Abstract
Organisms performing photosynthesis in the presence of oxygen have to cope with the formation of highly reactive singlet oxygen (1O2) and need to mount an adaptive response to photooxidative stress. Here we show that the alternative sigma factors RpoHI and RpoHII are both involved in the 1O2 response and in the heat stress response in Rhodobacter sphaeroides. We propose RpoHII to be the major player in the 1O2 response, whereas RpoHI is more important for the heat stress response. Mapping of the 5′ ends of RpoHII- and also RpoHI/RpoHII-dependent transcripts revealed clear differences in the −10 regions of the putative promoter sequences. By using bioinformatic tools, we extended the RpoHII regulon, which includes genes induced by 1O2 exposure. These genes encode proteins which are, e.g., involved in methionine sulfoxide reduction and in maintaining the quinone pool. Furthermore, we identified small RNAs which depend on RpoHI and RpoHII and are likely to contribute to the defense against photooxidative stress and heat stress.
Rhodobacter sphaeroides 2.4.1 induces the photosynthetic apparatus when the oxygen tension in the environment decreases in order to perform anoxygenic photosynthesis. In the presence of oxygen and light, R. sphaeroides still grows well even when photosynthetic pigments are highly abundant in the cell. Bacteriochlorophylls and their precursors can act as photosensitizers, causing the formation of highly toxic reactive singlet oxygen (1O2). Carotenoids prevent the formation of 1O2 by quenching excited bacteriochlorophyll molecules and by direct quenching of 1O2 (7, 10). However, R. sphaeroides needs to mount an adaptive response to 1O2 exposure that does not depend on carotenoids (2, 10).
Alternative sigma factors represent important regulatory systems, which control gene expression in bacteria under diverse stress conditions. It was shown earlier that the extracytoplasmic function (ECF) sigma factor RpoE is a major player in the 1O2 stress response in R. sphaeroides (2). Dissociation of RpoE from its anti-sigma factor ChrR leads to the activation of genes involved in the response to 1O2 (1, 2). Only recently we demonstrated that the expression of rpoHII, a gene encoding an alternative sigma factor of the σ32 family, directly depends on RpoE, forming a sigma factor cascade (22). RpoHII (σ38) is one of two alternative sigma factors in R. sphaeroides besides RpoHI (σ37), which are described as heat shock sigma factors. The two paralogs share 46% amino acid identity, and each sigma factor is able to complement a heat-sensitive rpoH deletion strain of Escherichia coli (12).
By proteome analysis of an rpoHII deletion strain, we showed that many of the RpoE-dependent proteins directly depend on RpoHII (22). Nevertheless, the synthesis of several proteins was reduced but not absent in the rpoHII deletion strain, and many other 1O2-triggered proteins do not even contain a putative RpoHII-specific promoter sequence. Therefore, other factors which lead to the induction of these genes by 1O2 exposure must exist. The earlier findings that the relative expression of rpoHI is also strongly induced under 1O2 stress (22) and that the hslO gene carries a heat-inducible promoter recognized by RpoHI and RpoHII (12) motivated us to analyze the role of RpoHI in the photooxidative stress response of R. sphaeroides.
For this purpose we constructed and characterized an rpoHI single-deletion strain and an rpoHI rpoHII double-deletion strain. We tested the sensitivity of these mutants to 1O2, methylglyoxal, and increased/decreased temperatures. Moreover, the 5′ ends of selected RpoHII and RpoHI/RpoHII-dependent transcripts were mapped by rapid amplification of 5′ cDNA ends (5′-RACE) to get more detailed information about the promoter specificity of these sigma factors. Based on those data, we performed a genome-wide search for putative RpoHII and RpoHI/RpoHII promoter sequences in close distance to open reading frames. In addition, we lately identified a set of small RNAs using high-throughput pyrosequencing (4). Here we demonstrate that expression of some small RNAs (sRNAs) is strongly dependent on RpoHI and RpoHII.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
R. sphaeroides strains were grown at 32°C in minimal salt medium containing malate as the carbon source (9). Aerobic growth conditions with a concentration of 160 to 180 μM dissolved oxygen were established by gassing cultures with air in flat glass bottles or by continuous shaking of Erlenmeyer flasks at 140 rpm with a culture volume of 20%. In semiaerobic cultures, a volume of 80% in Erlenmeyer flasks and shaking at 140 rpm lead to a dissolved oxygen concentration of approximately 25 μM. When necessary, kanamycin (25 μg ml−1), spectinomycin (10 μg ml−1), tetracycline (1.5 μg ml−1), or trimethoprim (50 μg ml−1) was added to liquid and solid growth media, which contained 1.6% agar. Antibiotics were omitted from precultures, cultures, and agar plates used for R. sphaeroides strains during stress experiments and inhibition zone assays (see below) to ensure identical culture conditions. Escherichia coli strains were grown at 37°C in LB medium under continuous shaking at 180 rpm or on solid growth media.
Construction of R. sphaeroides rpoHI deletion strains.
R. sphaeroides strains 2.4.1ΔrpoHI, 2.4.1ΔrpoHI ΔrpoHII, and 2.4.1TF18ΔrpoHI were generated by transferring the suicide plasmid pPHU2.4.1rpoHI::Km (Table 1) into R. sphaeroides 2.4.1, 2.4.1ΔrpoHII, and the rpoE chrR mutant strain TF18. Knockout candidates were screened for insertion of the kanamycin cassette into the chromosome by homologous recombination. Parts of the rpoHI gene of R. sphaeroides 2.4.1 together with upstream and downstream sequences were amplified by PCR using the oligonucleotides 2.4.1rpoHI_knockout-up_EcoRI, 2.4.1rpoHI_knockout-up_BamHI, 2.4.1rpoHI_knockout-down_BamHI, and 2.4.1rpoHI_knockout-down_SphI (see Table S1 in the supplemental material). The obtained PCR fragments were cloned into pPHU281 (15), using the appropriate restriction endonucleases. Then, the kanamycin cassette obtained from the plasmid pUC4K (34) was inserted into the BamHI restriction site to obtain the plasmid pPHU2.4.1rpoHI::Km. The plasmid pPHU2.4.1rpoHI::Km was transferred into E. coli strain S17-1 (27) and mobilized into R. sphaeroides strains by biparental conjugation. Conjugants were selected on malate minimal medium agar plates containing 25 μg kanamycin ml−1.
TABLE 1.
Strains and plasmids
| Strain/plasmid | Description | Source/reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17-1 | recA pro hsdR RP4-2-Tc::MuKm::Tn7 tra+ Kmr Spr | 27 |
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi (lac-proAB) | New England Biolabs |
| R. sphaeroides | ||
| 2.4.1 | Wild type | 33 |
| 2.4.1(pRK415) | Wild type harboring pRK415, Tcr | This study |
| TF18 | rpoE chrR mutation in 2.4.1, Tpr | 26 |
| TF18(pRK2.4.1rpoEchrR) | TF18 harboring pRK2.4.1rpoEchrR | This study |
| TF18(pRK415) | TF18 harboring pRK415, Tcr | This study |
| 2.4.1ΔrpoHII | 2.4.1rpoHII::ΩSpr/Smr | 22 |
| 2.4.1ΔrpoHII(pRK415) | 2.4.1ΔrpoHII harboring pRK415, Tcr | 26 |
| 2.4.1ΔrpoHII(pRK2.4.1rpoHII) | 2.4.1ΔrpoHI harboring pRK2.4.1rpoHII | 26 |
| 2.4.1ΔrpoHI | 2.4.1rpoHI::Kmr cassette | This study |
| 2.4.1ΔrpoHI(pRK415) | 2.4.1ΔrpoHI harboring pRK415, Tcr | This study |
| 2.4.1ΔrpoHI(pRK2.4.1rpoHI) | 2.4.1ΔrpoHI harboring pRK2.4.1rpoHI | This study |
| 2.4.1ΔrpoHI ΔrpoHII | 2.4.1ΔrpoHII rpoHI::Kmr cassette | This study |
| 2.4.1ΔrpoHI ΔrpoHII(pRK415) | 2.4.1ΔrpoHI ΔrpoHII harboring pRK415, Tcr | This study |
| 2.4.1ΔrpoHI ΔrpoHII (pRK2.4.1rpoHI) | 2.4.1ΔrpoHI ΔrpoHII harboring pRK2.4.1rpoHI | This study |
| TF18ΔrpoHI | TF18 rpoHI::Kmr cassette | This study |
| TF18ΔrpoHI(pRK415) | TF18ΔrpoHI harboring pRK415, Tcr | This study |
| TF18ΔrpoHI(pRK2.4.1rpoHI) | TF18ΔrpoHI harboring pRK2.4.1rpoHI | This study |
| Plasmids | ||
| pPHU281 | Suicide vector for R. sphaeroides, Tcr | 15 |
| pUC4K | Kmr, source of Kmr cassette | 25 |
| pRK415 | Tcr | 18 |
| pPHU2.4.1rpoHI::Kmr | pPHU281 with Kmr cassette, flanked by the upstream and downstream regions of rpoHI | This study |
| pRK2.4.1rpoHI | pRK415 harboring a 1.2-kb fragment containing rpoHI, flanked by the 239-bp upstream and 26-bp downstream regions | This study |
| pRK2.4.1rpoEchrR | pRK415 harboring a 1.6-kb fragment containing the rpoE-chrR operon, flanked by the 241-bp upstream and 158-bp downstream regions | This study |
| pDrive cloning vector | Apr Kmr | Qiagen |
PCR analysis of chromosomal DNA isolated from kanamycin-resistant and tetracycline-sensitive conjugants was carried out to confirm the double-crossover event of the kanamycin cassette into the R. sphaeroides chromosome. For this purpose, the whole rpoHI gene, including 6 bp of the upstream region and 365 bp of the downstream region, was PCR amplified using primers 2.4.1rpoHI-test-A and 2.4.1rpoHI-test-B (see Table S1 in the supplemental material). By insertion of the kanamycin cassette, 676 bp of the 897-bp R. sphaeroides rpoHI gene were deleted. For the double-crossover candidates, only one PCR fragment, with a size of 1,885 bp, consisting of the flanking regions of the rpoHI gene (592 bp) and the 1,293-bp kanamycin cassette, was obtained. For single-crossover candidates, the following two PCR fragments were obtained: a smaller fragment with the entire rpoHI gene, including the flanking regions (1,268 bp), which was also observed for the wild type, and a larger fragment with the deleted rpoHI gene, including the kanamycin cassette (1,885 bp).
