Abstract
Type III secretion (T3S) is important for the establishment and maintenance of a chlamydial infection. The genes encoding T3S components in Chlamydia are transcribed as separate temporal classes, but the mechanisms that regulate the timing of their expression are not understood. In this study, we demonstrate that promoters for 10 predicted T3S transcriptional units are each transcribed in vitro by the major form of chlamydial RNA polymerase but not by an alternative form of RNA polymerase containing σ28. Since changes in DNA supercoiling during chlamydial development have been proposed as a mechanism for temporal gene regulation, we examined the in vitro response of T3S promoters to altered superhelical density. Promoters for three T3S genes that are upregulated at mid times were activated in response to increased DNA supercoiling. In contrast, promoters for three late T3S genes were not sensitive to changes in superhelical density. This differential response to changes in DNA topology is similar to the pattern that has been reported for representative mid and late chlamydial genes that are unrelated to the T3S system. Based on these results, we propose that the temporal expression of T3S genes in Chlamydia is controlled by general mechanisms that regulate σ66-dependent gene expression during the developmental cycle. Our results are consistent with a model in which T3S genes that are upregulated in mid cycle are activated together with other mid genes in response to increased DNA supercoiling.
Gram-negative pathogenic bacteria utilize a type III secretion (T3S) system to deliver virulence factors into eukaryotic cells. The components of this specialized secretion machinery include structural proteins that are conserved among different bacteria, as well as specific effector proteins and regulatory chaperones. A wide range of effectors have been described with activities that modulate host cell functions to promote infection. For example, Salmonella secretes a T3S effector, SipA, into M cells to modulate actin dynamics and induce bacterial uptake (10). After entry, Salmonella secretes a different set of T3S effectors, such as SpiC, which inhibits the fusion of the Salmonella-containing vacuole with endosomes (31).
The Gram-negative pathogen Chlamydia utilizes a T3S system at different stages of its obligate intracellular infection (11). All Chlamydia species encode conserved T3S structural genes (5), and treatment with T3S inhibitors prevents intracellular chlamydial growth (15, 23, 33). Although there is no chlamydial T3S assay, the T3S machinery of other bacteria has been used to provide functional evidence that chlamydial T3S effectors can be secreted (4, 6, 26). An example of a chlamydial T3S effector is the translocated actin-recruiting phosphoprotein (TARP) that is secreted into host cells, where it induces actin recruitment and nucleation (3). These localized cytoskeletal rearrangements are necessary for chlamydial uptake into a membrane-bound cytoplasmic compartment, called the chlamydial inclusion, where chlamydiae replicate. Intracellular chlamydiae secrete T3S effectors to modify the inclusion membrane. For example, IncA is a chlamydial T3S effector that is translocated into the inclusion membrane where it plays an important role in the fusion of chlamydial inclusions (8, 26). These examples demonstrate that the T3S is involved in both the initiation and maintenance of an intracellular chlamydial infection (11).
There are several distinctive features about the organization and regulation of the genes that encode the T3S system in Chlamydia. Unlike other Gram-negative bacteria, in which T3S genes are located on a pathogenicity island, the chlamydial T3S genes are present on six loci dispersed throughout the genome (5). This difference has led to the hypothesis that the T3S genes originated in Chlamydia and then spread by horizontal gene transfer to other Gram-negative bacteria (13). In C. trachomatis, the structural T3S genes have been proposed to be transcribed as 10 operons by the major form of chlamydial RNA polymerase (9) on the basis of their putative promoter sequences, but this prediction has not been experimentally tested (21, 30). Thus, the chlamydial T3S does not appear to be regulated by an alternative form of RNA polymerase, as in the case of Salmonella enterica (18). Chlamydia spp. also do not encode orthologs of AraC-like transcriptional activators, such as Yersinia VirF, that regulate T3S genes in other bacteria (14).
Another unusual feature of chlamydial T3S genes is that they are transcribed at different times during the chlamydial developmental cycle (9, 19, 24). The CT863 operon is transcribed at early time points, soon after entry of the organism into the host cell. In contrast, many T3S genes are transcribed at higher levels at mid cycle when chlamydiae are actively growing and replicating by binary fission. Another subset of T3S genes is upregulated late in the developmental cycle, during conversion from the metabolically active intracellular form to an extracellular, infectious form. The mechanisms that regulate this temporal expression of T3S genes in Chlamydia have not been defined.
