Abstract
Enterococcus faecalis strains secrete multiple peptides representing different sex pheromones that induce mating responses by bacteria carrying specific conjugative plasmids. The pheromone cAM373, which induces a response by the enterococcal plasmid pAM373, has been of interest because a similar activity is also secreted by Streptococcus gordonii and Staphylococcus aureus. The potential to facilitate intergeneric DNA transfer from E. faecalis is of concern because of extensive multiple antibiotic resistance, including vancomycin resistance, that has emerged among enterococci in recent years. Here, we characterize the related pheromone determinant in S. gordonii and show that the peptide it encodes, gordonii-cAM373, does indeed induce transfer of plasmid DNA from E. faecalis into S. gordonii. The streptococcal determinant camG encodes a lipoprotein with a leader sequence, the last 7 residues of which represent the gordonii-cAM373 heptapeptide SVFILAA. Synthetic forms of the peptide had activity similar to that of the enterococcal cAM373 AIFILAS. The lipoprotein moiety bore no resemblance to the lipoprotein encoded by E. faecalis. We also identified determinants in S. gordonii encoding a signal peptidase and an Eep-like zinc metalloprotease (lspA and eep, respectively) similar to those involved in processing certain pheromone precursors in E. faecalis. Mutations generated in camG, lspA, and eep each resulted in the ablation of gordonii-cAM373 activity in culture supernatants. This is the first genetic analysis of a potential sex pheromone system in a commensal oral streptococcal species, which may have implications for intergeneric gene acquisition in oral biofilms.
Streptococcus gordonii is a member of the human commensal oral flora. The species is one of the primary colonizers of the salivary pellicle coating the tooth surface and as such plays an important role in determining the composition of the multispecies microbial community that composes the biofilm known as dental plaque (27). If S. gordonii enters the bloodstream, it can circulate and potentially colonize the cardiac endothelium to cause infective endocarditis (6).
Due to the close proximity of microbial cells in biofilms, the potential exists for DNA transfer among the multiple genera present. Acquisition of exogenous DNA by naturally competent oral streptococci can lead to genetic plasticity that may enhance their survival under conditions of environmental stress. Autoinducing peptides, such as the competence factors of S. gordonii, as well as Streptococcus mutans and Streptococcus pneumoniae, well known as signaling agents able to facilitate genetic exchange via transformation (15), are involved. In the case of S. gordonii, these peptides can exhibit strain specificity (30), and in S. gordonii CH1 (Challis), the related peptide has been shown to induce genome-wide transcriptional changes (62).
Another signaling system described in Gram-positive cocci that facilitates DNA acquisition is the sex pheromone response that causes aggregation of planktonic cells and conjugative-plasmid transfer through a genetically controlled response induced by sex pheromone peptides. This was first described for conjugative plasmids in Enterococcus faecalis (19, 20), and multiple genes that are involved in this response are well characterized (11). The ability of S. gordonii strains CH1 and G9B to secrete a peptide that induces a response in certain plasmid-containing E. faecalis strains similar to a pheromone-related response suggested that these oral streptococci might have the ability to signal intergeneric genetic exchange (10). An examination of the S. gordonii genome (NCBI GenBank accession number CP000725) revealed a number of DNA regions with abrupt variations in G-C content and codon usage, implying that the organism may have acquired foreign DNA through horizontal gene transfer from other species (M. M. Vickerman, unpublished observation).
E. faecalis is a member of the normal intestinal flora and an opportunistic pathogen notoriously involved in nosocomial infections (40, 47, 48). Sex pheromone-responding conjugative plasmids are ubiquitous in the species and frequently encode bacteriocins, antibiotic resistance, and/or virulence traits (14). The pheromone-like peptide produced by S. gordonii, designated gordonii-cAM373, with activities similar to cAM373 in E. faecalis, induces a response characteristic of that encoded by a family of conjugative plasmids represented by pAM373 (10) and the vancomycin-resistant element pAM368 (54). A similar activity is produced by most strains of Staphylococcus aureus (10). Whereas studies of pAM373 suggested that it could transfer into these nonenterococcal genera, the plasmid is not able to replicate in these organisms (10, 26). Since streptococci and staphylococci are presumed to lack the surface enterococcal binding substance (EBS), to which the pAM373-encoded aggregation substance (AS) binds in order to initiate the formation of mating aggregates in planktonic broth matings, intergeneric transfer appears to depend on the more passive contact resulting from matings on solid surfaces (14). Such conditions might resemble those involving multispecies populations occurring as biofilms. Previous studies indicated that although it is unable to replicate in nonenterococcal species, pAM373 may deliver a plasmid-borne transposon (10). It may also facilitate or enhance the mobilization of other, broader-host-range plasmids that have been shown to become established following transfer between E. faecalis and S. gordonii (12, 51) or persist in recipients as part of a cointegrate structure bearing a replicative component (26, 54).
The enterococcal sex pheromones are octa- or heptapeptides corresponding to carboxyl-terminal segments of the signal sequences of lipoprotein precursors (9) that have been specifically processed (1, 2). A cAM373-like peptide also originates as part of a lipoprotein signal sequence in S. aureus; however, the molecules are encoded by different genes, and therefore, the mature lipoproteins of S. aureus and E. faecalis bear no similarity to each other (24).
Cleavage by lipoprotein signal peptidase to release the lipoprotein from its signal sequence is thought to precede further processing that produces the active pheromone (7). For cases analyzed so far, the sites within the precursors that correspond to the carboxyl termini of the pheromones resemble classic lipoprotein signal peptidase target motifs (9, 24). In the case of the E. faecalis pheromone cAD1, a membrane metalloprotease designated Eep is involved in processing the cleaved signal sequence at what becomes the amino terminus of the active pheromone peptide (2). Eep also processes other sex pheromone precursors but does not appear to process the cAM373 precursor in E. faecalis (2). In this communication, we identify the S. gordonii determinant encoding cAM373-like activity and reveal additional determinants, including an eep-like determinant, that appear to be required for production of the peptide.
MATERIALS AND METHODS
Bacterial strains, media, and reagents.
The bacterial strains used in this study are listed in Table 1. Gram-positive strains were grown at 37°C in Todd-Hewitt broth (THB) (Becton Dickinson and Co.), with the addition of 1.5% agar when solid medium was required. E. faecalis and S. aureus were incubated aerobically; incubations involving S. gordonii, alone or with E. faecalis, were in a candle jar or 5% CO2 unless otherwise noted. Escherichia coli DH5α was grown in LB medium. Antibiotics were purchased from Sigma and used at the following concentrations for streptococci and enterococci: erythromycin (Em), 10 μg/ml; fusidic acid (Fa), 25 μg/ml; rifampin (Rf), 25 μg/ml; spectinomycin (Sp), 500 μg/ml; streptomycin (Sm), 1,000 μg/ml; and tetracycline (Tc), 10 μg/ml. Ampicillin (Ap) and Sp were used at 100 μg/ml and 50 μg/ml, respectively, in E. coli DH5α for cloning. Common laboratory chemicals and reagents were purchased from Fisher Scientific or Sigma.
TABLE 1.
