Abstract
The proline-rich homeodomain protein (PRH) plays multiple roles in the control of gene expression during embryonic development and in the adult. Vascular endothelial growth factor (VEGF) is a mitogen that stimulates cell proliferation and survival via cell surface receptors including VEGFR-1 and VEGFR-2. VEGF signaling is of critical importance in angiogenesis and hematopoiesis and is elevated in many tumors. Here we show that PRH binds directly to the promoter regions of the Vegf, Vegfr-1, and Vegfr-2 genes and that in each case PRH represses transcription. We demonstrate that overexpression or knockdown of PRH directly impinges on the survival of both leukemic and tumor cells and that the modulation of VEGF and VEGF receptor signaling by PRH mediates these effects. Our findings demonstrate that PRH is a key regulator of the VEGF signaling pathway and describe a mechanism whereby PRH plays an important role in tumorigenesis and leukemogenesis.
Tumor formation and progression to metastasis result from a multistep process that involves increased tumor cell survival or proliferation accompanied by the inhibition of differentiation, migration of tumor cells to sites favoring tumor growth (metastasis), increased tumor-related angiogenesis, and decreased immune surveillance. A critical signaling pathway involved in multiple processes that lead to tumor formation and progression to metastasis is triggered by vascular endothelial growth factor (VEGF). VEGF and its receptors VEGFR-1 (Flt-1) and VEGFR-2 (Flk-2/KDR) are essential for blood vessel formation (17, 23), and these proteins are involved in nearly all human tumors (23, 40, 54, 69). Several studies have demonstrated that many tumors exhibit increased survival as a result of a VEGF-dependent autocrine signaling pathway (24, 40, 75, 77). Increased expression of the VEGF receptors has also been observed in the tumor vasculature, underscoring the importance of VEGF signaling for tumor angiogenesis (7, 15). VEGF and VEGFR-1 are also essential for hematopoiesis (24, 32). VEGF is able to stimulate angiogenesis in tumors by recruiting bone marrow-derived VEGFR-1+ hematopoietic progenitor cells (HPCs) to the “premetastatic niche” (35), followed by recruitment of VEGFR-2+ circulating endothelial progenitors (CEPs) and perivascular VEGFR-1+ HPCs/progenitor cells (26, 31, 39). In addition, VEGF is involved in establishing the immune privilege of tumors by blocking dendritic cell differentiation (19, 20).
The Vegf gene is activated by a plethora of transcription factors and signaling pathways (38, 50). Physiological stress conditions such as hypoxia (25, 61) and hypoglycemia (51, 68) induce Vegf expression and thus contribute to tumor growth. Increased Vegf expression in response to hypoxia occurs as a result of transcription activation by hypoxia-inducible factor 1α (HIF1α) (59, 67) and also as a result of Vegf mRNA stabilization and increased translation (1, 5, 8, 60). The negative regulation of expression of the Vegf gene is less well characterized, although it has been noted that the tumor suppressor proteins p53 (56, 79), SMAD4/DPC4 (58), p16 (78), and von Hippel-Lindau (VHL) protein (41, 42) all downregulate angiogenesis and Vegf expression. Transcriptional activation and/or upregulation of the VEGF receptor genes under normoxia and hypoxia has also been investigated. The Vegfr-1 gene is activated by many transcription factors including CREB and ETS1 (74), HIF1α (22), ETS1 and HIF2α (14), and p53, together with estrogen receptors (45). The Vegfr-2 gene is activated by TFII-I at initiator elements in the promoter, and this activation is antagonized by TFII-IRD1 (33). Transcription of Vegfr-2 is also activated by SP1 (52), Ets1 in combination with HIF2α (16), and GATA-2, which binds in the 5′ untranslated region of this gene (47). Little is known about the negative regulation of Vegfr-1 and Vegfr-2.
The proline-rich homeodomain protein PRH/Hhex regulates hematopoiesis and vasculogenesis as well as many other processes in the developing embryo and adult (reviewed by Soufi and Jayaraman [62]). PRH is an oligomeric protein and has a high affinity for multiple clustered PRH binding sites (64, 76). PRH acts as a context-dependent transcription factor to activate or repress transcription depending on its target gene (62). When bound to DNA, PRH represses transcription by recruiting members of the Groucho/TLE family of corepressor proteins (27, 66), influencing both the phosphorylation and nuclear retention of these proteins (12). Microarray experiments in PRH−/− embryoid bodies have shown that many genes within the hematopoietic compartment are regulated by PRH expression, but as yet very few genes have been shown to be direct targets (28).
PRH functions as a negative regulator of cellular growth in hematopoietic cells (37, 71) and binds to the growth regulator and angiogenic inhibitor PML and eukaryotic translation initiation factor 4E (eIF4E) (70, 72). PRH can inhibit the mRNA transport activity of eIF4E, and this posttranscriptional activity of PRH blocks oncogenic transformation by eIF4E (71). High levels of PRH expression in hematopoietic cells generally lead to cell death (21, 34), although in mouse bone marrow transplantation experiments there is also outgrowth of T-cell leukemias (21). In contrast loss of PRH leads to increased cell proliferation in embryonic stem (ES) cell differentiation models (37) and in mice (29). Prh−/− knockout mice show a number of defects, including cardiac and vascular defects, and have elevated Vegf expression (29), and PRH−/− embryoid bodies have elevated Vegf mRNA levels, as determined using microarrays (28). Collectively, these findings strongly suggest that PRH negatively regulates vascular and hematopoietic cell growth and regulates Vegf expression. PRH also has a role in the expression of the VEGF receptors, as overexpression of PRH in human umbilical vein endothelial cells (HuVECs) leads to downregulation of the Vegfr-1 and Vegfr-2 genes as well as to downregulation of the VEGF coreceptor neuropilin-1 gene and some other proangiogenic genes (49). Furthermore, PRH has been shown to repress expression of the Vegfr-2 gene in endothelial cells by inhibiting the binding of GATA-2 to the Vegfr-2 promoter (46).
Here we demonstrate that PRH regulates the VEGF-VEGF receptor axis by repressing both the Vegf gene and the Vegfr-1 and Vegfr-2 receptor genes using a direct transcriptional mechanism. We show that regulation of the VEGF-VEGF receptor signaling pathway (VSP) by PRH exerts a profound influence on cell survival.
MATERIALS AND METHODS
Mammalian expression and reporter plasmids.
The expression vectors pMUG1-Myc-PRH, pMUG1-Myc-PRHF32E, and pCMV2-Flag-TLE1 are described in reference 66. pMUG1-Myc-PRHR188A,R189A, pMUG1-Myc-PRHN187A, and pEGFP-PRH are described in reference 12. A QuikChange kit (Stratagene) was used for the mutagenesis of pMUG1-Myc-PRH to produce pMUG1-Myc-PRHL23A,L24A. pCS2-Hex-VP16 (PRH-VP16) (6) was a generous gift from J. Brickman. pCMV2-TLE1 is a Flag-tagged TLE1 expression vector that has been previously described (66). The pGL2-1213Vegfr1-luc (R1 1.2), pGL2-694Vegfr1-luc (R1 0.7), and pGL2-200Vegfr1-luc (R1 0.2) reporter plasmids were constructed by cloning PCR fragments corresponding to sequences from −1213 to +154, −694 to +154, and −200 to +154 relative to the first exon in the Vegfr-1 gene between the BglII and HindIII sites of pGL2 (Promega). The pGL2-1326-Vegfr2-luc (R2 1.3) and pGL2-415-Vegfr-2-luc (R2 0.4) reporter plasmids were constructed by cloning PCR fragments corresponding to sequences from −1326 to +49 and −415 to +49 relative to the first exon in the Vegfr-2 gene, between the BglII and HindIII sites of pGL2. The pGL3 −1.8 Vegf reporter plasmid (A 1.2) was a gift from Bruce Spiegelman (2). The pGL3 −4.0 Vegf (A 4.0) and pGL3 −2.3 Vegf (A 2.3) reporter plasmids were constructed by cloning PCR fragments corresponding to sequences from 4.0 to 1.2 kbp and from 2.3 to 1.2 kbp, respectively, upstream of the first exon in Vegf into the KpnI site in the pGL3 −1.8 Vegf reporter (here referred to as A 1.2). All constructs were verified by DNA sequencing. pSV-lacZ (Promega), pTK-luciferase, and pTK-PRH-luciferase reporters have been described previously (27). PRH short hairpin RNA (shRNA) plasmids and green fluorescent protein (GFP) shRNA were from Origene. pBJ Flt-1, pBJ KDR, and pCDNA3 VEGF 165 are the expression plasmids for VEGFR-1, VEGFR-2 and VEGF, respectively, and were a kind gift from D. Bates.
Cell culture and transfections, transcription assays, and Western blotting.
Cell culture, transient transfections, transcription assays, and Western blotting were carried out as described in reference 12. VEGF antibody (VG76e) (18) was a gift from R. Bicknell. The anti-Myc mouse monoclonal antibody Myc9B11 was obtained from Cell Signaling Technology and the lamin A/C rabbit polyclonal antibody H-110 from Santa Cruz. Anti-PRH mouse polyclonal antibodies were produced in-house and have been described previously (63).
Quantitative reverse transcriptase-mediated PCR (qRT-PCR) and chromatin immunoprecipitation (ChIP).
Cells (1 × 107) were transfected with 10 μg pMUG1-PRH or PRH mutant plasmids for 48 h. In cotransfection experiments the PRH shRNA knockdown (KD) cells and shRNA control cells were transfected with 5 μg pMUG1-PRH and 2.5 μg pCMV2-TLE1 for 48 h. RNA was produced according to standard protocols. The quantitative PCR was performed in triplicate as described in reference 63 using primers listed below, and the data were analyzed using Rotorgene 6 software (Corbett Research; Rotorgene RG-3000). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as the internal control. Results for relative expression ratios were calculated according to the efficiency calibrated mathematical model (53).
The primers used were as follows: Vegfr-1 forward, 5′TGGCCATCACTAAGGAGCACTCC3′, and reverse, 5′GGAACTGCTGATGGCCACTGTG3′; Vegfr-2 forward, 5′TTAGTGACCAACATGGAGTCGTG3′, and reverse, 5′TAGTAAAGCCCTTCTTGCTGTCC3′; Vegf forward, 5′ATCAGCGCAGCTACTGCCATCC3′, and reverse, 5′TCTCCTATGTGCTGGCCTTGGTG3′; and GAPDH forward, 5′TGATGACATCAAGAAGGTGGTGAAG3′, and reverse, 5′TCCTTGGAGGCCATGTGGGCCAT3′.
