Abstract
In somatic cells, abscission, the physical separation of daughter cells at the completion of cytokinesis, requires CEP55, ALIX, and TSG101. In contrast, cytokinesis is arrested prior to abscission in differentiating male germ cells that are interconnected by TEX14-positive intercellular bridges. We have previously shown that targeted deletion of TEX14 disrupts intercellular bridges in all germ cells and causes male sterility. Although these findings demonstrate that intercellular bridges are essential for spermatogenesis, it remains to be shown how TEX14 and other proteins come together to prevent abscission and form stable intercellular bridges. Using a biochemical enrichment of male germ cell intercellular bridges, we identified additional bridge proteins, including CEP55. Although CEP55 is highly expressed in testes at the RNA level, there is no report of the presence of CEP55 in germ cells. We show here that CEP55 becomes a stable component of the intercellular bridge and that an evolutionarily conserved GPPX3Y motif of TEX14 binds strongly to CEP55 to block similar GPPX3Y motifs of ALIX and TSG101 from interacting and localizing to the midbody. Thus, TEX14 prevents the completion of cytokinesis by altering the destiny of CEP55 from a nidus for abscission to an integral component of the intercellular bridge.
Cytokinesis is the process by which a single cell separates into two genetically identical daughter cells (8). The events of cytokinesis begin shortly after the sister chromatids separate during mitotic anaphase. As the contractile ring narrows, the future daughter cells are connected by a narrow channel in which the evolutionarily conserved centralspindlin complex (a heterotetrameric complex of MKLP1 and MgcRacGap) localizes (16). Centrosomal 55-kDa protein (CEP55) is recruited from the centrosome to this centrally located complex and interacts with MKLP1 (6, 15, 22). Subsequently, ALIX (ALG-2 interacting protein X, also known as programmed cell death 6 interacting protein) and TSG101 (a component of the ESCRT-1 [endosomal sorting complex required for transport-1] complex) are recruited to the midbody through coiled-coil interactions with the CEP55 homodimer (3, 15, 17). Glycine (G)-proline (P)-proline (P)-X-X-X-tyrosine (Y) (GPPX3Y) motifs in ALIX and TSG101 are critical for this interaction with CEP55 (14, 17). Knockdown experiments in somatic cells have revealed that a deficiency of CEP55 leads to incomplete abscission and formation of multinucleated cells (3, 17). In addition, knockdown of either TSG101 or ALIX, known direct downstream interacting partners of CEP55, leads to a similar phenotype (3, 17). These interactions are essential for somatic cell abscission (2, 3, 17).
In contrast to these abscission events in somatic cells, differentiating germ cells do not complete cytokinesis and instead are linked together through 0.5- to 3-μm electron-dense “channels” called intercellular bridges (5, 7, 12). Intercellular bridges are evolutionarily conserved structures that are present in the gonads of essentially all multicellular organisms from fruit flies and hydra to marsupials, mice, and humans. In mammals, intercellular bridges play roles in synchronization of germ cells by passage of organelles and molecules between germ cells (especially important postmeiotically in haploid spermatids) (1, 19).
We have previously shown that testis expressed gene 14 (TEX14) localizes to male and female germ cell intercellular bridges (9, 11) and that the bridge forms through a direct interaction between TEX14 and the MKLP1-containing midbody protein complex (10). TEX14-positive intercellular bridges interconnect human and mouse spermatogonia as soon as spermatogonia begin to differentiate and continue to interconnect male germ cells up through formation of mature spermatozoa (11). Targeted deletion of TEX14 disrupts intercellular bridges in germ cells and causes sterility in male mice (11) but not in female mice (9). Furthermore, not only do MKLP1 and TEX14 interact in male germ cells, but MKLP1 and its centralspindlin complex partner, MgcRacGap, become stable components of the intercellular bridge (10). These results demonstrate that intercellular bridges are essential for spermatogenesis; however, until now, it was unclear how TEX14 participated in intercellular bridge formation to prevent abscission and the completion of cytokinesis in male germ cells. We demonstrate here that a TEX14-CEP55 interaction is critical for subverting abscission toward a stable intercellular bridge.
MATERIALS AND METHODS
Enrichment of intercellular bridges.
Intercellular bridge preparations were obtained from an 8-week-old wild-type mouse testis as previously described (10). The enriched intercellular bridge fractions were used for Western blot assays, and the final fraction PT was transferred to Superfrost/Plus microscope slides (Fisher Scientific) and allowed to air dry. After drying, the slides were lightly rinsed in TBS (100 mM Tris-HCl [pH 7.5]; 0.9%/150 mM NaCl) and used for immunofluorescence detection of mouse CEP55 and TEX14, as described below.
Production of anti-CEP55 and anti-MKLP1 antibodies.
Antibodies to full-length mouse CEP55 and MKLP1 protein were generated in guinea pigs using methods described previously (11). The antibodies were purified with the CEP55 or MKLP1 antigens, respectively, using the ProFound mammalian coimmunoprecipitation kit (Pierce).
Immunofluorescence analysis.
Mouse testes and ovaries were fixed overnight at 4°C, and cultured cells were fixed for 10 min at room temperature in 4% paraformaldehyde in TBS, followed by three washes in 70% ethanol and then overnight at 4°C in 70% ethanol. The testes and ovaries were processed and embedded by the Department of Pathology Core Services Laboratory (Baylor College of Medicine), and 4-μm sections were cut and prepared for immunostaining.
Samples were blocked in 3 or 5% bovine serum albumin (BSA)-TBS blocking buffer for 1 h at room temperature. Antibodies were diluted in 3 or 5% BSA-TBS blocking buffer and used for overnight incubation at 4°C at the following dilutions: rabbit or goat anti-TEX14, 1:500; guinea pig anti-CEP55, 1:500; and guinea pig anti-MKLP1, 1:200. Alexa 488- and Alexa 594-conjugated secondary antibodies were purchased from Molecular Probes. Samples were mounted with VectaShield mounting medium with DAPI (4′,6′-diamidino-2-phenylindole; Vector), covered with microscope coverslips (VWR Scientific), and examined by using an Axiovert 200 fluorescence microscope (Carl Zeiss). Fluorescence and differential interference contrast images were captured and processed using AxioVision release 4.6.
Generation of the N-terminal FLAG-tagged TEX14, MYC-tagged CEP55, mCherry-tagged TX or AAAX3A, and yellow fluorescent protein (YFP)-tagged full-length ALIX constructs.
The mouse TEX14 open reading frame (ORF) sequence was ligated into the BamHI and SalI sites of the pCMV-tag2 vector (Stratagene), which contains an N-terminal FLAG tag sequence. The mouse CEP55 ORF was cloned from testis cDNA and subcloned into the EcoRI and SalI sites of the pcDNA3 (Invitrogen) vector containing an N-terminal MYC tag sequence. The truncated and mutant TEX14 (TX, AAAX3A) and the full-length ALIX sequences were ligated into the BamHI and NotI or the NotI and XbaI sites, respectively, of the pcDNA3 vectors, while mCherry or YFP sequences were subcloned into the KpnI and BamHI sites. Purified plasmid DNA was obtained by using a QIAprep spin miniprep kit (Qiagen), and all constructs were sequenced for integrity.
Cell culture and transfection.
HEK293T or HeLa cells were maintained in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% fetal calf serum (SAFC Biosciences), 1% l-glutamine (Invitrogen), and penicillin-streptomycin (Invitrogen) and grown on poly-d-lysine (Sigma)-coated coverslips in culture plates at 37C° in a humidified 5% CO2 atmosphere. For immunoprecipitation and immunoblot experiments, cells were seeded at 50 to 80% confluence in 10-cm2 dishes (Corning) and transiently transfected using Fugene 6/HD transfection reagent (Roche) according to the manufacturer's instructions.
Coimmunoprecipitation and Western blot analysis.