Complementation of the R. sphaeroides rpoHI and rpoE deletion strains.
For complementation of rpoHI deletion strains, a 1,162-bp PCR fragment containing the entire rpoHI gene along with 239 bp of the upstream sequence and 26 bp of the downstream sequence was amplified using the oligonucleotides 2.4.1rpoHI-upstream and 2.4.1rpoHI-downstream (see Table S1 in the supplemental material). The obtained PCR fragment was cloned into the pDrive vector (Qiagen, Hilden, Germany). Digestion of the pDrive vector containing the insert with PstI and XbaI, followed by cloning with the same restriction sites into plasmid pRK415, obtained plasmid pRK2.4.1rpoHI. This plasmid was subsequently transformed in E. coli S17-1 and conjugated with strain 2.4.1ΔrpoHI, 2.4.1ΔrpoHI ΔrpoHII, and TF18ΔrpoHI to obtain the complemented strains 2.4.1ΔrpoHI(pRK2.4.1rpoHI), 2.4.1ΔrpoHI ΔrpoHII(pRK2.4.1rpoHI), and TF18ΔrpoHI(pRK2.4.1rpoHI).
For complementation of the rpoE chrR deletion strain TF18, a 1,595-bp PCR fragment containing the entire rpoE-chrR operon, along with 241 bp of the upstream sequence and 158 bp of the downstream sequence, was amplified using the oligonucleotides 2.4.1rpoEchrR-upstream and 2.4.1rpoEchrR-downstream (see Table S1 in the supplemental material). The obtained PCR fragment was cloned into the pDrive vector (Qiagen, Hilden, Germany). Digestion of the pDrive vector containing the insert with EcoRI, followed by cloning with the same restriction site into plasmid pRK415, obtained the plasmid pRK2.4.1rpoEchrR. This plasmid was subsequently transformed in E. coli S17-1 and conjugated with strain TF18 to obtain the complemented strain TF18(pRK2.4.1rpoEchrR).
Sensitivity to 1O2, methylglyoxal, and changes in temperature.
Measurement of sensitivity to 1O2 and methylglyoxal was performed as described previously (22). Exponential-phase cultures grown under semiaerobic conditions at 32°C were used to test the effect of temperature changes on R. sphaeroides 2.4.1 and the rpoHI and rpoHII deletion strains (Table 1). Cultures were diluted to an optical density at 660 nm (OD660) of 0.1, and for each strain, 5 μl of diluted culture was plated on agar plates, followed by incubation in the dark. Sensitivity to elevated temperatures was tested by incubating agar plates for 24 h at 42°C, followed by incubation at 32°C for 3 days to monitor growth. Growth was monitored also at 15°C for 21 days, and control agar plates were incubated at 32°C for 3 days.
High-light and photooxidative stress conditions.
High-light and photooxidative stress conditions were performed as described earlier (10). In brief, cultures were grown under semiaerobic conditions overnight to obtain pigmented cells. Cultures were diluted to an OD660 of 0.2 and allowed to double once under aerobic growth conditions in darkened flat glass bottles. High-light conditions were generated by illumination with 800 W m−2 white light. For photooxidative stress, 1O2-producing methylene blue was added to liquid cultures at a final concentration of 0.2 μM prior to illumination. Because these conditions are lethal for the rpoHI rpoHII double-deletion strain, illumination with 700 W m−2 and 0.2 μM methylene blue were used.
Radioactive labeling of proteins and gel-based proteome analysis.
Radioactive labeling and proteome analysis performed by two-dimensional (2D) gel electrophoresis followed the procedure described earlier (11). In brief, samples of 7 ml were retrieved from R. sphaeroides cultures, 15 μCi l-[35S]methionine (GE Healthcare, München, Germany) was added, and incubation was performed for 10 min under the experimental conditions described above. Samples were cooled on ice after incubation, and cells were harvested by centrifugation at 10,000 × g for 10 min at 4°C and stored at −20°C until further processing. For the extraction of soluble proteins, harvested cells were washed and disrupted by sonication. Intact cells and cell debris were removed by centrifugation, and the supernatant was used for ultracentrifugation at 100,000 × g. The radioactive label was quantified in the colorless supernatant by adding 10-μl aliquots to 1 ml Rotiszint scintillation cocktail (Roth, Karlsruhe, Germany) and measured in a Beckmann LS-6500 scintillation counter (Beckmann Coulter, Fullerton, CA). For 2D gel electrophoresis, protein samples containing 1.5 × 106 cpm were treated with RNase A (Qiagen) and RQ1 DNase I (Promega, Madison, WI) to remove nucleic acids. Proteins were precipitated using a final concentration of 10% trichloroacetic acid, and the protein pellets were dried and then solubilized in sample buffer (11). Then, samples were applied to immobilized pH gradient strips (ReadyStrip; Bio-Rad, Hercules, CA). After isoelectric focusing, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and gels were fixed, dehydrated, dried, and exposed to phosphorimaging screens for 48 h. Phosphorimages were read with a Molecular Imager FX (Bio-Rad) set to a resolution of 100 μm. Protein spots on digital phosphorimages were compared using the software Delta2D version 3.3 (Decodon, Greifswald, Germany).
RNA extraction, 5′-RACE, and Northern blot analysis.
RNA extraction for 5′-RACE and the 5′-RACE protocol was performed as described previously (22). For Northern blot analysis, samples from stress experiments were collected before (0 min) and 10 min after the onset of stress conditions. From these samples, total RNA was isolated by the hot phenol method (16) and quantified photometrically using a wavelength of 260 nm. Total RNA was separated on 10% polyacrylamide gels containing 7 M urea and then transferred onto Biodyne B 0.45-μm membranes (Pall) by semidry electroblotting. A total of 10 to 20 μg RNA per sample was loaded. Detection of the sRNAs RSs0680a and RSs2461 was carried out as described earlier (4). For detection of the newly identified sRNA RSs1543, the end-labeled oligodeoxynucleotide p-1543 (5′-ATG AAG CGG ACG AGA GAA CCC TC-3′) was used. Membranes were exposed on phosphorimaging screens (Bio-Rad) and analyzed with the Quantity One 1-D software (Bio-Rad).
Real-time RT-PCR.
The primers employed for analyzing relative expression of target genes using real-time reverse transcription-PCR (RT-PCR) are listed in Table S1 in the supplemental material. For normalization of mRNA levels, the rpoZ gene, which encodes the ω subunit of RNA polymerase of R. sphaeroides, was used (23). Conditions for real-time RT-PCR were described earlier in detail (10, 11). For real-time RT-PCR, a final concentration of 4 ng μl−1 of total RNA was applied in a one-step RT-PCR kit (Qiagen), and SYBR green I (Sigma-Aldrich) was added in a final dilution of 1:50,000 to the master mix to detect double-stranded DNA. Relative expression of target genes was calculated relative to expression of untreated samples and relative to rpoZ (24). PCR efficiency is 2.02 for rpoZ (10, 11), and further PCR efficiencies were determined experimentally using serial dilutions of RNA; respective values obtained are 2.12 for qxtA, 2.07 for RSP0150, 2.19 for RSP1948, and 1.9 for hslO.
