expression (Miller units) of: |
|||||
|---|---|---|---|---|---|---|
|
ttdA-lacZ fusion |
ttdR-lacZ fusion |
dcuB-lacZ fusion |
||||
| MC410 (wt) | IMW53 (dmlR) | IMW53 (wt) | IMW54 (dmlR) | MC410 (wt) | IMW53 (dmlR) | |
| None | 240 ± 46 | 160 ± 27 | 230 ± 5 | 220 ± 26 | 165 ± 27 | 138 ± 58 |
| l-Tartrate | 15,300 ± 4 | 11,400 ± 180 | 614 ± 14 | 520 ± 12 | 1,100 ± 157 | 1,010 ± 142 |
| d-malate | 558 ± 104 | 8,600 ± 474b | 504 ± 75 | 706 ± 33 | 1,340 ± 267 | 1,040 ± 273 |
| Fumarate | 125 ± 5 | 134 | 713 ± 31 | 711 ± 15 | 1,100 ± 210 | 1,080 ± 235 |
ttdA-′lacZ and dcuB-lacZ were provided on plasmids pMW322 and pMW99, and the ttdR-lacZ fusion was inserted into the chromosome. The expression was tested in strains with the corresponding reporter gene fusions in wild-type (wt) and dmlR backgrounds. The bacteria were grown in eM9 medium under anaerobic conditions in the presence of DMSO (20 mM), glycerol (50 mM), and the corresponding inducers (50 mM each) or without an inducer.
The value was 339 ± 15 Miller units for a ttdR dmlR double mutant.
Due to the link between DmlR and TtdR regulation, whether expression of ttdR coding for the regulator TtdR was changed in the dmlR mutant was tested (Table 3). Expression of ttdR-lacZ was stimulated slightly by l-tartrate, d-malate and fumarate (2.2- to 3.1-fold), similar to previous results (26), and the effects are known to depend on TtdR and DcuS-DcuR. In the mutant there was still stimulation by fumarate, whereas induction by l-tartrate and induction by d-malate were slightly decreased and increased, respectively. The expression of the DcuS-DcuR-regulated gene dcuB was stimulated by each of the effectors (Table 3) since DcuS responds to each type of C4-dicarboxylate (28). The little extra stimulation of dcuB-lacZ by d-malate was not observed after inactivation of dmlR, but otherwise had DmlR no effect on dcuB-lacZ expression with various effectors.
Overall, the expression of ttdA, ttdR, and dcuB exhibited minor (ttdR and dcuB) or clear (ttdA) changes in response to d-malate when dmlR was deleted. These findings can be explained by assuming that there is an indirect mechanism. d-Malate might accumulate in the dmlR mutant since d-malate degradation is inhibited, whereas uptake by the DmlR-independent DctA (or Dcu) transporters is not inhibited in the mutant. d-Malate can function as an effector of TtdR due to structural similarity to l- and meso-tartrate, and all of these compounds are C4-dicarboxylates with hydroxyl groups in the R configuration at the C-2 or C-3 position. This conclusion is supported by the results obtained for the dmlR ttdR double mutant, in which the level of expression of ttdA-lacZ was close to the background levels.
DISCUSSION
DmlR or d-malate regulon.
Along with the target gene dmlA encoding d-malate dehydrogenase, the regulator DmlR encoded by dmlR constitutes a regulon for d-malate metabolism that is separate from the general C4-dicarboxylate (DcuS-DcuR) and the l-tartrate (or TtdR) regulons. The gene products provide the capacity to grow aerobically on d-malate. This regulon responds to d-malate and the tartrate isomers, but only d-malate supports aerobic growth. Aerobic growth on l-tartrate like that described for E. coli W3110 and other “coliform” bacteria (38, 50) was not confirmed for the E. coli K-12 strains used in this study. DctA serves as the transporter for d-malate uptake, in agreement with previous reports (39), whereas the yeaV gene downstream of dmlA, which encodes a putative secondary carrier, is not involved in aerobic d-malate transport. The induction by d-malate and the tartrate isomers depends on the LysR-type regulator DmlR; DcuS-DcuR is required for induction of DctA and to some extent for regulation of DmlA. Overall, the DmlR and DmlA proteins in addition to enzymes of the central pathways are required for d-malate degradation.
