Abstract
SigF is an alternative sigma factor that is highly conserved among species of the genus Mycobacterium. In this study we identified the SigF regulon in Mycobacterium smegmatis using whole-genome microarray and promoter consensus analyses. In total, 64 genes in exponential phase and 124 genes in stationary phase are SigF dependent (P < 0.01, >2-fold expression change). Our experimental data reveal the SigF-dependent promoter consensus GTTT-N(15-17)-GGGTA for M. smegmatis, and we propose 130 potential genes under direct control of SigF, of which more than 50% exhibited reduced expression in a ΔsigF strain. We previously reported an increased susceptibility of the ΔsigF strain to heat and oxidative stress, and our expression data indicate a molecular basis for these phenotypes. We observed SigF-dependent expression of several genes purportedly involved in oxidative stress defense, namely, a heme-containing catalase, a manganese-containing catalase, a superoxide dismutase, the starvation-induced DNA-protecting protein MsDps1, and the biosynthesis genes for the carotenoid isorenieratene. Our data suggest that SigF regulates the biosynthesis of the thermoprotectant trehalose, as well as an uptake system for osmoregulatory compounds, and this may explain the increased heat susceptibility of the ΔsigF strain. We identified the regulatory proteins SigH3, PhoP, WhiB1, and WhiB4 as possible genes under direct control of SigF and propose four novel anti-sigma factor antagonists that could be involved in the posttranslational regulation of SigF in M. smegmatis. This study emphasizes the importance of this sigma factor for stationary-phase adaptation and stress response in mycobacteria.
The success of Mycobacterium tuberculosis as a pathogen can be attributed to its capacity to adapt to environmental changes throughout the course of infection. These changes include nutrient deprivation, hypoxia, various exogenous stress conditions, and the intraphagosomal environment. A large cohort of genes that facilitate this adaptation has been identified, and among them are many genes for transcriptional regulators such as sigma factors that modulate gene expression in response to different physiological cues. Sigma factors interact with the RNA polymerase to allow binding to specific promoter sequences and initiation of gene transcription. Mycobacteria harbor sigma factors of only the σ70 family, which fall into four different categories. SigA (group 1) is the essential primary sigma factor in mycobacteria, and SigB (group 2) is its nonessential paralog. SigF (group 3) and extracytoplasmic function (ECF) sigma factors (group 4) are alternative sigma factors, which allow adaptation to a wide range of internal and external stimuli. Alternative sigma factors vary considerably in type and numbers between species, mirroring their different requirements for stress response (15).
Thirteen sigma factors have been identified in M. tuberculosis (23, 29). Eleven of these are classified as alternative sigma factors, and many of them are recognized as virulence determinants. Loss of the alternative sigma factor SigF decreased the virulence of M. tuberculosis in mice (5) and disease-associated tissue damage in mice (13) as well as guinea pigs (20). Loss of SigF also leads to an altered cell wall composition due to a lack of virulence-related sulfolipids (13), and overexpression of sigF has been shown to affect the regulation of other cell wall-associated proteins involved in host-pathogen interaction (40).
SigF was originally thought to be absent in nonpathogenic, fast-growing mycobacteria such as Mycobacterium smegmatis (9). However, it has since become clear that SigF is well conserved among mycobacteria (30, 31) and regulates more than just virulence. While SigF is related to stress response and sporulation sigma factors in other bacteria (8), its role as a stress and stationary-phase sigma factor in M. tuberculosis is under debate (40).
In M. smegmatis, loss of sigF increases susceptibility to oxidative stress, acidic pH, and heat shock (12) and disables the synthesis of protective carotenoids (28). This suggests that SigF mediates a general stress response. However, there is still a paucity of basic knowledge pertaining to the SigF-regulated genes and how SigF fits into the regulatory network of sigma factors. Twenty-seven sigma factors have been proposed for M. smegmatis (35, 38). This is twice the number of sigma factors found in M. tuberculosis and possibly reflects the larger genome size and the more variable environments to which this species is required to adapt. Based on this observation, it has been suggested that the regulatory circuits involving SigF will differ between tuberculous and environmental mycobacteria given the different natures of their environments (31), but data on the regulation of SigF activity in M. smegmatis are lacking.
In this study we report on the SigF regulon of M. smegmatis during exponential- and stationary-phase growth. We propose a new SigF promoter consensus for M. smegmatis based on our experimental data, and we identify novel target genes under the direct control of this sigma factor. We provide a rationale for the phenotypes of the ΔsigF strain observed in previous stress challenge experiments and propose candidate genes involved in the posttranslational regulation of SigF in M. smegmatis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Mycobacterium smegmatis strain mc2155 (33) and an isogenic sigF (MSMEG_1804) deletion strain (12) were routinely grown in batch culture in Luria-Bertani broth supplemented with 0.05% (wt/vol) Tween 80 (LBT) at 37°C on a rotary shaker at 200 rpm. Gentamicin was added to a final concentration of 5 μg ml−1 where required. Culture growth was monitored by measuring the optical density at 600 nm (OD600). Samples were diluted in saline (8.5 g/liter NaCl) to bring the OD600 to below 0.5 when measured in cuvettes with a 1-cm light path length in a Jenway 6300 spectrophotometer. Starter cultures were grown to exponential phase (OD600 = 0.5 ± 0.2) and used immediately to inoculate 100 ml fresh LBT in 1-liter flasks to an initial OD600 of 0.005.
Cell harvest.
Batch cultures were harvested in exponential phase (μ = 0.26 ± 0.04 h−1) and stationary phase (μ = 0.07 ± 0.01 h−1) using cold glycerol-saline (36). In brief, cultures were mixed with 2 volumes of cold glycerol-saline (3:2 [vol/vol]; −20°C) and centrifuged for 20 min at 10,000 × g and −20°C. After centrifugation, the supernatant was decanted and the cells were resuspended in 1 ml glycerol-saline (1:1 [vol/vol]; −20°C), snap-frozen in a dry-ice/ethanol bath, and stored at −80°C.
RNA extraction.
Total RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA) following the manufacturer's recommendations. Frozen samples (−80°C) were centrifuged for 15 min at 13,000 × g and 4°C. Cells were resuspended in 1 ml TRIzol reagent and disrupted using 0.5-ml zirconium beads (0.1-mm diameter) in a Mini-BeadBeater (BioSpec Products, Bartlesville, OK) at 5,000 oscillations per min for three cycles of 30 s. The samples were chilled on ice for 30 s after each cycle. At the end of the extraction procedure, RNA pellets were air dried and redissolved in 50 μl diethyl pyrocarbonate (DEPC)-treated ultrapure water. Remaining DNA was removed with Turbo DNase (Applied Biosystems/Ambion, Austin, TX) following the manufacturer's instructions. RNA quality was assessed by electrophoresis on a standard 1% agarose gel, and RNA quantity was determined with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
Microarray resources.
Glass slide DNA microarrays with 7,736 unique 70-mer oligonucleotides (spotted in triplicates) representing every open reading frame (ORF) of the M. smegmatis mc2155 genome were acquired from the Pathogen Functional Genomics Resource Center (PFGRC) established by NIAID/JCVI (http://pfgrc.jcvi.org). Standard operating procedures (SOP) for RNA labeling and array hybridization as well as layout and annotation files for the microarray were downloaded from the PFGRC website. The open-source free TM4 software suite (www.tm4.org) was used for microarray analysis.
Synthesis/labeling of cDNA.
Mycobacterial RNA was aminoallyl (aa) labeled according to SOP M007 from PFGRC. In brief, cDNA was first reverse transcribed from 5 μg extracted total RNA using 3 μg random primers and SuperScript III reverse transcriptase (both from Invitrogen) with a 25 mM aa-dUTP labeling mix (2:3 aa-dUTP to dTTP). The 5-(3-aminoallyl)-dUTP was purchased from Sigma-Aldrich (St. Louis, MO). Synthesized cDNA was then coupled to either cyanine-3 or cyanine-5 (Cy-3/Cy-5) fluorescent dyes (GE Healthcare Bio-Sciences, Little Chalfont, United Kingdom) for 1 1/2 h. The concentration of cDNA and incorporation of dyes were measured with a NanoDrop ND-1000 spectrophotometer. Labeled probes were mixed and prepared as recommended for immediate hybridization to the microarray.
Microarray hybridization.
The microarrays were hybridized according to SOP M008 from PFGRC except that slides were handled individually in 50-ml conical tubes instead of Coplin jars. In brief, slides were blocked for at least 2 h and washed with filtered (0.22-μm) ultrapure water, followed by a final wash with isopropanol. Wet slides were centrifuged dry for 10 min at 800 × g at room temperature. Slides were then immediately hybridized with the prepared samples and incubated for at least 18 h. After hybridization, slides were washed, centrifuged dry as described above, and immediately scanned. Wash buffers were filtered (0.22 μm) prior to use. Hybridizations comparing wild-type and ΔsigF strains in both exponential and stationary phases were repeated in four biological replicates including dye swaps.
Image acquisition.
Slides were scanned using an Axon GenePix4000B microarray scanner (Molecular Devices, Sunnyvale, CA) at a 10-μm pixel size and autoadjusted photomultiplier (PMT) gain. Fluorescences at 532 nm (Cy3) and 635 nm (Cy5) were measured simultaneously and saved in separate 16-bit grayscale TIFF images, which were then analyzed with the TM4 programs Spotfinder, MIDAS, and MEV.
Data analysis.
Spots were identified with the fixed-circle segmentation method and quantified with 5% top background cutoff in Spotfinder (version 3.1.1). The spot signal intensities were normalized in MIDAS (version 2.19) using total array intensity and the LOWESS algorithm options. The gene expression ratio (n-fold change from ΔsigF strain to wild type) was calculated from the normalized signal intensities and averaged for each set of biological replicates. Ratios were tested for significance (P < 0.05 and P < 0.01) with a one-sample t test in MeV (version 4.3.02).
Quantitative real-time PCR.