The expression of genes in three temporal classes is a general feature of gene regulation in Chlamydia (2, 16, 22). Early genes are transcribed as soon as 1 h after infection, but the majority of chlamydial genes are not transcribed until mid times in the developmental cycle (2). We have proposed that the promoters of mid genes may be activated by the increased DNA supercoiling levels that we have measured during mid cycle (17). Furthermore, we have shown that representative mid-cycle promoters are transcribed at higher levels from more supercoiled templates in vitro (17), but we do not know if DNA supercoiling is a general mechanism for the activation of mid genes. Late genes are transcribed only at the end of the developmental cycle, and they appear to be regulated by two mechanisms. A subset is transcribed by the major chlamydial RNA polymerase, σ66 RNA polymerase, while another group of late genes is regulated by an alternative form of RNA polymerase containing σ28 instead of σ66 (34, 36). While the mechanism that regulates the σ66-dependent late genes has not been defined, it does not appear to involve DNA supercoiling since promoters for these genes were insensitive to changes in DNA supercoiling (17).
To understand how the T3S genes are regulated in Chlamydia, we examined if T3S promoters belonging to each temporal class are transcribed by different forms of chlamydial RNA polymerase and if transcription is affected by DNA topology. Our results indicate that there are similarities in the regulation of the promoters of T3S genes and non-T3S genes of the same temporal class. These results suggest that the temporal expression of T3S genes in Chlamydia may be regulated by general and not T3S-specific mechanisms.
MATERIALS AND METHODS
Construction of in vitro transcription plasmids.
Each promoter was cloned upstream of a promoterless G-less cassette transcription template as described previously (32). The promoter sequences were amplified by PCR from Chlamydia trachomatis serovar D genomic DNA and cloned into pMT1125 (32). All transcription plasmids and the primers used to clone the promoters contained therein are listed in Table 1.
TABLE 1.
Plasmids and cloning primers
| Plasmid | Promoter | Primer sequence | Region cloned |
|---|---|---|---|
| pMT1441 | cdsU | 5′-CTAGAATTCAAGAAAAAGCAAGATTAGTGCTTCA | −153 to +5 |
| 5′-AGGAATACTTCGCAAGTTACCG | |||
| pMT1442 | scc2 | 5′-CGCGAATTCTTGCAAGGCAACGACACACG | −144 to +5 |
| 5′-ATTATTAACAATATTAAATTCTAACGATCTGATTTAACAA | |||
| pMT1483 | CT863 | 5′-GGTTCTTTCGCACACCTTTC | −227 to +5 |
| 5′-AAAGGAGGGAACTTATCTAA | |||
| pMT1511 | cdsJ | 5′-AAAGAATTCGCGGATTCTGTTTTTGAAGC | −157 to +5 |
| 5′-TTTTTCACTCACCATAGCAAATG | |||
| pMT1512 | lpdA | 5′-ACCGAATTCGGCCAAAGAATCGCTCATAC | −169 to +5 |
| 5′-TATTTCCCGGGTTGTACAT | |||
| pMT1513 | cdsC | 5′-ACCGAATTCCCCCTAGAGATCCACGAACA | −146 to +5 |
| 5′-TTGTTTGGCAGAATATATCTATTT | |||
| pMT1514 | CT663 | 5′-CCCGAATTCAGTCGTTCGTGCCCGATT | −139 to +5 |
| 5′-ATTTTATGCTGAGTTATGCCAATAC | |||
| pMT1515 | CT665 | 5′-ACCGAATTCCGGTATCCAGGCCGTTATC | −170 to +5 |
| 5′-TAATTTTATATTTCAACCCTTCC | |||
| pMT1516 | fliF | 5′-CCCGAATTCCAGCTCTTTCGAGCTCATGTT | −154 to +5 |
| 5′-TTTTTGTCTTCTGTAACAAAAAGAGG | |||
| pMT1577 | incA | 5′-GTAGAATTCTTTCGTATCGCCAATGCAACA | −150 to +3 |
| 5′-TGAAACCAGATCCTATGGCTTTATATG |
Generation of transcription plasmid topoisomers.