The dmlA gene was also expressed at high levels during anaerobic growth, and the bacteria had high levels of of d-malate dehydrogenase activity. The high level of anaerobic induction might be due to the accumulation of d-malate after uptake without degradation, resulting in a high intracellular d-malate concentration. Under aerobic conditions d-malate is degraded, which prevents high intracellular levels from accumulating and causes moderate induction. The presence of d-malate dehydrogenase theoretically allows fermentation of d-malate according to the following equation: 2 d-malate → 1 ethanol + 1 acetate + 2 formate + 2 CO2.
The pathway uses the d-malate dehydrogenase DmlA, pyruvate-formate lyase, ethanol dehydrogenase AdhE, phosphotransacetylase (Pta), and common enzymes involved in mixed acid fermentation, including the acetate kinase AckA, resulting in a net yield of 1 mol of ATP per 2 mol of d-malate. However, the uptake of 2 mol of d-malate by the Dcu transporters (or DctA) during anaerobic growth requires 4 to 6 mol of H+ (11, 17), which is equivalent to consumption of 1 to 1.5 mol of ATP. Therefore, the overall reaction yields no ATP or there is a negative ATP yield, which explains the lack of growth.
In the presence of glycerol as an additional electron donor, d-malate is obviously used for fumarate respiration (despite the high activity of d-malate dehydrogenase), yielding 1 mol of succinate per mol of d-malate. Growth on d-malate plus glycerol by channeling d-malate into fumarate respiration and oxidizing glycerol to acetate, formate, and ethanol yields approximately 1.5 to 2 mol of ATP per mol of d-malate plus glycerol. d-Malate is presumably dehydrated by a side reaction of anaerobic fumarase B, which has also been observed for d-tartrate in d-tartrate fermentation by E. coli (25). This hypothesis is supported by the growth deficiency of the fumB mutant on glycerol plus d-malate. However, the fermentation balances suggest that small amounts of d-malate are degraded under anaerobic conditions by DmlA as well, and therefore, some acetate, ethanol, and formate are produced in addition to the amount produced from glycerol.
The d-malate dehydrogenase is a decarboxylating enzyme yielding pyruvate that was identified previously in E. coli (45). This enzyme is specific for d-malate and does not accept l-tartrate, in contrast to the regulator DmlR, which apparently responds to l- and meso-tartrate as well. The substrate specificity demonstrates the function of the enzyme in d-malate metabolism. Therefore, the previously described yeaU and yeaT genes and YeaU and YeaT proteins are renamed dmlA and dmlR and DmlA and DmlR, respectively. The expression of the genes is regulated essentially by d-malate and l- and meso-tartrate. The general metabolic regulators FNR, ArcB-ArcA, CRP, and DcuS-DcuR have only a limited effect, and it appears that d-malate metabolism is not fully integrated into the metabolic and regulatory system of E. coli, suggesting that d-malate metabolism plays a role only under rare and specific conditions.
The physiological source of d-malate is not known, but various bacteria are able to produce d-malate from maleate (2, 49) or from m-cresol (18). Despite the unknown origin of d-malate, growth on this substrate and the function of d-malate dehydrogenases have been described for various bacteria, including Rhodobacter capsulatus, Rhodopseudomonas sphaeroides Y, Pseudomonas fluorescens, and E. coli (22, 29, 32, 39).
Coordination of the d-malate-specific pathway with the general C4-dicarboxylate pathways: exogenous and endogenous regulation by C4-dicarboxylates.
In E. coli three regulatory systems are known for control of general C4-dicarboxylate metabolism (DcuS-DcuR), l-tartrate fermentation (TtdR), and aerobic d-malate catabolism (13, 22, 26, 36, 52). These pathways are differentially regulated but are coordinated by partially overlapping substrate specificities and by mutual control of regulator expression. Thus, TtdR responds to l- and meso-tartrate, which are C4-dicarboxylates with two hydroxyl groups at the C-2 or C-3 position, one of which has to be in the R configuration. DmlR, on the other hand, responds to d-malate and to l- and meso-tartrate, which are C4-dicarboxylates with one hydroxyl group at the C-2 or C-3 position that has to be in the R configuration. The second hydroxyl group of the tartrate isomers apparently is tolerated in any configuration. Binding of the effectors to DmlR and TtdR has not been demonstrated directly. DcuS, on the other hand, responds to all C4-dicarboxylates and tolerates a large number of groups at the C-2 or C-3 position, including hydroxyl, amino, and methyl groups, in the S or R configuration. Interaction and binding of the C4-dicarboxylates to DcuS have been demonstrated experimentally (7, 21, 28).