For ChIP K562 cells (108 cells per ChIP) were transiently transfected with 10 μg pMUG1-Myc-PRH. ChIP was carried out exactly as described in reference 76. Hot-start PCR (30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min) was performed using the following primers. For actin, the primers were FW, 5′CCGGCGGGGTCTTTGTCTGAGC3′, and REV, 5′GGGCCGGCCGCGTTATTACCA3′. For Vegfr-1, the primers were 1P1(FWD), 5′CCTTGGTGTGCAGCCCAGAAATG3′ (−1216 to −1194), and 1P1(REV), 5′CACCCGCCCAAGTCATTTCCTC3′ (−727 to −706); 1P2(FWD), 5′GCGCCTCAGTCCTCCGTGCCAAGAAC3′ (−481 to −456), and 1P2(REV), 5′CACTTCCTACCCCGGCACCTCCTTCTGG3′ (−72 to −45); and 1P3(FWD), 5′GAAGAGGGTAGGTGGGGAGGCGGATGA3′ (−123 to −97), and 1P3(REV), 5′CCCCAGCCGCGCCTCACCTGT3′ (+346 to +364). For Vegfr-2, the primers were 2P1(FWD), 5′TGCCTCTGCCAAAAGAAAAG3′ (−1322 to −1303), and 2P1(REV), 5′GAACTCCAATTCCTTCCTGACCACATTC3′ (−926 to −899); 2P2(FWD), 5′CCTTCTTGGGGCTAGGCAGGTCACTTCA3′ (−671 to −644), and 2P2(REV), 5′GATCTCCAGCTCCCCAAGCCCATTTA3′ (−148 to −123), and 2P3(FWD), 5′GTGTGGGGAAATGGGGAGATGTAAATGGG3′ (−169 to −141), and 2P3(REV), 5′CCTCTCCGCCCTCACCCGACCTGTC3′ (+401 to +424).
Thirty-five cycles of 94°C for 1 min, 49°C for 1 min, and 72°C for 1 min were performed using the following primers for Vegf: AP1(FWD), 5′AAAGACCCAACTCAAGTATCATCTCCAGG3′ (−4712 to −4684), and AP1(REV), 5′CACTCACTGTGTGGCCTTAGGTTATTCAAC3′ (−4296 to −4267); AP2(FWD), 5′TTTTGGGGTCAGACTTGGAGGAATAG3′ (−3966 to −3942), and AP2(REV), 5′TCTCTCTCCCCGCCTAGGTCTTTG3′ (−3536 to −3513); AP3(FWD), 5′TGGTCTTGTGATTGTTGTTTTGTGCTTT3′ (−2673 to −2646), and AP3(REV), 5′GTCCTCCATGGTGGTACCCAGCAA3′ (−2304 to −2280); AP4(FWD), 5′CTGGAGCGTTTTGGTTAAATTGAGGGA3′ (−1655 to −1629), and AP4(REV), 5′ATCAGCCCAAGCCCAGACTCATAGC3′ (−1232 to −1208); and AP5(FWD), 5′TCAGAAATAGGGGGTCCAGGAGCAAAC3′ (−671 to −645), and AP5(REV), 5′CACAGCCTGAAAATTACCCATCCG3′ (−237 to −214).
Knockdown experiments.
In PRH shRNA knockdown experiments, 5 × 106 cells were electroporated using 10 μg GFP shRNA (control) or 5 μg PRH49 and 5 μg PRH51 shRNA plasmids in combination. Cells were selected with 1 μg/ml puromycin for 1 week, and then 1 × 106 cells were harvested and used for RNA production or Western blotting. At least 5 independent knockdown experiments were performed with each cell line. Transient transfection of control and PRH KD cells was performed after 11 days of puromycin selection.
MTT assays and cumulative growth curves.
Around 2,000 K562 cells or 5,000 MCF-7 cells were placed in a 96-well plate. To treated cells, either 50 ng/ml VEGF165, 50 μg/ml VEGF antibody (VG76e) (18), 1 μM or 2 μM Sugen (SU1498), or 1 μM or 2 μM SU11248 was added. Control cells were treated with an equivalent volume of dimethyl sulfoxide (DMSO). After 72 h, cells were centrifuged at 200 × g and the media were removed. MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide) assays were carried out according to the method of Mosmann (48). SU1498 (Calbiochem) and VEGF antibody (VG76e) and SU11248 (LC Labs) were kind gifts from P. Hewett and R. Bicknell, respectively.
For cumulative growth curves, cells were set to 3 × 105 cells/ml daily or as required and viable cells were counted over 2 weeks using trypan blue exclusion staining. All dilutions of the cells were recorded, and the cumulative growth was calculated using the equation concentration × volume × dilution factor = total number of cells.
Flow cytometry and AV staining.
For cell cycle analysis ∼1 × 106 cells were washed twice in phosphate-buffered saline (PBS) and resuspended in RPMI media. Igepal (1%, final concentration) and propidium iodide (PI; 50 μg/ml, final concentration) were added immediately before data collection. Data (10,000 events/sample) were collected using a FACSCalibur apparatus (Becton Dickinson), and results were analyzed with Becton Dickinson Cell Quest software. For annexin V (AV) analysis, ∼1 × 106 cells were washed twice in PBS and resuspended in 1× buffer (10 mM HEPES-NaOH [pH 7.4], 0.14 M NaCl, 2.5 mM CaCl2). Cells were incubated with 5 μl annexin V-allophycocyanin (APC) (Becton Dickinson; 550474) and PI (5 μg/ml, final concentration) for 30 min at room temperature. Data were collected as described above. Results were analyzed for significance using the unpaired Student t test.
RESULTS
PRH represses Vegf, Vegfr-1, and Vegfr-2 expression.
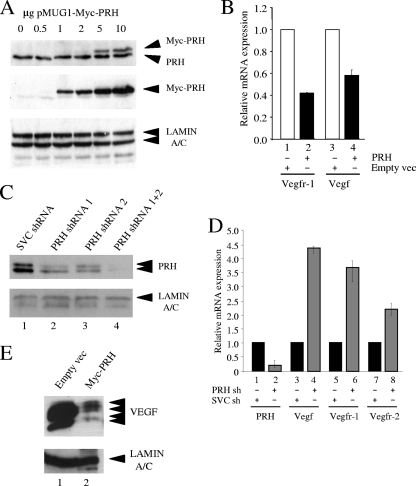
The K562 cell line is a chronic myeloid leukemia (CML) cell line in blast crisis (36) which expresses VEGF, VEGFR-1, and PRH and is therefore a useful cell type in which to examine the effects of PRH on the Vegf and Vegfr-1 genes (3, 13, 66). K562 cells were transiently transfected with a plasmid that expresses Myc-tagged PRH or with an equivalent amount of the empty vector. Titration experiments show that Myc-PRH expression levels in these experiments are roughly equivalent to the expression level of the endogenous PRH protein (Fig. 1A). Vegf and Vegfr-1 mRNA levels were then determined using quantitative reverse transcriptase-mediated PCR (qRT-PCR). PRH is able to repress the Vegfr-1 gene to approximately 40% of its unrepressed expression level and to repress expression of the Vegf gene to approximately 60% of its unrepressed expression level (Fig. 1B). Vegfr-2 mRNA is barely detectable in these cells (13), so it is not possible to determine the effect of PRH overexpression on Vegfr-2 mRNA levels.
FIG. 1.
PRH represses VSP genes. (A) Western blot of whole-cell extracts from K562 cells transiently transfected with increasing amounts of a plasmid expressing Myc-tagged PRH. Proteins were stained with a mouse anti-PRH antibody (top), the Myc9E10 monoclonal antibody (middle), and a lamin A/C antibody (bottom). (B) Vegfr-1 and Vegf mRNA levels in K562 cells 48 h after transfection with 10 μg pMUG1 or pMUG1-Myc-PRH. mRNA levels were determined by qRT-PCR using specific primers and compared to those of GAPDH. Values are means and standard deviations (SD) (n = 5). (C) Western blot of whole-cell extracts from K562 cells cotransfected with shRNA plasmids SVC shRNA, PRH shRNA 1, PRH shRNA 2, and PRH shRNA (1+2) (4). Proteins were stained with rabbit PRH antibody and lamin A/C antibody. (D) Prh, Vegf, Vegfr-1, and Vegfr-2 mRNA levels after shRNA (sh) cotransfection. Black bars represent SVC shRNA-targeted cells and gray bars PRH shRNA (1+2)-targeted cells. Values are means and SD (n = 5). (E) Western blot of whole-cell extract from K562 cells transfected with empty pMUG1 or pMUG1-MycPRH. Proteins were stained with VEGF antibody or lamin A/C antibody.
Knockdown experiments using shRNA constructs against the PRH transcript or scrambled control shRNA followed by Western blot analysis show that endogenous PRH levels are significantly reduced in cells expressing the PRH shRNA compared to cells expressing the control shRNA (Fig. 1C, top) and that equal amounts of protein are loaded (Fig. 1C, bottom). The combination of two different shRNAs against PRH gives the most potent knockdown (Fig. 1C, top), and subsequent knockdown experiments were carried out using this combination.
We next looked at the expression levels of the VSP genes in the PRH knockdown (KD) cells. The levels of relative mRNA expression of the Vegfr-1 and Vegf genes are increased by approximately 4.5-fold and 4-fold, respectively, in the PRH KD cells compared to the control cells (Fig. 1D). This suggests that PRH represses transcription of the Vegfr-1 and Vegf genes in these cells. Significantly, in the PRH KD cells the low level of Vegfr-2 mRNA is also increased compared to that in control cells. We infer that PRH also represses transcription of the Vegfr-2 gene in these cells. Western analysis confirms that VEGF protein levels are decreased when PRH is overexpressed (Fig. 1E). We were unable to examine the effects of PRH on the levels of the endogenous receptor proteins due to their low expression levels in K562 cells. Nevertheless, the results of these overexpression and knockdown studies confirm that the genes encoding VEGFR-1, VEGFR-2, and VEGF are all downregulated by PRH in K562 cells.
Repression by PRH requires DNA binding and recruitment of TLE.
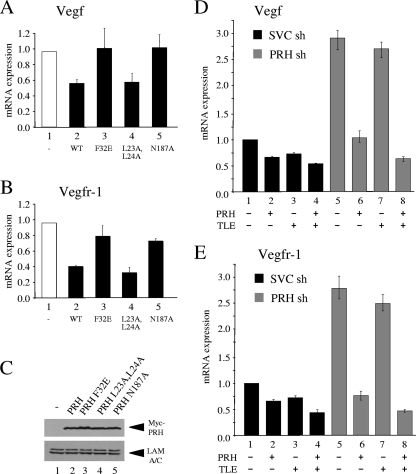
To determine whether the regulation of these genes requires the DNA binding activity of PRH and/or interaction with other proteins, we made use of mutated PRH proteins that have been described previously. PRH F32E is unable to bind to the corepressor protein TLE-1, and PRH N187A is unable to bind to DNA (12, 66). We also generated a mutant PRH protein that has previously been shown to be unable to bind to eIF4E (PRH L23A,L24A) and is consequently defective in posttranscriptional regulation of the cyclin D1 mRNA (71). K562 cells were transfected with the PRH expression vector described above or equivalent amounts of plasmids expressing the mutated proteins. Overexpression of wild-type PRH is able to repress expression of the Vegf gene to approximately 60% of its unrepressed level (Fig. 2A,). However, PRH F32E and PRH N187A are unable to repress the Vegf gene (bars 3 and 5), whereas PRH L23A,L24A represses expression to the same extent as wild-type PRH (bar 4). The mutated PRH proteins have the same effects on expression of the Vegfr-1 gene (Fig. 2B). Figure 2C shows that the mutated PRH proteins are expressed at roughly equal levels in the transfected cells.