Strains and plasmids used in this study
| Strain and plasmid | Description and relevant characteristicsa | Reference |
|---|---|---|
| E. faecalis | ||
| 39-5 | Carries pPD1 | 63 |
| JH2-2 | Rfr Far | 33 |
| OG1SS | Smr Spr | 28 |
| OG1X | Smr | 32 |
| S. aureus | ||
| 879R4RF | Rfr Far | 10 |
| S. gordonii | ||
| CH1S | Spontaneous Smr derivative of strain Challis | 61 |
| CH1 | Strain Challis | 55 |
| CH1811 | Deletion of camG signal sequence | This study |
| CH1967 | Nonpolar replacement of lspA with aad9; Spr | This study |
| CH1187 | Insertional inactivation of eep; Emr | This study |
| E. coli | ||
| DH5α | Cloning host | Invitrogen |
| Plasmids | ||
| pAD1 | Encodes response to pheromone cAD1 | 57 |
| pAM373 | Encodes response to pheromone cAM373 | 10 |
| pAM378 | pAM373 with Tn918 insert; Tcr | 10 |
| pAM8301 | Carries oriT373; Cmr | 26 |
| pAM4020 | pAM373 with Tn917lac insert; Emr | 17 |
| pAMS470 | Cloned oriT373 in pAMS749-12; Emr | This study |
| pAMS749-12 | Cloning vector; replicates in Enterococcus and Streptococcus; Emr | 25 |
| pCF10 | Encodes response to pheromone cCF10 | 18 |
| pPD1 | Encodes response to pheromone cPD1 | 63 |
| pVA891 | Integrative cloning vector; Emr | 37 |
| pVA8911 | Integrative cloning vector; Emr | 39 |
| pBluescript SK(+) | Cloning vector; Apr | Stratagene |
| pVA891ΔcamG | pVA891 carrying camG flanking regions; Emr | This study |
| pVA8911eep | pVA8911 carrying internal fragment of eep; Emr | This study |
| pBSΔlspA:Sp | pBluescript SK(+) carrying aad9 flanked by lspA fragments; Apr Spr | This study |
Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Emr, erythromycin resistant; Far, fusidic acid resistant; Rfr, rifampin resistant; Smr, streptomycin resistant; Spr, spectinomycin resistant; Tcr, tetracycline resistant.
Transformation of competent S. gordonii cells was done with horse serum as previously described (61). E. faecalis and E. coli were transformed by electroporation as previously described (23).
S. gordonii strain construction. (i) S. gordonii strain CH1811.
A search of the S. gordonii Challis CH1 genome sequence (GenBank accession number CP000725) for a sequence with homology to the cAM373 peptide AIFILAS and the staph-cAM373 peptide AIFILAA revealed a 351-bp open reading frame (ORF) (locus SGO_1242) encoding a lipoprotein precursor (116 amino acids long) containing the sequence SVFILAA, representing the last 7 residues of a 19-residue signal sequence containing a consensus signal sequence cleavage site predicted by Signal P (5) immediately preceding a characteristic cysteine residue (Fig. 1). A strain in which the nucleotides encoding this potential pheromone peptide were deleted from the chromosome was constructed using a nonreplicative-plasmid integration and excision approach. To construct strain CH1811, an 810-bp DNA fragment homologous to the S. gordonii chromosomal region located 6 bp upstream of the DNA encoding the SVFILAA sequence was directionally cloned into the HindIII and BamHI sites of the integrative plasmid pVA891 (37). A second, ∼830-bp DNA fragment homologous to the chromosomal sequence beginning 19 bp downstream from the DNA encoding the carboxyl amino acid of the SVFILAA peptide was cloned into the BamHI and EcoRI sites. Primers for PCR amplification of the cloned fragments used in this construction also had two in-frame translational stop codons. The resulting desired DNA fragment cloned into pVA891 carried a 1.64-kb HindIII-EcoRI fragment in which an ∼50-bp region of SGO_1242 that included the putative pheromone coding region was replaced with a BamHI site flanked by two in-frame translational stop codons. The construct, designated pVA891ΔcamG, was used to transform competent CH1 cells, and putative transformants were selected by growth on an erythromycin agar plate, suggesting that the plasmid had integrated by a single Campbell-type crossover into the chromosome. The resulting transformants were confirmed by Southern hybridization analysis to have the integrated pVA891 plasmid flanked by “duplicate” 1.64-kb regions. The selected transformants were grown in antibiotic-free broth medium to allow excision of the integrated plasmid by recombination of the flanking homologous DNA regions and plated on Todd-Hewitt agar. Colonies were then picked to erythromycin plates; colonies that did not grow were screened to confirm the loss of the integrated plasmid. Loss of the plasmid and the presence of either the 50-bp deletion or the parental sequence were confirmed by Southern hybridization and nucleotide sequence analysis. As expected from the central location of the engineered 50-bp deletion, approximately 50% of the erythromycin-sensitive colonies had the desired genotype. One of the erythromycin-sensitive revertant strains with the desired chromosomal sequence, in which SGO_1242 encoded a truncated protein (MKKIYTLA-stop) as the result of the desired 50-bp deletion, was designated strain CH1811 and selected for further characterization in these studies.
FIG. 1.
Sequence of gordonii-cAM373 peptide and associated lipoprotein. (A) Deduced amino acid sequence of CamG from S. gordonii Challis CH1. The solid arrow marks the putative site of lipoprotein signal peptidase cleavage of the prelipoprotein, which removes the 19-amino-acid signal sequence. The proposed site of cleavage by the Eep homologue at the amino-terminal end of the gordonii-cAM373 peptide is marked by the open arrow. The gordonii-cAM373 peptide (SVFILAA) is underlined. Amino acid residues for the lipoprotein precursor molecule, consisting of 116 amino acids, are numbered. (B) Comparison of the amino acid sequences of the peptide pheromone molecules cAM373 of E. faecalis, staph-cAM373 of S. aureus, and gordonii-cAM373 of S. gordonii. Positions in staph-cAM373 and gordonii-cAM373 showing variation from cAM373 are boxed.
(ii) S. gordonii strain CH1967.
Since pheromone peptide processing requires cleavage of the lipoprotein signal sequence in E. faecalis, an lspA mutant was constructed in S. gordonii to determine if similar processing is necessary for pheromone activity. The S. gordonii putative lspA gene was identified at genome locus SGO_1089 as the only gene in the genome with the type II signal peptidase signature (PFam 01252; Prosite 00855 [16]). Since this ORF is located between two ORFs with overlapping coding sequences and may be cotranscribed in an operon (Fig. 2B), the mutant strain was constructed by replacement of an ∼250-bp internal fragment of the 468-bp lspA gene, including the nucleotides encoding the 13-amino-acid SPII signature sequence, with an aad9 spectinomycin resistance gene (35) with no promoter or terminator, in order to minimize the potential for polar effects and allow readthrough to the downstream genes. The desired DNA fragments were amplified by PCR with primers listed in Table 2 and cloned sequentially into pBluescript KS(+) (Stratagene) in E. coli DH5α. The resulting 2,350-bp cloned linear double-stranded DNA fragment, with a 780-bp aad9 gene between segments of lspA flanking DNA, was released from the cloning vector by digestion with XhoI and BamHI, electrophoresed, gel purified, and used to transform competent CH1 cells. Putative transformants were screened on spectinomycin agar plates, confirming the expression of the aad9 gene.
FIG. 2.