The gene expression ratios detected by microarray analysis were confirmed by quantitative real-time PCR (qRT-PCR). Selected genes and primer pairs are listed in Table 1. Gene sequences were retrieved from JCVI/CMR (http://cmr.jcvi.org/tigr-scripts/CMR/GenomePage.cgi?database=gms). Total RNA (1 μg) from stationary-phase batch cultures of M. smegmatis mc2155 wild-type and ΔsigF strains was reverse transcribed with random primers (1 μg) and SuperScript III reverse transcriptase (both from Invitrogen) according to the manufacturer's protocol. Real-time PCR was performed using a SYBR green assay (Invitrogen) and optimized primer concentrations in a 7500 fast real-time PCR system (Applied Biosystems, Foster City, CA). Relative gene expression was determined from calculated threshold cycle (CT) values using MSMEG_2758 (sigA) as an internal normalization standard. n-fold changes were tested for significance with a Student t test.
TABLE 1.
Primer pairs for selected genes for real-time PCR
| Locus | Gene | Primer sequence (5′ → 3′) |
|---|---|---|
| MSMEG_1804 | sigF | GCTCAAGGAACTCCACTTGC (forward) |
| GATGGACAGCGTGTTGTACG (reverse) | ||
| MSMEG_2758 | sigA | GAAGACACCGACCTGGAACT (forward) |
| GACTCTTCCTCGTCCCACAC (reverse) | ||
| MSMEG_2927 | opuCB | TCTGTCGTTCCTCGCCTATC (forward) |
| AAACCGAAGAACACCAGCAT (reverse) | ||
| MSMEG_6213 | mcat | GGCAAGGACGAGATAATCCA (forward) |
| TCGTCGGTGAACTGTTTGAG (reverse) | ||
| MSMEG_6232 | katA | GCAGACCCATCTGGTCAAGT (forward) |
| AGTTCCCATTCCGGGTAGTC (reverse) | ||
| MSMEG_6467 | dps1 | ACAACGATCTGCATCTGACG (forward) |
| GTCACGCTCGACGGAGTAGT (reverse) | ||
| MSMEG_6515 | treS | GGCGACTTCTACGTCTGGAG (forward) |
| CGGGTTGTCGTAGTTGAGGT (reverse) |
Promoter search.
Of 134 SigF-dependent genes (microarray analysis under standard growth conditions; P < 0.01, gene expression ratio r < 0.5), the 400-bp regions immediately 5′ to the annotated start codons were scanned visually for sequences similar to the SigF-dependent promoter upstream of MSMEG_1802 identified in our previous study (12). Such a sequence was found in 49 of these regions. For use as training sets, three separate sets, each containing all 49 promoters, were generated by adjusting the spacing between the −10 and −35 elements to 15 bp, 16 bp, or 17 bp, respectively, by deleting bases before the −10 region or by inserting an “N.” These three sets were then used in separate analyses to create a custom position weight matrix (PWM) for a virtual footprint analysis of the M. smegmatis genome using PRODORIC (24) (http://prodoric.tu-bs.de). Search parameters were adjusted to ensure that a minimum of 80% of the training set promoters were recovered. For the 16-bp and 17-bp spacing sets, the settings were as follows: sensitivity = 0.7, core sensitivity = 0.6, and core size = 5. For the 15-bp spacing set, the settings were as follows: sensitivity = 0.8, core sensitivity = 0.6, and core size = 5. Only hits within 300 bp 5′ of an annotated start codon (JCVI/CMR) were considered. Of promoters identified by more than one of these analyses, the hits with the lower PWM score were removed, leaving a list of 130 genes. To generate a consensus sequence, all 130 identified promoters, plus the 12 promoters of the training set that were not recovered by the virtual footprint analysis, were adjusted to a spacing of 16 bp as before. This set was then used to create a sequence logo of the SigF promoter consensus using the WegLogo tool (7) (http://weblogo.berkeley.edu/logo.cgi).
Microarray data accession number.
All data have been deposited at the Gene Expression Omnibus (GEJO, NCBI) under accession number GSE19145.
RESULTS AND DISCUSSION
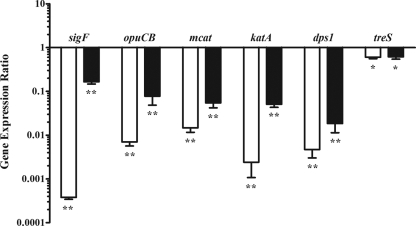
To determine the genes of M. smegmatis mc2155 that are regulated by the alternative sigma factor SigF, a genome-wide gene expression study was conducted for wild-type M. smegmatis mc2155 and an isogenic ΔsigF mutant (12) using microarray analysis. Comparing these two strains, 218 and 239 genes were differentially regulated (P < 0.01) in exponential- and stationary-phase cells, respectively (Table 2). Using a threshold value of a >2-fold difference in gene expression, the data revealed 65 genes in exponential-phase cells and 124 genes in stationary-phase cells under SigF control (Table 3). The majority of these genes showed reduced expression in the ΔsigF strain, in accordance with SigF as an initiator of gene transcription. Only 3 of the 65 genes in exponential phase and 2 of the 124 genes in stationary phase had a higher expression signal in the ΔsigF strain. Of the 124 SigF-dependent genes in stationary-phase cells, 73 genes were exclusively identified in this growth stage, while 51 genes showed reduced expression in exponential and stationary phases. Only 14 genes were unique to the exponential-phase SigF regulon. The entire expression data can be found in Data Set S1 in the supplemental material. To validate the microarray results, we performed real-time PCR on selected genes (Fig. 1). Expression ratios showed the same trend for all genes and were significantly different from 1 (P < 0.01).
TABLE 2.
SigF-dependent gene expression (P < 0.01) in M. smegmatis grown in LB-Tween batch cultures
| Mean growth rate μ, h−1, ± SD (OD600) | SigF-dependent genes (P < 0.01) | No. of genes with expression in ΔsigF straina: |
|
|---|---|---|---|
| Reduced (ratio of <0.5) | Increased (ratio of >2) | ||
| 0.25 ± 0.02 (0.4 ± 0.05) | 218 | 62 (11) | 3 (3) |
| 0.07 ± 0.01 (4.0 ± 0.2) | 239 | 122 (71) | 2 (2) |
The ratio is that of ΔsigF strain to wild-type gene expression. Genes unique to the growth phase are in parentheses.
TABLE 3.
SigF regulon (P < 0.01) in M. smegmatis grown in LB-Tween batch cultures
| Gene category | Expression in ΔsigF strain | Locus | Gene product |
|---|---|---|---|
| SigF-regulated genes exclusive to | Decreaseda | MSMEG_0266 | Arginine decarboxylase |
| stationary phase (μ = 0.07 ± | MSMEG_0267 | Esterase | |
| 0.01 h−1) | MSMEG_0536 | Intracellular protease, PfpI family protein | |
| MSMEG_0600 | Dehydrogenase | ||
| MSMEG_0637 | Iron-sulfur binding oxidoreductase | ||
| MSMEG_0670 | FAD-dependent oxidoreductase | ||
| MSMEG_0671 | S-(Hydroxymethyl)glutathione dehydrogenase | ||
| MSMEG_0684 | Aldehyde oxidase and xanthine dehydrogenase, molybdopterin binding | ||
| MSMEG_0963 | Hypothetical protein | ||
| MSMEG_1112 | Aconitate hydratase, putative | ||
| MSMEG_1315 | Small conductance mechanosensitive ion channel (MscS) family protein | ||
| MSMEG_1358 | Conserved hypothetical protein | ||
| MSMEG_1605 | Phosphate transport system regulatory protein PhoU | ||
| MSMEG_1766 | Conserved hypothetical protein | ||
| MSMEG_1767 | Conserved hypothetical protein | ||
| MSMEG_1768 | Conserved hypothetical protein | ||
| MSMEG_1769 | UsfY protein | ||
| MSMEG_1775 | Cytochrome P450 monooxygenase | ||
| MSMEG_1781 | Hypothetical protein | ||
| MSMEG_1783 | Hypothetical protein | ||
| MSMEG_1787 | RsbW protein | ||
| MSMEG_1792 | Conserved hypothetical protein | ||
| MSMEG_1794 | Dehydrogenase | ||
| MSMEG_1801 | Hypothetical protein | ||
| MSMEG_2160 | Hypothetical protein | ||
| MSMEG_2344 | Dehydrogenase | ||
| MSMEG_2345 | Lycopene cyclase | ||
| MSMEG_2346 | Phytoene synthase | ||
| MSMEG_2347 | Phytoene dehydrogenase | ||
| MSMEG_2376 | Conserved hypothetical protein | ||
| MSMEG_2913 | Hydrolase | ||
| MSMEG_2925 | Permease membrane component | ||
| MSMEG_2926 | Glycine betaine/carnitine/choline transport ATP binding protein OpuCA | ||
| MSMEG_3184 | Malto-oligosyltrehalose trehalohydrolase | ||
| MSMEG_3186 | Glycogen-debranching enzyme GlgX | ||
| MSMEG_3304 | Succinate semialdehyde dehydrogenase | ||
| MSMEG_3311 | Acyl carrier protein | ||
| MSMEG_3418 | Conserved hypothetical protein | ||
| MSMEG_3536 | Sugar transport protein | ||
| MSMEG_3541 | Cytochrome c biogenesis protein transmembrane region | ||
| MSMEG_3543 | Soluble secreted antigen MPT53 | ||
| MSMEG_3560 | Conserved hypothetical protein | ||
| MSMEG_3673 | 4-Alpha-glucanotransferase | ||
| MSMEG_4195 | Conserved hypothetical protein | ||