Genome-wide prediction of sigma factor target sites.
Based on the information on transcriptional start sites gained by 5′-RACE, putative sigma factor binding sites were identified. We then used those binding sites to create consensus sequences for promoters recognized only by RpoHII or by RpoHI and RpoHII. For a genome-wide prediction of putative sigma factor binding sites, those consensus sequences were used to search the R. sphaeroides 2.4.1 genome by using the DNA genome-scale DNA pattern software within the regulatory sequence analysis (RSA) tools (http://rsat.ulb.ac.be/rsat) (32). The whole chromosome was searched, and putative promoter positions were then analyzed with respect to their locations in intergenic regions or upstream of mRNA coding regions using ARTEMIS 11 software. Transcriptional start sites obtained previously by pyrosequencing of 1O2 and superoxide-treated R. sphaeroides cultures (4) were used to verify putative promoters obtained by the bioinformatic prediction.
Statistical analysis.
Statistical analysis for the comparison of threshold cycle values obtained for individual genes under different stress conditions using real-time RT-PCR was performed with Student's t test using Microsoft Excel 2003 (Microsoft, Redmond, WA). Significance levels (P of ≤0.1, ≤0.05, and ≤0.01) are indicated in the figure legends.
RESULTS
Deletion of rpoHI and rpoHII differently affects sensitivity to temperature, 1O2, and methylglyoxal.
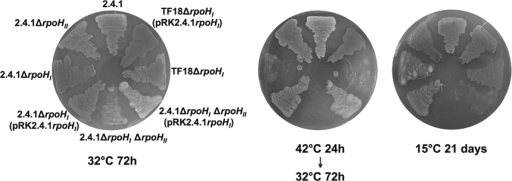
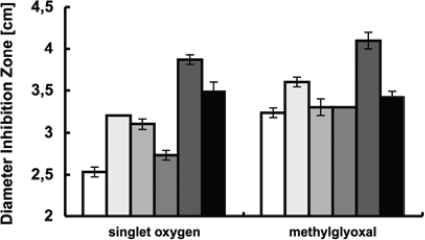
The construction of rpoHI deletion strains in the R. sphaeroides 2.4.1, 2.4.1rpoHII, and rpoE chrR mutant strain TF18 backgrounds yielded the strains 2.4.1ΔrpoHI, 2.4.1ΔrpoHI ΔrpoHII, and TF18ΔrpoHI as described in Materials and Methods. We tested the sensitivity of the rpoH deletion strains to changes in temperature (Fig. 1), 1O2, and methylglyoxal (Fig. 2). The rpoHI rpoHII deletion strain was most sensitive to all three stress factors compared to the other mutants and the wild-type strain. It was not growing on agar plates incubated at 15°C as well as after 24 h of incubation at 42°C. Inhibition zones obtained for 1O2 and methylglyoxal treatment were larger than those in all other strains tested (Fig. 2). Deletion of the rpoHI gene in strain TF18 was performed to test if dependency of rpoHII expression on RpoE leads to a phenotype similar to that of the rpoHI rpoHII deletion strain. Strain TF18ΔrpoHI exhibited a less sensitive phenotype for all three stress factors compared to that of the rpoHI rpoHII deletion strain but was more affected by 1O2 exposure than the parent strain TF18 (Fig. 2). Strain TF18 was not affected by changes in temperature, similar to the complemented rpoHI deletion strain TF18ΔrpoHI (Fig. 1). The less pronounced sensitivity of strain TF18ΔrpoHI compared to that of the rpoHI rpoHII deletion strain is in agreement with the finding that rpoHII expression is not entirely dependent on RpoE (22). The deletion of rpoHI in the wild-type and TF18 background generated a phenotype sensitive to increased temperature but not to decreased temperature (Fig. 1). In comparison, deletion of rpoHII in the wild type generated a phenotype more sensitive to 1O2 and methylglyoxal but not to temperature changes (22) (Fig. 1 and 2). Complementation with low-copy-number plasmid pRK415 harboring the rpoHI gene with its own promoter region restored the phenotype of the respective parental strains (Fig. 1; see also Table S2 in the supplemental material), except for strain 2.4.1ΔrpoHI ΔrpoHII(pRK2.4.1rpoHI) which showed weaker growth at 15°C compared to strain 2.4.1ΔrpoHII (Fig. 1). The presence of the plasmid pRK415 without the rpoHI gene did not influence the sensitivity of the strains to 1O2 and methylglyoxal (see Table S2) or to increased and decreased temperatures (data not shown).
FIG. 1.
Growth of R. sphaeroides wild-type and rpoHI and rpoHII deletion strains under different temperatures. The cultures were grown under semiaerobic conditions at 32°C to exponential phase and diluted to an OD660 of 0.1. For each strain, 5 μl of diluted culture was plated on minimal agar plates and incubated under the indicated temperatures in the dark. The agar plates incubated at 42°C were shifted to 32°C (optimal growth) after 24 h.
FIG. 2.
Inhibition of growth of the R. sphaeroides wild type, the TF18 strain, and the rpoHI and rpoHII deletion strains by 1O2 and methylglyoxal. Each point represents the mean result from three independent experiments. Error bars indicate the maximal deviations from three experiments. 2.4.1, open; 2.4.1ΔrpoHII, light gray (second bar); TF18(rpoE chrR mutant), gray (third bar); 2.4.1ΔrpoHI, dark gray (fourth bar); 2.4.1ΔrpoHI ΔrpoHII, dark gray (fifth bar); and TF18(rpoE chrR mutant)ΔrpoHI, filled.
Proteome analysis reveals overlapping RpoHI and RpoHII regulons.
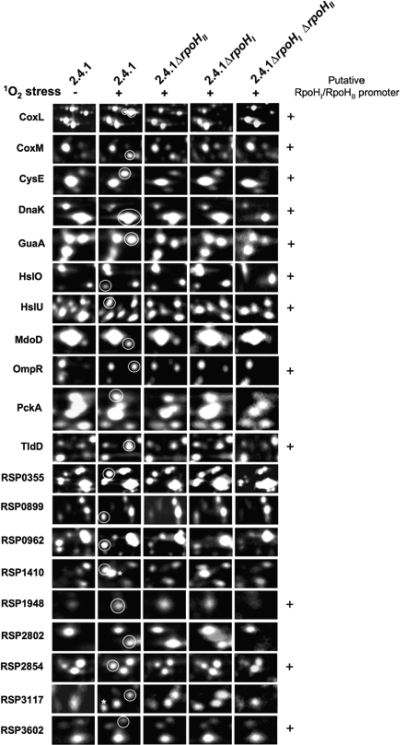
In a recent study we found that 25 soluble proteins induced by 1O2 exposure were strongly affected by the deletion of rpoHII (22). The majority of those proteins were not synthesized in the rpoHII deletion strain, but several proteins were decreased only in synthesis. In vitro studies suggested that RpoHI and RpoHII of R. sphaeroides 2.4.1 recognize the same set of heat-inducible promoters (12). Consequently, we assessed the role of potentially overlapping RpoHI and RpoHII regulons in the cellular defense against excess 1O2 generation. Patterns of soluble proteins obtained from the rpoHI and the rpoHI rpoHII deletion strains were compared with those obtained from the wild-type strain 2.4.1 and the rpoHII deletion strain (Fig. 3; see also Fig. S1 and S2 in the supplemental material) and revealed that RpoHI and RpoHII regulons indeed partially overlap.
FIG. 3.
Sections of inverted 2D gel images indicating differences in syntheses of several proteins between the wild-type strain 2.4.1 and strains 2.4.1ΔrpoHII, 2.4.1ΔrpoHI, and 2.4.1ΔrpoHI ΔrpoHII after 60 min of 1O2 exposure. Protein extracts were prepared from cells labeled in vivo with l-[35S]methionine during exponential growth in the dark or in the presence of methylene blue and high light. Proteins labeled with RSP numbers have hypothetical function, and those marked with asterisks were shown previously to be dependent on RpoHII (22).