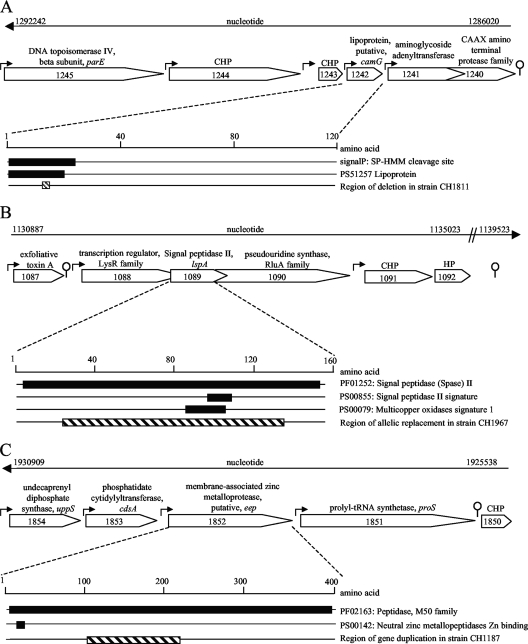
S. gordonii genome regions. The S. gordonii genome region is shown for each of the 3 genes of interest identified in these studies, SGO_1242 (camG) (A), SGO_1089 (lspA) (B), and SGO_1852 (eep) (C). The top line in each panel indicates the nucleotide numbers in the genome sequence in GenBank (accession number CP000725); the arrows at the end of each line show the ascending direction of the nucleotide numbers, indicating the orientation of the genome region pictured with respect to the chromosomal origin of replication. Beneath the genome nucleotide number line, each gene locus is indicated by the SGO number, which appears within an open arrow indicating the 5-prime-to-3-prime direction. The putative functional descriptions are based on the NCBI genome annotation and include conserved hypothetical (CHP) and hypothetical (HP) proteins. Putative bacterial promoters (forward arrows) and Rho-independent bacterial terminators (stem-loops) were identified in silico with the Bprom and FindTerm online software programs. Beneath camG, lspA, and eep are expansions that show the relevant features of the deduced CamG, LspA, and Eep proteins from the amino terminus (designated 1) to the carboxyl terminus. The filled blocks represent regions of conserved amino acids that share sequence homology to predicted functional domains identified in Prosite (PS) or pFam (PF) (22) or to signal sequences predicted by Signal P (41). The hatched bars represent the region of each encoded protein that was genetically manipulated in the construction of the isogenic mutant strains used in these studies (see Materials and Methods). (A) SGO_1242, which was designated camG, encodes a 116-amino-acid predicted lipoprotein with a processed signal sequence containing a peptide similar to cAM373. In silico analysis predicted that camG has its own promoter. Upstream of camG are two conserved hypothetical proteins, each with its own predicted promoter. Downstream of camG are two overlapping open reading frames with a single putative promoter; based upon sequence similarities, SGO_1241 encodes a 32.2-kDa protein similar to an aminoglycoside adenlytransferase (pFam04439) and SGO_1240 encodes a 27.2-kDa protein similar to CAAX amino-terminal proteases (pFam 02517) (42). No rho-independent terminators were detected in silico within the SGO_1244-through-SGO_1240 gene cluster. (B) SGO_1089, designated lspA, encodes a signal II peptidase signature motif (PF01252 and PS0085), as well as a conserved multicopper oxidase signature (PS00079). The lspA gene is located between two overlapping open reading frames with a single upstream promoter identified by both MacVector and Softberry promoter prediction software. In silico analysis of this operon indicated that the upstream gene (SGO_1088) encodes a putative transcriptional regulator of the LysR family. The downstream gene, SGO_1090, with its putative ribosomal binding site and start codon located within the lspA ORF, encodes a putative pseudouridine synthase. (C) SGO_1852, designated eep, encodes a 417-amino-acid protein with sequence similarity to E. faecalis Eep. The S. gordonii Eep carries a conserved zinc metalloprotease M-50 superfamily domain thought to be associated with the ability of a protein to cleave the transmembrane domains of substrate proteins, thereby transferring information and signals across bacterial membranes (38).
TABLE 2.
Oligonucleotide primers used in these studies
| Use | Primer name | Oligonucleotide primer sequence (5′ to 3′)a |
|---|---|---|
| Strain CH1967 construction | Xho1968UpF | ATCTCGAGGAATGGAGTTCTATGAACG |
| Hin3Stop1968UpR | ATAAGCTTAAGAGACAATAGCCCACTTAACCAAC | |
| Sma1966DownF | AACCCGGGTTGTCGATATGTTCCAATTTGAC | |
| Bam1966DownR | ATGGATCCGTTCCAAGACTTGGAAACGCG | |
| EcoRVplesSpF | ATGATATCTAGTGAGGAGGATATATTTGAATAC | |
| EcoR1plesSPR | ATGAATTCTTATAATTTTTTTAATCTGTTATT | |
| Probe construction to confirm lspA signature | LSPAdelF | AATAGGTGAAGTTAAGGGCT |
| LSPAdelR | CTTGGCGCAAGCGATCAATA | |
| Strain CH1811 construction | Hin1809F | GTAAGCTTGGAGACTTAACAATGTTTGAGTC |
| Bam1811R | TAGGATCCAAGCTATGCCAGTGTATAGAT | |
| Bam1811StopF | GCGGATCCTAGGCTTAATGGTACTTGGAAGG | |
| Eco1812R | TAGAATTCAGTCGCACTGGCTGGTGCCG | |
| Strain CH1187 construction | EcoR1StopEepF | ATGAATTCTAGACAGCCCTGCCAATGAATGTG |
| BamH1StopEepR | ATGGATCCTAACCAACCTTTAGAATCTGGTC | |
| Real-time PCR | SGO_1090L | CAGGACGAACTCATCAAATCC |
| SGO_1090R | CACCAGTTCGAGGATGGGTA | |
| SGO_1851L | ATGAGCGGGTTGGTGTTAAG | |
| SGO_1851R | CGATCAAGTTGTCTGCATGG | |
| SGO_1242L | TTGCTGAAAAAGGCGAAGTA | |
| SGO_1242R | CTCCATTAGTTGCAGAAATGC | |
| gyrA L | GAACGTCGAACGGAGCTTAT | |
| gyrA R | ACCTCCACGTTTTTGAGCTG |
Engineered 5′ restriction sites are underlined, and nucleotides modified to encode translational stop codons are italicized.
(iii) S. gordonii strain CH1187.
BLAST analysis of the S. gordonii genome identified only one locus that encoded a protein significantly similar (72% similar; 53% identical) to E. faecalis Eep. Gene locus SGO_1852 is a 1,254-bp ORF that, like the E. faecalis eep gene, encodes a membrane-associated zinc metalloprotease domain (Prosite 00142 [45]). To disrupt the eep homologue, a 370-bp internal fragment of the SGO_1852 ORF was flanked with engineered in-frame translational stop codons and cloned into the integrative plasmid pVA8911 (39). The resulting plasmid transformed CH1 competent cells and inserted into the chromosome via a single-crossover event. Putative transformants (Emr) were screened, and a transformant, designated strain CH1187, had the desired genotype with SGO_1852 disrupted by an integrated pVA8911 plasmid flanked by a 370-bp gene duplication.
(iv) Strain confirmation.
Mutant strains were confirmed by Southern hybridization and nucleotide sequence analysis of amplicons from chromosomal DNA. The strains were further confirmed to be derivatives of the parental strain CH1 by sequence analysis of the 16S rRNA gene using universal primers (4) and by comparison of sugar utilization patterns using inulin, salicin, mannitol, sorbitol, trehalose, melibiose, raffinose, lactose, sucrose, and glycerol as metabolic substrates.
(v) Quantitative real-time PCR.