FLAG-TEX14 and/or MYC-CEP55 (3 or 6 μg of DNA for double or single transfection, respectively) were overexpressed in HEK293T cells, as described above. At 48 h after transfections, cells were rinsed with phosphate-buffered saline (PBS) and lysed with M-PER mammalian protein extraction reagent (Pierce), and the lysates were sonicated and divided equally into three tubes. Lysates were incubated with anti-FLAG and anti-MYC antibodies and, as a control, protein G at 4°C for 1 h to overnight and then incubated with protein G-Sepharose 4B (Sigma) preblocked with 3% BSA-TBS. The immunoprecipitates were washed four times with PBS-1% Triton and once with PBS. The immunoprecipitates and total cell lysates were separated by 3 to 8% Tris-acetate gel (Invitrogen) and transferred onto a nitrocellulose membrane (Protran BA83). Western blot assays were performed using mouse anti-MYC monoclonal antibody (1:5,000; BD Biosciences) and mouse anti-FLAG monoclonal antibody (1:8,000; Sigma) as primary antibodies and horseradish peroxidase-conjugated anti-mouse IgG (1:10,000; Jackson Immunoresearch) as a secondary antibody. Western blot analyses of the enriched intercellular bridge fractions were performed using the affinity-purified goat anti-TEX14 antibody (1:500) and guinea pig anti-CEP55 antibody (1:500) as primary antibodies. Proteins were detected with chemiluminescence by SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) and exposed to BioMax XAR film (Eastman Kodak).
Yeast two-hybrid system and oxygen-biosensor assay.
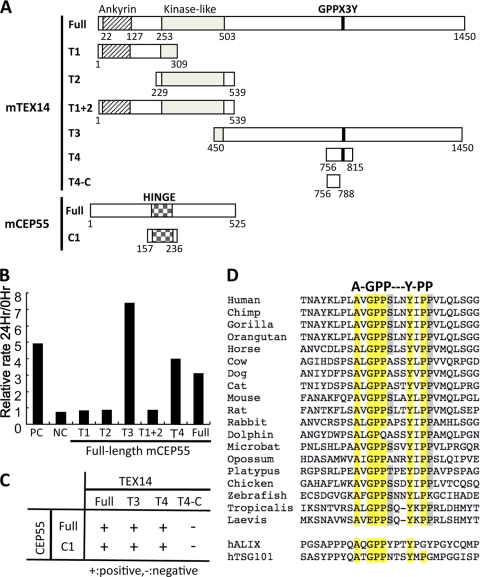
Protein-protein interactions were evaluated by using the Matchmaker Two-Hybrid System 3 (Clontech) as described previously (10). Mouse full-length TEX14 and MLKP1 were previously subcloned into the Matchmaker GAL4 two-hybrid pGBKT7 and pGADT7 vectors (10). The constructs of full-length mouse and human CEP55, human TEX14, truncated mouse CEP55-C1, and truncated mouse TEX14-T1, T2, T1+T2, T3, T4, and T4-C were made by using the Matchmaker GAL4 two-hybrid pGBKT7 bait vector and pGADT7 prey vectors. The constructs used are summarized in Fig. 5A and Table 1. Some Y2H interactions were examined with an oxygen biosensor assay (Clontech PT3584-1) that measured the fluorescence emitted by an oxygen-sensing platform that detects GAL4.
FIG. 5.
The region encoding the conserved TEX14 GPPX3Y motif interacts with the hinge region of CEP55. (A) The full-length and truncated regions of mouse TEX14 and CEP55 were cloned into the yeast two-hybrid vectors and used for the studies in panels B and C below. (B and C) Yeast two-hybrid oxygen biosensor between the full-length and truncated TEX14 proteins and the full-length CEP55 protein and/or selection plate analyses of the full-length and truncated TEX14 and CEP55 proteins. The terms “+” and “-” indicate positive and negative interactions, respectively. (D) Alignment of the GPPX3Y motif and flanking sequences from TEX14 orthologs and human ALIX and TSG101. Conserved amino acids are highlighted.
TABLE 1.
Constructs and results of yeast two-hybrid and biosensor experimentsa
| Assay | Figure | Species | Proteinb |
Interactionc | |
|---|---|---|---|---|---|
| pGADT7 | pGBKT7 | ||||
| Biosensor | 4B | Mouse | TEX14 | TEX14 | ++ |
| Mouse | MKLP1 | TEX14 | + | ||
| Mouse | CEP55 | TEX14 | ++ | ||
| 5B | Mouse | T1 | CEP55 | − | |
| Mouse | T2 | CEP55 | − | ||
| Mouse | T3 | CEP55 | +++ | ||
| Mouse | T1 + T2 | CEP55 | − | ||
| Mouse | T4 | CEP55 | ++ | ||
| Mouse | TEX14 | CEP55 | ++ | ||
| Y2H | 4C | Mouse | TEX14 | TEX14 | ++ |
| Mouse | TEX14 | CEP55 | ++ | ||
| Human | TEX14 | TEX14 | − | ||
| Human | TEX14 | CEP55 | ++ | ||
| 5C | Mouse | TEX14 | CEP55 | ++ | |
| Mouse | T3 | CEP55 | ++ | ||
| Mouse | T4 | CEP55 | ++ | ||
| Mouse | T4-C | CEP55 | − | ||
| Mouse | TEX14 | C1 | ++ | ||
| Mouse | T3 | C1 | ++ | ||
| Mouse | T4 | C1 | ++ | ||
| Mouse | T4-C | C1 | − | ||
This table summarizes the results of the biosensor and yeast two-hybrid assays in Fig. 4B and C and 5B and C. The source of the proteins that we used in these experiments is given in the species column.
TEX14, CEP55, and MKLP1 indicate full-length proteins, whereas T1 to T4, T4-C, or C1 denotes truncated proteins.
The terms “+” and “−” indicate positive and negative interactions, respectively, and the number of “+” symbols indicates the strength of the interaction.
Mammalian two-hybrid system.
A modified form of the CheckMate/Flexi vector mammalian two-hybrid system (Promega) was used to study the interactions of proteins in mammalian cells. A pGL4Cherry (mCherry/GAL4UAS/Hygro) vector was engineered from the pGL4.31 (luc2P/GAL4UAS/Hygro) vector by replacing the Luc2P firefly luciferase reporter sequence with the mCherry sequence. The pACT vector expresses a protein of interest, “X,” fused to a transcriptional activation domain (VP16-AD) and the pBIND vector expresses the other protein of interest, “Y,” fused to a DNA-binding domain (GAL4-BD) and Renilla luciferase by separate promoters. A pBIND-YFP vector was engineered from the pBIND vector by replacing Renilla luciferase sequence with the YFP sequence for experiments in Fig. 9.
FIG. 9.