The target genes of TtdR are limited to l-tartrate fermentation (genes ttdAB, ttdT, and ttdR) (26, 36), those for DmlR to d-malate degradation (dmlR and dmlA), those for DcuS-DcuR to fumarate respiration (dcuB, fumB, and frdABCD), and an aerobic C4-dicarboxylate transport gene (dctA) (1, 13, 52). Thus, l-tartrate, meso-tartrate, and d-malate induce only the genes of the peripheral pathways for l-tartrate fermentation and d-malate degradation. The same substrates induce the fumarate respiration genes via DcuS-DcuR, in accordance with the need for fumarate respiration when the substrates are metabolized anaerobically. In the presence of the “general” C4-dicarboxylates of the central pathways, like fumarate, l-malate, or aspartate, only the genes for central pathways of fumarate respiration (dcuB, fumB, and frdABCD) and C4-dicarboxylate degradation (dctA) are induced via DcuS-DcuR.
Coordination of the pathways is also achieved to some extent by mutual transcriptional regulation of the gene regulators. Thus, DcuS-DcuR controls the expression of the ttdR gene (26). In contrast, TtdR affects expression of DcuS-DcuR-regulated genes, like dcuB, but it is not clear whether this regulation is direct or indirect due to control of the expression of dcuS and dcuR. DmlR, on the other hand, regulates its own synthesis, but it is essentially independent of DcuS-DcuR and TtdR under most conditions.
It appears that the pathways for the general C4-dicarboxylates are regulated by exogenous induction using the DcuS-DcuR system, whereas the specific pathways for the uncommon C4-dicarboxylates d-malate and l-tartrate are regulated by endogenous induction using cytoplasmic LysR-type regulators. The use of different types of sensors or regulators is explained by the finding that extracellular (or exogenous) induction is observed for substrates that are also found in the central metabolic pathways, whereas intracellular (or endogenous) induction occurs for uncommon substrates (d-malate and l-tartrate) that are not intermediates in central metabolism. Substrates of this type can be perceived by intracellular regulators after uptake into the bacterial cell, since no interference from central metabolism is expected. Fumarate, succinate, and l-malate, on the other hand, are central metabolites under aerobic and anaerobic conditions. Therefore, extracellular sensing has to be used for differentiation of these compounds from intracellular metabolites. An early example of exogenous induction is regulation by glucose-6-phosphate (51). The extracellular sensing system responsible for this regulation was later identified as the UhpCBA two-component system (for a review, see reference 23).
Overall, it appears that C4-dicarboxylate metabolism and regulation in E. coli are complex, and the C4-dicarboxylates are versatile and important substrates under aerobic and anaerobic conditions. Recently, it was shown that in Bacillus subtilis l-malate is coutilized with glucose and that it is a second preferred carbon source (5, 14, 27), and it would be interesting to test whether C4-dicarboxylates are also preferred carbon sources in E. coli.
Acknowledgments
This work was supported by grants from Deutsche Forschungsgemeinschaft and the Innovationsstiftung Rheinland-Pfalz.
We are grateful to Katharina Eich for construction of plasmid pMW383 and to Patrick Scheu for construction of pMW323.
Footnotes
Published ahead of print on 16 March 2010.
REFERENCES
- 1.Abo-Amer, A. E., J. Munn, K. Jackson, M. Aktas, P. Golby, D. J. Kelly, and S. C. Andrews. 2004. DNA interaction and phosphotransfer of the C4-dicarboxylate-responsive DcuS-DcuR two-component regulatory system from Escherichia coli. J. Bacteriol. 186:1879-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asano, Y., M. Ueda, and H. Yamada. 1993. Microbial production of d-malate from maleate. Appl. Environ. Microbiol. 59:1110-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumer, C., A. Kleefeld, D. Lehnen, M. Heintz, U. Dobrindt, G. Nagy, K. Michaelis, L. Emödy, T. Polen, R. Rachel, V. F. Wendisch, and G. Unden. 2005. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology 151:3287-3298. [DOI] [PubMed] [Google Scholar]
- 4.Bongaerts, J., S. Zoske, U. Weidner, and G. Unden. 1995. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol. Microbiol. 16:521-534. [DOI] [PubMed] [Google Scholar]