For each of the following promoters cloned on a transcription plasmid (CT863, cdsC, fliF, incA, cdsJ, cdsU, and scc2), a series of topoisomers was generated using the method of Rhee et al. (20). For each plasmid, 10 μg of CsCl gradient-purified DNA was incubated for 3 h at 37°C with 5 U of wheat germ topoisomerase I (Promega) in a total of 40 μl containing 50 mM Tris-HCl (pH 7.6), 0.1 mM EDTA, 1 mM dithiothreitol, 50 mM NaCl, 10% glycerol, and various concentrations of ethidium bromide ranging from 0 to 48 μM. After incubation, the ethidium bromide was removed from the DNA by phenol-chloroform (1:1) extraction, followed by extraction with chloroform. The plasmid DNA was then recovered by ethanol precipitation and suspended in 30 μl of RNase-free TE buffer (10 mM Tris [pH 10], 1 mM EDTA). The purity and concentration of the DNA were assessed using a NanoDrop ND1000 spectrophotometer. The topoisomers were resolved on 1.4% agarose gels in 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA) with concentrations of ethidium bromide ranging from 0.02 to 0.16 μg/ml. Gel electrophoresis was performed at 3.5 V/cm with buffer circulation for 16 to 20 h at room temperature. The average difference in linking-number (ΔLK) for the topoisomers was determined by the band-counting method of Keller (12). The average superhelical density (σ) was calculated using the following formula: σ = −10.5(ΔLK/N), where N is the total number of base pairs in the plasmid.
Purification of chlamydial RNA polymerase.
L929 cells were infected with C. trachomatis serovar L2, and chlamydial σ66 RNA polymerase was partially purified at 21 h postinfection by heparin-agarose chromatography as previously described (29). Core enzyme lacking detectable σ66 activity was prepared from this partially purified RNA polymerase by gel filtration chromatography using a Superdex S200 10/300 GL column (GE Healthcare) on an AKTA Purifier 10 chromatography system (GE Healthcare). σ28 RNA polymerase was reconstituted by adding l μl of 15 nM recombinant chlamydial σ28 protein (34) to 4 μl of chlamydial core enzyme. Details of the transcription assays and the quantitation of promoter activity have been previously described (32, 34).
In vitro transcription.
Transcription experiments were performed as described previously (29), using 0.5 μl of chlamydial σ66 RNA polymerase or chlamydial σ28 RNA polymerase and 25 nM CsCl gradient-purified plasmid DNA or an individual plasmid topoisomer. The transcripts were resolved by electrophoresis on an 8 M urea-6% polyacrylamide gel, and the gels were fixed, dried, and exposed to a phosphor screen. The transcripts were visualized with a Personal FX phosphorimager (Bio-Rad). The amount of transcript produced by each promoter topoisomer was quantified using Quantity One software (Bio-Rad). Relative promoter activity was calculated by normalizing the amount of activity obtained from each topoisomer to the maximal promoter activity (defined as 100%) observed over the range of superhelical densities tested. In order to calculate a mean and standard deviation, three independent measurements of relative promoter activity were obtained for each set of promoter topoisomers.
RESULTS AND DISCUSSION
The T3S genes are transcribed by the major form of RNA polymerase in Chlamydia.
We first tested candidate promoters for 10 T3S operons in the C. trachomatis genome (Fig. 1) to determine if they were transcriptionally active. Nine of these promoters were predicted by Hefty and Stephens based on the mapping of in vivo transcription start sites (9). In addition we predicted a promoter for the T3S effector, incA, immediately upstream of a transcription start site mapped previously (26, 27). Each of these 10 candidate T3S promoters resembles the promoter transcribed by σ66 RNA polymerase (Table 2) (21, 28, 30) but not promoter sequences defined for Chlamydia σ28 RNA polymerase (35) or predicted for Chlamydia σ54 RNA polymerase (1).