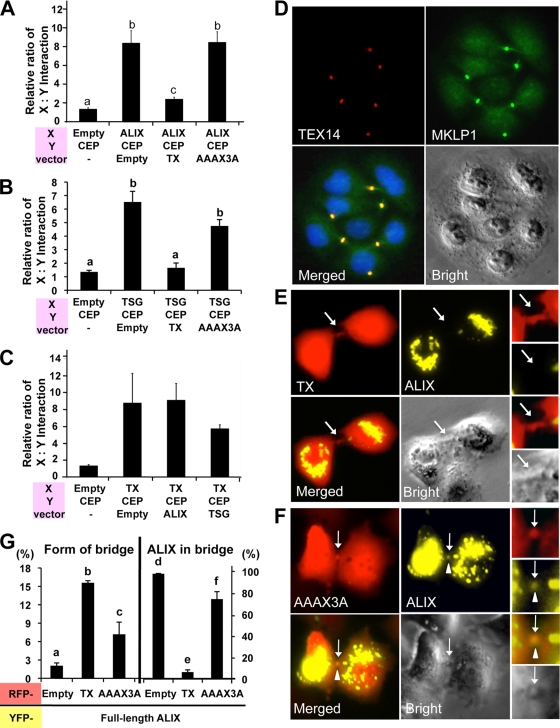
The GPPX3Y-containing TEX14 region interacts with CEP55 much more strongly than the equivalent regions of ALIX and TSG101 and inhibits the CEP55-ALIX and CEP55-TSG101 interactions. The pcDNA3 vectors lacking an insert (Empty) or containing the truncated TEX14 (TX), ALIX (ALIX), TSG101(TSG), and TEX14 mutant (AAAX3A) were cotransfected into HEK293T cells, along with VP16-AD-X, GAL4-BD-Y, and GAL4.3.1-mCherry vectors indicated at the bottom of each panel. The GAL4-BD-Y vector, containing the Renilla luciferase sequence, was replaced by yellow fluorescent protein (YFP) sequence. YFP expression was used for normalization of transfections instead of Renilla luciferase. The relative interaction of proteins X and Y is determined by the mCherry/YFP ratio. The interactions of chimeric VP16-AD-X and GAL4-BD-Y proteins in transfected HEK293T cells are shown (see Fig. 6B and Table 2 for additional details).
Sequences encoding the truncated TEX14, CEP55, ALIX, or TSG101 fragments were cloned into the multiple cloning regions of the pACT and pBIND vectors. The constructs used are summarized in Fig. 6A and B and Table 2 (procedure A). pACT (protein X), pBIND (protein Y), and pGL4Cherry (mCherry/GAL4UAS/Hygro) were transiently cotransfected into HEK293T cells using Fugene 6/HD transfection reagent, and the cells were examined 44 h later for red fluorescence by using an Axiovert 40 CFL microscope (Carl Zeiss). AxioVision release 4.6 was used for analysis of the image. The pACT-MyoD and pBIND-Id vectors served as positive controls, whereas the empty pACT and pBIND vectors served as negative controls for protein interaction. The cells were lysed by using Renilla luciferase assay reagent (Promega), and red fluorescence and Renilla luciferase were measured by using a Polar Star Omega microplate reader (BMG LabTech). The excitation and emission wavelengths were 580 and 610 nm, respectively, for red fluorescence (mCherry), 480 and 520 nm for yellow fluorescence (YFP), and luminescence for Renilla luciferase. Protein interactions were quantified by using the ratio of red fluorescence divided by Renilla luciferase or yellow fluorescence.
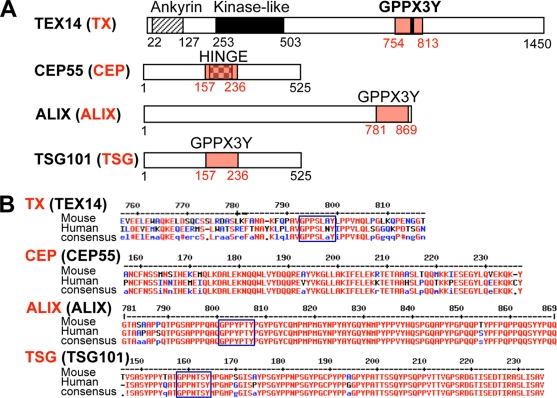
FIG. 6.
Essential motifs (A) and sequences (B) of TEX14, CEP55, ALIX, and TSG101. (A) The sizes and the domains/motif-containing regions of full-length TEX14, CEP55, ALIX, and TSG101 are shown. The domain regions highlighted in red are referred to as TX, CEP, ALIX, and TSG. (B) The corresponding amino acid sequences of these regions with the conserved consensus sequences in mouse and human are shown. These truncated proteins were used for the mammalian two-hybrid assays.
TABLE 2.
Constructs made for the mammalian two-hybrid assaysa
| Procedure | Figure | Human, mouse | pGAL4.3 | pACT (protein X) | pBIND (protein Y) | pcDNA3 (vector) | Interaction |
|---|---|---|---|---|---|---|---|
| A | 7C | Human, mouse | mCherry | Empty | CEP | − | |
| Human, mouse | mCherry | ALIX | CEP | ++ | |||
| Human, mouse | mCherry | TSG | CEP | + | |||
| Human, mouse | mCherry | TX | CEP | +++ | |||
| 7D | Human, mouse | mCherry | Empty | CEP | − | ||
| Human, mouse | mCherry | TX | CEP | +++ | |||
| Human, mouse | mCherry | Empty | ALIX | − | |||
| Human, mouse | mCherry | TX | ALIX | − | |||
| Human, mouse | mCherry | Empty | TSG | +, − | |||
| Human, mouse | mCherry | TX | TSG | +, − | |||
| 7E | Human, mouse | mCherry | Empty | TX | − | ||
| Human, mouse | mCherry | CEP | TX | +++ | |||
| Human, mouse | mCherry | Empty | TX-AAAX3A | − | |||
| Human, mouse | mCherry | CEP | TX-AAAX3A | − | |||
| 7F | Mouse | mCherry | Empty | CEP | − | ||
| Mouse | mCherry | TX | CEP | +++ | |||
| Mouse | mCherry | TX-APPX3Y | CEP | +++ | |||
| Mouse | mCherry | TX-GAPX3Y | CEP | − | |||
| Mouse | mCherry | TX-GPAX3Y | CEP | ++ | |||
| Mouse | mCherry | TX-GPPX3A | CEP | − | |||
| Mouse | mCherry | TX-AAAX3A | CEP | − | |||
| B | 8A and 9, left | Mouse, human | mCherry | Empty | CEP | NA | − |
| Mouse, human | mCherry | ALIX | CEP | Empty | ++ | ||
| Mouse, human | mCherry | ALIX | CEP | TX | + | ||
| Mouse, human | mCherry | ALIX | CEP | TX-AAAX3A | ++ | ||
| 8B and 9, center | Mouse | mCherry | Empty | CEP | NA | − | |
| Mouse | mCherry | TSG | CEP | Empty | + | ||
| Mouse | mCherry | TSG | CEP | TX | − | ||
| Mouse | mCherry | TSG | CEP | TX-AAAX3A | + | ||
| 8C and 9, right | Mouse | mCherry | Empty | CEP | NA | − | |
| Mouse | mCherry | TX | CEP | Empty | +++ | ||
| Mouse | mCherry | TX | CEP | ALIX | +++ | ||
| Mouse | mCherry | TX | CEP | TSG | +++ |
The table shows the species from which each protein is derived. In most cases the experiments were performed using both human and mouse TX, ALIX, TSG, and CEP. Boldfacing indicates experiments that are presented in Fig. 7C to F, Fig. 8A to C, and Fig. 9. “Empty” refers to the original pACT and pcDNA vector without insert. For procedure A, the two vectors were cotransfected with the pGL4-mCherry vector. For procedure B, the three vectors were cotransfected with the pGL4-mCherry vector. Under the “pcDNA3 (vector)” column, no additional vector is indicated by “NA.” A pBIND-YFP vector with CEP was used for mammalian two-hybrid analyses instead of a pBIND vector for Fig. 9. The terms “+” and “−” indicate positive and negative interactions, and the number of “+” symbols indicates the strength of the interaction.
Competition assays using the mammalian two-hybrid assay system.
The truncated TEX14 (TX), TEX14 AAAX3A mutant (AAAX3A), ALIX (ALIX), and TSG101 (TSG) pcDNA3 overexpression vectors were produced and expressed in HEK293T cells with mammalian two-hybrid vectors, pGL4Cherry (mCherry/GAL4UAS/Hygro), the VP16-AD-X and the GAL4-BD-Y vectors. Empty pcDNA3 vector was transfected as a control. The constructs used are summarized in Fig. 6A and B and Table 2, procedure B.
Alignment of the motif and flanking sequences from TEX14 orthologs.