| MSMEG_4562 | Conserved hypothetical protein | ||
| MSMEG_4993 | Hypothetical protein | ||
| MSMEG_5342 | Conserved hypothetical protein | ||
| MSMEG_5343 | Conserved hypothetical protein | ||
| MSMEG_5400 | Dehydrogenase | ||
| MSMEG_5542 | Transcriptional regulator, HTH_3 family protein | ||
| MSMEG_5590 | Carboxylate-amine ligase Nfa27300 | ||
| MSMEG_5606 | Cytochrome bd-I oxidase subunit II | ||
| MSMEG_5616 | Glyoxalase/bleomycin resistance protein/dioxygenase | ||
| MSMEG_5721 | Acetyl coenzyme A acetyltransferase | ||
| MSMEG_5826 | Pyruvate decarboxylase | ||
| MSMEG_5936 | Conserved hypothetical protein | ||
| MSMEG_6210 | Conserved hypothetical protein | ||
| MSMEG_6213 | Manganese-containing catalase | ||
| MSMEG_6232 | Catalase KatA | ||
| MSMEG_6305 | Conserved hypothetical protein | ||
| MSMEG_6354 | Serine esterase, cutinase family protein | ||
| MSMEG_6355 | Hypothetical protein | ||
| MSMEG_6501 | Hypothetical protein | ||
| MSMEG_6541 | Anti-sigma factor antagonist | ||
| MSMEG_6542 | B12 binding domain protein | ||
| MSMEG_6612 | ATPase, MoxR family protein | ||
| MSMEG_6615 | hypothetical protein | ||
| MSMEG_6616 | S-(Hydroxymethyl)glutathione dehydrogenase | ||
| MSMEG_6663 | C5-O-methyltransferase | ||
| MSMEG_6727 | Amino acid permease-associated region | ||
| MSMEG_6728 | Conserved hypothetical protein | ||
| MSMEG_6751 | Hypothetical protein | ||
| MSMEG_6767 | Mycocerosic acid synthase | ||
| Increasedb | MSMEG_3297 | Transcriptional regulator, CadC | |
| MSMEG_5934 | Conserved hypothetical protein | ||
| SigF-regulated genes exclusive to | Decreased | MSMEG_2343 | Methylesterase |
| exponential phase (μ = | MSMEG_3254 | RDD family protein, putative | |
| 0.25 ± 0.02 h−1) | MSMEG_3443 | Hypothetical protein | |
| MSMEG_5078 | Glucose-1-phosphate adenylyltransferase | ||
| MSMEG_5117 | Proline dehydrogenase | ||
| MSMEG_5119 | 1-Pyrroline-5-carboxylate dehydrogenase | ||
| MSMEG_5188 | CAAX amino protease family protein | ||
| MSMEG_5189 | Oxidoreductase | ||
| MSMEG_5335 | Formamidase | ||
| MSMEG_5336 | Amidate substrates transporter protein | ||
| MSMEG_5337 | Putative regulatory protein, FmdB family | ||
| Increased | MSMEG_2751 | Hypothetical protein | |
| MSMEG_3298 | Response regulator receiver domain protein | ||
| MSMEG_3299 | Putative oxidoreductase | ||
| SigF-regulated genes in both | Decreased | MSMEG_0451 | Oxidoreductase, FAD linked |
| exponential and stationary | MSMEG_0672 | Conserved hypothetical protein | |
| phases | MSMEG_0685 | Oxidoreductase, molybdopterin-binding subunit | |
| MSMEG_0696 | Alanine-rich protein | ||
| MSMEG_0697 | Integral membrane protein | ||
| MSMEG_1076 | Conserved hypothetical protein | ||
| MSMEG_1097 | Glycosyl transferase, group 2 family protein | ||
| MSMEG_1558 | Conserved hypothetical protein | ||
| MSMEG_1758 | Hypothetical protein | ||
| MSMEG_1770 | Conserved hypothetical protein | ||
| MSMEG_1771 | Methylase, putative | ||
| MSMEG_1772 | Conserved hypothetical protein | ||
| MSMEG_1773 | Conserved hypothetical protein | ||
| MSMEG_1774 | Conserved hypothetical protein | ||
| MSMEG_1777 | UsfY protein | ||
| MSMEG_1782 | Oxidoreductase, short-chain dehydrogenase/reductase family | ||
| MSMEG_1788 | Conserved hypothetical protein | ||
| MSMEG_1789 | Conserved hypothetical protein | ||
| MSMEG_1790 | Conserved hypothetical protein | ||
| MSMEG_1802 | ChaB protein | ||
| MSMEG_1804 | RNA polymerase sigma-F factor | ||
| MSMEG_1950 | Conserved hypothetical protein | ||
| MSMEG_1951 | Conserved domain protein | ||
| MSMEG_2112 | Secreted protein | ||
| MSMEG_2115 | Conserved hypothetical protein | ||
| MSMEG_2337 | Isopentenyl-diphosphate delta-isomerase, type 2 | ||
| MSMEG_2415 | Hemerythrin HHE cation binding region | ||
| MSMEG_2830 | ISMsm4, transposase | ||
| MSMEG_2924 | Permease binding protein component | ||
| MSMEG_2927 | ABC transporter, permease protein OpuCB | ||
| MSMEG_2958 | Conserved hypothetical protein | ||
| MSMEG_3022 | Transglycosylase-associated protein | ||
| MSMEG_3185 | Putative maltooligosyl trehalose synthase | ||
| MSMEG_3255 | DoxX subfamily protein, putative | ||
| MSMEG_3419 | Hypothetical protein | ||
| MSMEG_3439 | Hypothetical protein | ||
| MSMEG_4618 | Isochorismatase family protein | ||
| MSMEG_5402 | Dehydrogenase DhgA | ||
| MSMEG_5543 | Hypothetical protein | ||
| MSMEG_5550 | Protein-glutamate methylesterase | ||
| MSMEG_5617 | Immunogenic protein MPT63 | ||
| MSMEG_5722 | Conserved hypothetical protein | ||
| MSMEG_5799 | Nucleoside-diphosphate-sugar epimerase | ||
| MSMEG_6211 | Hypothetical protein | ||
| MSMEG_6212 | Hemerythrin HHE cation binding domain subfamily protein | ||
| MSMEG_6467 | Starvation-induced DNA-protecting protein | ||
| MSMEG_6500 | Conserved hypothetical protein | ||
| MSMEG_6579 | Conserved hypothetical protein | ||
| MSMEG_6610 | Protein of unknown function DUF58, putative | ||
| MSMEG_6665 | Integral membrane protein | ||
| MSMEG_6819 | Conserved domain protein |
ΔsigF strain/wild-type ratio of <0.5.
ΔsigF strain/wild-type ratio of >2.
FIG. 1.
Validation of microarray data with real-time PCR. Gene expression ratios (ΔsigF strain/wild type) of the six genes MSMEG_1804 (sigF), MSMEG_2927 (opuCB), MSMEG_6213 (mcat), MSMEG_6232 (katA), MSMEG_6467 (dps1), and MSMEG_6515 (treS) were determined by both microarray analysis (solid bars) and quantitative real-time PCR (open bars) for stationary-phase (μ = 0.07 ± 0.01) LBT batch cultures of M. smegmatis mc2155 wild-type and ΔsigF strains. Relative gene expression ratios were tested for significance (**, P ≤ 0.01; *, P ≤ 0.05). Results are shown as means ± standard deviations for four (solid bars) or three (open bars) biological replicates.
An improved SigF promoter consensus for M. smegmatis.
Previous studies have identified two genes directly regulated by SigF in M. smegmatis: chaB of the sigF operon (12) and crtI of the carotenoid synthesis gene cluster (28). The promoter motif preceding both genes is identical to the SigF consensus sequence of M. tuberculosis (13). A further 104 SigF-regulated genes have been proposed by an in silico analysis of the M. smegmatis genome based on the SigF consensus sequence of M. tuberculosis (28). However, only 12 genes of this theoretical regulon, in addition to chaB and crtI, proved to be SigF dependent (P < 0.01) in our study, which accounted for fewer than 4% of the identified genes.
We used our microarray data to conduct a promoter motif search tailored to M. smegmatis. Using the SigF-dependent promoter upstream of MSMEG_1802, mapped in our previous study (12), 400 bp upstream of the annotated start codon of 134 SigF-dependent genes (P < 0.01 and r < 0.5 only) were visually checked for sequence similarities. Forty-nine possible candidate genes were found. The spacing between the −10 and −35 elements varied between 14 and 19 bp, but the majority of promoters had a spacing of 15 to 17 bp (Fig. 2A). All 49 promoters were therefore adjusted to 15, 16, or 17 bp and used as separate training sets to create custom position weight matrices for virtual footprint analysis using the PRODORIC tool (24) as described in Materials and Methods. A total of 477 sequence hits were obtained, of which 153 were located within 300 bp 5′ of an annotated start codon. Of promoters identified more than once, the hits with the lower PWM score were removed, which led to a data set of 130 separate promoters. Of these, 62 had a spacing of 15 bp, 49 had a spacing of 16 bp, and 19 had a spacing of 17 bp (Table 4). Seventy percent of promoters were located within 100 bp of the annotated start codon (Fig. 2B).
FIG. 2.
Identification of the SigF promoter consensus. Visually identified promoter motifs upstream of 49 SigF-regulated genes (from microarray analysis) were used for a genome-wide virtual footprint analysis. (A) Promoter spacing variation between the −10 and −35 elements in the training set. (B) Number of promoters sorted into categories according to their distance to the start codon. (C) Derived SigF promoter consensus determined using the WebLogo tool.
TABLE 4.