In our previous study, some proteins were induced in the rpoHII deletion strain by 1O2 exposure but to a lower level than in the wild type (22). We observed that several proteins were fully synthesized upon 1O2 exposure in the wild-type strain and to the same or to a lower level in the rpoHI and rpoHII deletion strains, but those proteins were lacking in the rpoHI rpoHII deletion strain (Fig. 3). This observation was made for the protein CysE (serine O-acetyltransferase), which showed a decreased synthesis in the rpoHII deletion strain compared to that of the wild type (22) and was not synthesized in the rpoHI rpoHII deletion strain (Fig. 3). In a similar manner, CoxL (RSP2877, xanthine dehydrogenase, molybdenum binding subunit apoprotein), CoxM (RSP2876, putative carbon monoxide dehydrogenase, medium chain), HslO (putative Hsp33 protein), HslU (RSP1532, ATP-dependent protease ATP-binding subunit), MdoD (RSP3187, glucan biosynthesis protein D), OmpR (RSP0847, two-component transcriptional regulator), TldD (RSP1825, modulator of DNA gyrase), RSP0355 (serine protease), RSP0899 (thiol peroxidase [atypical 2-Cys peroxiredoxin]), RSP1410 (putative sulfite oxidase subunit), RSP1948 (hypothetical protein), RSP2802 (multidrug/cation efflux pump, membrane fusion protein subunit), RSP2854 (cation efflux transporter), and RSP3117 (hypothetical protein) were either not synthesized upon 1O2 exposure in the rpoHI rpoHII deletion strain or induction was lacking and the respective protein was synthesized in an amount similar to that in the nonexposed wild type (Fig. 3) and the rpoH single deletion strain (data not shown). The synthesis of the proteins DnaK (RSP1173, molecular chaperone), GuaA (RSP0005, GMP synthase), PckA (RSP1680, phosphoenolpyruvate carboxykinase), RSP0962 (dihydrolipoamide dehydrogenase), and RSP3602 (ABC efflux transporter, ATPase subunit) was not induced by 1O2 stress. These proteins were not synthesized or synthesized at lower levels in the rpoHI rpoHII deletion strain than in the wild type and the rpoHI and rpoHII deletion strains. This finding suggests a role of those proteins in basic cellular physiology and implies that they are not specific for the defense to stress generated by 1O2 exposure. Interestingly, HslU was the only protein observed in this study that was not synthesized in the rpoHI deletion strain and therefore seems to depend only on RpoHI under photooxidative stress.
Target sequences recognized by RpoHII and RpoHI/RpoHII.
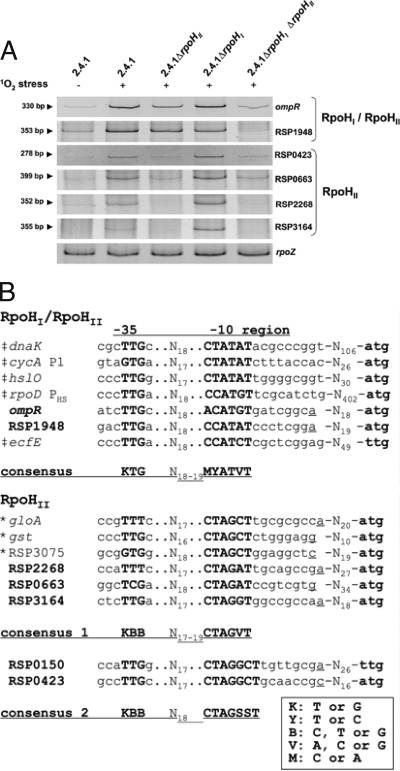
Our proteome data demonstrate that the synthesis of several proteins depends on both RpoHI and RpoHII. In order to identify sigma factor binding sites recognized by RpoHI and RpoHII, we analyzed the upstream region of the genes encoding OmpR and RSP1948 by 5′-RACE (Fig. 4). For both genes, putative sigma factor binding sites similar to the heat-inducible promoters upstream of rpoD and ecfE (12) were observed upstream of the determined 5′ ends. Both binding sites differ from those supposed to be recognized only by RpoHII (22). To find differences between RpoHII- and RpoHI/RpoHII-specific promoters, we also mapped the 5′ ends of further mRNA transcripts strictly depending on RpoHII, including RSP0423, RSP0663, RSP2268, and RSP3164.
FIG. 4.
(A) Separation of 5′-RACE products obtained from RNA extracts of wild-type and rpoHI and rpoHII deletion strains after 10 min of photooxidative stress. PCR products obtained after the second PCR (nested) were separated on a 10% polyacrylamide gel and stained with ethidium bromide. (B) Upstream of the 5′ ends of the sequences corresponding to the depicted DNA bands, putative RpoHII target sequences were found and displayed as an alignment. 5′ ends are underlined. ‡, RpoHI/RpoHII-dependent promoters verified by in vitro transcription (12); boldfaced letters, putative target sequences identified in this study; *, putative target sequences identified earlier (22). To match relative changes in mRNA levels of the respective genes, cDNA synthesis with all gene-specific RACE-1 primers in the same reaction was performed (see Table S1 in the supplemental material). We used a primer specific for rpoZ, a gene used for normalization in the real-time RT-PCR approach described earlier (23).
For all six selected mRNAs, we monitored relative changes in the intensity of 5′-RACE-PCR products obtained in R. sphaeroides 2.4.1, the rpoHII, the rpoHI, and the rpoHI rpoHII deletion strains (Fig. 4A). For rpoZ, which served as an internal control (36), PCR products from all strains were highly similar in intensity, which indicates that global transcript levels are not affected in the respective deletion strains or by 1O2 treatment. For all six other mRNAs, PCR products were amplified from wild-type cultures exposed to 1O2. The products were less abundant or missing using RNA extracts of the unstressed wild type compared to using the 1O2 stressed wild type. 5′-RACE for ompR and RSP1948 further supported the results obtained from proteome analysis. Expression of both genes depends on RpoHI and RpoHII, since strong PCR products were observed in all strains except for the rpoHI rpoHII deletion strain. PCR products were much weaker for ompR in the double-deletion strain than in all other strains exposed to 1O2, and products were lacking for RSP1948 (Fig. 4A). For RSP0423, RSP0663, RSP2268, and RSP3164, amplification of PCR products obtained from RNA extracts of the rpoHII and rpoHI rpoHII deletion strains were much weaker or missing in comparison to those of the wild type and the rpoHI deletion strain (Fig. 4A), underlining the strong dependence on only RpoHII (22).
We identified putative sigma factor target sequences directly upstream of the 5′ ends of all tested genes (Fig. 4B). The putative sigma factor target sequences identified upstream of the RpoHII-dependent genes RSP0423, RSP0663, RSP2268, and RSP3164 were highly similar to those identified earlier (22). By combining the information gained on the sigma factor target sequences, we identified a conserved consensus motif consisting of CTAGVT (V: A, C, or G) in the −10 region of RpoHII-dependent target sequences. Interestingly, the −10 region of the sigma factor target sequence upstream of RSP0423 exhibited an additional base pair in the hexameric motif CTAGVT, forming the heptameric −10 region CTAGGCT. In contrast, the −10 region also recognized by RpoHI consists of an MYATVT (M, C or A; Y, T or C) consensus motif. This motif shows high similarity to the RpoH-dependent promoter described for Caulobacter crescentus (TNNCNCCCTTGAA-N13-15-CCCCATNTA) (35) and to the E. coli RpoH promoter consensus sequence (CNCTTGAAA-N13-14-CCCCATNT) (13) (boldface shows similarities).
Genome-wide search for RpoHII and RpoHI/RpoHII target sequences.
In order to predict promoters exhibiting RpoHII or RpoHI/RpoHII target sites and thereby the respective regulons, we used the genome-scale DNA pattern software within the RSA tools (32). We used the −10 region consensus sequences recognized by RpoHII (Fig. 4B) and the respective consensus of the −35 region. Precisely, the R. sphaeroides 2.4.1 genome was searched for putative RpoHII-dependent promoters with the motif KBB-N17-19-CTAGVT (K: T or G; B: T, C, or G; V: A, C, or G), which revealed 47 highly conserved target sites not more than 400 bp upstream of translational start sites of protein coding genes (Table 2). Additionally, we searched for the motif KBB-N17-19-CTAGSST and identified 68 genes preceded by a putative RpoHII promoter with a heptameric −10 region (see Table S3 in the supplemental material). In a third search, the motif KTG-N(18-19)-MYATVT was used to identify putative RpoHI/RpoHII-dependent promoters, which gained 170 putative target genes (see Table S4 in the supplemental material). In all searches, we included only those target sites exhibiting at least one “T” in the −35 element.
TABLE 2.