By database mining using UCSC Genome Browser (http://genome.ucsc.edu/), NCBI (http://www.ncbi.nlm.nih.gov/), and Ensembl (http://www.ensembl.org), the protein sequences for TEX14 orthologs were obtained directly or deduced from ORFs of predicted cDNAs assembled from expressed sequence tags. Full-length Xenopus laevis and chicken (Gallus gallus) Tex14 cDNAs were cloned using 5′ and 3′ RACE (rapid amplification of cDNA ends), using the SMART RACE cDNA amplification kit (BD Biosciences). The following gene-specific primers were used: XlTex14-3RACE, 5′-CGTTGGTCAGCCCAGAAGTCATCA; XlTex14-5RACE, 5′-GGGCCCAGCAGTATGGTTTCCTACA; XlTex14-INT-F, 5′-CTTTTCTGGGAACCAGTGGAGT; XlTex14-INT-R, 5′-TTATGGCCAGAAATCACTGCAT; gTex14-5RACE, 5′-ACAGCGATGGTTAGGGTCTGAG; gTex14-3RACE, 5′-GGACCTAGACCACGAGCATTTA; gTex14-Ext-F, 5′-CGCTGTTAGGCCTTGGTAAGAT; and gTex14-Ext-R, 5′-TCTTCTTTGCTTCCACTTGCTG. The ORFs of deduced and cloned cDNAs were determined by using the EditSeq program of DNAStar (Madison, WI).
Statistical analysis.
Data were analyzed by analysis of variance according to the two-tailed Student t test. Differences between the mean values were considered to be statistically significant at P < 0.05.
RESULTS AND DISCUSSION
CEP55 is a component of stable intercellular bridge in testis and ovary.
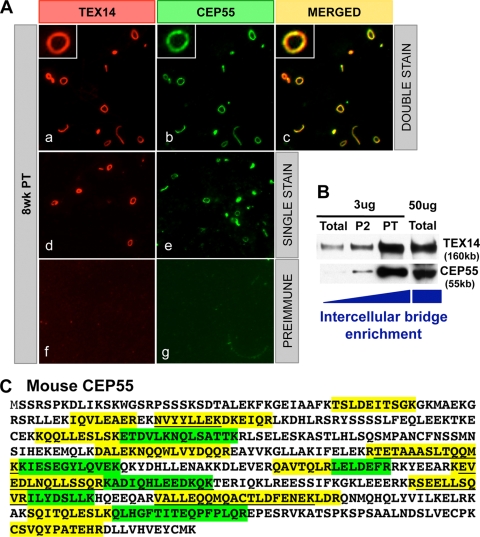
To identify components of the mammalian intercellular bridge and determine how the bridge forms, we developed a biochemical method to enrich intercellular bridges from mouse testes using TEX14 as a major marker protein (Fig. 1B) (10). This material was fractionated by size and subjected to liquid chromatography-tandem mass spectrometry proteomic analysis. In addition to TEX14, we isolated 19 proteins that have roles in cytokinesis, including the centralspindlin complex, MKLP1, and MgcRacGap (10). CEP55 mRNA was previously shown to be highly expressed in the testis (6, 15), and CEP55 protein was also identified with 24 peptides in our bridge preparation (Fig. 1C). To confirm that CEP55 was a component of the intercellular bridge and not a contaminant, we performed Western blot analyses of the enriched intercellular bridges and immunofluorescence of the purified intercellular bridge preparations. We discovered that CEP55 and TEX14 are coenriched by this biochemical method and colocalize in the purified intercellular bridges (Fig. 1A and B). To analyze the localization of CEP55 protein in immature and adult testes, we performed immunofluorescence analysis with antibodies generated to mouse TEX14 and mouse CEP55. Consistent with our purified bridge results, CEP55 and TEX14 perfectly colocalize as ring-shaped intercellular bridge structures in germ cells throughout the seminiferous tubules and at all stages of spermatogenesis (Fig. 2). The bridge diameter expands from the juvenile to the adult stage, suggesting that additional CEP55 and TEX14 are added to the intercellular bridge during spermatogenesis. In the female, CEP55 is expressed and colocalizes with TEX14 in embryonic day 18.5 mouse ovary (Fig. 3).
FIG. 1.
CEP55 is a component of the intercellular bridge. (A) Immunofluorescence was performed using custom-generated goat anti-mouse TEX14 and guinea pig anti-mouse CEP55 antibodies on isolated intercellular bridge preparations from an 8-week-old testis. The results from double staining (a to c), single staining (d and e), and preimmune using goat serum (f) or guinea pig serum (g) are shown as red, TEX14 or goat serum; green, CEP55 or guinea pig serum; and yellow, merged. (B) Western blot analysis of intercellular bridge enrichment using the anti-TEX14 and anti-CEP55 antibodies. (C) The 24 CEP55 peptides that were identified by proteomic analysis. The identified peptides are highlighted in yellow and green, with shorter overlapping peptides underlined.
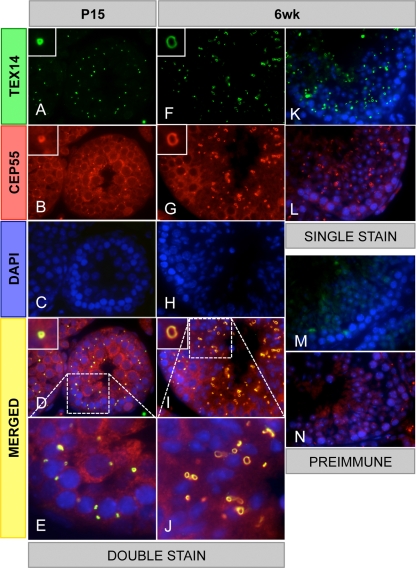
FIG. 2.
CEP55 colocalizes with TEX14 in testes. Immunofluorescence was performed with rabbit anti-TEX14 and guinea pig anti-CEP55 antibodies in postnatal 15-day-old and 6-week-old mouse testes. The results from double staining (A to J), single staining (K and L), and preimmune using rabbit serum (M) or guinea pig serum (N) are shown; green, TEX14 or rabbit serum; red, CEP55 or guinea pig serum; blue, DAPI; yellow, merged. High-magnification images (E and J) are derived from the boxed regions of panels D and I.
FIG. 3.