Genes of M. smegmatis with an identified SigF-dependent promoter consensus
| Expression category | Locus | Gene product | Microarray analysis |
PRODORIC promoter analysis |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Exponential phase |
Stationary phase |
Promoter sequence (5′ → 3′) | Positionb | PWM scorec | |||||
| Ratioa | P value | Ratio | P value | ||||||
| Strong SigF-dependent gene expressione in exponential and/or stationary phase | MSMEG_0266 | Arginine decarboxylase | 0.39 | 0.051 | 0.17 | 2.58E−04 | GTCG-N17-GGGAT | 160 | NF |
| MSMEG_0267 | Esterase | 0.32 | 0.05 | 0.14 | 4.86E−04 | GTTT-N15-GGGTA | 27 | 14.68 | |
| MSMEG_0451 | Oxidoreductase, FAD-linked | 0.12 | 1.12E−03 | 0.11 | 9.94E−05 | GTTC-N19-GGGCC | 49 | NF | |
| MSMEG_0670 | FAD-dependent oxidoreductase | 0.38 | 0.05 | 0.22 | 5.90E−03 | GGTT-N16-GGGTA | 9 | 12.99 | |
| MSMEG_0671 | S-(Hydroxymethyl)glutathione dehydrogenase | 0.29 | 0.049 | 0.07 | 1.96E−07 | GTTT-N15-GGGTA | 47 | 14.64 | |
| MSMEG_0672 | Conserved hypothetical protein | 0.19 | 2.29E−03 | 0.04 | 9.03E−06 | GTTT-N15-GGGTA | 50 | 14.58 | |
| MSMEG_0697 | Integral membrane protein | 0.18 | 4.43E−03 | 0.04 | 1.48E−06 | GTTT-N16-GGGAA | 58 | 13.79 | |
| MSMEG_1076 | Conserved hypothetical protein | 0.05 | 7.61E−06 | 0.03 | 4.16E−06 | GTTT-N16-GGGTA | 50 | 14.28 | |
| MSMEG_1097 | Glycosyl transferase, group 2 family protein | 0.05 | 4.74E−05 | 0.03 | 1.09E−06 | GTGT-N15-GGGTT | 11 | NF | |
| MSMEG_1112 | Aconitate hydratase | 0.36 | 0.06 | 0.19 | 6.42E−03 | GTTT-N16-GGGAA | 8 | 13.84 | |
| MSMEG_1131 | Tryptophan-rich sensory protein | 0.29 | 0.04 | 0.82 | 0.63 | GTGT-N16-GGGTA | 9 | 12.95 | |
| MSMEG_1315 | Small conductance mechanosensitive ion channel family protein | 0.46 | 0.026 | 0.29 | 1.59E−03 | GTTG-N17-GGGTA | 11 | 13.8 | |
| MSMEG_1758 | Hypothetical protein | 0.26 | 5.15E−03 | 0.09 | 1.06E−04 | GTTT-N16-GGGTA | 8 | 14.25 | |
| MSMEG_1766 | Conserved hypothetical protein | 0.57 | 0.1 | 0.15 | 3.66E−04 | GTTT-N16-GGGAA | 32 | 13.75 | |
| MSMEG_1770d | Conserved hypothetical protein | 0.04 | 1.28E−07 | 0.01 | 2.50E−07 | GTTT-N16-GGGCA | 64 | 13.74 | |
| MSMEG_1771 | Methylase, putative | 0.08 | 2.49E−04 | 0.04 | 3.18E−06 | GTTT-N15-GGGTA | 29 | 14.6 | |
| MSMEG_1773 | Conserved hypothetical protein | 0.19 | 9.80E−03 | 0.34 | 2.73E−03 | GTTT-N15-GGGAA | 11 | 14.13 | |
| MSMEG_1774 | Conserved hypothetical protein | 0.1 | 1.06E−03 | 0.03 | 5.55E−06 | GTTT-N16-GGGTA | 64 | 14.31 | |
| MSMEG_1775 | Cytochrome P450 monooxygenase | 0.34 | 0.07 | 0.09 | 1.07E−04 | GTTT-N15-GGGTA | 9 | 14.62 | |
| MSMEG_1777 | UsfY protein | 0.15 | 1.44E−03 | 0.05 | 1.13E−05 | GTTT-N16-GGGTA | 69 | 14.31 | |
| MSMEG_1782 | Oxidoreductase, short chain dehydrogenase/reductase | 0.19 | 9.73E−03 | 0.04 | 7.39E−06 | GTTT-N15-GGGTA | 221 | 14.16 | |
| MSMEG_1787 | RsbW protein | 0.5 | 0.04 | 0.45 | 8.18E−03 | GTTT-N17-GGGTA | 56 | 14.76 | |
| MSMEG_1792 | Conserved hypothetical protein | 0.44 | 0.11 | 0.16 | 5.01E−04 | GGGT-N14-GGGCA | 268 | NF | |
| MSMEG_1794 | Dehydrogenase | 0.31 | 0.047 | 0.09 | 7.03E−05 | GTGT-N17-GGGTA | 15 | 13.28 | |
| MSMEG_1801 | Hypothetical protein | 0.49 | 0.19 | 0.18 | 2.68E−03 | GGTG-N18-GGGAA | 173 | NF | |
| MSMEG_1802 | ChaB protein | 0.24 | 5.03E−03 | 0.1 | 8.96E−05 | GTTT-N16-GGGCA | 63 | 13.6 | |
| MSMEG_2112 | Secreted protein | 0.23 | 4.48E−03 | 0.31 | 3.52E−03 | GTTT-N15-GGGTA | 24 | 14.75 | |
| MSMEG_2337 | Isopentenyl-diphosphate delta-isomerase, type 2 | 0.21 | 3.50E−03 | 0.16 | 3.83E−04 | GGTG-N15-GGGTA | 67 | NF | |
| MSMEG_2347d | Phytoene dehydrogenase | 0.49 | 0.06 | 0.23 | 9.98E−04 | GTTT-N16-GGGTA | 97 | 14.4 | |
| MSMEG_2415 | Hemerythrin HHE cation binding region | 0.25 | 3.78E−03 | 0.08 | 1.12E−05 | GTTG-N15-GGGTA | 61 | 13.42 | |
| MSMEG_2830 | ISMsm4, transposase | 0.41 | 8.50E−03 | 0.54 | 5.29E−03 | GGTT-N16-GGGTG | 209 | NF | |
| MSMEG_2913 | Hydrolase | 0.36 | 0.042 | 0.1 | 1.36E−04 | GTTT-N15-GGGTA | 3 | 14.64 | |
| MSMEG_2927d | ABC transporter, permease protein OpuCB | 0.17 | 6.18E−04 | 0.11 | 3.14E−04 | GTTT-N16-GGGTA | 39 | 14.23 | |
| MSMEG_2958 | Conserved hypothetical protein | 0.17 | 3.78E−03 | 0.03 | 2.48E−06 | GTTC-N15-GGGTA | 24 | 13.34 | |
| MSMEG_3022d | Transglycosylase-associated protein | 0.05 | 5.77E−06 | 0.05 | 1.65E−05 | GTTT-N16-GGGTA | 30 | 14.34 | |
| MSMEG_3082 | Soul heme binding protein | 0.72 | 7.33E−03 | 0.44 | 5.67E−03 | GCTT-N16-GGGTA | 67 | 12.94 | |
| MSMEG_3141d | Conserved domain protein | 0.43 | 0.033 | 0.4 | 0.022 | GTGT-N16-GGGTA | 29 | 12.94 | |
| MSMEG_3255 | DoxX subfamily, putative | 0.02 | 6.01E−08 | 0.01 | 8.94E−08 | GTTT-N15-GGGAA | 36 | 14.09 | |
| MSMEG_3304 | Succinate semialdehyde dehydrogenase | 0.44 | 0.06 | 0.08 | 1.73E−07 | GTGT-N15-GGGTA | 25 | 13.24 | |
| MSMEG_3439 | Hypothetical protein | 0.15 | 4.30E−03 | 0.11 | 3.21E−04 | GTTT-N15-CGGTA | 59 | NF | |
| MSMEG_3443 | Hypothetical protein | 0.41 | 1.21E−03 | 0.47 | 0.026 | GTTT-N15-GGGAT | 45 | NF | |
| MSMEG_3536 | Sugar transport protein | 0.56 | 0.07 | 0.29 | 1.33E−05 | GTGG-N16-GGGTA | 134 | NF | |
| MSMEG_3543 | Soluble secreted antigen MPT53 | 0.28 | 0.015 | 0.09 | 1.14E−04 | GTTT-N16-GGGAA | 138 | 13.68 | |
| MSMEG_3673 | 4-Alpha-glucanotransferase | 0.55 | 0.08 | 0.38 | 3.26E−03 | GTTT-N16-GGGCA | 195 | 13.61 | |
| MSMEG_4195 | Conserved hypothetical protein | 0.58 | 0.017 | 0.42 | 9.23E−03 | GTTT-N15-GGGTA | 60 | 14.07 | |
| MSMEG_5189 | Oxidoreductase | 0.31 | 6.13E−03 | 0.42 | 0.013 | GGTT-N16-GGGTA | 25 | 12.93 | |
| MSMEG_5343 | Conserved hypothetical protein | 0.38 | 0.06 | 0.2 | 1.35E−03 | GTTT-N16-GGCTA | 35 | NF | |
| MSMEG_5402 | Dehydrogenase DhgA | 0.21 | 3.45E−03 | 0.18 | 9.24E−04 | GTTT-N15-GGGTA | 8 | 14.69 | |
| MSMEG_5543 | Hypothetical protein | 0.03 | 7.87E−07 | 0.02 | 9.61E−07 | GTTT-N17-GGGTA | 77 | 15.01 | |
| MSMEG_5550 | Protein-glutamate methylesterase | 0.29 | 3.07E−03 | 0.35 | 9.26E−03 | GTTT-N15-GGGTA | 20 | 14.58 | |
| MSMEG_5617 | Immunogenic protein MPT63 | 0.02 | 5.65E−11 | 0.04 | 4.26E−06 | GTTT-N15-GGGTA | 70 | 14.65 | |
| MSMEG_6211 | Hypothetical protein | 0.13 | 1.15E−03 | 0.06 | 4.79E−05 | GGTT-N15-GGGTA | 9 | 13.19 | |
| MSMEG_6212 | Hemerythrin HHE cation binding domain subfamily | 0.05 | 2.87E−05 | 0.02 | 3.65E−07 | GTTT-N15-GGGTA | 51 | 14.64 | |
| MSMEG_6213 | Manganese-containing catalase | 0.31 | 0.039 | 0.06 | 1.20E−05 | GTTT-N15-GGGTA | 40 | 14.57 | |
| MSMEG_6232 | Catalase KatA | 0.3 | 0.049 | 0.05 | 1.93E−06 | GTTT-N16-GGGAA | 67 | 13.78 | |