Conserved RpoHII target sequences upstream of protein coding genesa
| Target sequence | ATG distance (bp) | Locus tag | Annotated protein(s) | 5′ end |
|---|---|---|---|---|
| Energy metabolism | ||||
| TGT-N17-CTAGAT | 33 | RSP2785-2784 | CycF, cytochrome 554 | + |
| TTG-N17-CTAGAT | 35 | RSP3212-3210 | QxtA, quinol oxidase subunit I | |
| TCG-N19-CTAGAT | 82 | RSP3831 | Cox15, putative cytochrome oxidase assembly factor | |
| Transport | ||||
| TTG-N19-CTAGAT | 31 | RSP0367 | Preprotein translocase SecG subunit | |
| TGG-N19-CTAGCT | 40 | RSP0759 | Putative capsule polysaccharide exporter | + |
| TTG-N19-CTAGGT | 34 | RSP1053 | Signal recognition particle Ffh | |
| TGC-N18-CTAGCT | −28 | RSP1497 | Putative outer membrane lipoprotein carrier protein | + |
| TTG-N19-CTAGCT | 8 | RSP1803-1805 | CcmC, ABC heme transporter | + |
| TTC-N19-CTAGGT | −2 | RSP1843 | FtsY, signal recognition particle-docking protein | |
| TTG-N18-CTAGCT | 129 | RSP1882-1886 | ABC polyamine transporter, ATPase subunit | |
| Regulatory proteins | ||||
| TGG-N19-CTAGGT | 29 | RSP0269 | TspO, tryptophane-rich sensory protein | |
| TTG-N19-CTAGGT | 42 | RSP0762-0761 | Transcriptional regulator XRE family | |
| TGG-N19-CTAGAT | 83 | RSP2165 | PutR, transcriptional regulator, AsnC family | |
| TTT-N19-CTAGCT | 43 | RSP3217 | Cache sensor signal transduction histidine kinase | |
| TCG-N18-CTAGCT | 20 | RSP3430-3431 | Transcriptional regulator, winged helix family | |
| TTG-N19-CTAGCT | 8 | RSP3865 | Predicted transcriptional regulator | + |
| TTG-N19-CTAGGT | 133 | RSP4257-4258 | Anti-sigma factor antagonist STAS | + |
| Protein turnover and amino acid metabolism | ||||
| GTG-N19-CTAGCT | 49 | RSP0398 | GdhA, glutamate/leucine dehydrogenase | |
| TTT-N19-CTAGAT | 34 | RSP2627 | HisI, Phosphoribosyl-AMP cyclohydrolase | |
| TTT-N18-CTAGGT | 27 | RSP4043-4042 | Peptidylprolyl isomerase | + |
| TTG-N19-CTAGAT | 364 | RSP4294 | 16S rRNA | |
| TTG-N19-CTAGAT | 65 | RSP4330 | tRNA-Ala-GGC | |
| TTG-N19-CTAGAT | 364 | RSP4347 | 16S rRNA | + |
| TTG-N18-CTAGCT | 30 | RSP6044 | Serine/alanine racemase | |
| Stress response | ||||
| TTT-N19-CTAGCT | 29 | RSP0392 | GloA, probable lactothioglutathione synthetase | |
| TCG-N19-CTAGAT | 42 | RSP0663 | Formate-tetrahydrofolate ligase | |
| GTC-N19-CTAGAT | −2 | RSP0817-0816 | Aminodeoxychorismate synthase | |
| TTG-N17-CTAGCT | 18 | RSP1591 | Predicted glutathione S-transferase | + |
| TGG-N19-CTAGCT | 21 | RSP1869 | 2-octaprenyl-6-methoxyphenyl hydroxylase | + |
| TTT-N19-CTAGAT | 8 | RSP2092 | Putative uvrD/DNA helicase II | + |
| GGT-N18-CTAGCT | 202 | RSP2123 | Radical SAM domain protein and/or SplB, DNA repair photolyase domain | |
| TTG-N18-CTAGAT | 62 | RSP2617 | Peptide methionine sulfoxide reductase | + |
| TGT-N18-CTAGGT | 394 | RSP2647-2648 | Predicted SAM-dependent methyltransferase | + |
| TTG-N18-CTAGGT | 27 | RSP3164-3162 | Ferredoxin-like protein | + |
| TTG-N17-CTAGAT | 35 | RSP3212-3210 | QxtA, Quinol oxidase subunit I | |
| Hypothetical | ||||
| TTC-N19-CTAGCT | 29 | RSP0238 | Hypothetical protein | |
| TTC-N17-CTAGCT | 28 | RSP0855 | Hypothetical protein | + |
| TTG-N18-CTAGCT | 110 | RSP1366 | Hypothetical protein | |
| TGG-N19-CTAGAT | 30 | RSP1421 | Hypothetical protein | + |
| GTG-N19-CTAGCT | 27 | RSP3075-3076 | Hypothetical protein | + |
| TTG-N19-CTAGCT | 29 | RSP6001 | Hypothetical protein | + |
| TTC-N19-CTAGCT | 237 | RSP6037 | Hypothetical protein | |
| TTG-N19-CTAGCT | 31 | RSP6232 | Hypothetical protein | |
| Other functions | ||||
| GGT-N18-CTAGAT | 121 | RSP1502 | GAF domain protein | |
| TTT-N19-CTAGGT | 16 | RSP1896 | Guanine deaminase | |
| TTT-N18-CTAGAT | 36 | RSP2268 | Metallo-beta-lactamase family | |
| TCT-N19-CTAGAT | 70 | RSP2468-2470 | Putative portal protein | |
| TCG-N19-CTAGAT | 30 | RSP3261-3262 | BioA, aminotransferase |
The target sequences listed were searched using the RSA tools and the consensus sequence KBB-N17-19-CTAGVT. The distance to the translational start site is listed, and it is indicated if downstream genes might be cotranscribed and, hence, may form an operon. Negative distances to the translational start sites indicate that the start codon might have been annotated incorrectly. Positives (+) in the “5′ end” column indicate that primary transcripts were found using 454 pyrosequencing (4) and that the respective sigma factor target sequence is located directly upstream of the 5′ end.
The genome-wide location of putative RpoHII and RpoHI/RpoHII target sites was verified by comparison to that of primary transcripts obtained by 454 pyrosequencing that were aligned to the R. sphaeroides 2.4.1 genome (4). For almost one-third of our predicted promoters, we identified a 5′ end of the respective mRNA 5 to 10 bp downstream of the −10 region (Table 2; see also Tables S3 and S4 in the supplemental material).
Among those genes preceded by an RpoHII- or RpoHI/RpoHII-dependent promoter, several encode proteins typically involved in stress responses, but also proteins are encoded which are involved in basic cellular responses, e.g., energy metabolism, transport, protein turnover, and other functions like lipid metabolism. In our search for RpoHI/RpoHII-dependent promoters, we identified a number of genes encoding proteins known to be involved in the heat stress response, e.g., DnaK, GroES, GrpE, HslO, HrcA, HslV, HtpX, and IbpA (see Table S4 in the supplemental material).
Verification of RpoHII-dependent gene expression.
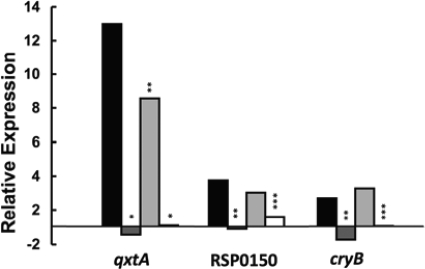
We tested the expression of three genes that were predicted to exhibit an RpoHII-dependent promoter by real-time RT-PCR. We focused on qxtA (RSP3212), encoding the quinol oxidase subunit I, with a predicted hexameric −10 region in the RpoHII target sequence (Table 2). In addition, we analyzed the following two genes preceded by heptameric, potentially RpoHII-specific −10 regions in the promoter sequence: RSP0150, a putative multi-sensor signal transduction histidine kinase related to bacteriophytochromes, and cryB (RSP3077), a bacterial cryptochrome (see Table S3 in the supplemental material) (16). After 1O2 exposure, mRNA levels were increased up to 4-fold for RSP0150, more than 12-fold for qxtA, and ∼3-fold for cryB in the wild type. For all three genes, mRNA levels did not increase in the rpoHII and the rpoHI rpoHII deletion strains, but fold changes in mRNA levels were increased in the rpoHI deletion strain (Fig. 5), indicating strong dependency on RpoHII. Mapping of the 5′ end of the RSP0150 mRNA then verified a putative promoter highly similar to the promoter upstream of RSP0423 (Fig. 4B). A similar promoter was also found to precede the cryB gene (see Table S3) (14).
FIG. 5.
