Abstract
Cellular senescence is an irreversible growth arrest and is presumed to be a natural barrier to tumor development. Like telomere shortening, certain defects in chromosome integrity can trigger senescence; however, the roles of centromere proteins in regulating commitment to the senescent state remains to be established. We examined chromatin structure in senescent human primary fibroblasts and found that CENP-A protein levels are diminished in senescent cells. Senescence-associated reduction of CENP-A is caused by transcriptional and posttranslational control. Surprisingly, forced reduction of CENP-A by short-hairpin RNA was found to cause premature senescence in human primary fibroblasts. This premature senescence is dependent on a tumor suppressor, p53, but not on p16INK4a-Rb; the depletion of CENP-A in p53-deficient cells results in aberrant mitosis with chromosome missegregation. We propose that p53-dependent senescence that arises from CENP-A reduction acts as a “self-defense mechanism” to prevent centromere-defective cells from undergoing mitotic proliferation that potentially leads to massive generation of aneuploid cells.
Cellular senescence is an irreversible growth arrest triggered by several types of stress, including DNA damage, oxidative stress, telomere shortening, and oncogene activation (7, 14, 15, 25, 55). While senescent cells maintain metabolic activity, cell cycle progression is permanently inhibited. The molecular basis of senescence has been studied intensively using normal diploid fibroblasts, melanocytes, and epithelial cells. In these studies, two tumor suppressor molecules, p53 and retinoblastoma protein (Rb), have been shown to play crucial roles in cell cycle arrest in senescent cells. In these cells, p53 effectively blocks cell cycle progression by upregulating its transcriptional target, p21CIP1. Rb is activated by p21CIP1 and p16INK4a, both of which are highly expressed in senescent cells (1, 7, 24). Activated Rb binds to E2F transcription factors to repress the expression of E2F target genes that promote cell proliferation (34). In contrast, p53 and p16INK4a-Rb pathways are often mutated in tumors (26, 53), and such tumor cells keep growing indefinitely without ever entering a senescent state. Senescence is therefore presumed to be a self-defense mechanism that prevents the uncontrolled proliferation of tumorigenic cells. Although it remains to be established how senescence, including the activation of the tumor suppressors, is initiated, certain defects in chromosome integrity, such as telomere shortening, can trigger it (7, 15). It was recently reported that BubR1-insufficient and Bub3/Rae1-haploinsufficient mice display an array of early aging-associated phenotypes (3-5) and Bub1 suppression in human fibroblasts activates a p53-dependent premature senescence response (22). Bub1, BubR1, and Bub3 are key players in the spindle assembly checkpoint (SAC) that blocks mitotic progression into anaphase in response to abnormalities in kinetochore-spindle interaction and/or kinetochore structure. These observations suggest that, like telomeres, kinetochores may also play a crucial role in regulating commitment to the senescent state.
Kinetochores are multiprotein complexes formed on a specialized region of each chromosome, designated the centromere. Kinetochore function is essential for the faithful segregation of chromosomes during mitosis and meiosis (13, 40). The centromere is composed of two domains, core centromeric chromatin and pericentric heterochromatin region. Numerous kinetochore-associated proteins have been identified to date, including centromere proteins (CENPs), Mis12, and SAC proteins (20, 23, 29, 40, 45, 47). CENP-A is an evolutionarily conserved centromere-specific histone H3 variant (8, 11, 18, 38, 49, 57, 59). As such, CENP-A represents an excellent candidate for an epigenetic marker of functional centromeres that could be monitored by senescence promoting networks. Studies of a variety of organisms have indicated that CENP-A plays a crucial role in organizing kinetochore chromatin for precise chromosome segregation; however, the impact of CENP-A loss upon proliferation varies widely in the context of species, cell types, and methods used to delete or deplete CENP-A (8, 23, 27, 51). CENP-B is another conserved centromere protein. CENP-B binds to a specific centromeric DNA sequence, the 17-bp “CENP-B box” in type I α-satellite repeats in human cells (19, 36). CENP-B is also important for proper organization of kinetochore chromatin. Although CENP-B is not essential for viability in higher eukaryotes (28, 30, 50), it is essential for heterochromatin formation of pericentromeres (41, 42, 48).