| MSMEG_6305 | Conserved hypothetical protein | 0.31 | 0.028 | 0.12 | 1.11E−04 | GTTT-N16-GGGCA | 8 | 13.55 | |
| MSMEG_6467 | Starvation-induced DNA-protecting protein | 0.04 | 2.65E−05 | 0.03 | 3.62E−06 | GTTC-N16-GGGCA | 100 | NF | |
| MSMEG_6541 | Anti-sigma factor antagonist | 0.8 | 0.033 | 0.5 | 2.45E−03 | GTTT-N15-GGGTA | 282 | 14.54 | |
| MSMEG_6665 | Integral membrane protein | 0.18 | 3.16E−03 | 0.13 | 1.88E−04 | GTTT-N15-GGGAA | 8 | 14.07 | |
| MSMEG_6751 | Hypothetical protein | 0.59 | 0.08 | 0.22 | 3.54E−04 | GGTT-N15-GGGTA | 9 | 13.17 | |
| MSMEG_6768 | Halogenase | 0.66 | 0.1 | 0.37 | 0.047 | GCTT-N16-GGGTA | 9 | 12.88 | |
| SigF-dependent gene expressionf in exponential and/or stationary phase | MSMEG_0686 | Oxidoreductase | 0.85 | 0.019 | 0.72 | 0.013 | GTTT-N15-GGGTA | 8 | 14.65 |
| MSMEG_1204d | 3-Oxoacyl-[acyl-carrier-protein] synthase 2 | 0.89 | 0.03 | 1.08 | 0.08 | GTTC-N15-GGGTA | 128 | 13.27 | |
| MSMEG_1360d | Endonuclease/exonuclease/phosphatase | 0.92 | 0.19 | 0.75 | 0.032 | GTGT-N15-GGGTA | 14 | 13.25 | |
| MSMEG_1477d | Major facilitator superfamily | 0.74 | 0.1 | 0.61 | 0.01 | GTGT-N16-GGGTA | 122 | 12.93 | |
| MSMEG_1807 | Acetyl-/propionyl-coenzyme A carboxylase alpha chain | 0.98 | 0.76 | 0.8 | 2.16E−03 | GGTT-N17-GGGTA | 294 | 13.33 | |
| MSMEG_1848 | Formate dehydrogenase-O, major subunit | 0.6 | 0.013 | 0.57 | 0.42 | GGTT-N15-GGGTA | 8 | 13.22 | |
| MSMEG_1853 | Na+/H+ antiporter NhaA | 0.69 | 0.026 | 0.66 | 0.023 | GTTT-N15-GGGTA | 99 | 14.73 | |
| MSMEG_2775 | Na+/H+ antiporter NhaA | 0.94 | 0.035 | 1.02 | 0.84 | GTTT-N16-GGGCA | 183 | 13.56 | |
| MSMEG_3046 | Carbamoyl-phosphate synthase, small subunit | 1.08 | 0.6 | 0.81 | 4.42E−03 | GTTT-N16-GGGAA | 206 | 13.72 | |
| MSMEG_3312 | Hemerythrin HHE cation binding domain subfamily | 0.74 | 0.08 | 0.56 | 0.025 | GTTT-N15-GGGTA | 26 | 14.58 | |
| MSMEG_3563d | Drug transporter | 1.14 | 0.83 | 0.61 | 5.64E−03 | GTTT-N16-GGGTA | 114 | 14.21 | |
| MSMEG_3621 | NADH dehydrogenase | 0.96 | 0.44 | 0.82 | 0.025 | GTTG-N15-GGGTA | 98 | 13.36 | |
| MSMEG_3689d | Sodium:solute symporter | 1.06 | 0.037 | 0.96 | 0.36 | GTTT-N16-GGGAA | 96 | 13.72 | |
| MSMEG_4831 | Transcriptional regulator, TetR family protein | 1.32 | 0.018 | 1.18 | 0.35 | GTTG-N16-GGGTA | 232 | 12.98 | |
| MSMEG_4918 | 1,4-Alpha-glucan branching enzyme | 0.6 | 0.027 | 0.52 | 0.022 | GGTT-N15-GGGTA | 172 | 13.2 | |
| MSMEG_5328 | Conserved hypothetical protein | 0.7 | 0.013 | 0.61 | 0.05 | GTTT-N16-GGGAA | 48 | 13.72 | |
| MSMEG_5554 | Antar domain protein | 0.9 | 0.3 | 1.11 | 9.33E−03 | GTTG-N15-GGGTA | 231 | 13.33 | |
| MSMEG_5661 | ABC transporter ATP binding protein | 1.1 | 0.81 | 0.64 | 0.028 | GTTT-N16-GGGAA | 39 | 13.7 | |
| MSMEG_5710 | Hypothetical protein | 0.9 | 0.015 | 0.93 | 0.43 | GTTT-N16-GGGCA | 211 | 12.99 | |
| MSMEG_6515d | Trehalose synthase | 0.68 | 3.70E−03 | 0.63 | 0.018 | GTGT-N16-GGGTA | 10 | 12.88 | |
| MSMEG_6676 | Probable conserved transmembrane protein | 0.54 | 9.69E−03 | 0.68 | 0.1 | GTTC-N16-GGGTA | 162 | 13.12 | |
| MSMEG_6739 | Hypothetical protein | 0.79 | 0.06 | 0.62 | 0.03 | GTTT-N16-GGGTA | 63 | 13.63 | |
| MSMEG_6822 | Beta-lactamase | 0.79 | 9.46E−03 | 0.55 | 0.039 | GTTT-N16-GGGTA | 46 | 14.2 | |
| No significant SigF-dependent gene expressiong observed under standard growth conditions | MSMEG_0191 | BadF/BadG/BcrA/BcrD ATPase family | 1.32 | 0.54 | 1.08 | 0.53 | GTTT-N17-GGGAA | 64 | 14.21 |
| MSMEG_0362 | Amidohydrolase 2 | 0.97 | 0.55 | 1.18 | 0.22 | GTTC-N17-GGGTA | 69 | 13.43 | |
| MSMEG_0430 | ISMsm4, transposase | No data | No data | GTTT-N16-GGGAA | 155 | 13.75 | |||
| MSMEG_0522 | pp24 protein | 1.01 | 0.86 | 0.78 | 0.09 | GTTT-N15-GGGTA | 34 | 14.61 | |
| MSMEG_0681 | P450 heme-thiolate protein | 1.03 | 0.56 | 1 | 0.99 | GTTT-N16-GGGCA | 45 | 13.02 | |
| MSMEG_0849 | Oxygenase | 1.26 | 0.6 | 0.99 | 0.62 | GTGT-N17-GGGTA | 102 | 13.29 | |
| MSMEG_1055 | Hexapeptide transferase family protein | No data | No data | GTTG-N17-GGGTA | 37 | 13.45 | |||
| MSMEG_1145 | Virulence factor Mce family protein | 0.97 | 0.22 | 1.15 | 0.43 | GTTT-N15-GGGGA | 214 | 12.9 | |
| MSMEG_1222 | ISMsm6, transposase | 1.07 | 0.77 | 1.09 | 0.36 | GTGT-N15-GGGTA | 8 | 13.11 | |
| MSMEG_1295 | Transthyretin | 0.76 | 0.06 | 0.8 | 0.08 | GTGT-N16-GGGTA | 160 | 12.86 | |
| MSMEG_1332 | Conserved hypothetical protein | 1.03 | 0.55 | 1.29 | 0.12 | GTTT-N17-GGGCA | 161 | 14.24 | |
| MSMEG_1435 | Ribosomal protein S10 | 1.01 | 0.94 | 0.86 | 0.5 | GTTC-N15-GGGTA | 257 | 13.24 | |
| MSMEG_1440 | Ribosomal protein S19 | 65.48 | 0.39 | 0.82 | 0.29 | GTGT-N15-GGGTA | 354 | 13.17 | |
| MSMEG_1698 | Putative ammonia monooxygenase superfamily | 1.93 | 0.33 | 1 | 0.96 | GTTT-N15-GGGTA | 8 | 14.54 | |
| MSMEG_1742 | Oxidoreductase | 0.73 | 0.19 | 0.9 | 0.08 | GTTT-N15-GGGTA | 89 | 14.63 | |
| MSMEG_1784 | Type I topoisomerase | 0.9 | 0.22 | 0.85 | 0.12 | GTGT-N16-GGGTA | 183 | 12.91 | |
| MSMEG_1844 | Conserved hypothetical protein | 0.85 | 0.35 | 0.88 | 0.44 | GTTT-N15-GGGTA | 24 | 14.6 | |
| MSMEG_1919 | Transcription factor WhiB | 0.85 | 0.08 | 0.86 | 0.37 | GCTT-N16-GGGTA | 199 | 12.95 | |
| MSMEG_2200d | Formyltetrahydrofolate deformylase | 1 | 0.91 | 0.97 | 0.71 | GTTT-N15-GGGTA | 237 | 14.6 | |
| MSMEG_2335 | Hexapeptide transferase family protein | 1.02 | 0.53 | 1.08 | 0.1 | GTTG-N17-GGGTA | 37 | 13.45 | |
| MSMEG_2425 | Ammonium transporter | 1.33 | 0.22 | 1.02 | 0.76 | GTTC-N15-GGGTA | 238 | 13.25 | |
| MSMEG_2466 | Glutaryl coenzyme A dehydrogenase | 1.17 | 0.17 | 0.96 | 0.79 | GTTC-N17-GGGTA | 174 | 13.44 | |
| MSMEG_2780 | Uroporphyrinogen decarboxylase | 1.05 | 0.6 | 1.02 | 0.71 | GTTT-N15-GGGCA | 92 | 13.91 | |
| MSMEG_2804 | Two-component system sensor kinase | 0.92 | 0.07 | 1.13 | 0.18 | GTTG-N16-GGGTA | 39 | 13.13 | |
| MSMEG_2837 | Nitrate reductase NarB | 0.86 | 0.2 | 0.8 | 0.73 | GTTT-N16-GGGTA | 42 | 14.26 | |
| MSMEG_2938 | Acyl coenzyme A thioesterase II | 1.04 | 0.49 | 1.18 | 0.35 | GTCT-N15-GGGTA | 60 | 13.09 | |
| MSMEG_3026 | Conserved hypothetical protein | 1.12 | 0.08 | 1 | 1 | GTTC-N15-GGGTA | 51 | 13.35 | |
| MSMEG_3205 | Histidinol dehydrogenase | 1.13 | 0.07 | 1.13 | 0.47 | GCTT-N15-GGGTA | 201 | 13.15 | |
| MSMEG_3289 | gp61 protein | 0.82 | 0.23 | 0.68 | 0.12 | GTTT-N15-GGGTA | 29 | 14.68 | |
| MSMEG_3610 | Conserved hypothetical protein | 1.03 | 0.26 | 1.29 | 0.13 | GTTG-N15-GGGTA | 73 | 13.29 | |
| MSMEG_3611 | d-Xylulose 5-phosphate/d-fructose 6-phosphate phosphoketolase | 1.01 | 0.97 | 0.73 | 0.05 | GGTT-N15-GGGTA | 68 | 13.29 | |