FIG. 2.
PRH represses VSP genes in conjunction with TLE proteins. (A) Vegf mRNA levels in K562 cells 48 h after transfection with 10 μg pMUG1 (−) or pMUG1 vectors expressing PRH (wild type [WT], PRH F32E, PRH L23A,L24A, or PRH N187A). mRNA levels were determined as for Fig. 1. Values are means and SD (n = 3). (B) As for panel A except that relative levels of expression of Vegfr-1 are shown. Values are means and SD (n = 3). (C) Western blot of Myc-PRH expression in K562 cells 48 h after transfection with the vectors described for panel A. Whole-cell extracts were run on a sodium dodecyl sulfate-polyacrylamide gel (SDS-PAG) and stained with Myc and lamin A/C antibodies. (D) Vegf mRNA levels in K562 shRNA (sh) control cells and PRH shRNA cells 48 h after transfection with pMUG1-MycPRH (5 μg), pCMV2-TLE1(2.5 μg), or both plasmids were determined as for Fig. 1. Values are means and SD (n = 3). (E) As for panel D except that relative levels of expression of Vegfr-1 are shown. Values are means and SD (n = 3).
Given that interaction between PRH and TLE appears to be required for repression of these genes, we next examined the effects of overexpressing TLE-1 and PRH in control cells and PRH KD cells. Overexpression of PRH alone represses expression of the Vegf mRNA to about 60% of its unrepressed level (Fig. 2D, bar 2), and overexpression of TLE-1 alone represses this gene to approximately 75% of its unrepressed level (Fig. 2D, bar 3). Coexpression of both PRH and TLE-1 results in approximately 50% repression (Fig. 2D, bar 4). As expected, in PRH KD cells Vegf mRNA levels are elevated compared to those in control cells (compare bars 1 and 5). Overexpression of PRH in these cells results in strong repression of the Vegf gene (Fig. 2D, bar 6). In contrast, overexpression of TLE1 in PRH KD cells does not result in a significant change in Vegf mRNA levels (Fig. 2D, bar 7). However, coexpression of PRH and TLE-1 in these cells results in a more potent repression of the Vegf gene than overexpression of PRH alone (compare bars 6 and 8). These data show that PRH and TLE-1 together cause additional repression of the Vegf gene and that the effects of TLE on Vegf expression in control cells are dependent upon endogenous PRH. Corepression by TLE-1 and PRH is also observed for the Vegfr-1 mRNA (Fig. 2E). Taken together with the data shown above, these results strongly suggest that repression of these genes by PRH does not involve interaction with eIF4E but rather occurs through the regulation of transcription in conjunction with TLE proteins.
PRH is present at the Vegf, Vegfr-1, and Vegfr-2 promoters.
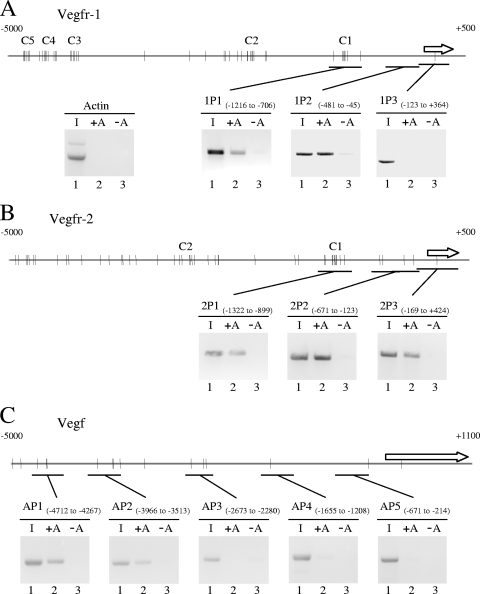
SELEX (systematic evolution of ligands by exponential enrichment) experiments with random oligonucleotides and a truncated protein consisting of the PRH homeodomain and C-terminal region have identified a consensus DNA binding sequence 5′C/tA/tATTAAA/g3′ (lowercase letters indicate base pairs found less frequently) (9). Unlike the truncated PRH protein used in the SELEX experiments, full-length PRH forms oligomers and binds with high affinity to promoter DNA containing multiple 5′ATTA3′ sequences that do not match the consensus sequence (64, 76). To assess whether PRH is likely to bind to sites within the regulatory regions of the Vegfr-1, Vegfr-2, and Vegf genes, we analyzed the sequence 5,000 bp upstream of the first exon for the presence of core 5′ATTA3′ sequences. As expected, the Vegf, Vegfr-1, and Vegfr-2 genes all contain multiple potential binding sites for PRH as defined by the core 5′ATTA3′ motif. The Vegfr-1 promoter region contains 5 clusters of potential PRH sites located approximately 0.98 kb, 2.2 kb, 4.2 kb, 4.6 kb, and 4.8 kb upstream of the first exon (Fig. 3A). In addition there are several nonclustered potential PRH sites. At the Vegfr-2 promoter there are 2 major clusters of potential PRH binding sites located approximately 1.1 kb and 2.8 kb upstream of the first exon (Fig. 3B). Both of these clusters contain 7 potential PRH sites within 250 bp of DNA. Again, there are many additional nonclustered potential PRH sites. However, like the Vegfr-1 promoter, the Vegfr-2 promoter region does not contain an exact match to the consensus PRH site defined by SELEX. At the Vegf promoter there are 14 potential PRH sites located within 5,000 bp upstream of the first exon, including a consensus PRH binding site at 4.77 kb upstream; however, the sites appear to be less closely clustered than those present in the Vegfr-1 and Vegfr-2 promoters (Fig. 3C).
FIG. 3.
PRH binds to the Vegf, Vegfr-1, and Vegfr-2 promoters in cells. (A) Schematic of the human Vegfr-1 promoter (top line), with the positions of potential PRH binding sites indicated as bars. C1 to C5 indicate clusters of potential sites. The arrow corresponds to the first exon. Each inset panel shows the results of PCRs using the following templates: (1) input DNA, (2) DNA precipitated from cells expressing Myc-PRH with beads plus Myc antibody, (3) DNA precipitated from cells expressing Myc-PRH with beads plus normal IgG. The amplicons are indicated by solid bars. In the same experiment, primers within the actin promoter did not produce a product (left inset). (B) As for panel A except that the schematic and ChIP results are for the Vegfr-2 promoter. (C) As for panel A except that the schematic and ChIP results are for the Vegf promoter.
We next carried out chromatin immunoprecipitation (ChIP) assays using Myc-PRH and a Myc antibody. Endogenous PRH could not be assessed in these experiments, as the available antibodies do not immunoprecipitate PRH sufficiently robustly. PCR was performed with primer pairs that specifically amplify sequences within the promoter region of each gene. Primer pairs specific for sequences present in the actin promoter were used as a negative control in this experiment and all subsequent ChIP experiments. In the presence of exogenous PRH, there is no specific product from within the actin promoter (Fig. 3A, left). At the Vegfr-1 promoter, primer pair 1P3 (−123 to +364 relative to the start of the first exon) did not produce a PCR product after ChIP (Fig. 3A). However, amplicons 1P2 (−481 to −45) and 1P1 (−1216 to −706) both produced specific PCR products (Fig. 3A). In the presence of normal IgG there is relatively little or no product visible with these primer pairs. These experiments thus demonstrate that Myc-PRH is associated with the Vegfr-1 promoter. The region between −1216 and −706 contains multiple potential PRH sites (1P1). The association of PRH with DNA from −481 to −45 (1P2) could be the result of PRH binding to nonconsensus sites within this region or could simply be a consequence of the intrinsic low resolution of semiquantitative ChIP experiments. To address this point, we carried out electrophoretic mobility shift assay (EMSA) experiments with the PRH homeodomain and DNA fragments derived from regions 1P1 and 1P2. In all cases the PRH homeodomain formed a ladder of PRH-DNA complexes, confirming that PRH can bind to multiple sites within 1P1 and suggesting that PRH also binds to multiple nonconsensus binding sites in 1P2 (data not shown). We conclude that PRH binding at the Vegfr-1 promoter includes but is not limited to “ATTA” sequences.
At the Vegfr-2 promoter, primer pairs that amplify sequences located approximately 1,322 bp to 899 bp (2P1) and 671 bp to 123 bp (2P2) upstream of the first exon, as well as primers located between −169 and +424 (2P3), all produced specific PCR products after ChIP with the Myc antibody (Fig. 3B). In all cases, in the presence of normal IgG antibody no PCR product was produced. These data suggest that PRH could be associated with the cluster of potential PRH binding sites present around 1.1 kb upstream of the Vegfr-2 promoter as well as with the more promoter-proximal PRH binding sites. EMSA experiments show that PRH is capable of binding to DNA fragments derived from 2P1 and 2P2 in vitro (data not shown). We conclude that PRH also binds to multiple binding sites within the Vegfr-2 promoter that include ATTA and nonconsensus sequences.
At the Vegf promoter three sets of primer pairs that amplify sequences across the region from 2,673 to 214 bp upstream of the first exon did not produce strong PCR products after ChIP with the Myc antibody (Fig. 3C). However, in the same experiment, primer pairs located from 4,712 to 4,267 bp (AP1) and from 3,966 to 3,500 bp (AP2) upstream of the first exon both produced a specific PCR product in the presence of the Myc antibody (Fig. 3C). These data show that PRH is most strongly associated with DNA in the far-distal promoter region of the Vegf gene. EMSA experiments confirm that PRH is capable of binding to promoter sequences between kbp −3.9 and −3.4 (data not shown).
PRH directly represses the Vegf, Vegfr-1, and Vegfr-2 promoters.