Selected functional genes depending on RpoHII for expression, predicted by genome-wide search for putative RpoHII target sequences. Relative expression of qxtA, RSP0150, and cryB in strains 2.4.1 (black bars), 2.4.1ΔrpoHII (dark gray bars), 2.4.1ΔrpoHI (light gray bars), and 2.4.1ΔrpoHI ΔrpoHII (white bars) was investigated under photooxidative stress. Relative expression was determined by real-time RT-PCR by using threshold cycle values obtained from RNA samples before and after 7 min of singlet oxygen exposure. For normalization, mRNA levels for rpoZ were used as an internal standard. Levels of significance are indicated as follows: *, P ≤ 0.01; **, P ≤ 0.05; and ***, P ≤ 0.1.
RpoHI/RpoHII-dependent sRNAs are induced by 1O2 and heat stress.
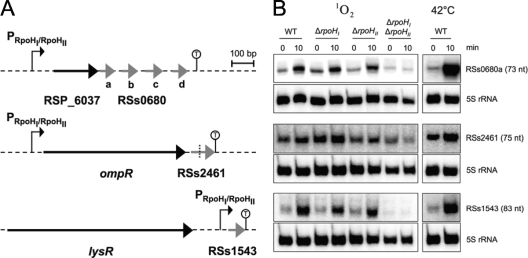
In a genome-wide search for primary transcripts using a 454 pyrosequencing approach, four abundant sRNAs were affected by 1O2 exposure (4). Two of these sRNAs, RSs0680a and RSs2461, were supposed to be transcribed from an RpoHI/RpoHII promoter. The 454 pyrosequencing data revealed the presence of a third sRNA, RSs1543, with a length of 83 nucleotides (nt), that is triggered by photooxidative stress. RSs0680a and RSs2461 are cotranscribed with upstream genes and lack their own promoters (4). In detail, RSs0680a is cotranscribed with RSP6037 (hypothetical protein) from the putative promoter TTG-N19-CCATGT-N75-ATG. The sRNA RSs2461 is cotranscribed with the ompR gene and thereby transcribed from the promoter upstream of ompR (Fig. 4B; see also Table S4 in the supplemental material). In contrast, RSs1543 is directly preceded by a promoter (Fig. 6a) with high similarity to the putative RpoHI/RpoHII consensus motif, consisting of TTG-N19-CTAAAT. Northern blot analysis revealed that all three sRNAs depend on both RpoHI and RpoHII. The two sRNAs, RSs0680a and RSs1543, were strongly induced after 10 min of 1O2 exposure in the wild type and the rpoHI and rpoHII single-deletion strains but not in the rpoHI rpoHII double-deletion strain (Fig. 6B). For the third sRNA, RSs2461, the result was similar, but the induction was less pronounced. We also tested if the three sRNAs are inducible under heat stress conditions (Fig. 6B). An induction was observed for all three sRNAs 10 min after the shift from 32°C to 42°C, which fits into the observation that genes transcribed from an RpoHI/RpoHII promoter are induced under 1O2 as well as heat stress. Our findings clearly showed that the overlapping RpoHI and RpoHII regulons include noncoding sRNAs. This extends the functional role of genes controlled by the two RpoH sigma factors and suggests that sRNAs may be involved in posttranscriptional gene regulation in response to 1O2 and heat in R. sphaeroides.
FIG. 6.
Induction of RSs0680a, RSs2461, and RSs1543 under 1O2 and heat stress depends on both RpoHI and RpoHII. (A) The genetic localization of RSs0680a, RSs2461, and RSs1543 is depicted. RSs0680a and RSs2461 are transcribed together with the upstream gene from an RpoHI/RpoHII promoter. RSs0680a is followed by three homologous sRNAs (RSs0680b to -d). RSs2461 is further processed after cotranscription (4), and the respective processing site is indicated by a dotted line. In contrast, RSs1543 is directly preceded by an RpoHI/RpoHII promoter. A terminating structure is indicated by a stem-loop with a “T” in the loop. (B) Northern blot analysis of RSs0680a, RSs2461, and RSs1543. All three sRNAs are induced after 10 min of 1O2 exposure in the wild-type (WT) and the rpoHI and rpoHII single-deletion strains. This induction is not observed in the rpoHI rpoHII deletion strain. After 10 min of heat stress, all three sRNAs are upregulated. In the case of RSs2461, only the 75-nt fragment is shown, which originates from a 116-nt precursor (4).
DISCUSSION
RpoHI and RpoHII control the heat and photooxidative stress responses.
Increased temperatures in surface layers of, e.g., aquatic photic zone environments, microbial mats, and soils, are usually caused by high solar radiation. Hence, both stresses, heat and photooxidative stress, as caused by chlorophyll-mediated 1O2 formation, are likely to occur simultaneously and may therefore be tightly linked in photosynthetic organisms. The idea that the regulatory response to 1O2 and heat stress may overlap in R. sphaeroides is based on the findings that (i) the deletion of the gene encoding the heat shock-like sigma factor RpoHII leads to sensitivity against 1O2 exposure (22) and that (ii) RpoHI and RpoHII recognize similar promoters in vitro (12). Our idea is supported by the severe phenotype of the rpoHI rpoHII deletion strain found under both stress conditions. Furthermore, we found that expression of several genes, e.g., ompR, RSP1948, and hslO, as well as the sRNAs RSs0680a, RSs2461, and RSs1543, depends on both RpoHI and RpoHII and that those genes are induced by photooxidative stress and heat.
Exclusive roles of RpoHI and RpoHII.
Despite the fact that both RpoHI and RpoHII contribute to the response to heat and 1O2, each sigma factor has its exclusive role. After induction by RpoE, RpoHII is the main factor of the 1O2 stress response, controlling at least 115 genes with specific binding sites and potentially more than 170 genes that are also recognized by RpoHI. RpoHII-specific target sites are not activated even after 10 min of heat stress, in contrast to genes preceded by a promoter also recognized by RpoHI (data not shown). The important role of RpoHII for the photooxidative stress response is also confirmed by the 1O2-sensitive phenotype of the rpoHII deletion strain (22) and the failure of RpoHI to fully compensate the lack of RpoHII. In contrast, RpoHII is able to compensate the lack of RpoHI under 1O2 stress, as the rpoHI deletion strain shows no significant change in 1O2 sensitivity compared to that of the wild type. This indicates that most promoters recognized by RpoHI are also recognized by RpoHII. The rpoHI rpoHII deletion strain is more sensitive to 1O2 than the rpoHII deletion strain (Fig. 2); hence, RpoHI also contributes to the 1O2 stress response but has most likely a minor role.
Our data demonstrate that RpoHI controls the heat stress response of R. sphaeroides, in contrast to earlier results showing that the deletion of rpoHI does not lead to a heat-sensitive phenotype in liquid cultures growing photosynthetically or by aerobic or anaerobic respiration (17). We now demonstrate that the rpoHI deletion strain is sensitive to heat when grown on malate minimal agar plates (Fig. 1). The rpoHII deletion strain was not affected and therefore, RpoHI, but not RpoHII, controls mainly the heat shock response in R. sphaeroides.
RpoHII target promoters differ from those recognized by both RpoHI and RpoHII.
In earlier work, we mapped the transcriptional start sites of selected RpoHII-dependent genes to identify the corresponding putative target sequences. We found the highly conserved −10 element CTAGCT (22) that clearly differs from the respective elements in heat-inducible promoters recognized in vitro by both RpoHI and RpoHII (12).
To unravel what makes a target sequence specific for RpoHII, we mapped the 5′ ends of further RpoHII-dependent genes and additionally the 5′ ends of the ompR and RSP1948 mRNA, whose products are clearly dependent on both sigma factors. Obviously, the G in the CTAGVT motif allows recognition by only RpoHII, whereas a T at this position leads to recognition by both factors. Interestingly, a variation of the −10 region motif comprised of CTAGSST was observed. Together with the finding that the MYATVT consensus is also recognized by RpoHII, this leads to the conclusion that RpoHII is more flexible and recognizes a wider variety of −10 regions than RpoHI. It is conceivable that distinct differences in the amino acid sequences of the region 2.4 of RpoHI and RpoHII (12), which is involved in the recognition of the −10 element, are responsible for the difference in promoter selectivity.
Redundancy in promoter elements of the regulatory response to photooxidative stress and heat.