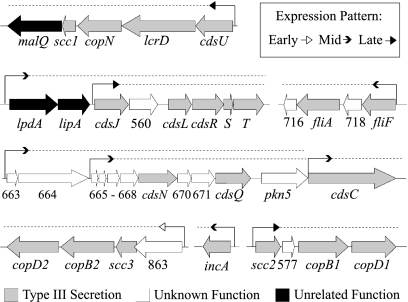
FIG. 1.
Transcriptional organization and temporal expression of T3S operons. For each operon, the promoter location is indicated by a bent arrow, followed by a dashed line above each gene in the operon, and the arrowheads indicate the temporal expression patterns of the first gene in each operon, as noted on the figure. Genes or open reading frames are identified by name or number, respectively, below the large filled arrows, which are color coded according to functional class. An unrelated function is one not involved in T3S.
TABLE 2.
Promoter sequences of chlamydial T3S operons
| Operona | Promoter sequenceb | Temporal class | Discriminatorc G/C content (%) |
|---|---|---|---|
| cdsU (CT091) | TTGAGAAAAACATTTATATACGGTAACTTGCCAAGT*A | Late | 50 |
| lpdA (CT557) | TTGAGATTTTATCCACCCAGATGTACAACCCGGG*A | Mid | 83 |
| cdsJ (CT559) | TTGGCACTAATCTCCCCATTTCGTATGGTGAGTG*A | Late | 50 |
| scc2 (CT576) | TTGTTAAATCAGATCGTTAGAATTTAATATTGTTAGT*A | Late | 25 |
| CT663 | TCTTTTTAAAGTAGGTATTGGCATAACTCAGCATA*A | Mid | 29 |
| CT665 | TTGTATCTTTTTAGAACGGGAAGGGTTGAAATATA*A | Mid | 0 |
| cdsC (CT674) | TTGCAAGATAGAGGGCAAATAGATATATTCTGCCA*A | Mid | 57 |
| fliF (CT719) | TTGTTTTTAATAGCCTCTTTTTGTTACAGAAGAC*A | Mid | 33 |
| CT863 | TTGCATGAAAAATACTTTTTAGATAAGTTCCCT*C | Early | 80 |
| incA (CT119) | TAGAATTTTTATCATATAAAGCCATAGGATCTGGT*T | Mid | 50 |
Each operon is identified by the name and/or the open reading frame identification number of its first gene.
Transcription start sites (*) were mapped by Hefty and Stephens (9), with the exception of that for incA, which was mapped by Suchland et al. (27) Promoter elements (−35 and −10 elements, underlined) were predicted by Hefty and Stephens (9), with the exception of the incA promoter, which was predicted in this study.
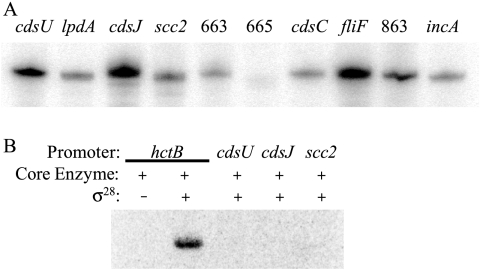
We tested these putative C. trachomatis T3S promoters for transcriptional activity with in vitro assays (32) since an experimental genetic system to test in vivo promoter activity in Chlamydia is not yet available. All transcription plasmids and the primers used to clone the promoters contained therein are listed in Table 1. We found that all 10 candidate T3S promoters were transcribed by σ66 RNA polymerase, which is the major form of chlamydial RNA polymerase (Fig. 2A). We also examined whether T3S promoters can be transcribed by σ28 RNA polymerase since this alternative RNA polymerase regulates a subset of late genes in Chlamydia (34, 36). However, σ28 RNA polymerase did not transcribe the promoters for three late T3S operons, cdsU, cdsJ, and scc2 (Fig. 2B). We were unable to test for σ54-dependent promoter activity because there is no assay for chlamydial σ54 RNA polymerase at this time. Taken together, these results indicate that the main T3S genes in Chlamydia are transcribed by the major form of RNA polymerase and do not support a role for the late regulator, σ28 RNA polymerase, in the expression of T3S genes. Additional mechanisms must exist, however, to explain how the T3S genes are transcribed by σ66 RNA polymerase as three temporal groups during the chlamydial developmental cycle (24).