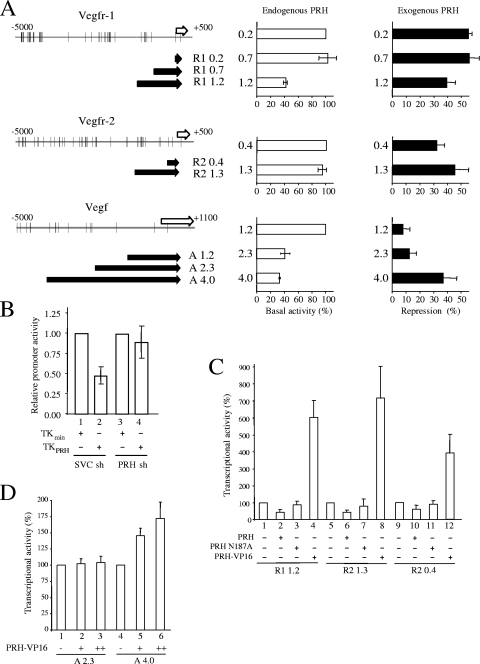
To determine whether the promoter regions identified using ChIP are responsible for the regulation of each gene by PRH, promoter sequences were cloned upstream of the luciferase reporter gene in pGL2 (Fig. 4A). These reporter constructs were transiently cotransfected into K562 cells either alone or together with a PRH expression vector. For the Vegfr-1 gene, reporter constructs R1 1.2 (bp −1213 to +154 relative to the start of exon 1), R1 0.7 (−694 to +154), and R1 0.2 (−200 to +154) were transiently transfected into K562 cells. Figure 4A (top middle) shows that these reporters have very different basal activities; the R1 1.2 construct has low basal promoter activity relative to the R1 0.7 and R1 0.2 promoter derivatives. This suggests that endogenous PRH represses the R1 1.2 construct. Over a range of R1 1.2 reporter plasmid amounts, reporter activity increases in a linear fashion as increasing amounts of reporter plasmid are transfected into the cells (data not shown). Therefore, the plasmid DNA does not titrate out endogenous PRH under these conditions, and the R1 0.2 and R1 0.7 reporters are not more active than R1 1.2 because they have titrated away PRH. In keeping with this view, the same reporters show no significant difference in basal activity in HeLa cells that do not express PRH (data not shown). The difference in basal activity is similar to that observed in K562 cells for the minimal thymidine kinase (TK) promoter compared to the minimal TK promoter containing multiple upstream PRH binding sites (Fig. 4B, bars 1 and 2). However, in PRH KD cells the minimal TK promoter and the minimal TK promoter containing PRH binding sites have the same basal activity (Fig. 4B, bars 3 and 4). We conclude that repressive PRH-dependent elements are located between kbp −1.2 and −0.7 and that endogenous PRH binds to these sites to repress promoter activity. This is in agreement with the ChIP data which show that PRH binds to sequences within this region (Fig. 3A).
FIG. 4.
PRH directly represses the Vegf, Vegfr-1, and Vegfr-2 promoters. (A, left) Schematic representations of the Vegfr-1, Vegfr-2, and Vegf promoters indicating the position of the first exon for each gene (white arrows) and the promoter derivatives (black arrows) used in this study. (Middle) Relative promoter activity found in K562 cell extracts following transfection with Vegfr-1, Vegfr-2, or Vegf reporters. Five micrograms of each reporter and 5 μg of the β-galactosidase plasmid (pSV-lacZ) were cotransfected into cells. Relative promoter activity was determined 24 h posttransfection as the luciferase activity normalized for transfection efficiency using the β-galactosidase activity. Values are means and SD (n = 3). (Right) Percent repression by PRH for each reporter construct. Cells were transfected and reporter assays were carried out as described above except that cells were cotransfected with 1 μg pMUG1-Myc-PRH. Percent repression is promoter activity in the presence of PRH as a percentage of reporter activity in the absence of PRH. (B) K562 cells were transfected with PRH shRNA (sh) or control shRNA and grown in selection for 10 days. The cells were then retransfected with 5 μg of pSV-lacZ and 5 μg of the minimal TK reporter (TKmin; bars 1 and 3) or the minimal TK reporter containing PRH binding sites (TKPRH; bars 2 and 4). Twenty-four hours posttransfection, relative promoter activity was determined as described in the text. Values are means and SD (n = 3). (C) Cells were transfected and assayed for reporter activity as for panel A. The graph shows the activity of the R1 1.2, R2 1.3, and R2 0.4 reporters cotransfected into cells with 1 μg pMUG1-PRH, 1 μg pMUG1-PRH N187A, or 3 μg pCS2-Hex-VP16. (D) Cells were transfected and assayed for reporter activity as for panel A. The graph shows the activity of the A 2.9 and A 4.0 Vegf reporters with 3 μg or 6 μg pCS2-Hex-VP16.
We next examined the effects of PRH overexpression on the activity of these promoter constructs. Overexpression of PRH represses the activity of all three reporter constructs (Fig. 4A, top right). Since the R1 0.7 and R1 0.2 reporters are repressed to very similar degrees, this suggests that PRH-responsive elements are also present within the minimal R1 0.2 promoter region (the R1 0.2 promoter overlaps sequences that bind PRH in ChIP).
To confirm that the PRH-responsive elements in the R1 1.2 promoter require DNA binding by PRH for the observed repression, we examined the effects of overexpressing wild-type PRH and the PRH protein defective in binding to DNA (PRH N187A). We also made use of a PRH-VP16 fusion protein, which is able to strongly activate transcription when bound to PRH sites. As expected, PRH brings about repression of the R1 1.2 reporter (Fig. 4C, bar 2). In contrast, PRH N187A is not able to significantly repress the activity of this reporter (bar 3), whereas PRH-VP16 is able to strongly activate this reporter activity (bar 4). These data confirm that the R1 1.2 reporter is responsive to PRH and show that the DNA binding activity of PRH is required for repression.
To examine the regulation of the Vegfr-2 promoter by PRH reporter constructs, R2 1.3 (−1326 to +49) and R2 0.4 (−415 to +49) were transfected into K562 cells (Fig. 4C, middle). In contrast to the Vegfr-1 reporters, the Vegfr-2 reporter constructs have very similar basal activities (Fig. 4A, middle center). Cotransfection of these reporters with a PRH expression vector has a repressive effect on the activity of the R2 0.4 and R2 1.3 reporters (Fig. 4A, middle right). Figure 4C confirms that PRH represses the activity of both reporters and shows that PRH N187A has little effect. Furthermore, expression of the PRH-VP16 fusion protein described above strongly activates the R2 1.3 and R2 0.4 reporters (Fig. 4C). These data suggest that PRH binds to the Vegfr-2 promoter-proximal region in order to bring about the repression of transcription.
ChIP experiments suggest that PRH does not bind strongly to the promoter-proximal region of the Vegf promoter, and for this reason we made Vegf reporter constructs that extend up to 4 kbp upstream of the first exon. Equivalent molar amounts of Vegf reporters A 1.2, A 2.3, and A 4.0 (Fig. 4A, bottom) were transfected into K562 cells. These reporters have different basal activities in these cells: the A 1.2 reporter has high basal activity, while the A 2.3 reporter and the A 4.0 reporter have lower basal promoter activities. In contrast, the basal activities of these promoters are not significantly different in cells that do not expresses PRH. This suggests that repressive elements are located between kbp −1.2 and −2.3 and also between kbp −2.3 and −4.0 in the Vegf promoter. However, only the more distal promoter regions are strongly associated with PRH in ChIP experiments. To determine whether PRH overexpression has any effect on the activity of these reporters, they were cotransfected into K562 cells with a PRH expression plasmid. Figure 4A (bottom right) shows that PRH represses the activity of the A 4.0 reporter but has only a small repressive effect on the activities of the A 1.2 and A 2.3 reporters. This suggests that critical PRH-responsive elements are located between approximately kbp −4.0 and −2.3 in this promoter, in agreement with the ChIP data. In order to confirm this conclusion, we looked at the effects of PRH-VP16 on the activities of the A 4.0 and A 2.3 reporters. PRH-VP16 activates the A 4.0 reporter in a dose-dependent fashion, whereas this fusion protein has no effect on the activity of the A 2.3 reporter (Fig. 4D, bottom right). PRH N187A does not significantly repress the activity of either reporter, indicating that direct binding of PRH is required for repression (data not shown). Together, these reporter data support the idea that PRH binding sites located between kbp −4.0 and −2.3 are important for repression by PRH. It is noteworthy that these binding sites respond to PRH even in the absence of the upstream consensus sequence CAATTAA, located at kbp −4.77.
PRH influences cell survival.
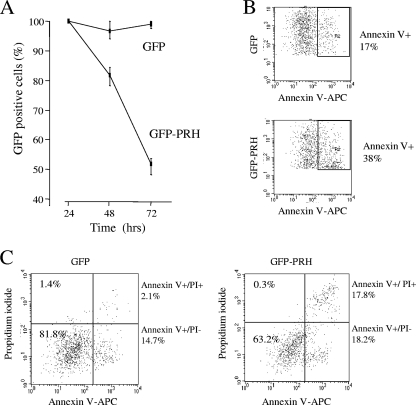
Since we have shown that PRH coordinately regulates at least three genes in the VEGF signaling pathway, we next set out to determine whether PRH expression influences cell proliferation and/or cell survival via this pathway. We cotransfected K562 cells with a plasmid expressing GFP or a GFP-PRH fusion protein and determined the percentages of green GFP+ cells and GFP-PRH+ cells over a 72-h time course using flow cytometry. Figure 5A shows that the percentage of GFP-PRH+ cells declines to around 50% whereas the percentage of GFP+ cells is roughly maintained over the same time period. Cell death in the transfected (GFP+) population was measured by flow cytometric analysis of green fluorescent cells costained with fluorescent antibodies to annexin V (AV). Figure 5B shows a representative analysis of the amount of AV staining of the GFP+ and GFP-PRH+ cell populations. Around 20% of GFP+ cells are AV positive, whereas around 40% of GFP-PRH+ cells are AV positive. To confirm these results, we repeated the experiment and costained cells with the fluorescent DNA binding propidium iodide (PI) stain and fluorescent antibodies to AV. PI staining provides a measure of the percentage of dying cells, and cells that are PI positive and AV positive can be considered to be in late apoptosis. In the control GFP+ population, around 15% of the cells are AV+ whereas only 2% are both PI+ and AV+ (Fig. 5C). In contrast, in the GFP-PRH+ population around 18% of cells are AV+ while around 18% are both PI+ and AV+. Thus, there is a small increase in the early apoptotic cell population and a dramatic increase in the late apoptotic cell population on expression of GFP-PRH. This suggests that PRH induces apoptosis in these cells.
FIG. 5.
PRH overexpression induces apoptosis in K562 cells. (A) Cells transfected with pEGFP alone or pEGFP and pMUG1-Myc-PRH. The percentage of GFP-expressing cells 24 h posttransfection was set as 100%, and the change from this was tracked over 72 h. Values are means and SD (n = 3). (B) Cells from panel A were analyzed by flow cytometry. The percentage of GFP-positive/annexin V (AV)-positive cells was determined 24 h posttransfection. One representative dot plot of 3 is shown. (C) Cells from panel A were dual stained with propidium iodide (PI)/AV (APC antibody) and analyzed by flow cytometry. The dot plot shows the percentages of live cells (PI− AV−), necrotic cells (PI+), early apoptotic cells (AV+), and late apoptotic cells (AV+ PI+) after gating for GFP+ cells. One representative dot plot of 3 is shown.