- 5.Brückner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. [DOI] [PubMed] [Google Scholar]
- 6.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, J., and W. A. Hendrickson. 2008. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J. Biol. Chem. 283:30256-30265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in E. coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, S. J., P. Golby, D. Omrani, S. A. Broad, V. L. Harrington, J. R. Guest, D. J. Kelly, and S. C. Andrews. 1999. Inactivation and regulation of the aerobic C4-dicarboxylate transport (dctA) gene of Escherichia coli. J. Bacteriol. 181:5624-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Easton, J. A., P. Thompson, and M. W. Crowder. 2006. Time-dependent translational response of Escherichia coli to excess Zn(II). J. Biomol. Tech. 17:303-307. [PMC free article] [PubMed] [Google Scholar]
- 11.Engel, P., R. Krämer, and G. Unden. 1994. Transport of C4-dicarboxylates by anaerobically grown Escherichia coli: energetics and mechanism of exchange, uptake and efflux. Eur. J. Biochem. 222:605-614. [DOI] [PubMed] [Google Scholar]
- 12.Giffhorn, F., and A. Kuhn. 1983. Purification and characterization of a bifunctional l-(+)-tartrate dehydrogenase-d-(+)-malate dehydrogenase (decarboxylating) from Rhodopseudomonas sphaeroides Y. J. Bacteriol. 155:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golby, P., S. Davies, D. J. Kelly, J. R. Guest, and S. C. Andrews. 1999. Identification and characterization of a two-component sensor-kinase and response regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli. J. Bacteriol. 181:1238-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Görke, B., and J. Stülke. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613-624. [DOI] [PubMed] [Google Scholar]
- 15.Guest, J. R., J. Green, A. S. Irvine, and S. Spiro. 1996. The FNR modulon and FNR regulated gene expression, p. 317-342. In E. C. C Lin and A. S. Lynch (ed.), Regulation of gene expression in Escherichia coli. Chapman and Hall, New York, NY.
- 16.Gunsalus, R. P. 1992. Control of electron flow in Escherichia coli: coordinated transcription of respiratory pathway genes. J. Bacteriol. 174:7069-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutowski, S. J., and H. Rosenberg. 1975. Succinate uptake and related proton movements in Escherichia coli K-12. Biochem. J. 152:647-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopper, D. J., P. J. Chapman, and S. Dagley. 1968. Enzymic formation of d-(+)-malate. Biochem. J. 110:798-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopper, J. D., J. P. Chapman, and S. Dagley. 1970. Metabolism of l-malate and d-malate by a species of Pseudomonas. J. Bacteriol. 104:1197-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iuchi, S., and E. C. Lin. 1993. Adaptation of Escherichia coli to redox environments by gene expression. Mol. Microbiol. 9:9-15. [DOI] [PubMed] [Google Scholar]
- 21.Janausch, I. G., I. Garcia-Moreno, and G. Unden. 2002. Function of DcuS from Escherichia coli as a fumarate-stimulated histidine protein kinase in vitro. J. Biol. Chem. 277:39809-39814. [DOI] [PubMed] [Google Scholar]
- 22.Janausch, I. G., E. Zientz, Q. H. Tran, A. Kröger, and G. Unden. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39-56. [DOI] [PubMed] [Google Scholar]
- 23.Kadner, R. J., M. D. Island, J. L. Dahl, and C. A. Webber. 1994. A transmembrane signalling complex controls transcription of the Uhp sugar phosphate transport system. Res. Microbiol. 381-387. [DOI] [PubMed]
- 24.Kim, O. B., and G. Unden. 2007. The l-Tartrate/succinate antiporter TtdT (YgjE) of l-tartrate fermentation in Escherichia coli. J. Bacteriol. 189:1597-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, O. B., S. Lux, and G. Unden. 2007. Anaerobic growth of Escherichia coli on d-tartrate is independent of d- or l-tartrate specific transporters and enzymes. Arch. Microbiol. 188:583-589. [DOI] [PubMed] [Google Scholar]
- 26.Kim, O. B., J. Reimann, H. Lukas, U. Schumacher, J. Grimpo, P. Dünnwald, and G. Unden. 2009. Regulation of tartrate metabolism by TtdR and relation to the DcuS-DcuR regulated C4-dicarboxylate metabolism of Escherichia coli. Microbiology 155:3632-3640. [DOI] [PubMed] [Google Scholar]