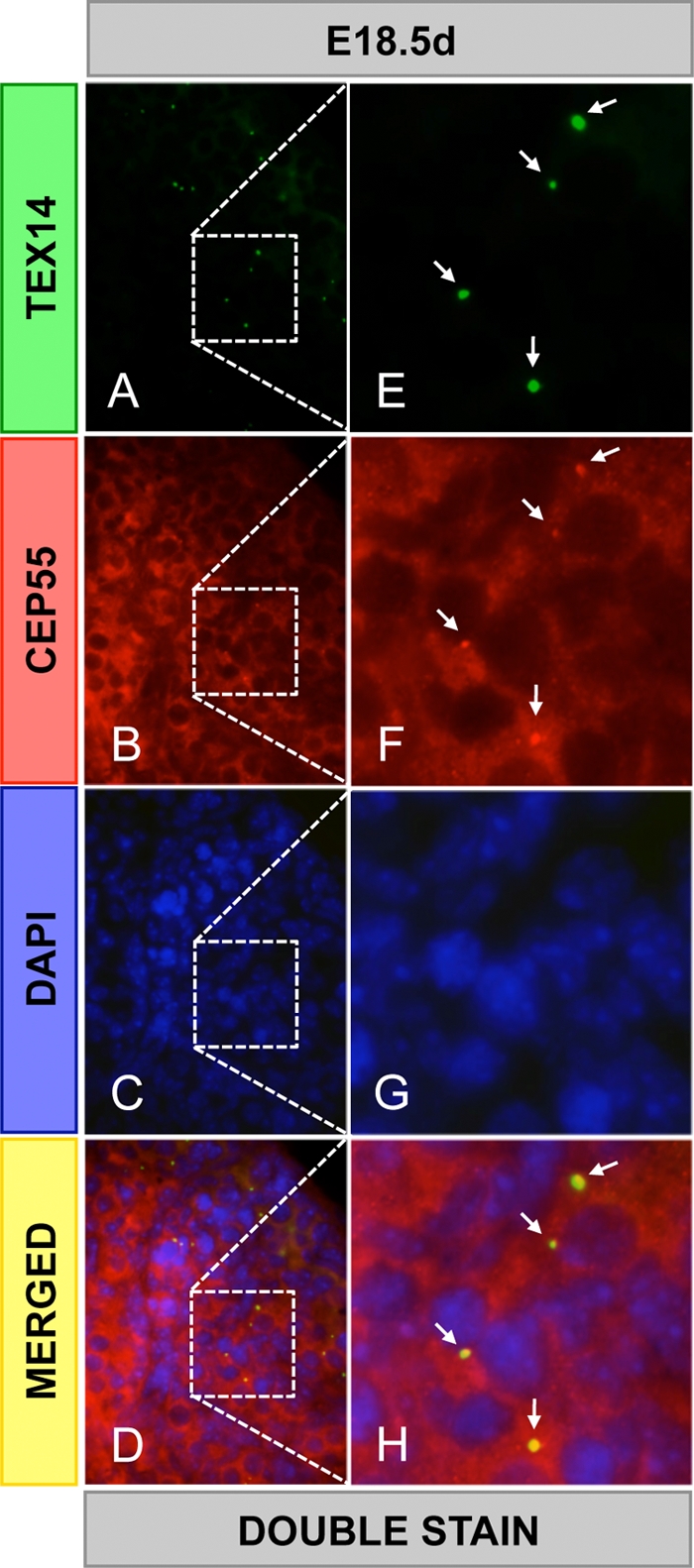
CEP55 colocalizes with TEX14 in the ovary. Immunofluorescence was performed using goat anti-TEX14 and guinea pig anti-CEP55 antibodies in embryonic day 18.5 mouse ovaries. The results from double staining (A to H): green, TEX14; red, CEP55; blue, DAPI; and yellow, merged. High-magnification images (E to H) are derived from the boxed regions in panels A to D, respectively. Arrows, intercellular bridges.
TEX14 interacts with CEP55.
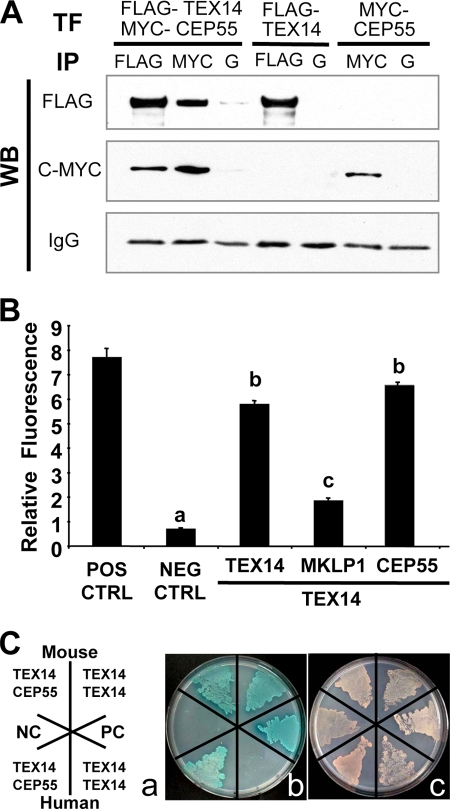
Since both CEP55 and TEX14 interact with MKLP1 (10, 22), we wanted to know whether CEP55 and TEX14 are components of the intercellular bridge as direct or indirect interactors. We first overexpressed full-length FLAG-TEX14 and MYC-CEP55 constructs in HEK293T cells (Fig. 4A). Immunoprecipitation with anti-FLAG antibody, followed by Western blot analysis with anti-FLAG or anti-MYC antibodies, reveals that we not only immunoprecipitate FLAG-TEX14 but also MYC-CEP55. Likewise, when we immunoprecipitate with anti-MYC antibody, we immunoprecipitate both MYC-CEP55 and FLAG-TEX14. When we express only FLAG-TEX14 and immunoprecipitate with anti-FLAG antibody, the anti-MYC antibody does not cross-react and conversely when we only express MYC-CEP55 and immunoprecipitate with anti-MYC antibody, the anti-FLAG antibody does not cross-react. Thus, by coimmunoprecipitation, we are able to show that TEX14 and CEP55 form a complex.
FIG. 4.
TEX14 interacts with CEP55. (A) Immunoprecipitation (IP) of FLAG-TEX14 and/or MYC-CEP55 from HEK293T cells, followed by Western blot analysis with the antibodies as shown. Immunoprecipitation of protein G (lanes G) is the control. (B) Yeast two-hybrid analyses using vectors encoding full-length mouse TEX14, MKLP1, CEP55, and positive and negative controls. The relative ratios of TEX14-TEX14, TEX14-MKLP1, and TEX14-CEP55 interactions were determined by using an oxygen-biosensor system. (C) Yeast two-hybrid interactions of mouse and human full-length TEX14 and CEP55. (a) Yeast were stably transformed with vectors and plated for yeast two-hybrid analysis as depicted. SV40 T antigen with p53 and SV40 T antigen with lamin C were used as positive and negative controls, respectively. (b) Interactions between the two-hybrid proteins are evident by colony growth and blue color on selection plates. (c) A nonselection plate shows that all of the transformed yeast are capable of growing.
To determine the intensity of the interactions of TEX14 and CEP55, we first performed yeast two-hybrid assays with full-length mouse or human TEX14 and CEP55. Mouse TEX14 interacts strongly with itself and CEP55 and more weakly with MKLP1 (Fig. 4B and C and Table 1). Human TEX14 and CEP55 also interact, but, oddly, human TEX14 does not interact with itself in this assay (Fig. 4C). Thus, mammalian orthologs of TEX14 and CEP55 interact strongly.
TEX14 has a GPPX3Y motif.
To define the core regions of TEX14 and CEP55 that interact, we constructed yeast two-hybrid vectors to express full-length and truncated mouse TEX14 and CEP55 proteins (Fig. 5A). TEX14 has three ankyrin repeats, a kinaselike domain, and a C-terminal leucine zipper dimerization motif (21) (Fig. 5A). Whereas full-length TEX14 interacts with full-length CEP55, N-terminal truncations of TEX14 that contained the ankyrin repeats, the kinaselike domains, or both failed to interact with full-length CEP55 (Fig. 5B and Table 1). However, the long TEX14 C-terminal domain that contains several coiled-coil domains interacts with full-length CEP55 (Fig. 5B and Table 1).
In somatic cells, CEP55 dimerizes through coiled-coil domains at the N terminus (17, 22) (see Fig. 11, left). During cytokinesis, GPPX3Y-containing coiled-coil domains of ALIX and TSG101 bind to the “hinge” region of CEP55 (14, 17) (Fig. 6A and B and see Fig. 11, left). These interactions are essential for somatic cell abscission (2, 3, 17). Because the C-terminal region of TEX14 contains coiled-coil domains and interacts with full-length CEP55 (Fig. 5B and C and Table 1), we searched for similar GPPX3Y domains in TEX14 that could be mimicking the ALIX and TSG101 interactions with CEP55. In addition to previously published mouse and human TEX14 orthologs (20, 21), we cloned the Xenopus laevis and Gallus gallus orthologs and uncovered an additional 15 full-length or partial TEX14 sequences in the public database. Although there is significant divergence of these orthologs in their C-terminal regions compared to the ankyrin repeats and kinaselike domains in their N termini, 17 of the 19 proteins share a similar GPPX3Y motif in their C termini, with the exceptions being the Xenopus laevis and dolphin orthologs (Fig. 5D). Analysis of the TEX14 sequences in and around the GPPX3Y motif demonstrated a conserved alanine upstream of the motif that is also shared with the human ALIX and TSG101 sequences, a conserved serine/threonine/alanine within the motif, and two conserved prolines downstream, one of which is also present in TSG101.