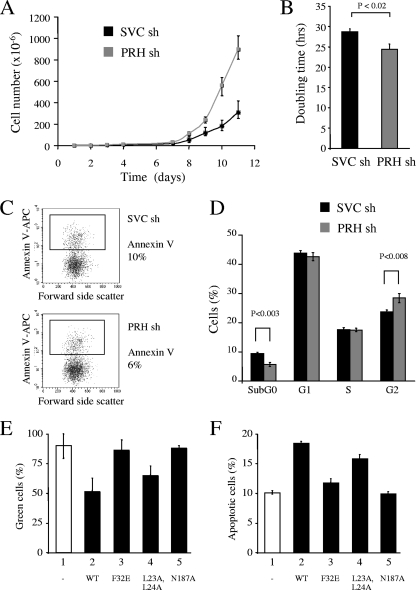
To confirm and extend these results, we examined the growth and survival of the PRH KD cells described above. Control cells and PRH KD cells were maintained at a constant density over a 2-week period and counted daily using a trypan blue exclusion assay to identify viable cells. Cumulative growth curves for each population show that PRH knockdown results in increased cell numbers (Fig. 6A) and that PRH KD cells have a significantly shorter doubling time (Fig. 6B). To examine the amount of apoptosis in the PRH KD cells, the cells were costained with PI and AV antibodies. On average PRH KD results in around 6% apoptotic cells whereas in the control population there are around 10% apoptotic cells, and this small decrease in cell death appears to be statistically significant (P < 0.004) (Fig. 6C). To examine cell proliferation in the PRH KD cell population, we used PI staining and flow cytometry. There is little difference in the percentages of cells in the G1 and S phases of the cell cycle in the PRH KD cells compared to the control cells (Fig. 6D). However in the PRH KD cells on average there is a small increase in the number of cells in G2 (28% compared to 24%) and a decrease in the number of cells in the sub-G0 (dead) population (4%) compared to the control (9%). Over time these differences may account for the increased growth of these cells. Taken together, these data suggest that the removal of PRH promotes cell growth via a decrease in cell death.
FIG. 6.
PRH KD increases cell proliferation. (A) Cumulative growth curves for K562 cells transfected with SVC shRNA (sh) or PRH shRNA (1+2). Cells were selected with puromycin 24 h posttransfection. After 7 days, 3 × 106 cells of each cell type were plated out and counted over 11 days. (B) Graph of the doubling time for K562 cells transfected with SVC shRNA or PRH shRNA (1+2) (gray bar). Values are means and SD (n = 3). (C) Cells from panel A were dual stained with PI/AV (APC antibody) and analyzed by flow cytometry. The dot plots show percentages of live cells (AV−) and percentages of apoptotic cells (AV+). (D) Percent distribution of cells in each stage of the cell cycle. PI staining of K562 cells transfected with SVC shRNA or PRH shRNA (1+2) is shown (n = 3). (E) Percentages of GFP+ cells in cotransfection experiments. K562 cells were transfected with pEGFP and either pMUG1 (bar 1) or pMUG1 vectors expressing PRH (bar 2; WT, wild type), PRH F32E (bar 3), PRH L23A,L24A (bar 4), or PRH N187A (bar 5), and the percentage of GFP+ cells was measured 72 h posttransfection. Values are means and SD (n = 3). (F) K562 cells were transfected as for panel E and dual stained with PI/AV antibody 24 h posttransfection for analysis by flow cytometry (n = 3).
DNA binding and recruitment of TLE are required for increased cell death.
To determine whether the effects of PRH on the survival of these cells require DNA binding and/or binding to TLE or eIF4E, we coexpressed PRH and the mutated PRH proteins with GFP and determined the percentage of green GFP+ cells with time. Coexpression of PRH with GFP causes a significant decrease in the number of GFP+ cells over 72 h (Fig. 6E). In contrast PRH N187A and PRH F32E have little effect on the number of GFP+ cells. PRH L23A,L24A is able to decrease the number of GFP+ cells with time, although it is not as effective as wild-type PRH (Fig. 6E, bar 4). We also measured the amount of cell death in the GFP+ transfected cell populations. Myc-PRH brings about an increase in cell death, from approximately 10% to 20% of the transfected cells (Fig. 6F, bars 1 and 2). Although, expression of the N187A and F32E mutants has little effect, expression of the L23A L24A mutant brings about an increase in cell death (Fig. 6F, bar 4). We conclude that the mutant PRH proteins that do not bind TLE or DNA and that are defective for transcriptional repression do not induce cell death. This suggests that the cell death caused by PRH in these experiments requires the ability of PRH to bind DNA and corepress transcription with TLE. Since the PRH L23A,L24A protein shows a somewhat decreased ability to induce cell death, we infer that the interaction of PRH with eIF4E may also contribute to the effects of PRH on the survival of these cells.
The effects of PRH on cell survival are mediated by the VSP.
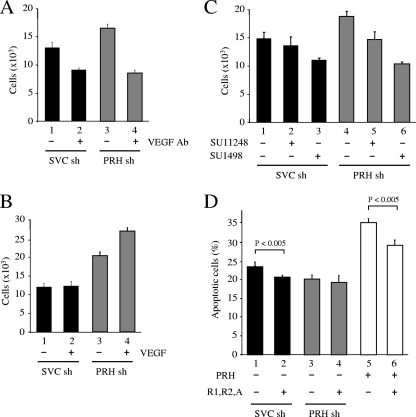
To investigate whether the effect of PRH on cell survival is mediated by VEGF and the VEGF receptors, equal numbers of control cells and PRH KD cells were resuspended in growth media either with no other additions or with a VEGF blocking antibody (VG76e) (18). Cells were grown for 48 h, and the effects of the blocking antibody on cell growth were assessed using MTT assays. PRH KD cells show enhanced cell growth after 48 h compared to control cells (Fig. 7A, compare bars 1 and 3). The VEGF blocking antibody inhibits the growth of the control cells, indicating that these cells are dependent upon VEGF (Fig. 7A, bar 2). The VEGF blocking antibody abolishes the increased growth seen in the PRH KD cells (Fig. 7A, bar 4). We conclude that the higher expression of VEGF receptors in the PRH KD cells allows the concomitantly elevated VEGF to bring about robust paracrine signaling that in turn stimulates cell proliferation. It is important to note that autocrine regulation by VEGF would not be influenced by the external VEGF blocking antibody. In keeping with this view, enhanced growth is observed when PRH KD cells are incubated with exogenous VEGF (Fig. 7B, bar 4). In contrast, the addition of VEGF has no effect on growth of the control cells (Fig. 7B, bar 2). We conclude that additional VEGF has a stimulatory effect on cell growth in PRH KD cells because there are higher levels of VEGF receptor proteins in these cells than in control cells.
FIG. 7.
PRH KD cells are sensitive to VEGF inhibition. (A) K562 cells were transfected with SVC shRNA (sh) or PRH shRNA and then incubated with 50 μg/ml anti-VEGF antibody (bars 2 and 4) or an equal volume of DMSO (bars 1 and 3) for 72 h. An MTT assay was then used to calculate cell numbers. Values are means and SD (n = 5). (B) As for panel A except that cells were incubated with 50 ng/ml VEGF. Values are means and SD (n = 5). (C) As for panel A except that cells were incubated with DMSO (bars 1 and 4), 2 μM SU11248 (bars 2 and 5), or 2 μM SU1498 (bars 3 and 6). Values are means and SD (n = 3). (D) K562 cells transfected with SVC shRNA (black bars) or PRH shRNA (gray bars) or not shRNA transfected (white bars) were then transfected with a vector expressing PRH (3 μg) (bars 5 and 6) and/or vectors expressing VEGF, VEGFR-1, and VEGFR-2 (bars 2, 4, and 6) (total, 3 μg). Twenty-four hours posttransfection the cells were dual stained with PI/AV antibody for analysis by flow cytometry. The graph shows the means and SD (n = 3).
SU11248 (sunitinib) selectively inhibits the activity of a subgroup of tyrosine kinase receptors which includes the VEGF family of receptors (44). As expected, PRH KD cells show enhanced growth compared to control cells (Fig. 7C, compare bars 4 and 1). The addition of SU11248 has no significant inhibitory effect on growth of control cells (Fig. 7C, compare bars 1 and 2). In contrast, SU11248 abolishes the enhanced growth of the PRH KD cells (Fig. 7C, bar 5). We conclude that the KD cells are more sensitive to the effects of SU11248 than the control cells.
The addition of the VEGFR-2 receptor kinase inhibitor SU1498 (65) inhibits the growth of the control cells (20 to 30% reduction in cell number) (Fig. 7C, bars 1 and 3). SU1498 is known to block extracellular signal-regulated kinase (ERK) signaling, which might contribute to its inhibitory effect on control cells (4). However, the addition of SU1498 to PRH KD cells abolishes the growth-stimulatory effect of PRH KD and reduces the number of KD cells to around the same level seen after treatment of the control cells (Fig. 7C, compare bars 4 and 6). The simplest explanation for this difference in sensitivity is that the PRH KD cells express higher levels of the VEGFR-2 receptor and that as a consequence the VEGF signaling pathway is more active in these cells and more strongly inhibited by SU1498.
The results of the inhibitor experiments described above, combined with the data obtained with the VEGF antibody and via addition of VEGF, strongly suggest that the effects of PRH on cell survival are a result of the repression of VSP genes. If this is the case, overexpression of VSP proteins using a PRH-independent promoter should overcome the effects of overexpressing PRH. In order to test this hypothesis, we cotransfected control cells and PRH KD cells with vectors expressing VEGF, VEGFR-1, and VEGFR-2 under the control of the cytomegalovirus (CMV) promoter and determined the number of apoptotic cells 24 h posttransfection (Fig. 7D). Overexpression of VEGF, VEGFR-1, and VEGFR-2 brings about a small but significant decrease in apoptosis in the control cells (Fig. 7D, bars 1 and 2). However, overexpression of VEGF, VEGFR-1, and VEGFR-2 in PRH KD cells has no effect on the level of apoptosis (Fig. 7D, bars 3 and 4). This suggests that endogenous PRH represses the endogenous VSP genes, resulting in increased cell death, and that the overexpressed VSP proteins overcome the effects of this repression. In cells overexpressing PRH there is increased apoptosis, and this is abrogated by overexpression of the VSP proteins (Fig. 7D, bars 5 and 6). These data confirm that the effects of PRH on cell death are mediated in large part at least by regulation of the VSP.
PRH influences the survival of breast cancer cells.
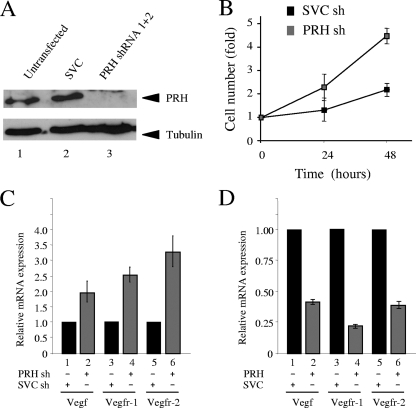
To determine whether regulation of the VSP by PRH plays a more generic role in the control of tumor cell proliferation, we next looked at the effects of PRH KD and overexpression on the growth and survival of breast cancer cells. MCF-7 breast cancer cells express PRH, but the subcellular localization is aberrant, with strong staining of the nucleolus (55). Using the PRH shRNA constructs described above, we knocked down PRH in these cells (Fig. 8A). PRH KD results in an increase in the cumulative growth rate of these cells, suggesting that some endogenous PRH is active despite the aberrant localization. This difference in growth rate can be clearly seen in MTT assays in which equal numbers of cells are plated out 5 days after selection and their growth is monitored over a further 48 h (Fig. 8B). qRT-PCR shows that Vegf, Vegfr-1, and Vegfr-2 are significantly upregulated in PRH KD cells (Fig. 8C). Moreover, overexpression of PRH in MCF-7 cells represses the expression of each of these genes (Fig. 8D). We infer that PRH represses the expression of all three of these genes in MCF-7 cells and that PRH KD results in their derepression and in increased cell growth.