Several alphaproteobacteria that belong to the Rhizobiales and Rhodobacterales contain two or even three rpoH homologs in their genomes (12). Most likely, the additional heat shock sigma factors have been evolved by a gene duplication event and then diverged. Obviously, RpoHII of R. sphaeroides has been recruited for the response to 1O2 and is controlled directly by RpoE, whereas RpoHI remained the major factor for the heat stress response. Rhizobium etli also harbors two heat shock-like sigma factors, RpoH1 and RpoH2. As in R. sphaeroides, they are able to complement a heat-sensitive rpoH deletion strain of E. coli and have different functions in R. etli (12, 20). RpoH1 is involved mainly in the heat shock and oxidative stress response, whereas RpoH2 participates in osmotic tolerance. In contrast to the rpoH1 deletion strain, the rpoH2 deletion strain is not negatively affected by heat shock, which is similar to R. sphaeroides. The rpoH1 rpoH2 double-deletion strain shows the most severe phenotype to heat shock (20), which is in agreement with our studies for R. sphaeroides. The picture emerges that in the ancestors of the Rhodobacterales and Rhizobiales lineages, one of the rpoH copies was recruited for more specific functions. Together, the rpoH homologs appear to provide a balanced response to heat, oxidative and photooxidative stress, and changes in osmolarity and are involved in the interaction with other organisms (12, 20, 22). Other bacterial lineages also use redundancy in promoter elements to provide a balanced response to environmental changes. For example, in E. coli, the molecular basis of selective promoter recognition is known in detail for RpoD (σ70) and RpoS (29). Here, a combination of exchanges in the single nucleotides in the −10 region and the nucleotides located between the −10 and −35 region that form either AT-rich stretches or an extended −10 element are important for the selectivity of the promoter. Furthermore, AT-rich stretches upstream of the −35 element bind further transcriptional regulators that affect sigma factor binding (30).
Bioinformatically predicted RpoHII-dependent genes.
In our bioinformatic search, we identified 115 genes preceded by a putative RpoHII promoter with a hexameric (CTAGVT) and/or a heptameric (CTAGSST) −10 element. We do not exclude the possibility that further genes preceded by very similar promoters may depend on RpoHII. For example, in the upstream regions of the genes gloB (RSP0799) and gpx (RSP2389), we identified putative promoter sequences similar to those of the RpoHII promoter, with a heptameric motif in the −10 element. The putative promoter sequence CTG-N18-CTATGAT is located 27 bp upstream of the translation start site of RSP0799, and a very similar sequence (TTG-N18-CTATGCT) was identified 74 bp upstream of the translation start site of gpx. The gloB and gpx genes were shown previously to be dependent on RpoHII (22). In addition, we identified in our pyrosequencing approach several 5′ ends 5 to 10 bp downstream of putative −10 elements of promoters highly similar to those upstream of gloB and gpx. We therefore assume that the heptameric −10 element recognized by RpoHII is more variable than the hexameric −10 element. Also, the putative RpoHI/RpoHII promoter upstream of the sRNA RSs1543, consisting of TTG-N19-CTAAAT, shows a discrepancy to the RpoHI/RpoHII promoter consensus motif KTG-N(18-19)-MYATVT. The RpoHI/RpoHII regulon is likely to include even more genes, as identified by our bioinformatic search.
New functions in the response to 1O2 exposure in R. sphaeroides.
The response to 1O2 requires DNA repair (2, 5), quinol oxidation, reduction of glutathione peroxide (5, 11), detoxification of methylglyoxal, 1O2 scavenging (11, 22), glutathione (GSH)-dependent defense systems, protein turnover, export of small toxic waste products of biomolecules (ABC transporters), and scavenging of divalent metal ions (11).
Some of the genes identified by our bioinformatic prediction encode proteins which extend the functional response of R. sphaeroides to photooxidative stress known so far. Those proteins are involved in methionine sulfoxide reduction and aerobic respiration; homologs of RSP2617, a peptide methionine sulfoxide reductase, are involved in protection against oxidative stress in almost all organisms. Peptide methionine sulfoxide reductases catalyze the thioredoxin-dependent reduction of methionine sulfoxide (6, 19), which is generated by the reaction of methionine with reactive oxygen species, including 1O2 (8).
Some genes induced by 1O2 exposure in an RpoHII-dependent manner encode the following proteins that are involved in establishing and maintaining the respiratory system in the cell: QxtA (Rsp3213), the quinol oxidase I subunit; Cox15 (RSP3831), a putative cytochrome oxidase assembly factor; and Abc1 (RSP3305), a putative ubiquinol-cytochrome c reductase assembly protein belonging to the ABC1 family. The function of the respiratory chain is linked to the maintenance of an appropriate redox environment in the periplasm (3, 28). Induction of components of the respiratory pathway under photooxidative stress will keep the respiratory system functional after damages by 1O2; otherwise, uncontrolled electron transfer from dehydrogenases to O2 could lead to the formation of superoxide and hydrogen peroxide.
QxtA can counteract oxidative stress due to the maintenance of the quinone pool and its ability to limit accumulation of superoxide and peroxide by rapidly abstracting electrons from upstream dehydrogenases and transferring them to the oxidases (28). It was shown earlier that qxtA expression responds to redox alterations (21).
Furthermore, regulatory proteins encoded by the bioinformatically predicted genes could contribute to the photooxidative stress response. In this study and in earlier studies (11, 22), we observed proteins which are altered in synthesis upon 1O2 exposure, but the respective genes are not preceded by a putative promoter recognized by RpoE, RpoHII, or rather RpoHI and RpoHII. The RpoHI/RpoHII-dependent regulatory proteins may contribute to the expression of those proteins and thereby extend the functional response to 1O2.
Stress-inducible sRNAs extend the RpoHI and RpoHII regulons.
The sRNAs RSs0680a, RSs2461, and RSs1543 are induced under 1O2 and heat stress in an RpoHI/RpoHII-dependent manner. While we confirmed cotranscription of RSs0680a and RSs2461 together with their upstream genes, the sRNA RSs1543 is directly preceded by a conserved RpoHI/RpoHII binding site. Similar to its paralog RSs2461, also RSs1543 is located next to a transcriptional regulator, maybe implying combined regulatory action. For RSs2461, which is cotranscribed with ompR, it has to be explored if this sRNA somehow regulates expression of ompR or if OmpR-regulated genes are regulated in concert with RSs2461. The low but detectable levels of ompR mRNA and RSs2461 sRNA in the rpoHI rpoHII deletion strain obtained with 5′-RACE and Northern blot experiments indicate recognition by an additional sigma factor such as, e.g., RpoD. In enterobacteria like E. coli or Salmonella, OmpR itself induces sRNAs like, e.g., MicC or MicF, which are regulators of the outer membrane protein composition (31). Targets of the RpoHI/RpoHII-dependent sRNAs of R. sphaeroides are not identified to date, and no function can be assigned. In our proteome analysis of the 1O2-stressed rpoHI rpoHII deletion strain, we observed some protein spots, which are not present or less abundant in the stressed wild type. Somehow, the synthesis or stability of these proteins or the transcription of the respective genes is negatively affected by RpoHI/RpoHII. It is conceivable that the repression is due to (i) an RpoHI/RpoHII-dependent repressor or (ii) the action of the identified sRNAs.
Conclusion.
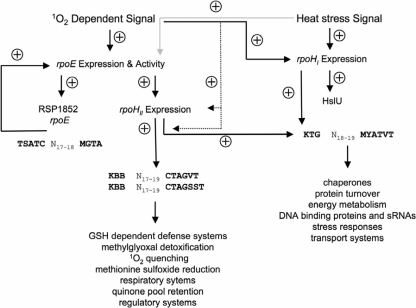
Our current model (Fig. 7) shows in which manner the response to photooxidative stress and heat overlap in R. sphaeroides and thereby substantially extends previous models (22, 37). In brief, 1O2 is recognized as a stress signal and increases the expression of rpoE and of those genes exhibiting a conserved RpoE promoter, including rpoHII. Heat stress also triggers rpoE gene expression at a very low level. RpoHII controls (i) genes specific to photooxidative stress and (ii) genes needed in the response to heat exposure by recognizing three different consensus target sites. Only one of those sites is also recognized by RpoHI, which very likely regulates the heat response in concert with RpoHII. Besides the RpoE promoter, the rpoHII gene putatively exhibits a second promoter recognized by itself and by RpoHI (see Table S4 in the supplemental material). This explains the low induction of the rpoHII gene in the rpoE chrR mutant strain TF18. More important, exposure to 1O2 and heat triggers the expression of several regulatory factors, including DNA binding proteins and sRNAs. We assume that those factors are important for the regulatory control of genes not exhibiting an RpoE, RpoHII, or RpoHI/RpoHII promoter.
FIG. 7.