RNA was prepared from mid-log-phase S. gordonii cells grown in THB, and real-time PCR was performed as previously described (43). Briefly, cultures were extracted with hot acid-phenol, and precipitated RNA was resuspended in diethyl pyrocarbonate (DEPC) water and purified via an RNeasy Midi spin column (Qiagen). After DNase treatment, the RNA was repurified with a second RNeasy Midi spin column, eluted in RNase-free H2O with 0.4 Units RNaseOUT (Invitrogen) per microliter, and stored in aliquots at −80°C. Primer3 software (http://frodo.wi.mit.edu/primer3/input.htm) (46) was used to design primers for real-time PCR (Table 2). The input parameters for primer pairs were 150-bp target amplicon size, 20-bp primer sequence length, a G-C content of 40 to 60% for the primer sequences, and an annealing-temperature range of 59 to 61°C. All primers were checked against the S. gordonii genome sequence to confirm their specificity. RNA preparations from strains CH1, CH1967, CH1187, and CH1811 were used as templates in real-time PCR to determine the expression of SGO_1090 located downstream of SGO_1089, disrupted in strain CH1967 (Fig. 2B), and SGO_1851 located downstream of SGO_1852, disrupted in strain CH1187 (Fig. 2C). The primers for camG expression were located within SGO_1242 downstream of the deleted nucleotides corresponding to the signal sequence (Fig. 2A). Expression of gyrA was measured in each strain for normalization. The iScript One-Step RT-PCR Kit with SYBR green (Bio-Rad Laboratories) was used according to the manufacturer's directions for reverse transcription and labeling of cDNA. Real-time data were collected using the MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad Laboratories). Data were analyzed using the Livak (2−ΔΔCT) method (36), and expression of SGO_1090, SGO_1851, or SGO_1242 was reported as a fold change normalized to gyrA expression in each strain.
E. faecalis strain construction.
A mobilizable replicon was created by subcloning oriT373 from pAM8301 (26) on a 0.5-kb XbaI-BamHI fragment into the erythromycin-resistant vector pAMS749-12 (25) digested with XbaI and BamHI, creating pAMS470, which replicates and expresses erythromycin resistance in both E. faecalis and S. gordonii. Subsequently, pAMS749-12 and pAMS470 were separately introduced into JH2-2/pAM378 by electroporation. Pheromone-sensing and transfer functions were active for pAM378 (12), which corresponds to pAM373 with an insert of Tn918 providing selectable tetracycline resistance. Agarose gel analyses with various restriction enzymes (data not shown) confirmed that the expected plasmids were intact and independent in each of the new donor strains, JH2-2/pAM378/pAMS749-12 and JH2-2/pAM378/pAMS470.
Pheromones and clumping assay.
Peptides were synthesized by the University of Michigan Biomedical Research Core Protein Structure facility and resuspended in dimethyl sulfoxide (DMSO) at a concentration of 1 mg/ml. Appropriate dilutions were made in THB. Activity was tested through the range of 0.05 to 1,000 ng/ml using a microtiter 2-fold dilution assay (20) essentially as previously described (24). Clumping of the responder strain was evaluated by eye, with additional verification in some cases using ×1,000 phase-contrast magnification.
Pheromone activity secreted into the culture medium was assayed in the following way. Fresh THB was inoculated with a 0.01-volume inoculum from an overnight culture and incubated at 37°C, either with shaking at 100 rpm for E. faecalis or S. aureus or stationary for S. gordonii. At late log stage (optical density at 650 nm [OD650], ∼0.85), the cultures were placed briefly on ice and pelleted. The supernatants were filtered through a 0.22-μm filter (Millipore; Millex PES membrane) to remove cells, placed in a boiling water bath for 15 min, and stored at 4°C. Activity in the culture filtrate was measured by utilizing a microtiter 2-fold dilution assay as described above, but using culture filtrate instead of diluted peptide. The titer was calculated as the highest dilution factor that still caused clumping of responder cells.
Conjugative plasmid transfer and analysis.
Filter matings were done essentially as previously described (24). Donors were incubated for 90 min in the presence of synthetic peptide at a concentration of 50 ng/ml for induction or in an equal volume of THB. After mating, the cells were resuspended by vortexing the filter in THB, diluted, and plated on selective antibiotic plates to determine the viable count (CFU/ml) of donors (Rf plus Fa), recipients (Sm), and transconjugants (Sm plus Em, or Sm plus Tc). For experiments using S. gordonii recipients, transconjugants were confirmed by their inability to grow on medium containing bile and sodium azide (Enterococcosel Agar; Becton Dickinson and Co.), a medium that inhibits growth of S. gordonii.
Plasmid DNA was purified by cell lysis using lysozyme and Sarkosyl, followed by centrifugation to equilibrium in a cesium chloride-ethidium bromide gradient essentially as previously described (24). Restriction enzymes were purchased from New England Biolabs. Agarose gel electrophoresis was as previously described (25).
RESULTS
Identification of the gordonii-cAM373 precursor determinant in S gordonii.
S. gordonii camG was detected by a visual search of all the encoded lipoproteins in the S. gordonii genome for predicted signal peptides that shared sequence similarity immediately upstream from their lipoprotein signal peptidase processing site to the cAM373 peptide AIFILAS and the staph-cAM373 peptide AIFILAA (24). SGO_1242 revealed an open reading frame encoding a putative lipoprotein precursor with the sequence SVFILAA, representing the last 7 residues of a 19-residue signal sequence immediately preceding a characteristic cysteine residue (Fig. 1 and 2A). The encoded 13.2-kDa lipoprotein, subsequently designated CamG, has no significant similarities to any bacterial proteins in the NCBI databases based on BLAST searches. Like that of the E. faecalis CamE lipoprotein (24), the CamG function is not known.
A synthetic form of the heptameric peptide SVFILAA, designated gordonii-cAM373, was generated and tested for cAM373-like activity using E. faecalis OG1X/pAM373 as a responder in a microtiter dilution and clumping assay routinely used to quantitate pheromone activity (20). The presence of a sex pheromone peptide induces visual clumping of cells that carry a conjugative plasmid encoding a response to that specific peptide. The gordonii-cAM373 peptide induced OG1X/pAM373 responder cell clumping at a concentration as low as 0.1 ng/ml. This was similar to the concentration of the S. aureus staph-cAM373 peptide (AIFILAA) but about 2-fold more than the amount of E. faecalis cAM373 needed to induce a response (24). The peptide appeared highly specific in that it had no detectable activity when tested similarly (in the range of 0.05 to 1,000 ng/ml) against strains of E. faecalis containing plasmids that respond to different pheromones: OG1X/pAD1 (responds to cAD1), OG1SS/pCF10 (responds to cCF10), and 39-5 (carries pPD1 and responds to cPD1).
To confirm that the pheromone-like response was due to processing of CamG to release gordonii-cAM373 peptide into the culture supernatant, S. gordonii strain CH1811 was constructed with a 42-bp deletion within camG that removed the SVFILAA coding region and replaced it with a premature translation stop codon. Although the S. gordonii parental strain CH1 culture supernatant could elicit a clumping response from OG1X carrying either pAM373 or pAM4020 (17), a pAM373 derivative that responds to cAM373, similar preparations from strain CH1811 were not able to generate responder cell clumping within the limits of detection of this assay (Table 3).
TABLE 3.
Absence of cAM373-like activity in S. gordonii mutants
| Culture filtrate straina | Titer using responder E. faecalisb |
|
|---|---|---|
| OG1X/pAM373 | OG1X/pAM4020 | |
| E. faecalis JH2-2 | 64-128 | 32-64 |
| S. aureus 879R4RF | 16-32 | 16 |
| S. gordonii | ||
| CH1 | 16-32 | 8 |
| CH1811 | <2 | <2 |
| CH1967 | <2 | <2 |
| CH1187 | <2 | <2 |
| THB broth control | <2 | <2 |
Culture filtrates were prepared as described in Materials and Methods.
Based on a microtiter dilution assay, the titer is the highest culture filtrate dilution factor still resulting in clumping of the responder. Titers are reported as the range in replicate tests (n ≥ 3). In this assay, a titer of 2 represents the lower limit of detection.
Lipoprotein signal processing is necessary for camG-related peptide activity.