| MSMEG_3672 | Transporter, small multidrug resistance (SMR) family | 1.12 | 0.41 | 1 | 0.96 | GTTC-N17-GGGTA | 62 | 13.43 | |
| MSMEG_3811 | Universal stress protein family, putative | 0.85 | 0.21 | 1.15 | 0.26 | GTTG-N15-GGGTA | 1 | 13.38 | |
| MSMEG_3822 | Regulatory protein GntR, HTH | 1.01 | 0.92 | 1.08 | 0.63 | GTCT-N15-GGGTA | 28 | 13.22 | |
| MSMEG_3918 | Hypothetical protein | 0.97 | 0.58 | 1.19 | 0.13 | GTTT-N17-GGGTA | 75 | 14.57 | |
| MSMEG_4070d | Transcriptional regulator, TetR family, putative | 0.98 | 0.87 | 1.18 | 0.35 | GTGT-N16-GGGTA | 69 | 12.94 | |
| MSMEG_4232 | UDP-N-acetylmuramoylalanyl-d-glutamate-2,6-diaminopimelate ligase | 1.16 | 0.06 | 1.1 | 0.32 | GTCT-N16-GGGTA | 232 | 12.87 | |
| MSMEG_4270 | Adenosine kinase | 1.1 | 0.14 | 1.2 | 0.13 | GTTG-N16-GGGTA | 246 | 13.08 | |
| MSMEG_4405 | Putative ECF sigma factor RpoE1 | 1.03 | 0.68 | 0.99 | 0.94 | GCTT-N15-GGGTA | 234 | 13.1 | |
| MSMEG_4427 | Transmembrane efflux pump | 1.11 | 0.47 | 1.15 | 0.35 | GTTC-N15-GGGAA | 279 | 12.87 | |
| MSMEG_4441 | Cupin domain protein | 1.58 | 0.35 | 1 | 0.99 | GTCT-N17-GGGTA | 147 | 13.3 | |
| MSMEG_4633 | Peptidase S9, prolyl oligopeptidase | 1.33 | 0.12 | 1.04 | 0.81 | GTTT-N16-GGGTA | 95 | 14.16 | |
| MSMEG_4707 | Nonheme bromoperoxidase BPO-A2 | 0.47 | 0.06 | 0.9 | 0.87 | GTTT-N15-GGGTA | 35 | 13.94 | |
| MSMEG_4737 | Conserved hypothetical protein | 1.1 | 0.45 | 1.27 | 0.3 | GTTT-N15-GGGCA | 97 | 13.39 | |
| MSMEG_5011 | Hypothetical protein | 1.06 | 0.53 | 1.18 | 0.19 | GGTT-N15-GGGTA | 42 | 13.22 | |
| MSMEG_5376 | Conserved hypothetical protein | 1.59 | 0.28 | 1.3 | 0.23 | GTGT-N17-GGGTA | 99 | 13.34 | |
| MSMEG_5434 | Hypothetical protein | 0.95 | 0.61 | 0.85 | 0.57 | GGTT-N15-GGGTA | 119 | 13.16 | |
| MSMEG_5499 | Conserved hypothetical protein | 1.26 | 0.32 | 1.46 | 0.35 | GTTT-N16-GGGCA | 197 | 13.53 | |
| MSMEG_5559 | Metabolite/sugar transport protein | 0.87 | 0.43 | 0.84 | 0.08 | GTTT-N16-GGGTA | 39 | 14.12 | |
| MSMEG_5580d | Ku protein | 1 | 0.98 | 1.15 | 0.18 | GTTT-N16-GGGTA | 9 | 13.67 | |
| MSMEG_5754 | gp41 protein | 1.02 | 0.34 | 1 | 0.98 | GTTG-N15-GGGAA | 301 | 12.87 | |
| MSMEG_5773 | Fatty acid desaturase | 0.97 | 0.49 | 1.09 | 0.32 | GTGT-N17-GGGTA | 177 | 13.63 | |
| MSMEG_5872 | DNA binding response regulator PhoP | 1 | 0.96 | 1.2 | 0.05 | GTTT-N17-GGGTA | 71 | 14.75 | |
| MSMEG_5907 | Acyl coenzyme A dehydrogenase | 1.09 | 0.29 | 1.12 | 0.28 | GGTT-N15-GGGTA | 181 | 13.21 | |
| MSMEG_6091 | Negative regulator of genetic competence ClpC/MecB | 1.01 | 0.82 | 1.12 | 0.18 | GTTG-N15-GGGTA | 136 | 13.42 | |
| MSMEG_6199 | Transcription factor WhiB | 1.23 | 0.1 | 1.29 | 0.19 | GCTT-N15-GGGTA | 23 | 13.14 | |
| MSMEG_6427 | Superoxide dismutase (Mn) | 0.91 | 0.24 | 1.02 | 0.45 | GGTT-N17-GGGTA | 120 | 13.3 | |
| MSMEG_6847 | Conserved hypothetical protein | 0.93 | 0.49 | 1.17 | 0.22 | GTTG-N15-GGGAA | 280 | 12.84 | |
ΔsigF strain/wild-type gene expression ratio.
Base pairs between end of −10 promoter element and annotated start codon.
PWM, position weight matrix. NF, not found.
SigF promoter site previously proposed by Provvedi et al. (28).
P < 0.05, >2-fold difference between wild type and ΔsigF strain.
P < 0.05.
P > 0.05.
A position weight matrix of the identified promoters was created and a sequence logo for the resulting SigF consensus generated using the WebLogo tool (7) as described in Materials and Methods. This consensus for SigF-dependent promoters in M. smegmatis was identified as GTTT-N(15-17)-GGGTA (Fig. 2C).
Of the 130 genes identified here, 20 had previously been proposed by Provvedi and colleagues (28) to be directly recognized by SigF, but for 6 of these the predicted promoter site differs from the site predicted by our analysis. Our consensus is supported by experimental data: 72 of the 130 genes show a significant reduction in expression (P < 0.05) in the ΔsigF strain in exponential- and/or stationary-phase cells. The remaining 58 genes carry the identified SigF promoter consensus but were not differentially expressed. They are most likely part of specific SigF-regulated stress response regulons or could represent false-positive hits.
SigF regulates genes with purported roles in oxidative stress response and pigment production.
A phenotypic characteristic well established for the M. smegmatis ΔsigF strain is its pronounced sensitivity to hydrogen peroxide (12, 28). The protection against reactive oxygen species in pathogenic mycobacteria has attracted much attention due to its implication in survival within the host. The main detoxifying enzymes, catalase-peroxidase, KatG, and the alkyl hydroperoxide reductase AhpC, are conserved and well studied across the genus (16). However, none of these enzymes appear to be involved in the SigF-mediated hydrogen peroxide resistance (12). Accordingly, the katG and ahpC genes were SigF independent in the present study.
Our analysis revealed a number of alternative genes that could play a role in oxidative stress resistance in M. smegmatis. SigF regulates the expression of two potential H2O2-detoxifying enzymes: the heme-containing catalase KatA (MSMEG_6232) and a manganese-containing catalase (MSMEG_6213). Expression of both genes was 20-fold decreased in stationary-phase cells (P < 0.01) and 3-fold decreased in exponential-phase cells (P < 0.05) in the ΔsigF strain relative to the wild type. Neither of these catalases is found in other mycobacteria, except for a homolog of MSMEG_6213 in M. avium. Both enzymes supply M. smegmatis with an alternative route of hydrogen peroxide degradation, which is not available to other mycobacteria. Additionally, we found the SigF promoter upstream of sodA (MSMEG_6427), encoding a superoxide dismutase, which is highly conserved in mycobacteria.
MSMEG_6467, encoding a probable starvation-induced DNA binding protein, MsDps1, exhibited reduced expression in the ΔsigF strain in both exponential phase (r = 0.04, P < 0.01) and stationary phase (r = 0.03, P < 0.01). Dps proteins have been linked with oxidative stress resistance in bacteria (1). MsDps1 was first identified in carbon-starved M. smegmatis cultures (17) and is preceded by promoter motifs recognized by the sigma factors SigF and SigH (6). A homolog of this gene can be found in other environmental mycobacteria (e.g., M. avium, M. avium paratuberculosis, or M. kansasii). In B. subtilis stress-induced production of Dps is controlled by SigB, which is the functional equivalent to mycobacterial SigF (2).