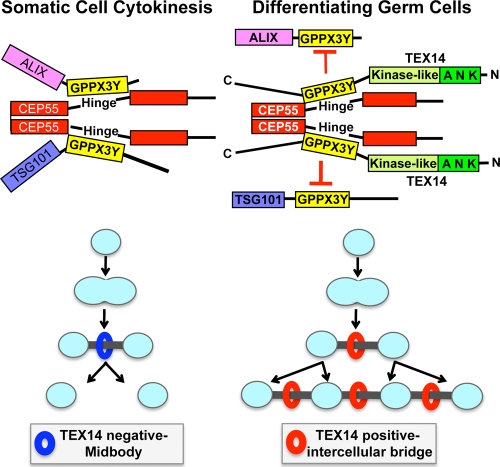
FIG. 11.
Models for cytokinesis and intercellular bridge formation. (Left) Model of somatic cell abscission. CEP55 is essential in the recruitment of additional proteins (e.g., TSG101 and ALIX) that are required for abscission of the midbody (2, 3, 17). The regions of TSG101 and ALIX containing GPPX3Y motifs interact with the hinge region of CEP55 (14, 17) to complete cytokinesis. (Right) Model of the intercellular bridge in differentiating germ cells. The conserved GPPX3Y motif of TEX14 interacts strongly with the hinge region of CEP55 in differentiating germ cells to block CEP55 interactions with TSG101 and ALIX, resulting in formation of a stable intercellular bridge.
The region including the GPPX3Y motif of mouse and human TEX14 interacts with the hinge of CEP55.
To determine the relevance of this GPPX3Y motif as it relates to the interaction of TEX14 with CEP55, we performed yeast two-hybrid assays with mouse TEX14 amino acids 756 to 815 that contain the GPPX3Y motif (Fig. 5A). These 60 amino acids interacted with full-length CEP55 (Fig. 5B and C and Table 1). Conversely, an 80-amino-acid fragment of CEP55 (C1) that contains the hinge region interacted with full-length TEX14, the C terminus (T3), and GPPX3Y-containing fragment (T4) but not a shorter sequence that lacked the GPPX3Y motif (T4-C) (Fig. 5A to C and Table 1). Thus, similar GPPX3Y-containing regions of TEX14, ALIX, and TSG101 interact with the hinge region of CEP55.
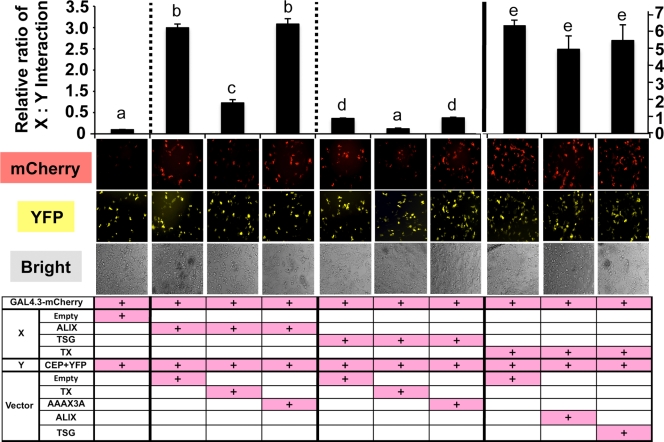
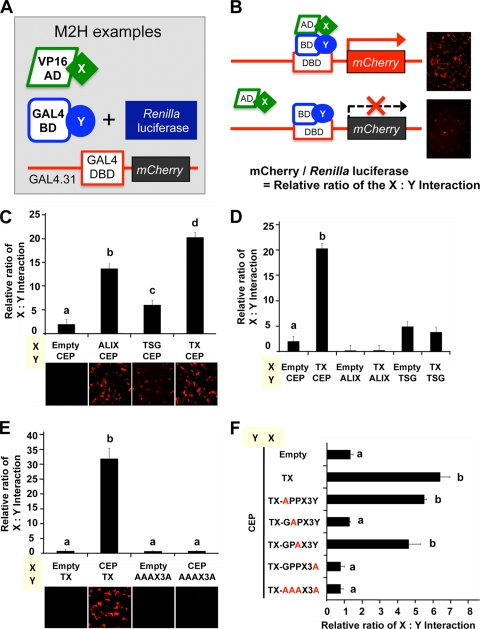
Based on these expression, colocalization, and yeast two-hybrid interaction data, we hypothesized that the conserved GPPX3Y motif of TEX14 (Fig. 5D) interacts strongly with the hinge region of CEP55 and blocks the interaction of CEP55 with TSG101 and ALIX (see Fig. 11, right). To test this hypothesis, we developed several modified mammalian two-hybrid assays to study these protein-protein interactions in a more natural mammalian cell setting. When our expressed proteins “X” and “Y” interact, the mCherry protein can be detected as red fluorescence (Fig. 7A and B). The GAL4-BD (“Y”) vector expresses Renilla reniformis luciferase (Fig. 7C to F and Fig. 8A to C) or yellow fluorescence (YFP) (Fig. 9) under the control of the simian virus 40 (SV40) promoter, normalizing for differences in transfection efficiency. Key regions of mouse and human TEX14 (TX), CEP55 (CEP), ALIX (ALIX), and TSG101 (TSG) (Fig. 6A and B) were cloned in frame into these vectors and cotransfected into HEK293T cells. Similar to previous reports (14, 17), we confirmed that the regions including the GPPX3Y motifs of human TSG101 and ALIX interact with the hinge region of human CEP55 (Fig. 7C and Table 2). Similar to our yeast two-hybrid data, the GPPX3Y region of human TEX14 (TX) (the 60 amino acids from 754 to 813, Fig. 6A and B) strongly binds to the hinge region of human CEP55 in our mammalian two-hybrid assay (Fig. 7C and Table 2). In all three cases, intracellular red fluorescence was also detected (Fig. 7C, bottom). In contrast, TEX14 does not interact with ALIX and, in the case of TSG101, TEX14 binding is not significantly different than the TSG101 interaction with empty pACT vector as a control (Fig. 7D and Table 2).
FIG. 7.
The GPPX3Y motif of TEX14 is essential for binding to the hinge region of CEP55. (A and B) Summary of the modified mammalian-two-hybrid assays (A) Three kinds of transfection vectors were made. One protein coding sequence (“X”) was fused to a transcriptional activation domain sequence (VP16-AD), and the other protein coding sequence (“Y”) was fused to a DNA-binding domain sequence (GAL4-BD). (B) When proteins “X” and “Y” interact, transcriptional activation of the mCherry gene occurs, which is detected as red fluorescence (B, top right). The GAL4-BD-Y vector expresses the Renilla reniformis luciferase, allowing for normalization of transfections. The relative interaction of protein X and Y is determined by the mCherry/Renilla reniformis luciferase ratio. (C to F) Mammalian-two-hybrid interactions of chimeric VP16-AD-X and GAL4-BD-Y proteins in transfected HEK293T cells is shown (see Fig. 6 and Table 2 for additional details).
FIG. 8.