Since in E. faecalis production of pheromone peptides requires cleavage of the lipoprotein signal sequence by the lipoprotein-specific signal peptidase, processing of gordonii-cAM373 was investigated in S. gordonii by constructing a strain in which the lipoprotein signal peptidase was inactivated. SGO_1089 (Fig. 2B) is the only S. gordonii strain CH1 gene locus encoding a signal peptidase II signature motif protein; during the genome annotation process, this gene was designated lspA. The lspA open reading frame encodes a ca. 17.4-kDa protein with hydrophobicity data (not shown) consistent with the predicted location of LspA as an integral membrane protein. The signal II peptidase family of integral membrane proteins processes lipoprotein precursors by cleaving the leader peptide adjacent to a conserved cysteine residue (44). Although signal peptidase is essential for cell viability in E. coli (64), mutants lacking this gene have been constructed in Bacillus subtilis and Lactococcus lactis (56, 60). Indeed, a nonpolar mutant S. gordonii strain, designated CH1967, was constructed which had doubling times similar to that of the parental CH1 strain in THB (data not shown). Like strain CH1811, culture supernatants of strain CH1967 were unable to induce clumping in cAM373 responder cells (Table 3), implying that lspA is involved in the processing of the gordonii-cAM373 precursor.
S. gordonii eep is required for pheromone processing.
In E. faecalis, the Eep protease has been previously reported to play an essential role in cAD1 pheromone synthesis by cleaving adjacent to what becomes the amino terminus site of the cAD1 peptide (2). Therefore, the S. gordonii genome was examined for a gene encoding a similar protease that might play a role in processing gordonii-cAM373. SGO_1852 was identified as the only S. gordonii locus that encodes a protein with sequence similarity to Eep (Fig. 2C). Like E. faecalis eep, S. gordonii SGO_1852 encodes a putative zinc metalloprotease. The ca. 45.6-kDa S. gordonii-encoded protein contains several predicted transmembrane regions with no predicted signal sequence (Signal P analysis [41]), consistent with Eep's predicted membrane-associated location necessary for signal processing. SGO_1852 was disrupted, and the resulting isogenic mutant (CH1187) was tested for the ability of the culture supernatant to induce clumping in responder cells. S. gordonii strain CH1187 was unable to elicit a clumping response from E. faecalis cAM373 responder cells (Table 3), indicating that SGO_1852 is important in gordonii-cAM373 processing; accordingly, this S. gordonii determinant was designated eep.
The loss of pheromone-like induction of responder cell clumping by each of the three isogenic mutants (Table 3) appears to be gene specific. Quantitative-PCR data verified that none of the S. gordonii mutant strains had decreased transcriptional expression downstream of the gene deletions or disruptions (Table 4).
TABLE 4.
Quantitative-PCR analyses to rule out potential polar effects of gene disruptions in S. gordonii strains
| ΔΔCT analysis of: | Strain | Avg CT | gyrA avg CT | ΔCT | ΔΔCT | Fold change (2−ΔΔCT)c |
|---|---|---|---|---|---|---|
| camG, SGO_1242 | CH1 | 19.12 | 17.03 | 2.09 | 0.00 | 1.00 |
| CH1811 | 17.21 | 15.65 | 1.56 | −0.52 | 1.44 | |
| SGO_1090a | CH1 | 19.13 | 16.98 | 2.15 | 0.00 | 1.00 |
| CH1967 | 19.14 | 16.99 | 2.14 | −0.01 | 1.01 | |
| SGO_1851b | CH1 | 16.69 | 17.03 | −0.35 | 0.00 | 1.00 |
| CH1187 | 15.49 | 16.71 | −1.21 | −0.87 | 1.82 |
Open reading frame immediately downstream from lspA (SGO_1089).
Open reading frame immediately downstream from eep (SGO_1852).
Relative expression normalized to gyrA; expression in mutant strains with disrupted SGO_1242 (camG), SGO_1089 (lspA), or SGO_1852 (eep) compared to strain CH1.
The S. gordonii camG-related peptide induces transfer of DNA from E. faecalis to S. gordonii.
Since pAM373 does not replicate in S. gordonii, we could not directly examine the ability of pAM373 to transfer from E. faecalis to S. gordonii. We therefore constructed a mobilizable plasmid, based on the vector pAMS749-12, that replicated in S. gordonii as well as E. faecalis. A segment of DNA containing the origin of transfer of pAM373 (oriT373) (26) was spliced into pAMS749-12. The resulting construct, pAMS470, which carried an erythromycin resistance determinant, was introduced by electroporation into E. faecalis strain JH2-2/pAM378 to generate JH2-2/pAM378/pAMS470 (pAM378 is a derivative of pAM373 with an insertion of the tetracycline resistance transposon Tn918 [closely related to Tn916] and exhibits a normal pheromone response to cAM373 [10]). When used as a donor, E. faecalis JH2-2/pAM378/pAMS470 was able to mobilize pAMS470 very efficiently to E. faecalis OG1X (which secretes cAM373) in 4-h filter matings (>100 transconjugants per donor); however, in the absence of pAM378, no transfer of pAMS470 was detected (<3.0 × 10−8 transconjugants per donor) (Table 5).
TABLE 5.
Intergeneric gordonii-cAM373-induced plasmid transfer
| Donora | Donor inductionb | Recipient | Filter mating time | Selection | Frequencyc | Cotransfer of unselected marker (%)d |
|---|---|---|---|---|---|---|
| JH2-2/pAM378/pAMS470 | No | OG1X | 4 h | Em | 7.8 × 100 | 30 |
| Tc | 4.4 × 100 | 86 | ||||
| JH2-2/pAMS470 | No | OG1X | 4 h | Em | < 3.0 × 10−8 | NA |
| JH2-2/pAM378/pAMS470 | No | CH1S | 30 min | Em | 3.8 × 10−7 | ND |
| Tc | 1.1 × 10−5 | ND | ||||
| JH2-2/pAM378/pAMS470 | Yes | CH1S | 30 min | Em | 7.0 × 10−4 | 0 |
| Tc | 1.1 × 10−4 | 7 | ||||
| JH2-2/pAM378/pAMS749-12 | No | CH1S | 30 min | Em | < 3.6 × 10−8 | NA |
| Tc | 4.8 × 10−6 | 0 | ||||
| JH2-2/pAM378/pAMS749-12 | Yes | CH1S | 30 min | Em | < 4.3 × 10−8 | NA |
| Tc | 7.3 × 10−6 | 0 | ||||
| JH2-2/pAMS470 | No | CH1S | 4 h | Em | < 6.5 × 10−8 | NA |
| JH2-2/pAM378 | No | CH1S | 4 h | Tc | 1.2 × 10−5 | NA |
pAM378 is a pAM373::Tn918 Tc-resistant derivative responsive to pheromone. pAMS749-12 is a cloning vector carrying Em resistance. pAMS470 is pAMS749-12 carrying oriT373.
Induction involved 90-min exposure of the donor to 50 ng/ml of the synthetic peptide gordonii-cAM373 before donor and recipient were mixed.
Conjugation frequencies are given as transconjugants per donor. Frequencies of conjugation to recipient CH1S are mean averages of 3 independent experiments. High rates for pheromone-induced conjugative transfer for positive-control E. faecalis recipients are consistent with published results (11).
To determine the cotransfer rate, 200 transconjugants in each case were screened for resistance to the unselected marker (Em or Tc). NA, not applicable; ND, not determined.