FIG. 2.
In vitro transcription of candidate C. trachomatis T3S promoters. (A) Transcription of 10 candidate promoters with chlamydial σ66 RNA polymerase. The promoter for each operon is indicated by the name or the open reading frame number of its first gene. The promoters for the CT663, CT665, and CT863 operons are abbreviated as 663, 665, and 863, respectively. (B) Three late T3S promoters were not transcribed by chlamydial σ28 RNA polymerase. The hctB promoter is a known σ28-regulated promoter that was used as a positive control (33).
T3S promoters show different responses to changes in superhelical density.
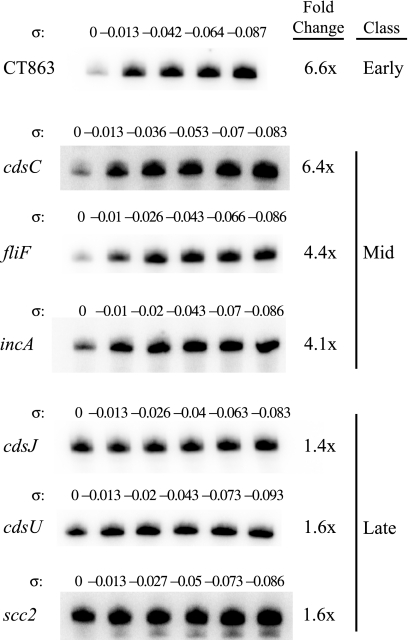
Since DNA supercoiling has been proposed as a mechanism to regulate mid-cycle genes but not late genes in Chlamydia (17), we examined whether T3S promoters are sensitive to alterations in DNA supercoiling. We performed a supercoiling-sensitivity transcription assay to test promoter activity at different supercoiling levels using plasmid topoisomers that differ only in superhelical density (17). We tested promoters for three T3S operons that are first expressed at mid times in the chlamydial developmental cycle (cdsC, fliF, and incA) and another three T3S operons that are not transcribed until late time points (cdsJ, cdsU, and scc2) (2, 9, 24). We also tested the promoter for the CT863 operon as a putative early T3S promoter based on detection of the CT863 transcript as early as 1.5 h postinfection (24). We cloned each T3S promoter on a transcription plasmid and generated a panel of topoisomers for each plasmid over a range of superhelical densities (σ) from 0 (completely relaxed) to >−0.08 (highly negatively supercoiled) (20) to encompass the physiologic range of supercoiling (−0.028 to −0.077) that we have measured in Chlamydia (17).
Four of the seven T3S promoters tested were transcribed in a supercoiling-responsive manner (Fig. 3). The CT863, cdsC, fliF, and incA promoters were upregulated by 4.1- to 6.6-fold in response to increased superhelical density. In contrast, the cdsJ, cdsU, and scc2 promoters were insensitive to changes in DNA supercoiling since they showed only 1.4-, 1.6-, and 1.6-fold respective changes in promoter activity across the range of superhelical densities tested. None of these T3S promoters was transcribed at higher levels from completely relaxed DNA.
FIG. 3.
Supercoiling sensitivity transcription assay. For each T3S promoter cloned on a transcription plasmid, a series of plasmid topoisomers was generated over a range of superhelical densities (σ) from 0 (completely relaxed DNA) to >−0.08 (highly negatively supercoiled). Each topoisomer was transcribed by σ66 RNA polymerase in an in vitro transcription reaction to measure the promoter activity at different superhelical densities. The greatest difference in promoter activity over this range of superhelical densities was calculated as fold change and is reported as the average of three independent experiments. For each promoter, the temporal expression class (early, mid, or late) is listed.
It has been proposed that the sequence of a chlamydial promoter may determine whether it is responsive to DNA supercoiling and that supercoiling-sensitive promoters contain a high proportion of G or C (G/C) sequence (17). The two most supercoiling-responsive promoters, CT863 and cdsC, have a high G/C content (80% and 57%, respectively) in the discriminator region, which is the sequence between the −10 promoter element and the transcription start site (Table 2). However, we found no clear correlation between the supercoiling responsiveness and the G/C content of the other T3S promoters.