FIG. 8.
PRH represses VSP genes in MCF-7 cells. (A) Western analysis of whole-cell extracts from untransfected MCF-7 cells or cells cotransfected with SVC shRNA or PRH shRNA (1+2). Extracts were stained with rabbit PRH antisera (top) and tubulin antibody (bottom). (B) Growth curves for MCF-7 cells transfected with SVC shRNA (sh) or PRH shRNA (1+2). Cells were selected with puromycin 24 h posttransfection, and after 5 days in selection, MTT assays were performed over 3 days. Results shown are representative of the results from 6 independent experiments. (C) Vegf, Vegfr-1, and Vegfr-2 mRNA levels in MCF-7 cells after shRNA cotransfection (as above). Levels of mRNA were determined by qRT-PCR using specific primers and compared to that for GAPDH. Black bars represent the SVC shRNA-targeted cells and gray bars the PRH shRNA (1+2)-targeted cells. Values are means and SD (n = 5). (D) Vegfr-1, Vegfr-2, and Vegf mRNA levels in MCF-7 cells 48 h after transfection with pMUG1 (empty vector) or pMUG1-Myc-PRH. mRNA levels were determined as above. Values are means and SD (n = 3).
DISCUSSION
In this study we have shown through manipulation of PRH expression that there is an inverse relationship between the levels of PRH protein and the expression of the Vegf, Vegfr-1, and Vegfr-2 mRNAs. We have also shown that PRH requires the ability to bind to DNA and its corepressor TLE to bring about robust repression of the Vegf and Vegfr-1 genes, suggesting that the repression of these genes occurs predominantly through a transcriptional mechanism. ChIP experiments reveal that PRH is associated with the Vegf, Vegfr-1, and Vegfr-2 promoters, supporting the idea that PRH is a direct transcriptional repressor of these genes. Although the Vegfr-1, Vegfr-2, and Vegf promoters are all regulated by PRH, they differ considerably in architecture. The Vegfr-1 promoter contains a TATA box, a potential PRH site within the first exon of the gene, and a cluster of potential PRH sites located around 1 kb upstream of the first exon. The precise mapping of PRH binding sites is not possible using semiquantitative ChIP. However, the combination of reporter assays and ChIP data together suggests that PRH binds to distal sequences at around kbp −1 as well as to more-proximal DNA sequences. This arrangement of PRH sites, namely, a distal cluster and one or more promoter-proximal sites, is similar to that observed at another PRH target gene, Goosecoid (76). The Vegfr-2 promoter lacks a TATA box but contains initiator elements and downstream GATA elements. PRH has previously been shown to block the DNA binding activity of GATA-2 and thereby function as an indirect repressor of the Vegfr-2 gene (46). However, in our reporter constructs, the downstream GATA element is not present, suggesting that repression of Vegfr-2 by PRH also involves direct transcriptional regulation. The combination of ChIP and reporter assays suggests that PRH represses the Vegfr-2 promoter by binding to sequences present in the proximal promoter. In marked contrast to both of these promoters, truncated versions of the Vegf promoter are not regulated by PRH until sequences located approximately 2 to 4 kbp upstream of the proximal promoter are present. This far-distal promoter region contains several potential nonconsensus PRH sites that bind PRH in ChIP. Although these sites are not as closely clustered as the sites in the Goosecoid, Vegfr-1, and Vegfr-2 promoters, they nevertheless allow repression by PRH. Further work will be required to determine to what extent the preference of the oligomeric PRH protein for clustered PRH sites determines promoter selection and regulation. It is possible that PRH also utilizes different as yet unidentified binding partners in these different promoters.
In this study we demonstrate that overexpression of PRH in K562 cells brings about increased apoptosis and, conversely, that knockdown of PRH results in increased cell survival and decreased apoptosis. A central question then is whether PRH causes these effects on cell survival and apoptosis through the repression of the VEGF signaling pathway or through an unrelated mechanism. VEGF blocking antibodies and VEGF kinase inhibitors show that PRH KD cells are more sensitive to depletion of VEGF or inhibition of VEGF receptor activity than control cells. Furthermore, overexpression of VEGF and its receptors from PRH-independent promoters counteracts the effects of PRH overexpression. The simplest explanation for these results is that PRH KD cells are dependent on the VEGF signaling pathway for their increased survival. This conclusion is supported by the fact that the PRH KD cells show enhanced growth upon the addition of exogenous VEGF, whereas control cells do not respond to exogenous VEGF. The inability of mutated PRH proteins that are defective in transcriptional repression to bring about apoptosis provides further support for the idea that there is a direct antagonism between the prosurvival functions of the VEGF signaling pathway and the expression of PRH. In summary, our results show that a normal function of PRH is to limit cell proliferation and that an important mechanism that PRH uses to influence cell proliferation occurs via coordinate repression of genes in the VEGF signaling pathway. The coordinate transcriptional regulation of the growth factor and the receptor proteins may be particularly apposite but not limited to autocrine signaling. It is clear that repression of the VEGF signaling pathway is not the only mechanism that PRH uses to bring about decreased cell proliferation. The interaction of PRH with eIF4E is known to lead to decreased transport of mRNAs that have specific secondary structures (71), and, as the Vegf mRNA is regulated posttranscriptionally (1, 8), it remains possible that PRH might regulate Vegf expression posttranscriptionally as well as transcriptionally.
K562 cells represent a useful model for understanding the role of PRH in the leukemic stem cell, as these cells derive from a patient with chronic myeloid leukemia in blast crisis (CML-BC) (10, 36). BCR-ABL signaling is known to result in the constitutive expression of VEGF in CML cells (43). VEGFR-2 appears to be required for the proliferation and migration of leukemic cells (13, 57). Significantly, although both VEGFR-1 and VEGFR-2 are expressed at high levels in bone marrow from patients with CML, it is VEGFR-2 expression that correlates with decreased patient survival (73). Our results suggest that PRH levels play a central role in influencing the survival and growth of K562 leukemic cells. Although K562 cells do not normally express VEGFR-2, knockdown of PRH results in increased VEGFR-2 expression. Inhibition of the VEGFR-2 receptor in the KD cells decreases the survival advantage of the KD cells compared to control K562 cells, suggesting that VEGFR-2 expression contributes to enhanced cell survival. PRH expression and subcellular localization are aberrant in both primary CML cells in blast crisis and also in the M4/M5 subset of acute myeloid leukemia (AML) cells (72) although it is not known whether these cells also have elevated VEGF signaling. It is likely that loss of PRH repression activity may be a feature of many leukemias that exhibit elevated VEGF and VEGF receptor expression.
The results of PRH KD and overexpression in MCF-7 breast cancer cells suggest that PRH is a critical regulator of cell growth outside the hematopoietic compartment and that it may play a role in the regulation of cell survival in many cellular contexts that were not identified in studies of embryonic development. PRH subcellular localization is aberrant in some breast tumors, including MCF-7 cells, and in thyroid tumors (11, 55). In addition, the product of the PRH-repressed gene Goosecoid promotes tumor cell malignancy and is strongly upregulated in breast tumors (30). Deregulation of PRH activity and the consequent deregulation of VSP and Goosecoid expression may be involved in the development of a variety of tumors. It is tempting to speculate that deregulated PRH activity in vascular stem cells or in hematopoietic migratory cells could contribute to tumor angiogenesis and tumor metastasis via upregulation of the VSP. Further studies will be required to determine the roles that PRH plays in these processes.
Acknowledgments
We are grateful to P. Hewett, C. Bunce, and R. Bicknell (University of Birmingham, United Kingdom) for reagents and useful discussions. We thank B. Spiegelman and Z. Arany (Dana-Farber Cancer Institute and Harvard Medical School), J. Brickman (University of Edinburgh, United Kingdom), and D. Bates (University of Bristol, United Kingdom) for plasmids used in this study.
P.N. is grateful to the University of Birmingham for a Ph.D. Studentship, H.W. is grateful to the BBSRC for a Committee Strategic Research Ph.D. Studentship, and A.S. is grateful to the Royal Thai Government for a Ph.D. Studentship. P.-S.J. and K.G. are grateful to The Wellcome Trust and Breast Cancer Campaign for grant funding.
Footnotes
Published ahead of print on 22 February 2010.