To determine if the gordonii-cAM373 peptide could induce mating functions in E. faecalis JH2-2/pAM378/pAMS470 that would facilitate transfer to S. gordonii, donors were exposed to 50 ng/ml of the peptide for 90 min prior to a 30-min filter mating with S. gordonii recipient CH1S (resistant to streptomycin). Transconjugants acquiring pAMS470 (Em resistant) were generated at a frequency 3 orders of magnitude higher than for donors not exposed to the peptide (7.0 × 10−4 per donor versus 3.8 × 10−7 per donor, respectively [Table 5]). Transfer of the Em resistance marker was not detected when the donor carried the vector pAMS749-12 instead of pAMS470, showing that mobilization depends on the presence of oriT373. In addition, pAMS470 was not able to transfer when pAM378, which supplies conjugation functions, was not coresident in the donor. For the JH2-2/pAM378/pAMS470 donor, the nonlinked (≤7% cotransfer) transfer of tetracycline resistance to S. gordonii is related to the efficient excision of Tn918 and insertion into the recipient chromosome, which is characteristic of conjugative transposons in the Tn916 family (13). Analysis by agarose gel electrophoresis (Fig. 3) of plasmid DNA in transconjugants from the induced mating of JH2-2/pAM378/pAMS470 with CH1S confirmed the presence of pAMS470 in Em-resistant transconjugants but showed no evidence of autonomously replicating pAM378 in Tc-resistant transconjugants. Overall, the data clearly indicate that gordonii-cAM373 can induce plasmid transfer from E. faecalis to S. gordonii.
FIG. 3.
Donor, recipient, and transconjugant plasmid analyses. Plasmid preparations from donor, recipient, and nine representative transconjugant strains were digested with EcoRI and analyzed by agarose gel electrophoresis. Lane 1, “1 Kb Plus DNA Ladder” molecular size standard (Invitrogen); lane 2, donor E. faecalis JH2-2/pAM378/pAMS470; lane 3, recipient S. gordonii CH1S (plasmid free); lanes 4 to 6, transconjugants resistant to erythromycin; lanes 7 to 9, transconjugants resistant to tetracycline; lanes 10 to 12, transconjugants resistant to erythromycin and tetracycline. The expected EcoRI digestion fragments from pAM378 were 25.5, 13.3, 10.0, 4.8, and 0.9 kb; those from pAMS470 were 4.9, 0.46, and 0.05 kb.
DISCUSSION
The production of a sex pheromone-like activity has previously been reported for S. gordonii strains Challis and G9B (10). In the present study, we identified a determinant, camG, in S. gordonii that encodes a lipoprotein precursor for which a heptapeptide derived from the processed signal sequence has an activity similar to that of the cAM373 pheromone produced by E. faecalis. The deduced gordonii-cAM373 amino acid sequence differs at three positions (Fig. 1) from that of the cAM373 peptide of E. faecalis. It has a serine and alanine at the amino and carboxyl termini, respectively, whereas the reverse is the case for E. faecalis cAM373. The other difference is at position 2, where a valine is present in the S. gordonii sequence rather than an isoleucine. The gordonii-cAM373 peptide differs from staph-cAM373 of S. aureus by only 2 residues, those at positions 1 and 2 (Fig. 1). The activity of the gordonii-cAM373 peptide is similar to that of staph-cAM373, as determined by cell-clumping assays (data not shown), which is about 50% that of the E. faecalis peptide (24), implying that the differences at positions 1 and 2 have little if any further effect. This seems consistent with the structural conservation of the corresponding amino acid residues.
The gordonii-cAM373 peptide appears to be derived from processing of a lipoprotein precursor by both a lipoprotein signal peptidase at the carboxyl terminus and an Eep homologue at the amino terminus; disruption of either lspA or eep ablated the clumping of E. faecalis in response to S. gordonii culture supernatant. It is noteworthy that while the previously reported Eep protein of E. faecalis processes the precursors of cAD1, cPD1, and cCF10 pheromones, it is not essential for processing the cAM373 precursor (2); another activity presumed to be directed toward the latter remains to be identified in E. faecalis. Whereas Eep is not required for expression of cAM373 in E. faecalis, an S. gordonii mutant with eep disrupted was unable to secrete the pheromone-like activity (strain CH1187 in Table 3), implying that S. gordonii Eep plays a role in gordonii-cAM373 processing. The reason for the difference is not clear. It is possible, however, that it is related to the major differences in the amino acid sequences that precede the pheromone moieties of cAM373 and gordonii-cAM373. Other than the first 4, the residues are quite different (24). Supportive of this view is a report by Chandler and Dunny (8) showing that a hybrid DNA construct in which the region corresponding to the cCF10 pheromone peptide was fused to DNA encoding the region upstream of cAM373 (from E. faecalis) expressed cCF10 with no dependence on Eep.
It has been proposed (20) that the peptide sex pheromones in E. faecalis may have functions other than signaling the availability of plasmid-free cells as potential recipients; however, any additional functions remain to be identified. It is possible, if not likely, that certain plasmids, such as pAM373, have evolved in such a way as to take advantage of specific secreted peptides as mating signals. Accordingly, it is likely that a sex pheromone role is not the primary function of gordonii-cAM373. One possibility is that the peptides constitute regulatory (e.g., feedback) signals that relate to expression of the associated lipoproteins encoded by the precursors. However, while mutations downstream of the encoded pheromones in the cognate lipoprotein moieties of cAM373 and cAD1 in E. faecalis did not eliminate secretion of the peptides, no change in the physiological behavior of the cells has yet been identified. Mutants unable to produce pheromone peptides exhibited normal growth, and there was no effect on the donor potential of such enterococcal variants carrying their respective peptide-responding plasmids (1, 24). In E. faecalis, the CamE lipoprotein from which cAM373 is processed is encoded by camE, which is flanked by genes encoding an upstream ABC transporter and a downstream cryptic protein and convergent thioredoxin reductase. In the case of staph-cAM373, the related camS is the fourth gene in an operon that includes a DNA helicase and a DNA ligase gene (24); a functional role for the CamS lipoprotein is unknown. Similarly, in S. gordonii, a function for the CamG lipoprotein has not been identified, and the chromosomal locations of camE, camS, and camG are unrelated.
While sex pheromone-related conjugative systems have not been described within bacterial genera unrelated to enterococci, the presence of cAM373-like activity in an oral streptococcal species is intriguing. The fact that the related lipoprotein precursors are processed by an Eep-like activity suggests a purpose for the resulting peptide, unless it simply represents a step in the degradation of the signal peptide. It is noteworthy that certain enterococcal pheromone-related peptides, including cAM373, have been reported to exhibit neutrophil chemotaxis activities (21, 49); however, such activities have not been examined in the case of the nonenterococcal cAM373-related peptides. It is possible that nonenterococcal peptides are also able to influence/modulate neutrophils or perhaps other mammalian host defense assets.
The fact that gordonii-cAM373 is able to induce a mating response encoded by pAM373-like plasmids in E. faecalis and has now been shown to mobilize an erythromycin resistance plasmid carrying the pAM373 oriT from E. faecalis to S. gordonii implies that the peptide may play a significant role in facilitating the transmission of DNA and possibly antibiotic resistance (e.g., a vancomycin resistance trait, such as that carried by the cAM373-responsive pAM368) to S. gordonii. Furthermore, since enterococci have been found in the oral environment (50, 52, 53), the usual location for S. gordonii (27), and conversely, oral streptococci have been identified in the gut environment (31), routinely inhabited by E. faecalis, the opportunity for cohabitation of the two organisms further supports the potential for genetic exchange.