The response of T3S promoters to alterations in DNA supercoiling correlates with their temporal expression pattern.
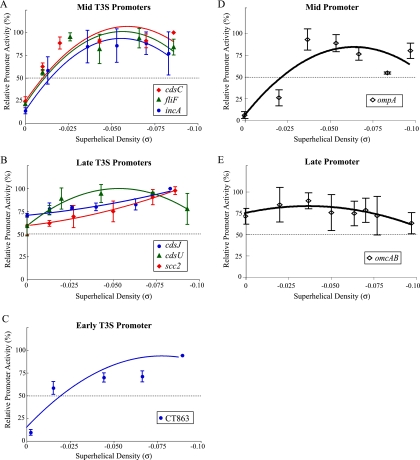
We next examined if there is a relationship between this differential response to DNA supercoiling and the temporal expression of T3S genes. For each promoter, we calculated the relative promoter activity by defining the maximal level of transcription at any superhelical density as 100% and normalizing other transcription levels to this value. To compare the promoters, we graphed the effects of superhelical density on the relative activity of each promoter. The promoters for the three mid-cycle T3S operons were transcribed at low levels from a relaxed template and at much higher levels from more supercoiled templates. Transcription of the cdsC, fliF, and incA promoters was highest at a superhelical density (σ) of approximately −0.06 and leveled off or decreased slightly for a σ of >−0.06 (Fig. 4A). These superhelical density optima are close to the in vivo superhelicity of −0.063 to −0.077 that we measured for the chlamydial plasmid in the reticulate body (RB) stage of the developmental cycle (17). In addition, this response to DNA supercoiling closely mirrors the supercoiling response pattern for promoters of two other non-T3S chlamydial mid genes, ompA (Fig. 4D) and pgk (17). ompA encodes the major outer membrane protein (7), and pgk is the gene for phosphoglycerol kinase (25); neither is known to be associated with the T3S system in Chlamydia.
FIG. 4.
Graphs showing the relationship between promoter activity and superhelical density for promoters of T3S genes belonging to mid (A), late (B), and early (C) temporal classes. For comparison, the graphs for mid (D) and late (E) promoters of non-T3S genes are shown (adapted from reference 17). Relative promoter activity was calculated as a percentage of the maximal promoter activity over the range of superhelical densities (σ) tested. A relative promoter activity of 50% is indicated by a dashed horizontal line on each graph. All reactions were performed as three independent experiments, and error bars represent standard deviations.
In contrast, the promoters for three late T3S operons, cdsJ, cdsU, and scc2, were not affected by alterations in DNA supercoiling. Transcript levels from each late promoter did not change over the range of superhelical densities, and the relative promoter activity was above 50% at all superhelical densities tested (Fig. 4B). Thus, the promoters for late T3S genes show a lack of response to DNA supercoiling that is similar to promoters for the late genes omcAB (Fig. 4E), hctA, and ltuB (17), which have no known connection to the T3S system.
The promoter for the single early T3S operon showed a supercoiling-responsive transcription pattern. The CT863 promoter was transcribed at low levels from a relaxed template and at higher levels with increasing superhelical density (Fig. 4C). The maximal activity was at the highest superhelical density tested (σ of −0.087), but unlike the three mid T3S promoters, the CT863 promoter did not reach a supercoiling optimum prior to this supraphysiologic level of DNA supercoiling. The CT863 promoter is the first early chlamydial promoter whose supercoiling response has been characterized, and thus it remains to be seen whether its supercoiling dependence is typical of early promoters. In summary, promoters of T3S genes show differential responses to changes in DNA supercoiling that correlate with their temporal patterns of gene expression. We found that three mid T3S promoters were transcribed in a supercoiling-dependent manner that is similar to the pattern described for two other mid chlamydial promoters (17). These in vitro findings are consistent with a model in which the T3S mid genes, together with other supercoiling-responsive mid genes, are activated by increased DNA supercoiling during mid cycle. In contrast, we demonstrated that three late T3S promoters were transcribed in a supercoiling-insensitive manner by σ66 RNA polymerase but were not transcribed by σ28 RNA polymerase. Thus, the late T3S genes appear to belong to the subset of supercoiling-independent, σ66-regulated late genes that includes omcAB, which encode two outer membrane proteins that are specific for elementary bodies, and hctA, the gene for the histone-like protein, Hc1, that condenses chlamydial DNA. The ability of late promoters to be well transcribed from relaxed templates is consistent with the low DNA supercoiling levels that we have measured at late time points in the chlamydial developmental cycle (17). Our results indicate that the temporal expression of T3S genes in Chlamydia appears to be controlled by general mechanisms that regulate developmental gene expression in this pathogenic bacterium.