REFERENCES
- 1.Akiri, G., D. Nahari, Y. Finkelstein, S. Y. Le, O. Elroy-Stein, and B. Z. Levi. 1998. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene 17:227-236. [DOI] [PubMed] [Google Scholar]
- 2.Arany, Z., S. Y. Foo, Y. Ma, J. L. Ruas, A. Bommi-Reddy, G. Girnun, M. Cooper, D. Laznik, J. Chinsomboon, S. M. Rangwala, K. H. Baek, A. Rosenzweig, and B. M. Spiegelman. 2008. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature 451:1008-1012. [DOI] [PubMed] [Google Scholar]
- 3.Bess, K. L., T. E. Swingler, J. Rivett, K. Gaston, and P.-S. Jayaraman. 2003. The transcriptional repressor protein PRH interacts with the proteasome. Biochem. J. 374:667-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boguslawski, G., P. W. McGlynn, K. A. Harvey, and A. T. Kovala. 2004. SU1498, an inhibitor of vascular endothelial growth factor receptor 2, causes accumulation of phosphorylated ERK kinases and inhibits their activity in vivo and in vitro. J. Biol. Chem. 279:5716-5724. [DOI] [PubMed] [Google Scholar]
- 5.Bornes, S., L. Prado-Lourenco, A. Bastide, C. Zanibellato, J. S. Iacovoni, E. Lacazette, A. C. Prats, C. Touriol, and H. Prats. 2007. Translational induction of VEGF internal ribosome entry site elements during the early response to ischemic stress. Circ. Res. 100:305-308. [DOI] [PubMed] [Google Scholar]
- 6.Brickman, J. M., C. M. Jones, M. Clements, J. C. Smith, and R. S. Beddington. 2000. Hex is a transcriptional repressor that contributes to anterior identity and suppresses Spemann organiser function. Development 127:2303-2315. [DOI] [PubMed] [Google Scholar]
- 7.Chan, A. S., S. Y. Leung, M. P. Wong, S. T. Yuen, N. Cheung, Y. W. Fan, and L. P. Chung. 1998. Expression of vascular endothelial growth factor and its receptors in the anaplastic progression of astrocytoma, oligodendroglioma, and ependymoma. Am. J. Surg. Pathol. 22:816-826. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, T., D. Nahari, L. W. Cerem, G. Neufeld, and B. Z. Levi. 1996. Interleukin 6 induces the expression of vascular endothelial growth factor. J. Biol. Chem. 271:736-741. [DOI] [PubMed] [Google Scholar]
- 9.Crompton, M. R., T. J. Bartlett, A. D. MacGregor, G. Manfioletti, E. Buratti, V. Giancotti, and G. H. Goodwin. 1992. Identification of a novel vertebrate homeobox gene expressed in haematopoietic cells. Nucleic Acids Res. 20:5661-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daley, G. Q. 2004. Chronic myeloid leukemia: proving ground for cancer stem cells. Cell 119:314-316. [DOI] [PubMed] [Google Scholar]
- 11.D'Elia, A. V., G. Tell, D. Russo, F. Arturi, F. Puglisi, G. Manfioletti, V. Gattei, D. L. Mack, P. Cataldi, S. Filetti, C. Di Loreto, and G. Damante. 2002. Expression and localization of the homeodomain-containing protein HEX in human thyroid tumors. J. Clin. Endocrinol. Metab. 87:1376-1383. [DOI] [PubMed] [Google Scholar]
- 12.Desjobert, C., P. Noy, T. E. Swingler, H. Williams, K. Gaston, and P.-S. Jayaraman. 2009. The PRH/Hex repressor protein causes nuclear retention of Groucho/TLE corepressors. Biochem. J. 417:121-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dias, S., K. Hattori, Z. Zhu, B. Heissig, M. Choy, W. Lane, Y. Wu, A. Chadburn, E. Hyjek, M. Gill, D. J. Hicklin, L. Witte, M. A. Moore, and S. Rafii. 2000. Autocrine stimulation of VEGFR-2 activates human leukemic cell growth and migration. J. Clin. Invest. 106:511-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutta, D., S. Ray, J. L. Vivian, and S. Paul. 2008. Activation of the VEGFR1 chromatin domain: an angiogenic signal-ETS1/HIF-2alpha regulatory axis. J. Biol. Chem. 283:25404-25413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dvorak, H. F., M. Detmar, K. P. Claffey, J. A. Nagy, L. van de Water, and D. R. Senger. 1995. Vascular permeability factor/vascular endothelial growth factor: an important mediator of angiogenesis in malignancy and inflammation. Int. Arch. Allergy Immunol. 107:233-235. [DOI] [PubMed] [Google Scholar]
- 16.Elvert, G., A. Kappel, R. Heidenreich, U. Englmeier, S. Lanz, T. Acker, M. Rauter, K. Plate, M. Sieweke, G. Breier, and I. Flamme. 2003. Cooperative interaction of hypoxia-inducible factor-2alpha (HIF-2alpha) and Ets-1 in the transcriptional activation of vascular endothelial growth factor receptor-2 (Flk-1). J. Biol. Chem. 278:7520-7530. [DOI] [PubMed] [Google Scholar]
- 17.Ferrara, N., H. P. Gerber, and J. LeCouter. 2003. The biology of VEGF and its receptors. Nat. Med. 9:669-676. [DOI] [PubMed] [Google Scholar]
- 18.Fraser, H. M., S. E. Dickson, S. F. Lunn, C. Wulff, K. D. Morris, V. A. Carroll, and R. Bicknell. 2000. Suppression of luteal angiogenesis in the primate after neutralization of vascular endothelial growth factor. Endocrinology 141:995-1000. [DOI] [PubMed] [Google Scholar]
- 19.Gabrilovich, D., T. Ishida, T. Oyama, S. Ran, V. Kravtsov, S. Nadaf, and D. P. Carbone. 1998. Vascular endothelial growth factor inhibits the development of dendritic cells and dramatically affects the differentiation of multiple hematopoietic lineages in vivo. Blood 92:4150-4166. [PubMed] [Google Scholar]
- 20.Gabrilovich, D. I., H. L. Chen, K. R. Girgis, H. T. Cunningham, G. M. Meny, S. Nadaf, D. Kavanaugh, and D. P. Carbone. 1996. Production of vascular endothelial growth factor by human tumors inhibits the functional maturation of dendritic cells. Nat. Med. 2:1096-1103. [DOI] [PubMed] [Google Scholar]
- 21.George, A., H. C. Morse III, and M. J. Justice. 2003. The homeobox gene Hex induces T-cell-derived lymphomas when overexpressed in hematopoietic precursor cells. Oncogene 22:6764-6773. [DOI] [PubMed] [Google Scholar]
- 22.Gerber, H. P., F. Condorelli, J. Park, and N. Ferrara. 1997. Differential transcriptional regulation of the two vascular endothelial growth factor receptor genes. Flt-1, but not Flk-1/KDR, is up-regulated by hypoxia. J. Biol. Chem. 272:23659-23667. [DOI] [PubMed] [Google Scholar]
- 23.Gerber, H. P., and N. Ferrara. 2003. The role of VEGF in normal and neoplastic hematopoiesis. J. Mol. Med. 81:20-31. [DOI] [PubMed] [Google Scholar]
- 24.Gerber, H. P., A. K. Malik, G. P. Solar, D. Sherman, X. H. Liang, G. Meng, K. Hong, J. C. Marsters, and N. Ferrara. 2002. VEGF regulates haematopoietic stem cell survival by an internal autocrine loop mechanism. Nature 417:954-958. [DOI] [PubMed] [Google Scholar]
- 25.Goldberg, M. A., and T. J. Schneider. 1994. Similarities between the oxygen-sensing mechanisms regulating the expression of vascular endothelial growth factor and erythropoietin. J. Biol. Chem. 269:4355-4359. [PubMed] [Google Scholar]
- 26.Grunewald, M., I. Avraham, Y. Dor, E. Bachar-Lustig, A. Itin, S. Jung, S. Chimenti, L. Landsman, R. Abramovitch, and E. Keshet. 2006. VEGF-induced adult neovascularization: recruitment, retention, and role of accessory cells. Cell 124:175-189. [DOI] [PubMed] [Google Scholar]
- 27.Guiral, M., K. Bess, G. Goodwin, and P.-S. Jayaraman. 2001. PRH represses transcription in hematopoietic cells by at least two independent mechanisms. J. Biol. Chem. 276:2961-2970. [DOI] [PubMed] [Google Scholar]
- 28.Guo, Y., R. Chan, H. Ramsey, W. Li, X. Xie, W. C. Shelley, J. P. Martinez-Barbera, B. Bort, K. Zaret, M. Yoder, and R. Hromas. 2003. The homeoprotein Hex is required for hemangioblast differentiation. Blood 102:2428-2435. [DOI] [PubMed] [Google Scholar]
- 29.Hallaq, H., E. Pinter, J. Enciso, J. McGrath, C. Zeiss, M. Brueckner, J. Madri, H. C. Jacobs, C. M. Wilson, H. Vasavada, X. Jiang, and C. W. Bogue. 2004. A null mutation of Hhex results in abnormal cardiac development, defective vasculogenesis and elevated Vegfa levels. Development 131:5197-5209. [DOI] [PubMed] [Google Scholar]
- 30.Hartwell, K. A., B. Muir, F. Reinhardt, A. E. Carpenter, D. C. Sgroi, and R. A. Weinberg. 2006. The Spemann organizer gene, Goosecoid, promotes tumor metastasis. Proc. Natl. Acad. Sci. U. S. A. 103:18969-18974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hattori, K., S. Dias, B. Heissig, N. R. Hackett, D. Lyden, M. Tateno, D. J. Hicklin, Z. Zhu, L. Witte, R. G. Crystal, M. A. Moore, and S. Rafii. 2001. Vascular endothelial growth factor and angiopoietin-1 stimulate postnatal hematopoiesis by recruitment of vasculogenic and hematopoietic stem cells. J. Exp. Med. 193:1005-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hattori, K., B. Heissig, Y. Wu, S. Dias, R. Tejada, B. Ferris, D. J. Hicklin, Z. Zhu, P. Bohlen, L. Witte, J. Hendrikx, N. R. Hackett, R. G. Crystal, M. A. Moore, Z. Werb, D. Lyden, and S. Rafii. 2002. Placental growth factor reconstitutes hematopoiesis by recruiting VEGFR1(+) stem cells from bone-marrow microenvironment. Nat. Med. 8:841-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson, T. A., H. E. Taylor, D. Sharma, S. Desiderio, and S. K. Danoff. 2005. Vascular endothelial growth factor receptor-2: counter-regulation by the transcription factors, TFII-I and TFII-IRD1. J. Biol. Chem. 280:29856-29863. [DOI] [PubMed] [Google Scholar]
- 34.Jayaraman, P.-S., J. Frampton, and G. Goodwin. 2000. The homeodomain protein PRH influences the differentiation of haematopoietic cells. Leuk. Res. 24:1023-1031. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan, R. N., R. D. Riba, S. Zacharoulis, A. H. Bramley, L. Vincent, C. Costa, D. D. MacDonald, D. K. Jin, K. Shido, S. A. Kerns, Z. Zhu, D. Hicklin, Y. Wu, J. L. Port, N. Altorki, E. R. Port, D. Ruggero, S. V. Shmelkov, K. K. Jensen, S. Rafii, and D. Lyden. 2005. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 438:820-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koeffler, H. P., and D. W. Golde. 1980. Human myeloid leukemia cell lines: a review. Blood 56:344-350. [PubMed] [Google Scholar]
- 37.Kubo, A., V. Chen, M. Kennedy, E. Zahradka, G. Q. Daley, and G. Keller. 2005. The homeobox gene HEX regulates proliferation and differentiation of hemangioblasts and endothelial cells during ES cell differentiation. Blood 105:4590-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loureiro, R. M., and P. A. D'Amore. 2005. Transcriptional regulation of vascular endothelial growth factor in cancer. Cytokine Growth Factor Rev. 16:77-89. [DOI] [PubMed] [Google Scholar]