Recent determinations of genome sequences further support the idea that commensal bacteria may act as reservoirs for potential virulence factors or ecologically disruptive genes that are selected for and expressed under conditions of environmental stress or antibiotic selective pressures. Clinical acquisition of antibiotic resistance determinants is thought to occur between species that are closely related phylogenetically (3). Genes encoding signaling molecules may be similarly exchanged among related species. Gene order plots comparing sequenced chromosomes of several streptococcal species (Fig. 4) suggest that the region between the parE and parC genes, encoding the DNA topoisomerase IV beta and alpha subunits, respectively, may be a hot spot for DNA recombination and integration. Indeed, the possibility of genetic recombination in this region has been noted because of its potential for transmitting fluoroquinolone resistance due to recombination among the parE and parC genes in S. pneumoniae and viridans streptococci (34, 59). In S. gordonii, camG is one of several genes located between parE and parC (Fig. 4). A multigene region between parE and parC is also seen in the other oral streptococcal species, Streptococcus sanguinis and S. mutans. BLAST searches and sequence comparisons of some of the genes suggest that they may have arisen from the remnants of a lactococcal phage (29, 58). The genomic-comparison data suggest that S. gordonii may have acquired camG through horizontal transfer. PCR analyses with camG-specific primers and chromosomal DNA of S. gordonii strain G9B (data not shown), which also elicits an aggregation response in pAM373-containing E. faecalis cells (10), suggest that camG is not unique to strain CH1. It is tempting to speculate that the acquisition and maintenance of camG in S. gordonii has the potential to signal additional horizontal gene transfer events involving other organisms in its ecological niche, further contributing to the genetic plasticity of this oral commensal bacterium.
FIG. 4.
Comparison of parC-parE chromosomal regions of streptococcal species. Comparison of genomic regions of representative streptococcal species indicated that the region flanking S. gordonii camG is potentially a hot spot for genetic integration and/or recombination. The top line is a nucleotide scale in kb for all the genomic regions. The parE and parC genes, encoding DNA topoisomerase IV beta and alpha subunits, respectively, and their immediate flanking genes and intergenic regions are shown for six representative streptococcal species: S. gordonii strain CH1 (GenBank CP000725; nucleotides 1293043 to 1282321), S. mutans strain UA159 (GenBank AE014133; nucleotides 1153799 to 1142879), S. sanguinis strain SK36 (GenBank CP000387; nucleotides 1257686 to 1247679); S. pneumoniae strain R6 (GenBank AE007317; nucleotides 749110 to 755873), S. pyogenes strain M1 GAS (GenBank AE004092; nucleotides 751576 to 757996), and S. agalactiae strain 2603 V/R (GenBank AE009948; nucleotides 1161100 to 1154670). The arrows indicate the size and direction of each ORF. The number beneath each ORF is the gene locus number. The name of each gene or annotation description is given above each arrow. Open arrows represent the genes with conserved gene order among all six species. The ORFs between parE and parC in the oral streptococcal species (S. gordonii, S. mutans, and S. sanguinis) were compared to each other by BLAST search at the amino acid and nucleotide levels. ORFs that showed no significant similarity to any of the ORFs in the intergenic regions of any of the three oral streptococcal species are shaded black. ORFs that showed significant similarity to each other are shaded gray (S. gordonii SGO_1243, S. mutans SMU.1208, and S. sanguinis SSA_1230) or hatched (SGO_1241 and SSA_1227). The region between parE and parC contains genes that suggest a phage or plasmid origin. The hypothetical protein (HP) encoded by SGO_1244 has amino acid regions with significant similarity to the Abi family proteins (PF07751) involved in bacteriophage resistance mediated by abortive infection in lactococcal species (29). The fic gene of S. mutans, which despite its location has no homology to camG, encodes a putative mobilization protein similar to that in lactococcal plasmid pNZ4000 (58).
Acknowledgments
This work was supported by Public Health Service grant DE11090 from the National Institute of Dental and Craniofacial Research to M.M.V., the University of Michigan Office of the Vice President for Research, the University of Michigan Horace H. Rackham Graduate School, and the University of Michigan School of Dentistry.
Footnotes
Published ahead of print on 16 March 2010.
REFERENCES
- 1.An, F. Y., and D. B. Clewell. 2002. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 184:1880-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, F. Y., M. C. Sulavik, and D. B. Clewell. 1999. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 181:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andremont, A. 2003. Commensal flora may play key role in spreading antibiotic resistance. ASM News 69:601-607. [Google Scholar]
- 4.Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541-555. [DOI] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.Beynon, R. P., V. K. Bahl, and B. D. Prendergast. 2006. Infective endocarditis. BMJ 333:334-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391-398. [DOI] [PubMed] [Google Scholar]
- 8.Chandler, J. R., and G. M. Dunny. 2008. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J. Bacteriol. 190:1172-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clewell, D. B., F. Y. An, S. E. Flannagan, M. Antiporta, and G. M. Dunny. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol. Microbiol. 35:246-247. [DOI] [PubMed] [Google Scholar]
- 10.Clewell, D. B., F. Y. An, B. A. White, and C. Gawron-Burke. 1985. Streptococcus faecalis sex pheromone (cAM373) also produced by Staphylococcus aureus and identification of a conjugative transposon (Tn918). J. Bacteriol. 162:1212-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clewell, D. B., and G. Dunny. 2002. Conjugation and genetic exchange in enterococci, p. 265-300. In M. S. Gilmore (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC.
- 12.Clewell, D. B., G. F. Fitzgerald, L. Dempsey, L. E. Pearce, F. Y. An, B. A. White, and C. Gawron-Burke. 1985. Streptococcal conjugation: plasmids, sex pheromones, and conjugative transposons, p. 194-203. In S. E. Mergenhagen and B. Rosan (ed.), Molecular basis of oral microbial adhesion. American Society for Microbiology, Washington, DC.
- 13.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 14.Clewell, D. B., and M. V. Francia. 2004. Conjugation in gram-positive bacteria, p. 227-256. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, DC.
- 15.Cvitkovitch, D. G. 2001. Genetic competence and transformation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:217-243. [DOI] [PubMed] [Google Scholar]
- 16.Dalbey, R. E., and G. Von Heijne. 1992. Signal peptidases in prokaryotes and eukaryotes—a new protease family. Trends Biochem. Sci. 17:474-478. [DOI] [PubMed] [Google Scholar]
- 17.De Boever, E. H., D. B. Clewell, and C. M. Fraser. 2000. Enterococcus faecalis conjugative plasmid pAM373: complete nucleotide sequence and genetic analyses of sex pheromone response. Mol. Microbiol. 37:1327-1341. [DOI] [PubMed] [Google Scholar]
- 18.Dunny, G., M. Yuhasz, and E. Ehrenfeld. 1982. Genetic and physiological analysis of conjugation in Streptococcus faecalis. J. Bacteriol. 151:855-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. U. S. A. 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunny, G. M., R. A. Craig, R. L. Carron, and D. B. Clewell. 1979. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid 2:454-465. [DOI] [PubMed] [Google Scholar]
- 21.Ember, J. A., and T. E. Hugli. 1989. Characterization of the human neutrophil response to sex pheromones from Streptococcus faecalis. Am. J. Pathol. 134:797-805. [PMC free article] [PubMed] [Google Scholar]