Current model that displays the role of RpoE, RpoHI and RpoHII in the response to 1O2 and heat stress in R. sphaeroides. Solid black arrows indicate strong positive effects in the regulatory cascade triggered by 1O2 and heat stress. Solid gray arrows indicate less pronounced positive effects in the regulatory cascade. Dashed arrows indicate the function of hypothetical expression factors involved in the RpoE-RpoHII-dependent gene induction. Representative promoter sequences for RpoE (2, 11) and the putative promoter consensus sequences for RpoHII or rather RpoHI/RpoHII (Fig. 4B) are depicted. K: T or G; Y: T or C; B: T, C, or G; V: A, C, or G; M: C or A.
Supplementary Material
Acknowledgments
We thank Monica Zobawa and Friedrich Lottspeich for identification of proteins by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), Ulrike Ruppert for deletion of the rpoHI gene, and Johannes Schwarz for proteome analysis of the rpoHI deletion strains.
This project is funded by DFG grant Kl 563/20-1.
Footnotes
Published ahead of print on 19 March 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anthony, J. R., J. D. Newman, and T. J. Donohue. 2004. Interactions between the Rhodobacter sphaeroides ECF sigma factor, σE, and its anti-sigma factor, ChrR. J. Mol. Biol. 341:345-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, J. R., K. L. Warczak, and T. J. Donohue. 2005. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc. Natl. Acad. Sci. U. S. A. 102:6502-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bader, M., W. Muse, D. P. Ballou, C. Gassner, and J. C. Bardwell. 1999. Oxidative protein folding is driven by the electron transport system. Cell 98:217-227. [DOI] [PubMed] [Google Scholar]
- 4.Berghoff, B., A. J. Glaeser, C. Sharma, M. J. Vogel, and G. Klug. 2009. Photooxidative stress-induced and abundant small RNAs in Rhodobacter sphaeroides. Mol. Microbiol. 74:1497-1512. [DOI] [PubMed] [Google Scholar]
- 5.Braatsch, S., O. V. Moskvin, G. Klug, and M. Gomelsky. 2004. Responses of the Rhodobacter sphaeroides transcriptome to blue light under semiaerobic conditions. J. Bacteriol. 186:7726-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brot, N., and H. Weissbach. 2000. Peptide methionine sulfoxide reductase: biochemistry and physiological role. Biopolymers 55:288-296. [DOI] [PubMed] [Google Scholar]
- 7.Cogdell, R. J., T. D. Howard, R. Bittl, E. Schlodder, I. Geisenheimer, and W. Lubitz. 2000. How carotenoids protect bacterial photosynthesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:1345-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies, M. J. 2005. The oxidative environment and protein damage. Biochim. Biophys. Acta 1703:93-109. [DOI] [PubMed] [Google Scholar]
- 9.Drews, G. 1983. Mikrobiologisches Praktikum. Springer Verlag, Heidelberg, Germany.
- 10.Glaeser, J., and G. Klug. 2005. Photo-oxidative stress in Rhodobacter sphaeroides: protective role of carotenoids and expression of selected genes. Microbiology 151:1927-1938. [DOI] [PubMed] [Google Scholar]
- 11.Glaeser, J., M. Zobawa, F. Lottspeich, and G. Klug. 2007. Protein synthesis patterns reveal a complex regulatory response to singlet oxygen in Rhodobacter. J. Proteome Res. 6:2460-2471. [DOI] [PubMed] [Google Scholar]
- 12.Green, H. A., and T. J. Donohue. 2006. Activity of Rhodobacter sphaeroides RpoHII, a second member of the heat shock sigma factor family. J. Bacteriol. 188:5712-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross, C. A. 1996. Function and regulation of the heat shock proteins, p. 1382-1399. In N. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Rily, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 14.Hendrischk, A. K., S. W. Frühwirth, J. Moldt, R. Pokorny, S. Metz, G. Kaiser, A. Jager, A. Batschauer, and G. Klug. 2009. A cryptochrome-like protein is involved in the regulation of photosynthesis genes in Rhodobacter sphaeroides. Mol. Microbiol. 74:990-1003. [DOI] [PubMed] [Google Scholar]
- 15.Hübner, P., J. C. Willison, P. M. Vignais, and T. A. Bickle. 1991. Expression of regulatory nif genes in Rhodobacter capsulatus. J. Bacteriol. 173:2993-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janzon, L., S. Lofdahl, and S. Arvidson. 1986. Evidence for a coordinate transcriptional control of alpha-toxin and protein-a synthesis in Staphylococcus aureus. FEMS Microbiol. Lett. 33:193-198. [Google Scholar]
- 17.Karls, R. K., J. Brooks, P. Rossmeissl, J. Luedke, and T. J. Donohue. 1998. Metabolic roles of a Rhodobacter sphaeroides member of the sigma 32 family. J. Bacteriol. 180:10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 19.Levine, R. L., J. Moskovitz, and E. R. Stadtman. 2000. Oxidation of methionine in proteins: roles in antioxidant defense and cellular regulation. IUBMB Life 50:301-307. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Salazar, J. M., M. Sandoval-Calderon, X. Guo, S. Castillo-Ramirez, A. Reyes, M. G. Loza, J. Rivera, X. Alvarado-Affantranger, F. Sanchez, V. Gonzalez, G. Davila, and M. A. Ramirez-Romero. 2009. The Rhizobium etli RpoH1 and RpoH2 sigma factors are involved in different stress responses. Microbiology 155:386-397. [DOI] [PubMed] [Google Scholar]
- 21.Mouncey, N. J., E. Gak, M. Choudhary, J. Oh, and S. Kaplan. 2000. Respiratory pathways of Rhodobacter sphaeroides 2.4.1: identification and characterization of genes encoding quinol oxidases. FEMS Microbiol. Lett. 192:205-210. [DOI] [PubMed] [Google Scholar]
- 22.Nuss, A. M., J. Glaeser, and G. Klug. 2009. RpoHII activates oxidative-stress defense systems and is controlled by RpoE in the singlet oxygen-dependent response in Rhodobacter sphaeroides. J. Bacteriol. 191:220-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pappas, C. T., J. Sram, O. V. Moskvin, P. S. Ivanov, R. C. Mackenzie, M. Choudhary, M. L. Land, F. W. Larimer, S. Kaplan, and M. Gomelsky. 2004. Construction and validation of the Rhodobacter sphaeroides 2.4.1 DNA microarray: transcriptome flexibility at diverse growth modes. J. Bacteriol. 186:4748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prentki, P., A. Binda, and A. Epstein. 1991. Plasmid vectors for selecting IS1-promoted deletions in cloned DNA: sequence analysis of the omega interposon. Gene 103:17-23. [DOI] [PubMed] [Google Scholar]
- 26.Schilke, B. A., and T. J. Donohue. 1995. ChrR positively regulates transcription of the Rhodobacter sphaeroides cytochrome c2 gene. J. Bacteriol. 177:1929-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon, R., M. O'Connell, M. Labes, and A. Puhler. 1986. Plasmid vectors for the genetic analysis and manipulation of Rhizobia and other gram-negative bacteria. Methods Enzymol. 118:640-659. [DOI] [PubMed] [Google Scholar]
- 28.Soballe, B., and R. K. Poole. 2000. Ubiquinone limits oxidative stress in Escherichia coli. Microbiology 146(Pt. 4):787-796. [DOI] [PubMed] [Google Scholar]
- 29.Typas, A., G. Becker, and R. Hengge. 2007. The molecular basis of selective promoter activation by the sigma(S) subunit of RNA polymerase. Mol. Microbiol. 63:1296-1306. [DOI] [PubMed] [Google Scholar]
- 30.Typas, A., S. Stella, R. C. Johnson, and R. Hengge. 2007. The −35 sequence location and the Fis-sigma factor interface determine sigmas selectivity of the proP (P2) promoter in Escherichia coli. Mol. Microbiol. 63:780-796. [DOI] [PubMed] [Google Scholar]
- 31.Urban, J. H., and J. Vogel. 2007. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 35:1018-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Helden, J., B. Andre, and J. Collado-Vides. 2000. A web site for the computational analysis of yeast regulatory sequences. Yeast 16:177-187. [DOI] [PubMed] [Google Scholar]
- 33.van Niel, C. B. 1944. The culture, general physiology, morphology, and classification of the non-sulfur purple and brown bacteria. Bacteriol. Rev. 8:1-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 35.Wu, J., and A. Newton. 1996. Isolation, identification, and transcriptional specificity of the heat shock sigma factor sigma32 from Caulobacter crescentus. J. Bacteriol. 178:2094-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeller, T., and G. Klug. 2004. Detoxification of hydrogen peroxide and expression of catalase genes in Rhodobacter. Microbiology 150:3451-3462. [DOI] [PubMed] [Google Scholar]
- 37.Ziegelhoffer, E. C., and T. J. Donohue. 2009. Bacterial responses to photo-oxidative stress. Nat. Rev. Microbiol. 7:856-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.