Acknowledgments
We thank Christopher Rosario for providing σ28 RNA polymerase and Christine Sütterlin, Johnny Akers, Allan Chen, Eric Cheng, Kirsten Johnson, and Karen Kelley for critical reading of the manuscript and helpful advice.
This work was supported by a grant from the NIH (AI 71104 to E.M.P.). M.T. was supported by an NIH Independent Scientist Award (AI 057563).
Footnotes
Published ahead of print on 16 March 2010.
REFERENCES
- 1.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acids Res. 27:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belland, R. J., G. Zhong, D. D. Crane, D. Hogan, D. Sturdevant, J. Sharma, W. L. Beatty, and H. D. Caldwell. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U. S. A. 100:8478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clifton, D. R., K. A. Fields, S. S. Grieshaber, C. A. Dooley, E. R. Fischer, D. J. Mead, R. A. Carabeo, and T. Hackstadt. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U. S. A. 101:10166-10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fields, K. A., and T. Hackstadt. 2000. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol. 38:1048-1060. [DOI] [PubMed] [Google Scholar]
- 5.Fields, K. A., and T. Hackstadt. 2006. The Chlamydia type III secretion system: structure and implications for pathogenesis, p. 219-234. In P. Bavoil and P. Wyrick (ed.), Chlamydia: genomics and pathogenesis. Horizon Bioscience, Wymondham, United Kingdom.
- 6.Fields, K. A., D. J. Mead, C. A. Dooley, and T. Hackstadt. 2003. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 48:671-683. [DOI] [PubMed] [Google Scholar]
- 7.Frost, E. H., S. Deslandes, S. Veilleux, and D. Bourgaux-Ramoisy. 1991. Typing Chlamydia trachomatis by detection of restriction fragment length polymorphism in the gene encoding the major outer membrane protein. J. Infect. Dis. 163:1103-1107. [DOI] [PubMed] [Google Scholar]
- 8.Hackstadt, T., M. A. Scidmore-Carlson, E. I. Shaw, and E. R. Fischer. 1999. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell Microbiol. 1:119-130. [DOI] [PubMed] [Google Scholar]
- 9.Hefty, P. S., and R. S. Stephens. 2007. Chlamydial type III secretion system is encoded on ten operons preceded by σ70-like promoter elements. J. Bacteriol. 189:198-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higashide, W., S. Dai, V. P. Hombs, and D. Zhou. 2002. Involvement of SipA in modulating actin dynamics during Salmonella invasion into cultured epithelial cells. Cell Microbiol. 4:357-365. [DOI] [PubMed] [Google Scholar]
- 11.Jamison, W. P., and T. Hackstadt. 2008. Induction of type III secretion by cell-free Chlamydia trachomatis elementary bodies. Microb. Pathog. 45:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller, W. 1975. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc. Natl. Acad. Sci. U. S. A. 72:4876-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, J. F. 2001. Revisiting the chlamydial type III protein secretion system: clues to the origin of type III protein secretion. Trends Genet. 17:65-69. [DOI] [PubMed] [Google Scholar]
- 14.Lambert de Rouvroit, C., C. Sluiters, and G. R. Cornelis. 1992. Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol. Microbiol. 6:395-409. [PubMed] [Google Scholar]
- 15.Muschiol, S., S. Normark, B. Henriques-Normark, and A. Subtil. 2009. Small molecule inhibitors of the Yersinia type III secretion system impair the development of Chlamydia after entry into host cells. BMC Microbiol. 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholson, T. L., L. Olinger, K. Chong, G. Schoolnik, and R. S. Stephens. 2003. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J. Bacteriol. 185:3179-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niehus, E., E. Cheng, and M. Tan. 2008. DNA supercoiling-dependent gene regulation in Chlamydia. J. Bacteriol. 190:6419-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osborne, S. E., and B. K. Coombes. 