Carotenoids are able to scavenge reactive oxygen species (ROS) (42). M. smegmatis produces the yellow carotenoid isorenieratene under light exposure and nutrient starvation (28). The synthesis of this carotenoid was shown to be SigF dependent in M. smegmatis ATCC 607, and the authors suggested that this is a unique feature of that strain (28). However, our study reveals that strain mc2155 also produces this carotenoid (Fig. 3) and that the expression of the corresponding biosynthesis genes MSMEG_2343 to MSMEG_2347 is SigF dependent (Table 3). All genes of the cluster showed decreased expression in the ΔsigF strain in both growth stages (see Data Set S1 in the supplemental material). Supporting our expression data, a difference in pigmentation was observed for the mc2155 wild-type and ΔsigF strains when grown on LBT agar plates under illumination (Fig. 3). The wild-type colonies developed a distinct yellow color over the course of a week, whereas the ΔsigF strain retained its white color (Fig. 3). Complementation of the ΔsigF strain restored the original phenotype (data not shown).
FIG. 3.
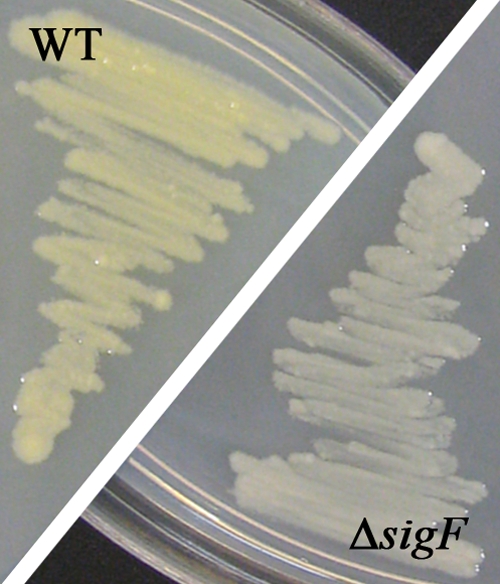
SigF-dependent pigmentation of bacterial colonies. M. smegmatis mc2155 wild-type (WT) and ΔsigF deletion strains were grown on LBT agar under standard fluorescent light at 37°C for 5 days.
Trehalose biosynthesis, osmoprotection, and heat stress.
The microarray data suggested that seven glycosidases involved in the metabolism of trehalose and glycogen are under SigF control. Trehalose is found in a number of bacteria, where it is usually accumulated as an osmoprotectant or stored as an additional carbon source in response to stress (3, 11). In mycobacteria, trehalose is essential for growth (25, 41), and trehalose-containing glycolipids are components of their waxy, highly impenetrable cell wall (3, 11). Three different trehalose biosynthesis pathways (OtsAB, TreYZ, and TreS) have been characterized in M. tuberculosis (10) and in M. smegmatis (41). Two of these systems, TreS (MSMEG_6515) and TreYZ (MSMEG_3186-MSMEG_3185), showed reduced expression in the ΔsigF strain, and our promoter analysis further identified the TreS pathway, but not the TreYZ pathway, to be under direct control of SigF. Two genes encoding the glycogen-debranching enzyme GlgX, MSMEG_3186 (in an operon with TreYZ) and MSMEG_6507, exhibited lower expression in the ΔsigF strain. Glycogen is converted to trehalose by trehalose synthase (TreS). The genes of the OtsA-OtsB pathway were SigF independent in both exponential- and stationary-phase cells (see Data Set S1 in the supplemental material). Trehalose-containing glycolipids are important for cell wall integrity (26), and it has been shown that trehalose synthesis is a prerequisite for the survival of M. smegmatis at elevated temperatures (41). Considering this, the heat-sensitive phenotype that we observed previously for the ΔsigF strain (12) could be caused by its decreased ability to synthesize trehalose as a thermoprotectant.
Furthermore, we could identify a potential system for the uptake of osmoregulatory compounds under direct control of SigF. The genes MSMEG_2927 to MSMEG_2924 are annotated as components of an ATP-binding cassette (ABC) transporter homologous to the OpuC transporter of Bacillus subtilis for the uptake of glycine betaine/l-proline/carnitine/choline, which is controlled by the functionally equivalent sigma factor, SigB (18, 19, 37).
Potential SigF-dependent regulators.
Our promoter analysis predicted a SigF-dependent promoter sequence upstream of sigH3 (MSMEG_0573), whiB1 (MSMEG_1919), whiB4 (MSMEG_6199), and phoP (MSMEG_5872). SigH and its paralogs have been shown to be upregulated under heat and oxidative stress in M. smegmatis (32). WhiB proteins are regulatory proteins unique to actinomycetes (34). In M. tuberculosis expression of whiB4 has been observed at elevated temperatures (14), and WhiB1 has been similarly linked with heat and oxidative stress resistance in Corynebacterium glutamicum (21). Inactivation of phoP in M. tuberculosis led to an altered cell envelope composition and stress responses in vitro, as well as an attenuation of the pathogen in vivo (39). Direct control of phoP by SigF has been predicted previously (28), but the sequence motif we identified here is located at a different site.
Regulation of SigF. (i) Expression of the genomic region surrounding sigF.
In M. smegmatis, sigF is part of an operon with its anti-sigma factor RsbW encoded upstream (designated UsfX in M. tuberculosis) and a ChaB family protein encoded upstream of rsbW. This arrangement of the sigF gene with its anti-sigma factor is conserved in mycobacteria (30). The microarray analysis showed a reduced expression of chaB in the ΔsigF strain at both time points (r < 0.24, P < 0.005), whereas rsbW was less compromised (r < 0.7, P < 0.02). This result is in accordance with our previous study, where we reported that sigF is transcribed from two different promoters: a SigF-dependent promoter preceding chaB and a SigF-independent promoter preceding rsbW (12). The lack of expression from the promoter upstream of chaB would have a more pronounced effect on chaB expression than on rsbW expression, as the latter is also transcribed from the SigF-independent promoter.
MSMEG_1806 (encoding a conserved hypothetical protein), directly downstream of sigF, was the only gene in the study with a strong increase in expression in the ΔsigF strain for both exponential- and stationary-phase cells (r = 12.9 and r = 21.2, respectively; P < 0.01). Most likely, the upregulation of MSMEG_1806 is due to a polar effect of the inserted gentamicin cassette (aacC-1) into the adjacent sigF gene. However, all phenotypic characteristics of the ΔsigF strain, which were observed in previous growth and stress challenge experiments in our laboratory (12), were reversible by complementation of the deleted sigF gene, rendering the increased expression of MSMEG_1806 without a noticeable impact.
(ii) Posttranslational regulation of SigF.
There is little evidence of transcriptional regulation of the rsbW-sigF operon in M. smegmatis under standard growth conditions and for most stress conditions applied in vitro (12, 31). It therefore appears likely that SigF is regulated mainly at the posttranslational level. In M. tuberculosis, posttranslational regulation of SigF is highly complex, involving the anti-sigma factor UsfX as well as five anti-sigma factor antagonists, RsfA, RsfB, Rv1364c, Rv1904, and Rv2638 (4, 27). The latter are not part of the usfX-sigF operon but are dispersed across the genome. To date no anti-sigma factor antagonists have been described for SigF in M. smegmatis.
Close inspection of our microarray data to identify potential candidates for the regulatory cascade of SigF revealed a region immediately preceding the SigF locus, where 20 of the 29 genes from MSMEG_1766 to MSMEG_1794 were affected by the sigF deletion (r < 0.5 and P < 0.01 in stationary phase). Furthermore, 11 of these genes were found to contain a SigF promoter. The region contains an anti-sigma factor (MSMEG_1787) with similarity to M. smegmatis RsbW (43% identities) and three UsfY proteins. In M. tuberculosis, the usfY gene is part of the sigF locus (8), but no function has been assigned to it. The close proximity of usfY genes to sigF in both M. smegmatis and M. tuberculosis indicates that the protein is involved in the control of SigF activity, although experimental evidence is missing to date.
Control of SigF activity in M. tuberculosis involves five anti-sigma factor antagonists. A SMART (Simple Modular Architecture Research Tool) (22) search revealed that these proteins share a STAS domain (sulfate transporter and anti-sigma factor antagonist domain). We identified four proteins with such a domain in M. smegmatis, either by homology to the M. tuberculosis anti-sigma factor antagonist RsfA (MSMEG_1786) or RsfB (MSMEG_6127) or by their annotation as STAS domain proteins (MSMEG_0586 and MSMEG_5551). A fifth STAS domain protein, MSMEG_6541, was identified from the microarray data, which showed reduced expression in the ΔsigF strain in stationary phase (r < 0.5, P < 0.01) and possesses a SigF-dependent promoter. None of the other four STAS domain proteins mentioned showed a change in expression in the ΔsigF strain. Strikingly, MSMEG_1786 (RsfA) is located in the 29-gene region upstream of sigF, and MSMEG_5551 is located immediately downstream of a gene under direct control of SigF (MSMEG_5550) (Table 3).
Our analysis has identified four candidate proteins as anti-SigF antagonists in M. smegmatis: MSMEG_1786 (based on homology to M. tuberculosis RsfA and the location of its gene), MSMEG_6127 (based on homology to M. tuberculosis RsfB), MSMEG_6541 (based on SigF-dependent expression), and MSMEG_5551 (based on the location of its gene). Together with the presence of a possible second anti-sigma factor for SigF, MSMEG_1787, this would suggest careful fine-tuning of SigF activity, allowing the cell to exert close control over the large SigF regulon identified in this study. Future work will be required to elucidate which function each of these proteins has as part of the SigF cascade in M. smegmatis.
Conclusions.
In this communication we report on the SigF regulon of M. smegmatis mc2155. We present 138 candidate genes under direct or indirect control of SigF, among them several catalase genes, the biosynthesis gene for the pigment isorenieratene, and genes for two of three trehalose-generating pathways in M. smegmatis. We describe a promoter consensus for 130 genes, of which more than 50% showed a reduced expression in a ΔsigF strain in either exponential- or stationary-phase batch cultures. We further report indications for a posttranslational regulatory cascade of SigF, as predicted previously (12), and propose possible anti-SigF antagonists. In summary, this study has revealed an array of novel SigF-dependent genes which could be involved in defense against oxidative stress, heat stress, and osmotic stress in M. smegmatis, suggesting SigF as a key player for stationary-phase adaptation and stress response in mycobacteria.