- 27.Kleijn, R. J., J. M. Buescher, L. LeChat, M. Jules, S. Aymerich, and W. Sauer. 2010. Metabolic fluxes during strong carbon catabolite repression by malate in Bacillus subtilis. J. Chem. Biol. 285:1587-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kneuper, H., I. G. Janausch, V. Vijayan, M. Zweckstetter, V. Bock, C. Griesinger, and G. Unden. 2005. The nature of the stimulus and of the fumarate binding site of the fumarate sensor DcuS of Escherichia coli. J. Biol. Chem. 280:20596-20603. [DOI] [PubMed] [Google Scholar]
- 29.Knichel, W., and F. Radler. 1982. d-Malic enzyme of Pseudomonas fluorescens. Eur. J. Biochem. 123:547-552. [DOI] [PubMed] [Google Scholar]
- 30.Latour, D. J., and J. H. Weiner. 1987. Investigation of Escherichia coli fumarate reductase subunit function using transposon Tn5. J. Gen. Microbiol. 133:597-607. [DOI] [PubMed] [Google Scholar]
- 31.Lehnen, D., C. Blumer, T. Polen, B. Wackwitz, V. F. Wendisch, and G. Unden. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521-532. [DOI] [PubMed] [Google Scholar]
- 32.Martínez-Luque, M., F. Castillo, and R. Blasco. 2001. Assimilation of d-malate by Rhodobacter capsulatus E1F1. Curr. Microbiol. 43:154-157. [DOI] [PubMed] [Google Scholar]
- 33.Miles, J. S., and J. R. Guest. 1987. Molecular genetic aspects of the citric acid cycle of Escherichia coli, p. 45-66. In J. Kay and P. D. J. Weitzman (ed.), Krebs' citric acid cycle. The Biochemical Society, London, UK. [PubMed]
- 34.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 35.Nimmo, H. G. 1987. The tricarboxylic cycle and anaplerotic reactions, p. 156-159. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. ASM Press, Washington, DC.
- 36.Oshima, T., and F. Biville. 2006. Functional identification of ygiP as a positive regulator of the ttdA-ttdB-ygjE operon. Microbiology 152:2129-2135. [DOI] [PubMed] [Google Scholar]
- 37.Rabin, R. S., and V. Stewart. 1993. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 175:3259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reaney, S. K., C. Begg, S. J. Bungard, and J. R. Guest. 1993. Identification of the l-tartratase genes (ttdA and ttdB) of Escherichia coli and evolutionary relationship with the class I fumarase genes. J. Gen. Microbiol. 139:1523-1530. [DOI] [PubMed] [Google Scholar]
- 39.Reed, J. L., T. R. Patel, K. H. Chen, A. R. Joyce, M. K. Applebee, C. D. Herring, O. T. Bui, E. M. Knight, S. S. Fong, and B. O. Palsson. 2006. Systems approach to refining genome annotation. Proc. Natl. Acad. Sci. U. S. A. 103:17480-17484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Sauer, U., and B. J. Eikmanns. 2005. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29:765-794. [DOI] [PubMed] [Google Scholar]
- 42.Sawers, G. 1993. Specific transcriptional requirements for positive regulation of the anaerobically inducible pfl operon by ArcA and FNR. Mol. Microbiol. 10:737-747. [DOI] [PubMed] [Google Scholar]
- 43.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 44.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Stern, J. R., and C. S. Hegre. 1966. Inducible d-malic enzyme in Escherichia coli. Nature 212:1611-1612. [DOI] [PubMed] [Google Scholar]
- 46.Stewart, V. 1993. Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol. Microbiol. 9:425-434. [DOI] [PubMed] [Google Scholar]
- 47.Unden, G., and J. Bongaerts. 1997. Alternative respiratory pathway of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1321:217-234. [DOI] [PubMed] [Google Scholar]
- 48.Unden, G., and A. Kleefeld. 30 July 2004, posting date. Chapter 3.4.5, C4-dicarboxylate degradation in aerobic and anaerobic growth. In R. Curtiss III et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org. [DOI] [PubMed]
- 49.Van der Werf, M. J., W. J. van der Tweel, and S. Hartmans. 1992. Screening for microorganisms producing d-malate from maleate. Appl. Environ. Microbiol. 58:2854-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaughn, R. H., G. L. Marsch, T. C. Stadtmann, and B. C. Cantino. 1946. Decomposition of tartrates by the coliform bacteria. J. Bacteriol. 52:311-325. [PMC free article] [PubMed] [Google Scholar]
- 51.Winkler, H. H. 1971. Kinetics of exogenous induction of the hexose-6-phosphate transport system of Escherichia coli. J. Bacteriol. 107:74-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zientz, E., J. Bongaerts, and G. Unden. 1998. Fumarate regulation of gene expression in Escherichia coli by the DcuSR (dcuSR) two-component regulatory system. J. Bacteriol. 180:5421-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]