The GPPX3Y motif of TEX14 inhibits the CEP55-ALIX and CEP55-TSG101 interactions and the entry of ALIX to the midbody, resulting in formation of stable intercellular bridges. (A to C) pcDNA3 vectors lacking an insert (Empty) or containing the truncated TEX14 (TX), ALIX (ALIX), TSG101(TSG), and TEX14 mutant (AAAX3A) were cotransfected into HEK293T cells along with VP16-AD-X, GAL4-BD-Y, and GAL4.31-mCherry vectors indicated at the bottom of each panel (see Fig. 6B and Table 2 for additional details). The relative ratios of the interactions of protein X and protein Y are shown. (D) Transfection of the full-length TEX14 vectors into HeLa cells. Immunofluorescence using goat anti-TEX14 and guinea pig anti-MKLP1 antibodies was performed: red, TEX14; green, MKLP1; blue, DAPI; and yellow, merged. (E and F) Cotransfection of pcDNA-YFP-full-length ALIX overexpression vector with pcDNA-mCherry-truncated GPPX3Y TEX14 (TX) (E) and TEX14 mutant AAAX3A (F) overexpression vectors into HeLa cells. The localization patterns of ALIX were microscopically examined for yellow fluorescence within a background of cells expressing TX or AAAX3A (red fluorescence). Arrow, midbody; arrowhead, ALIX. (G) Quantification of the experiment in panels E and F. The graphs were made by analyzing 1,000 double-positive cells with RFP and YFP from 11 to 13 separate experiments. The graphs show the percentage of the number of bridge containing cells/the number of double-positive RFP and YFP cells (left) and the number of ALIX localized in midbody/the number of RFP positive bridges in double-positive RFP and YFP cells (right).
The GPPX3Y domain of TEX14 and the first P and Y of GPPX3Y are essential for binding with the hinge of CEP55.
To define how the TEX14 GPPX3Y motif functions during the binding of TEX14 with CEP55, we generated the alanine mutant, AAAX3A. The TEX14 mutant AAAX3A is unable to bind to CEP55 (Fig. 7E and F and Table 2). To precisely define the amino acids in the TEX14 GPPX3Y motif that are most required for the interaction with CEP55, we mutated individual glycine (G), proline (P), and tyrosine (Y) residues to alanines. Specific mutations in the first proline (i.e., GAPX3Y) and the tyrosine (i.e., GPPX3A) of the motif are sufficient to abolish TEX14 interaction with CEP55 (Fig. 7F and Table 2). Consistent with the lack of conservation of the glycine in the Xenopus laevis ortholog of TEX14 (i.e., a G-to-E substitution) (Fig. 5D), mutation of this glycine to alanine did not alter the interaction of human TEX14 with CEP55. It is unclear whether the presence of the glutamine (Q) in the dolphin ortholog (i.e., GPQX3Y) functions like a proline in interactions with CEP55 or is a result of a genomic sequencing error.
The GPPX3Y motif of TEX14 inhibits the interaction of truncated CEP55 with ALIX or TSG101.
Based on our model (see Fig. 11, right), we hypothesize that TEX14 competes more efficiently for the CEP55 hinge region than either ALIX or TSG101. To test this hypothesis, we first analyzed the ALIX-CEP55 and TSG101-CEP55 interactions when TEX14 was overexpressed. Whereas ALIX and TSG101 demonstrate a strong interaction with CEP55 in our mammalian two-hybrid assay when an empty pcDNA vector is cotransfected, overexpression of TEX14 (TX) in this setting suppressed both the ALIX-CEP and the TSG-CEP interactions (Fig. 8A and B, Fig. 9 [left and center], and Table 2). However, overexpression of the TEX14 mutant AAAX3A does not block either the ALIX-CEP or the TSG-CEP interaction (Fig. 8A and B, Fig. 9 [left and center], and Table 2), confirming that the GPPX3Y motif is important for the interaction with CEP55 and also essential for the antagonism of ALIX and TSG101 interactions with CEP55. In contrast, overexpression of ALIX and TSG fails to disrupt the TX-CEP interaction (Fig. 8C, Fig. 9 [right], and Table 2). In addition, the TX-CEP interaction was typically stronger than the ALIX-CEP or TSG-CEP interactions (Fig. 9 and Table 2).
Full-length TEX14 localizes to the midbody and blocks cell abscission.
When full-length TEX14 is overexpressed in HeLa cells, many interconnected cells are visualized with anti-TEX14 and anti-MKLP1 antibodies by immunofluorescence (Fig. 8D). These data suggest that overexpressed TEX14 binds with CEP55 and blocks endogenous HeLa cell-produced ALIX and TSG101 from interacting with CEP55, resulting in inhibition of the completion in cytokinesis and stabilization of a transient intercellular bridge.
The GPPX3Y motif of TEX14 inhibits entry of full-length ALIX into the midbody.
In the testes of mice lacking TEX14, male germ cells continue to divide and enter meiosis, independent of the intercellular bridges. However, these germ cells quickly die at the pachytene spermatocyte stage (11). To study how the expression of TEX14 prevents these germ cells from dividing and stabilizes the intercellular connection, we transfected our various constructs into mammalian cells. We focused on ALIX because of its strong interactions with CEP55 in our assays (Fig. 7C, Fig. 8A, and Fig. 9, left) and on the truncated TEX14 to confirm that the GPPX3Y motif of TEX14 inhibits the interaction of ALIX:CEP55. We generated pcDNA-mCherry-truncated TEX14 (wild-type TX or mutant AAAX3A) and pcDNA-YFP full-length ALIX overexpression vectors and cotransfected them into HeLa cells. The truncated TEX14 and AAAX3A mutant are overexpressed throughout the cells, including the midbody, but do not specifically localize to the midbody like full-length TEX14 (Fig. 8D) because the truncated TEX14 and AAAX3A mutant are only 60 amino acids and do not have the other critical domains except the GPPX3Y motif (Fig. 6). The truncated GPPX3Y TEX14 (TX) inhibits full-length ALIX localization to the midbody (Fig. 8E and G, right), and several interconnecting cells were visualized in the TX transfected cells similar to the full-length TEX14-transfected cells in Fig. 8D (Fig. 8G, left). In contrast, full-length ALIX continues to localize to the midbody when the truncated TEX14 mutant (AAAX3A) construct is cotransfected (Fig. 8F and G, right), similar to the control experiment when the pcDNA-mCherry empty vector is transfected along with ALIX (data not shown). Thus, TEX14 blocks the ability of ALIX to interact with endogenous CEP55, preventing abscission, and instead TEX14-CEP55 complexes contribute to the formation of stable intercellular bridges (Fig. 8D and E).
Conclusions and implications.
In summary, our studies show that the GPPX3Y containing region of TEX14 binds to the hinge region of CEP55 and inhibits the interactions of ALIX and TSG101 with CEP55 (Fig. 7C to F, Fig. 8A and B, and Fig. 9). Our studies involving mutations in the GPPX3Y motif and disruption of the interactions between CEP55 and ALIX or TSG101 argue strongly for this to be a direct interaction. In our model (see Fig. 11), the steps involved in abscission normally are MKLP1 → CEP55 → ALIX/TSG101 →∼→ abscission. However, in differentiating male (and presumably female) germ cells, the steps involved appear to be MKLP1 → TEX14 → CEP55 → intercellular bridge. When GPPX3Y containing TEX14 fragments are present (Fig. 7E and F), ALIX does not enter the midbody, suggesting that TEX14 prevents the completion of cytokinesis, including the localization of ALIX, by altering the fate of CEP55 from a midbody organizer protein that recruits proteins for abscission to the midbody to a major component in the stable intercellular bridge. One additional mechanism by which TEX14 could more effectively compete for CEP55 is through its interaction with MKLP1. Our previous results (10) show that TEX14 appears early in the process of cytokinesis by binding MKLP1 and “filling” the intercellular bridge. At a later point, MKLP1, MgcRacGap, and TEX14 are overlapping in a single structure. We believe it is at this point that CEP55 approaches the midbody and is bound tightly and simultaneously by both MKLP1 and TEX14, never allowing the recruitment of ALIX or TSG101 for abscission.