- 22.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flannagan, S. E., and D. B. Clewell. 1991. Conjugative transfer of Tn916 in Enterococcus faecalis: trans activation of homologous transposons. J. Bacteriol. 173:7136-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flannagan, S. E., and D. B. Clewell. 2002. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol. Microbiol. 44:803-817. [DOI] [PubMed] [Google Scholar]
- 25.Flannagan, S. E., D. B. Clewell, and C. M. Sedgley. 2008. A “retrocidal” plasmid in Enterococcus faecalis: passage and protection. Plasmid 59:217-230. [DOI] [PubMed] [Google Scholar]
- 26.Francia, M. V., and D. B. Clewell. 2002. Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: identification of the pAD1 nic site, a specific relaxase and a possible TraG-like protein. Mol. Microbiol. 45:375-395. [DOI] [PubMed] [Google Scholar]
- 27.Frandsen, E. V., V. Pedrazzoli, and M. Kilian. 1991. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol. Immunol. 6:129-133. [DOI] [PubMed] [Google Scholar]
- 28.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garvey, P., G. F. Fitzgerald, and C. Hill. 1995. Cloning and DNA sequence analysis of two abortive infection phage resistance determinants from the lactococcal plasmid pNP40. Appl. Environ. Microbiol. 61:4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Havarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi, H., R. Takahashi, T. Nishi, M. Sakamoto, and Y. Benno. 2005. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J. Med. Microbiol. 54:1093-1101. [DOI] [PubMed] [Google Scholar]
- 32.Ike, Y., R. A. Craig, B. A. White, Y. Yagi, and D. B. Clewell. 1983. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc. Natl. Acad. Sci. U. S. A. 80:5369-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janoir, C., I. Podglajen, M. D. Kitzis, C. Poyart, and L. Gutmann. 1999. In vitro exchange of fluoroquinolone resistance determinants between Streptococcus pneumoniae and viridans streptococci and genomic organization of the parE-parC region in S. mitis. J. Infect. Dis. 180:555-558. [DOI] [PubMed] [Google Scholar]
- 35.LeBlanc, D. J., L. N. Lee, and J. M. Inamine. 1991. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1804-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 37.Macrina, F. L., R. P. Evans, J. A. Tobian, D. L. Hartley, D. B. Clewell, and K. R. Jones. 1983. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25:145-150. [DOI] [PubMed] [Google Scholar]
- 38.Marchler-Bauer, A., J. B. Anderson, F. Chitsaz, M. K. Derbyshire, C. DeWeese-Scott, J. H. Fong, L. Y. Geer, R. C. Geer, N. R. Gonzales, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, A. Tasneem, N. Thanki, R. A. Yamashita, D. Zhang, N. Zhang, and S. H. Bryant. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205-D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mechold, U., M. Cashel, K. Steiner, D. Gentry, and H. Malke. 1996. Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J. Bacteriol. 178:1401-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray, B. E. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 42.Pei, J., and N. V. Grishin. 2001. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem. Sci. 26:275-277. [DOI] [PubMed] [Google Scholar]
- 43.Petersen, H. J., C. Keane, H. F. Jenkinson, M. M. Vickerman, A. Jesionowski, J. C. Waterhouse, D. Cox, and S. W. Kerrigan. 2010. Human platelets recognize a novel surface protein PadA on Streptococcus gordonii through a unique interaction involving fibrinogen receptor GPIIbIIIa. Infect. Immun. 78:413-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi, H. Y., K. Sankaran, K. Gan, and H. C. Wu. 1995. Structure-function relationship of bacterial prolipoprotein diacylglyceryl transferase: functionally significant conserved regions. J. Bacteriol. 177:6820-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rawlings, N. D., and A. J. Barrett. 1995. Evolutionary families of metallopeptidases. Methods Enzymol. 248:183-228. [DOI] [PubMed] [Google Scholar]
- 46.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 47.Ruoff, K. L., L. de la Maza, M. J. Murtagh, J. D. Spargo, and M. J. Ferraro. 1990. Species identities of enterococci isolated from clinical specimens. J. Clin. Microbiol. 28:435-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahm, D. F., H. K. Marsilio, and G. Piazza. 1999. Antimicrobial resistance in key bloodstream bacterial isolates: electronic surveillance with the Surveillance Network Database—USA. Clin. Infect. Dis. 29:259-263. [DOI] [PubMed] [Google Scholar]
- 49.Sannomiya, P., R. A. Craig, D. B. Clewell, A. Suzuki, M. Fujino, G. O. Till, and W. A. Marasco. 1990. Characterization of a class of nonformylated Enterococcus faecalis-derived neutrophil chemotactic peptides: the sex pheromones. Proc. Natl. Acad. Sci. U. S. A. 87:66-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sedgley, C., A. Nagel, G. Dahlen, C. Reit, and A. Molander. 2006. Real-time quantitative polymerase chain reaction and culture analyses of Enterococcus faecalis in root canals. J. Endod. 32:173-177. [DOI] [PubMed] [Google Scholar]
- 51.Sedgley, C. M., E. H. Lee, M. J. Martin, and S. E. Flannagan. 2008. Antibiotic resistance gene transfer between Streptococcus gordonii and Enterococcus faecalis in root canals of teeth ex vivo. J. Endod. 34:570-574. [DOI] [PubMed] [Google Scholar]
- 52.Sedgley, C. M., S. L. Lennan, and D. B. Clewell. 2004. Prevalence, phenotype and genotype of oral enterococci. Oral Microbiol. Immunol. 19:95-101. [DOI] [PubMed] [Google Scholar]
- 53.Sedgley, C. M., A. C. Nagel, C. E. Shelburne, D. B. Clewell, O. Appelbe, and A. Molander. 2005. Quantitative real-time PCR detection of oral Enterococcus faecalis in humans. Arch. Oral Biol. 50:575-583. [DOI] [PubMed] [Google Scholar]
- 54.Showsh, S. A., E. H. De Boever, and D. B. Clewell. 2001. Vancomycin resistance plasmid in Enterococcus faecalis that encodes sensitivity to a sex pheromone also produced by Staphylococcus aureus. Antimicrob. Agents Chemother. 45:2177-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sulavik, M. C., G. Tardif, and D. B. Clewell. 1992. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J. Bacteriol. 174:3577-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tjalsma, H., V. P. Kontinen, Z. Pragai, H. Wu, R. Meima, G. Venema, S. Bron, M. Sarvas, and J. M. van Dijl. 1999. The role of lipoprotein processing by signal peptidase II in the Gram-positive eubacterium Bacillus subtilis. Signal peptidase II is required for the efficient secretion of alpha-amylase, a non-lipoprotein. J. Biol. Chem. 274:1698-1707. [DOI] [PubMed] [Google Scholar]
- 57.Tomich, P. K., F. Y. An, S. P. Damle, and D. B. Clewell. 1979. Plasmid-related transmissibility and multiple drug resistance in Streptococcus faecalis subsp. zymogenes strain DS16. Antimicrob. Agents Chemother. 15:828-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Kranenburg, R., M. Kleerebezem, and W. M. de Vos. 2000. Nucleotide sequence analysis of the lactococcal EPS plasmid pNZ4000. Plasmid 43:130-136. [DOI] [PubMed] [Google Scholar]
- 59.Varon, E., and L. Gutmann. 2000. Mechanisms and spread of fluoroquinolone resistance in Streptococcus pneumoniae. Res. Microbiol. 151:471-473. [DOI] [PubMed] [Google Scholar]
- 60.Venema, R., H. Tjalsma, J. M. van Dijl, A. de Jong, K. Leenhouts, G. Buist, and G. Venema. 2003. Active lipoprotein precursors in the Gram-positive eubacterium Lactococcus lactis. J. Biol. Chem. 278:14739-14746. [DOI] [PubMed] [Google Scholar]
- 61.Vickerman, M. M., D. G. Heath, and D. B. Clewell. 1993. Construction of recombination-deficient strains of Streptococcus gordonii by disruption of the recA gene. J. Bacteriol. 175:6354-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vickerman, M. M., S. Iobst, A. M. Jesionowski, and S. R. Gill. 2007. Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J. Bacteriol. 189:7799-7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yagi, Y., R. E. Kessler, J. H. Shaw, D. E. Lopatin, F. An, and D. B. Clewell. 1983. Plasmid content of Streptococcus faecalis strain 39-5 and identification of a pheromone (cPD1)-induced surface antigen. J. Gen. Microbiol. 129:1207-1215. [DOI] [PubMed] [Google Scholar]
- 64.Yamagata, H., C. Ippolito, M. Inukai, and M. Inouye. 1982. Temperature-sensitive processing of outer membrane lipoprotein in an Escherichia coli mutant. J. Bacteriol. 152:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]