2009. RpoE fine tunes expression of a subset of SsrB-regulated virulence factors in Salmonella enterica serovar Typhimurium. BMC Microbiol. 9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouellette, S. P., Y. M. Abdelrahman, R. J. Belland, and G. I. Byrne. 2005. The Chlamydia pneumoniae type III secretion-related lcrH gene clusters are developmentally expressed operons. J. Bacteriol. 187:7853-7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhee, K. Y., M. Opel, E. Ito, S. Hung, S. M. Arfin, and G. W. Hatfield. 1999. Transcriptional coupling between the divergent promoters of a prototypic LysR-type regulatory system, the ilvYC operon of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96:14294-14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaumburg, C. S., and M. Tan. 2003. Mutational analysis of the Chlamydia trachomatis dnaK promoter defines the optimal −35 promoter element. Nucleic Acids Res. 31:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw, E. I., C. A. Dooley, E. R. Fischer, M. A. Scidmore, K. A. Fields, and T. Hackstadt. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol. Microbiol. 37:913-925. [DOI] [PubMed] [Google Scholar]
- 23.Slepenkin, A., P. A. Enquist, U. Hagglund, L. M. de la Maza, M. Elofsson, and E. M. Peterson. 2007. Reversal of the antichlamydial activity of putative type III secretion inhibitors by iron. Infect. Immun. 75:3478-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slepenkin, A., V. Motin, L. M. de la Maza, and E. M. Peterson. 2003. Temporal expression of type III secretion genes of Chlamydia pneumoniae. Infect. Immun. 71:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 26.Subtil, A., C. Parsot, and A. Dautry-Varsat. 2001. Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol. Microbiol. 39:792-800. [DOI] [PubMed] [Google Scholar]
- 27.Suchland, R. J., B. M. Jeffrey, M. Xia, A. Bhatia, H. G. Chu, D. D. Rockey, and W. E. Stamm. 2008. Identification of concomitant infection with Chlamydia trachomatis IncA-negative mutant and wild-type strains by genomic, transcriptional, and biological characterizations. Infect. Immun. 76:5438-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan, M. 2006. Regulation of gene expression, p. 103-132. In P. Bavoil and P. Wyrick (ed.), Chlamydia: genomics and pathogenesis. Horizon Bioscience, Wymondham, United Kingdom.
- 29.Tan, M., and J. N. Engel. 1996. Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatis rRNA P1 promoter. J. Bacteriol. 178:6975-6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan, M., T. Gaal, R. L. Gourse, and J. N. Engel. 1998. Mutational analysis of the Chlamydia trachomatis rRNA P1 promoter defines four regions important for transcription in vitro. J. Bacteriol. 180:2359-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uchiya, K., M. A. Barbieri, K. Funato, A. H. Shah, P. D. Stahl, and E. A. Groisman. 1999. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18:3924-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson, A. C., and M. Tan. 2002. Functional analysis of the heat shock regulator HrcA of Chlamydia trachomatis. J. Bacteriol. 184:6566-6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf, K., H. J. Betts, B. Chellas-Gery, S. Hower, C. N. Linton, and K. A. Fields. 2006. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol. Microbiol. 61:1543-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu, H., and M. Tan. 2003. σ28 RNA polymerase regulates hctB, a late developmental gene in Chlamydia. Mol. Micro. 50:577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, H. H. Y., E. G. Di Russo, M. A. Rounds, and M. Tan. 2006. Mutational analysis of the promoter recognized by Chlamydia and Escherichia coli σ28 RNA polymerase. J. Bacteriol. 188:5524-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, H. H. Y., D. Kibler, and M. Tan. 2006. In silico prediction and functional validation of σ28-regulated genes in Chlamydia and Escherichia coli. J. Bacteriol. 188:8206-8212. [DOI] [PMC free article] [PubMed] [Google Scholar]