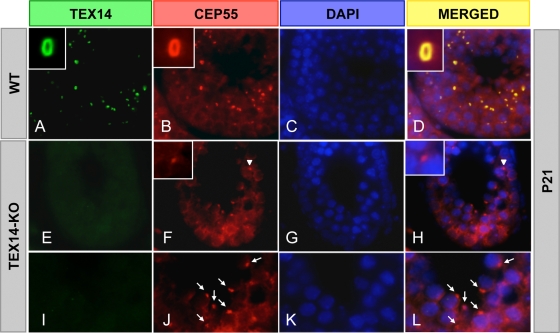
Thus, in male (and presumably female) germ cells, through a combination of a high local concentration of TEX14 in the transient bridge and a stronger TEX14-CEP55 interaction relative to the CEP55-ALIX or CEP55-TSG101 interactions (Fig. 9), ALIX and TSG101 are not recruited to the midbody and are unable to bind with CEP55 to complete cytokinesis (see Fig. 11, right). In the testes of TEX14 knockout mice, CEP55 localizes to the midbody and, in the absence of local TEX14 to interact, ALIX and TSG101 are permitted access to CEP55 to complete cytokinesis, disrupting the transient intercellular bridge and resulting in an eventual failure of spermatogenesis (Fig. 10 and Fig. 11, left) (11).
FIG. 10.
TEX14 inhibits the completion of cytokinesis in the testis. Immunofluorescence of wild-type (WT) and TEX14 knockout (KO) testes from 3-week-old mice. TEX14 and CEP55 colocalize at the intercellular bridge in WT mouse testis (A to D). In TEX14 knockout testes (E to L), CEP55 localizes to midbodies (F and H, arrowhead) and abscissed midbodies (J and L, arrow). Green, TEX14; red, CEP55; blue, DAPI; yellow, merged.
CEP55 mRNA and protein are increased in a number of tumor cell lines and carcinoma tissues (13, 18), and CEP55 is part of a 70-gene signature of chromosomal instability that is predictive of decreased survival in several cancer types (4). These findings suggest that interfering with CEP55 function in tumor cells may be a novel therapeutic strategy for targeting multiple different types of cancer. Further studies are warranted to determine whether small peptides containing the GPPX3Y motif of TEX14 could function as antineoplastic agents, capable of suppressing cancer cell proliferation by binding with CEP55 and generating stable intercellular bridges.
Acknowledgments
These studies were supported in part by National Institutes of Health grants R01HD057880, U01HD060496, and U54HD07495 (to M.M.M.). M.A.E. is supported by T32HD007165. M.A.E. and M.P.G. were supported by T32GM07330 and Baylor Research Advocates for Student Scientists.
We thank Yi-Nan Lin for technical advice; Julio Agno for genotyping mice; and Shirley Baker, Roopa Nalam, and Gregory Buchold for help with manuscript preparation.
Footnotes
Published ahead of print on 22 February 2010.
REFERENCES
- 1.Braun, R. E., R. R. Behringer, J. J. Peschon, R. L. Brinster, and R. D. Palmiter. 1989. Genetically haploid spermatids are phenotypically diploid. Nature 337:373-376. [DOI] [PubMed] [Google Scholar]
- 2.Carlton, J. G., M. Agromayor, and J. Martin-Serrano. 2008. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. U. S. A. 105:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlton, J. G., and J. Martin-Serrano. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316:1908-1912. [DOI] [PubMed] [Google Scholar]
- 4.Carter, S. L., A. C. Eklund, I. S. Kohane, L. N. Harris, and Z. Szallasi. 2006. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38:1043-1048. [DOI] [PubMed] [Google Scholar]
- 5.Dym, M., and D. W. Fawcett. 1971. Further observations on the numbers of spermatogonia, spermatocytes, and spermatids connected by intercellular bridges in the mammalian testis. Biol. Reprod. 4:195-215. [DOI] [PubMed] [Google Scholar]
- 6.Fabbro, M., B. B. Zhou, M. Takahashi, B. Sarcevic, P. Lal, M. E. Graham, B. G. Gabrielli, P. J. Robinson, E. A. Nigg, Y. Ono, and K. K. Khanna. 2005. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev. Cell 9:477-488. [DOI] [PubMed] [Google Scholar]
- 7.Fawcett, D. W., S. Ito, and D. Slautterback. 1959. The occurrence of intercellular bridges in groups of cells exhibiting synchronous differentiation. J. Biophys. Biochem. Cytol. 5:453-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glotzer, M. 2005. The molecular requirements for cytokinesis. Science 307:1735-1739. [DOI] [PubMed] [Google Scholar]
- 9.Greenbaum, M. P., N. Iwamori, J. E. Agno, and M. M. Matzuk. 2009. Mouse TEX14 is required for embryonic germ cell intercellular bridges but not female fertility. Biol. Reprod. 80:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenbaum, M. P., L. Ma, and M. M. Matzuk. 2007. Conversion of midbodies into germ cell intercellular bridges. Dev. Biol. 305:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenbaum, M. P., W. Yan, M. H. Wu, Y. N. Lin, J. E. Agno, M. Sharma, R. E. Braun, A. Rajkovic, and M. M. Matzuk. 2006. TEX14 is essential for intercellular bridges and fertility in male mice. Proc. Natl. Acad. Sci. U. S. A. 103:4982-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huckins, C. 1978. Spermatogonial intercellular bridges in whole-mounted seminiferous tubules from normal and irradiated rodent testes. Am. J. Anat. 153:97-121. [DOI] [PubMed] [Google Scholar]
- 13.Inoda, S., Y. Hirohashi, T. Torigoe, M. Nakatsugawa, K. Kiriyama, E. Nakazawa, K. Harada, H. Takasu, Y. Tamura, K. Kamiguchi, H. Asanuma, T. Tsuruma, T. Terui, K. Ishitani, T. Ohmura, Q. Wang, M. I. Greene, T. Hasegawa, K. Hirata, and N. Sato. 2009. Cep55/c10orf3, a tumor antigen derived from a centrosome residing protein in breast carcinoma. J. Immunother. 32:474-485. [DOI] [PubMed] [Google Scholar]
- 14.Lee, H. H., N. Elia, R. Ghirlando, J. Lippincott-Schwartz, and J. H. Hurley. 2008. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science 322:576-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Garay, I., A. Rustom, H. H. Gerdes, and K. Kutsche. 2006. The novel centrosomal associated protein CEP55 is present in the spindle midzone and the midbody. Genomics 87:243-253. [DOI] [PubMed] [Google Scholar]
- 16.Mishima, M., S. Kaitna, and M. Glotzer. 2002. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell 2:41-54. [DOI] [PubMed] [Google Scholar]
- 17.Morita, E., V. Sandrin, H. Y. Chung, S. G. Morham, S. P. Gygi, C. K. Rodesch, and W. I. Sundquist. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakai, M., T. Shimokawa, T. Kobayashi, S. Matsushima, Y. Yamada, Y. Nakamura, and Y. Furukawa. 2006. Elevated expression of C10orf3 (chromosome 10 open reading frame 3) is involved in the growth of human colon tumor. Oncogene 25:480-486. [DOI] [PubMed] [Google Scholar]
- 19.Ventela, S., J. Toppari, and M. Parvinen. 2003. Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: mechanisms of haploid gene product sharing. Mol. Biol. Cell 14:2768-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, P. J., J. R. McCarrey, F. Yang, and D. C. Page. 2001. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 27:422-426. [DOI] [PubMed] [Google Scholar]
- 21.Wu, M. H., A. Rajkovic, K. H. Burns, W. Yan, Y. N. Lin, and M. M. Matzuk. 2003. Sequence and expression of testis-expressed gene 14 (Tex14): a gene encoding a protein kinase preferentially expressed during spermatogenesis. Gene Expr. Patterns 3:231-236. [DOI] [PubMed] [Google Scholar]
- 22.Zhao, W. M., A. Seki, and G. Fang. 2006. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol. Biol. Cell 17:3881-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]