Supplementary Material
Acknowledgments
This work was funded by New Zealand Lottery Health and the University of Otago, Health Research Council NZ, Maurice Wilkins Centre for Molecular Biodiscovery, Swiss National Science Foundation, and National Institute of Allergy and Infectious Diseases.
We thank Les McNoe from the Otago Genomics Facility for technical assistance. We also thank Thorsten Mascher for expert advice on the in silico promoter analysis and for critical reading of the manuscript.
Footnotes
Published ahead of print on 16 March 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Almiron, M., A. J. Link, D. Furlong, and R. Kolter. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646-2654. [DOI] [PubMed] [Google Scholar]
- 2.Antelmann, H., S. Engelmann, R. Schmid, A. Sorokin, A. Lapidus, and M. Hecker. 1997. Expression of a stress- and starvation-induced dps/pexB-homologous gene is controlled by the alternative sigma factor sigmaB in Bacillus subtilis. J. Bacteriol. 179:7251-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arguelles, J. C. 2000. Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch. Microbiol. 174:217-224. [DOI] [PubMed] [Google Scholar]
- 4.Beaucher, J., S. Rodrigue, P. E. Jacques, I. Smith, R. Brzezinski, and L. Gaudreau. 2002. Novel Mycobacterium tuberculosis anti-sigma factor antagonists control SigmaF activity by distinct mechanisms. Mol. Microbiol. 45:1527-1540. [DOI] [PubMed] [Google Scholar]
- 5.Chen, P., R. E. Ruiz, Q. Li, R. F. Silver, and W. R. Bishai. 2000. Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF. Infect. Immun. 68:5575-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdhury, R. P., S. Gupta, and D. Chatterji. 2007. Identification and characterization of the dps promoter of Mycobacterium smegmatis: promoter recognition by stress-specific extracytoplasmic function sigma factors SigmaH and SigmaF. J. Bacteriol. 189:8973-8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeMaio, J., Y. Zhang, C. Ko, and W. R. Bishai. 1997. Mycobacterium tuberculosis sigF is part of a gene cluster with similarities to the Bacillus subtilis sigF and sigB operons. Tuber. Lung Dis. 78:3-12. [DOI] [PubMed] [Google Scholar]
- 9.DeMaio, J., Y. Zhang, C. Ko, D. B. Young, and W. R. Bishai. 1996. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 93:2790-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Smet, K. A., A. Weston, I. N. Brown, D. B. Young, and B. D. Robertson. 2000. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology 146:199-208. [DOI] [PubMed] [Google Scholar]
- 11.Elbein, A. D., Y. T. Pan, I. Pastuszak, and D. Carroll. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology 13:17R-27R. [DOI] [PubMed] [Google Scholar]
- 12.Gebhard, S., A. Hümpel, A. D. McLellan, and G. M. Cook. 2008. The alternative sigma factor SigF of Mycobacterium smegmatis is required for survival of heat shock, acidic pH and oxidative stress. Microbiology 154:2786-2795. [DOI] [PubMed] [Google Scholar]
- 13.Geiman, D. E., D. Kaushal, C. Ko, S. Tyagi, Y. C. Manabe, B. G. Schroeder, R. D. Fleischmann, N. E. Morrison, P. J. Converse, P. Chen, and W. R. Bishai. 2004. Attenuation of late-stage disease in mice infected by the Mycobacterium tuberculosis mutant lacking the SigF alternate sigma factor and identification of SigF-dependent genes by microarray analysis. Infect. Immun. 72:1733-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geiman, D. E., T. R. Raghunand, N. Agarwal, and W. R. Bishai. 2006. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 50:2836-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57:441-466. [DOI] [PubMed] [Google Scholar]
- 16.Gupta, S., and D. Chatterji. 2005. Stress responses in mycobacteria. IUBMB Life 57:149-159. [DOI] [PubMed] [Google Scholar]
- 17.Gupta, S., S. B. Pandit, N. Srinivasan, and D. Chatterji. 2002. Proteomics analysis of carbon-starved Mycobacterium smegmatis: induction of Dps-like protein. Protein Eng. 15:503-512. [DOI] [PubMed] [Google Scholar]
- 18.Holtmann, G., and E. Bremer. 2004. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of Opu transporters. J. Bacteriol. 186:1683-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kappes, R. M., B. Kempf, and E. Bremer. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karls, R. K., J. Guarner, D. N. McMurray, K. A. Birkness, and F. D. Quinn. 2006. Examination of Mycobacterium tuberculosis sigma factor mutants using low-dose aerosol infection of guinea pigs suggests a role for SigC in pathogenesis. Microbiology 152:1591-1600. [DOI] [PubMed] [Google Scholar]
- 21.Kim, T. H., J. S. Park, H. J. Kim, Y. Kim, P. Kim, and H. S. Lee. 2005. The whcE gene of Corynebacterium glutamicum is important for survival following heat and oxidative stress. Biochem. Biophys. Res. Commun. 337:757-764. [DOI] [PubMed] [Google Scholar]
- 22.Letunic, I., T. Doerks, and P. Bork. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229-D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manganelli, R., R. Provvedi, S. Rodrigue, J. Beaucher, L. Gaudreau, and I. Smith. 2004. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 186:895-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Münch, R. H., K. A. Grote, M. Scheer, J. Klein, M. Schobert, and D. Jahn. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187-4189. [DOI] [PubMed] [Google Scholar]
- 25.Murphy, H. N., G. R. Stewart, V. V. Mischenko, A. S. Apt, R. Harris, M. S. McAlister, P. C. Driscoll, D. B. Young, and B. D. Robertson. 2005. The OtsAB pathway is essential for trehalose biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 280:14524-14529. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen, L., S. Chinnapapagari, and C. J. Thompson. 2005. FbpA-dependent biosynthesis of trehalose dimycolate is required for the intrinsic multidrug resistance, cell wall structure, and colonial morphology of Mycobacterium smegmatis. J. Bacteriol. 187:6603-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parida, B. K., T. Douglas, C. Nino, and S. Dhandayuthapani. 2005. Interactions of anti-sigma factor antagonists of Mycobacterium tuberculosis in the yeast two-hybrid system. Tuberculosis (Edinb.) 85:347-355. [DOI] [PubMed] [Google Scholar]
- 28.Provvedi, R., D. Kocincova, V. Dona, D. Euphrasie, M. Daffe, G. Etienne, R. Manganelli, and J. M. Reyrat. 2008. SigF controls carotenoid pigment production and affects transformation efficiency and hydrogen peroxide sensitivity in Mycobacterium smegmatis. J. Bacteriol. 190:7859-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigue, S., R. Provvedi, P. E. Jacques, L. Gaudreau, and R. Manganelli. 2006. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 30:926-941. [DOI] [PubMed] [Google Scholar]
- 30.Sechi, L. A., G. E. Felis, N. Ahmed, D. Paccagnini, D. Usai, S. Ortu, P. Molicotti, and S. Zanetti. 2007. Genome and transcriptome scale portrait of sigma factors in Mycobacterium avium subsp. paratuberculosis. Infect. Genet. Evol. 7:424-432. [DOI] [PubMed] [Google Scholar]
- 31.Singh, A. K., and B. N. Singh. 2008. Conservation of Sigma F in mycobacteria and its expression in Mycobacterium smegmatis. Curr. Microbiol. 56:574-580. [DOI] [PubMed] [Google Scholar]
- 32.Singh, A. K., and B. N. Singh. 2009. Differential expression of sigH paralogs during growth and under different stress conditions in Mycobacterium smegmatis. J. Bacteriol. 191:2888-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 34.Soliveri, J. A., J. Gomez, W. R. Bishai, and K. F. Chater. 2000. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology 146:333-343. [DOI] [PubMed] [Google Scholar]
- 35.Staron, A., H. J. Sofia, S. Dietrich, L. E. Ulrich, H. Liesegang, and T. Mascher. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 74:557-581. [DOI] [PubMed] [Google Scholar]
- 36.Villas-Boas, S. G., and P. Bruheim. 2007. Cold glycerol-saline: the promising quenching solution for accurate intracellular metabolite analysis of microbial cells. Anal. Biochem. 370:87-97. [DOI] [PubMed] [Google Scholar]
- 37.von Blohn, C., B. Kempf, R. M. Kappes, and E. Bremer. 1997. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 25:175-187. [DOI] [PubMed] [Google Scholar]
- 38.Waagmeester, A., J. Thompson, and J. M. Reyrat. 2005. Identifying sigma factors in Mycobacterium smegmatis by comparative genomic analysis. Trends Microbiol. 13:505-509. [DOI] [PubMed] [Google Scholar]
- 39.Walters, S. B., E. Dubnau, I. Kolesnikova, F. Laval, M. Daffe, and I. Smith. 2006. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 60:312-330. [DOI] [PubMed] [Google Scholar]
- 40.Williams, E. P., J. H. Lee, W. R. Bishai, C. Colantuoni, and P. C. Karakousis. 2007. Mycobacterium tuberculosis SigF regulates genes encoding cell wall-associated proteins and directly regulates the transcriptional regulatory gene phoY1. J. Bacteriol. 189:4234-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodruff, P. J., B. L. Carlson, B. Siridechadilok, M. R. Pratt, R. H. Senaratne, J. D. Mougous, L. W. Riley, S. J. Williams, and C. R. Bertozzi. 2004. Trehalose is required for growth of Mycobacterium smegmatis. J. Biol. Chem. 279:28835-28843. [DOI] [PubMed] [Google Scholar]
- 42.Ziegelhoffer, E. C., and T. J. Donohue. 2009. Bacterial responses to photo-oxidative stress. Nat. Rev. Microbiol. 7:856-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.