- 39.Lyden, D., K. Hattori, S. Dias, C. Costa, P. Blaikie, L. Butros, A. Chadburn, B. Heissig, W. Marks, L. Witte, Y. Wu, D. Hicklin, Z. Zhu, N. R. Hackett, R. G. Crystal, M. A. Moore, K. A. Hajjar, K. Manova, R. Benezra, and S. Rafii. 2001. Impaired recruitment of bone-marrow-derived endothelial and hematopoietic precursor cells blocks tumor angiogenesis and growth. Nat. Med. 7:1194-1201. [DOI] [PubMed] [Google Scholar]
- 40.Masood, R., J. Cai, T. Zheng, D. L. Smith, D. R. Hinton, and P. S. Gill. 2001. Vascular endothelial growth factor (VEGF) is an autocrine growth factor for VEGF receptor-positive human tumors. Blood 98:1904-1913. [DOI] [PubMed] [Google Scholar]
- 41.Maxwell, P. H., and P. J. Ratcliffe. 2002. Oxygen sensors and angiogenesis. Semin. Cell Dev. Biol. 13:29-37. [DOI] [PubMed] [Google Scholar]
- 42.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 43.Mayerhofer, M., P. Valent, W. R. Sperr, J. D. Griffin, and C. Sillaber. 2002. BCR/ABL induces expression of vascular endothelial growth factor and its transcriptional activator, hypoxia inducible factor-1alpha, through a pathway involving phosphoinositide 3-kinase and the mammalian target of rapamycin. Blood 100:3767-3775. [DOI] [PubMed] [Google Scholar]
- 44.Mendel, D. B., A. D. Laird, X. Xin, S. G. Louie, J. G. Christensen, G. Li, R. E. Schreck, T. J. Abrams, T. J. Ngai, L. B. Lee, L. J. Murray, J. Carver, E. Chan, K. G. Moss, J. O. Haznedar, J. Sukbuntherng, R. A. Blake, L. Sun, C. Tang, T. Miller, S. Shirazian, G. McMahon, and J. M. Cherrington. 2003. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin. Cancer Res. 9:327-337. [PubMed] [Google Scholar]
- 45.Menendez, D., A. Inga, J. Snipe, O. Krysiak, G. Schonfelder, and M. A. Resnick. 2007. A single-nucleotide polymorphism in a half-binding site creates p53 and estrogen receptor control of vascular endothelial growth factor receptor 1. Mol. Cell. Biol. 27:2590-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minami, T., T. Murakami, K. Horiuchi, M. Miura, T. Noguchi, J. Miyazaki, T. Hamakubo, W. C. Aird, and T. Kodama. 2004. Interaction between hex and GATA transcription factors in vascular endothelial cells inhibits flk-1/KDR-mediated vascular endothelial growth factor signaling. J. Biol. Chem. 279:20626-20635. [DOI] [PubMed] [Google Scholar]
- 47.Minami, T., R. D. Rosenberg, and W. C. Aird. 2001. Transforming growth factor-beta 1-mediated inhibition of the flk-1/KDR gene is mediated by a 5′-untranslated region palindromic GATA site. J. Biol. Chem. 276:5395-5402. [DOI] [PubMed] [Google Scholar]
- 48.Mosmann, T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65:55-63. [DOI] [PubMed] [Google Scholar]
- 49.Nakagawa, T., M. Abe, T. Yamazaki, H. Miyashita, H. Niwa, S. Kokubun, and Y. Sato. 2003. HEX acts as a negative regulator of angiogenesis by modulating the expression of angiogenesis-related gene in endothelial cells in vitro. Arterioscler. Thromb. Vasc. Biol. 23:231-237. [DOI] [PubMed] [Google Scholar]
- 50.Pages, G., and J. Pouyssegur. 2005. Transcriptional regulation of the vascular endothelial growth factor gene—a concert of activating factors. Cardiovasc. Res. 65:564-573. [DOI] [PubMed] [Google Scholar]
- 51.Park, S. H., K. W. Kim, Y. S. Lee, J. H. Baek, M. S. Kim, Y. M. Lee, M. S. Lee, and Y. J. Kim. 2001. Hypoglycemia-induced VEGF expression is mediated by intracellular Ca2+ and protein kinase C signaling pathway in HepG2 human hepatoblastoma cells. Int. J. Mol. Med. 7:91-96. [PubMed] [Google Scholar]
- 52.Patterson, C., Y. Wu, M. E. Lee, J. D. DeVault, M. S. Runge, and E. Haber. 1997. Nuclear protein interactions with the human KDR/flk-1 promoter in vivo. Regulation of Sp1 binding is associated with cell type-specific expression. J. Biol. Chem. 272:8410-8416. [DOI] [PubMed] [Google Scholar]
- 53.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Podar, K., and K. C. Anderson. 2005. The pathophysiologic role of VEGF in hematologic malignancies: therapeutic implications. Blood 105:1383-1395. [DOI] [PubMed] [Google Scholar]
- 55.Puppin, C., F. Puglisi, L. Pellizzari, G. Manfioletti, M. Pestrin, M. Pandolfi, A. Piga, C. Di Loreto, and G. Damante. 2006. HEX expression and localization in normal mammary gland and breast carcinoma. BMC Cancer 6:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin, G., R. Kishore, C. M. Dolan, M. Silver, A. Wecker, C. N. Luedemann, T. Thorne, A. Hanley, C. Curry, L. Heyd, D. Dinesh, M. Kearney, F. Martelli, T. Murayama, D. A. Goukassian, Y. Zhu, and D. W. Losordo. 2006. Cell cycle regulator E2F1 modulates angiogenesis via p53-dependent transcriptional control of VEGF. Proc. Natl. Acad. Sci. U. S. A. 103:11015-11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santos, S. C., and S. Dias. 2004. Internal and external autocrine VEGF/KDR loops regulate survival of subsets of acute leukemia through distinct signaling pathways. Blood 103:3883-3889. [DOI] [PubMed] [Google Scholar]
- 58.Schwarte-Waldhoff, I., O. V. Volpert, N. P. Bouck, B. Sipos, S. A. Hahn, S. Klein-Scory, J. Luttges, G. Kloppel, U. Graeven, C. Eilert-Micus, A. Hintelmann, and W. Schmiegel. 2000. Smad4/DPC4-mediated tumor suppression through suppression of angiogenesis. Proc. Natl. Acad. Sci. U. S. A. 97:9624-9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Semenza, G. L. 1999. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 15:551-578. [DOI] [PubMed] [Google Scholar]
- 60.Shima, D. T., U. Deutsch, and P. A. D'Amore. 1995. Hypoxic induction of vascular endothelial growth factor (VEGF) in human epithelial cells is mediated by increases in mRNA stability. FEBS Lett. 370:203-208. [DOI] [PubMed] [Google Scholar]
- 61.Shweiki, D., A. Itin, D. Soffer, and E. Keshet. 1992. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature 359:843-845. [DOI] [PubMed] [Google Scholar]
- 62.Soufi, A., and P.-S. Jayaraman. 2008. PRH/Hex: an oligomeric transcription factor and multifunctional regulator of cell fate. Biochem. J. 412:399-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soufi, A., P. Noy, M. Buckle, A. Sawasdichai, K. Gaston, and P.-S. Jayaraman. 2009. CK2 phosphorylation of the PRH/Hex homeodomain functions as a reversible switch for DNA binding. Nucleic Acids Res. 37:3288-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soufi, A., C. Smith, A. R. Clarke, K. Gaston, and P.-S. Jayaraman. 2006. Oligomerisation of the developmental regulator proline rich homeodomain (PRH/Hex) is mediated by a novel proline-rich dimerisation domain. J. Mol. Biol. 358:943-962. [DOI] [PubMed] [Google Scholar]
- 65.Strawn, L. M., G. McMahon, H. App, R. Schreck, W. R. Kuchler, M. P. Longhi, T. H. Hui, C. Tang, A. Levitzki, A. Gazit, I. Chen, G. Keri, L. Orfi, W. Risau, I. Flamme, A. Ullrich, K. P. Hirth, and L. K. Shawver. 1996. Flk-1 as a target for tumor growth inhibition. Cancer Res. 56:3540-3545. [PubMed] [Google Scholar]
- 66.Swingler, T. E., K. L. Bess, J. Yao, S. Stifani, and P.-S. Jayaraman. 2004. The proline-rich homeodomain protein recruits members of the Groucho/Transducin-like enhancer of split protein family to co-repress transcription in hematopoietic cells. J. Biol. Chem. 279:34938-34947. [DOI] [PubMed] [Google Scholar]
- 67.Tang, N., L. Wang, J. Esko, F. J. Giordano, Y. Huang, H. P. Gerber, N. Ferrara, and R. S. Johnson. 2004. Loss of HIF-1alpha in endothelial cells disrupts a hypoxia-driven VEGF autocrine loop necessary for tumorigenesis. Cancer Cell 6:485-495. [DOI] [PubMed] [Google Scholar]
- 68.Textor, B., M. Sator-Schmitt, K. H. Richter, P. Angel, and M. Schorpp-Kistner. 2006. c-Jun and JunB are essential for hypoglycemia-mediated VEGF induction. Ann. N. Y. Acad. Sci. 1091:310-318. [DOI] [PubMed] [Google Scholar]
- 69.Toi, M., T. Matsumoto, and H. Bando. 2001. Vascular endothelial growth factor: its prognostic, predictive, and therapeutic implications. Lancet Oncol. 2:667-673. [DOI] [PubMed] [Google Scholar]
- 70.Topcu, Z., D. L. Mack, R. A. Hromas, and K. L. Borden. 1999. The promyelocytic leukemia protein PML interacts with the proline-rich homeodomain protein PRH: a RING may link hematopoiesis and growth control. Oncogene 18:7091-7100. [DOI] [PubMed] [Google Scholar]
- 71.Topisirovic, I., B. Culjkovic, N. Cohen, J. M. Perez, L. Skrabanek, and K. L. Borden. 2003. The proline-rich homeodomain protein, PRH, is a tissue-specific inhibitor of eIF4E-dependent cyclin D1 mRNA transport and growth. EMBO J. 22:689-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Topisirovic, I., M. L. Guzman, M. J. McConnell, J. D. Licht, B. Culjkovic, S. J. Neering, C. T. Jordan, and K. L. Borden. 2003. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol. Cell. Biol. 23:8992-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verstovsek, S., S. Lunin, H. Kantarjian, T. Manshouri, S. Faderl, J. Cortes, F. Giles, and M. Albitar. 2003. Clinical relevance of VEGF receptors 1 and 2 in patients with chronic myelogenous leukemia. Leuk. Res. 27:661-669. [DOI] [PubMed] [Google Scholar]
- 74.Wakiya, K., A. Begue, D. Stehelin, and M. Shibuya. 1996. A cAMP response element and an Ets motif are involved in the transcriptional regulation of flt-1 tyrosine kinase (vascular endothelial growth factor receptor 1) gene. J. Biol. Chem. 271:30823-30828. [DOI] [PubMed] [Google Scholar]
- 75.Wang, E. S., J. Teruya-Feldstein, Y. Wu, Z. Zhu, D. J. Hicklin, and M. A. Moore. 2004. Targeting autocrine and paracrine VEGF receptor pathways inhibits human lymphoma xenografts in vivo. Blood 104:2893-2902. [DOI] [PubMed] [Google Scholar]
- 76.Williams, H., P.-S. Jayaraman, and K. Gaston. 2008. DNA wrapping and distortion by an oligomeric homeodomain protein. J. Mol. Biol. 383:10-23. [DOI] [PubMed] [Google Scholar]
- 77.Wu, Y., A. T. Hooper, Z. Zhong, L. Witte, P. Bohlen, S. Rafii, and D. J. Hicklin. 2006. The vascular endothelial growth factor receptor (VEGFR-1) supports growth and survival of human breast carcinoma. Int. J. Cancer 119:1519-1529. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, J., A. Lu, D. Beech, B. Jiang, and Y. Lu. 2007. Suppression of breast cancer metastasis through the inhibition of VEGF-mediated tumor angiogenesis. Cancer Ther. 5:273-286. [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang, L., D. Yu, M. Hu, S. Xiong, A. Lang, L. M. Ellis, and R. E. Pollock. 2000. Wild-type p53 suppresses angiogenesis in human leiomyosarcoma and synovial sarcoma by transcriptional suppression of vascular endothelial growth factor expression. Cancer Res. 60:3655-3661. [PubMed] [Google Scholar]