Despite the extensive studies of centromere-associated proteins, it remains unclear whether these proteins are involved in the control of cell proliferation; previous studies focused on the roles of centromere proteins in chromosome segregation and were mainly conducted in immortalized cell lines, such as HeLa. In HeLa cells, p53 and Rb are known to be inactivated due to the integration of the human papillomavirus that leads to their immortalization. Although it is essential to use primary human cells to uncover the regulatory roles of centromere proteins in cell proliferation, such studies have not been done. In our exploration of senescence-associated alterations in nuclear structure using primary human cells, we found that CENP-A levels were markedly reduced in the senescent cells. Furthermore, we showed that short-hairpin RNA (shRNA)-mediated depletion of CENP-A induces senescence in primary human fibroblast TIG3 cells but not HeLa cells. Inactivation of p53 in TIG3 cells depleted of CENP-A restores the proliferation, leading to an increase in the number of cells exhibiting aberrant chromosome behavior. These results indicate that the reduction of CENP-A drives normal human diploid fibroblasts into a senescent state in a p53-dependent manner. The senescence that arises from CENP-A depletion may be a self-defense mechanism to suppress the otherwise catastrophic impact upon genome integrity that would arise from kinetochore dysfunction following certain types of stress.
MATERIALS AND METHODS
Cell culture, vectors, and retroviral gene transfer.
Primary human diploid lung fibroblasts, TIG3 cells, were purchased from Health Science Research Resources Bank (Japan). When grown in vitro under standard conditions, TIG3 cells proliferated for a limited time and ceased dividing at 86 population doubling levels (PDLs) in our laboratory. The quiescent cells were produced by a combination of contact inhibition and 0.5% serum. Phoenix ampho and Phoenix eco cells were provided by G. P. Nolan (Stanford University, Stanford, CA). TIG3, HeLa, and Phoenix cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and penicillin-streptomycin at 37°C.
The following plasmids were used for generation of retroviruses: pWZL-neo encoding ecotropic receptor; pBabe-puro or pBabe-hygro encoding oncogenic ras, triple FLAG-tagged human CENP-A (FLAG-CENP-A) (see Fig. 5A), or histone H2B-EGFP; and pRetro Super-puro or pRetro Super-hygro (a gift from R. Agami, Netherlands Cancer Institute) encoding shRNA directed against CENP-A, p16INK4a, p53, or control. Retrovirus-shRNAs were generated as described previously (9). The target sequences for silencing CENP-A, p16INK4a (62), p53 (10), and the control are listed in Table 1. Human CENP-A cDNA (provided by M. Ikeno, School of Medicine, Keio University, Keio, Japan) and human histone H2B cloned by reverse transcription-PCR (RT-PCR) from total RNA of TIG3 cells were used as templates to generate the retroviral vectors according to standard molecular biology procedures.
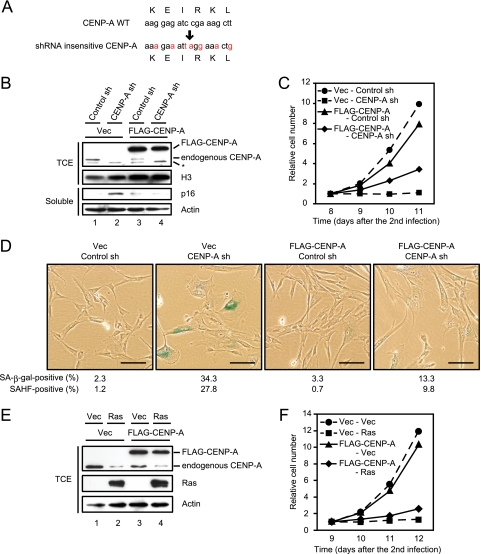
FIG. 5.
The expression of FLAG-tagged shRNA insensitive CENP-A rescues senescencelike phenotypes caused by shRNA to CENP-A. (A) A mutated shRNA cleavage site in an shRNA insensitive CENP-A cDNA is shown. (B) Immunoblotting of TIG3 cells expressing vector control (Vec) or FLAG-tagged shRNA insensitive CENP-A (FLAG-CENP-A) (first infection) in combination with control or CENP-A shRNA (second infection). TCE and soluble proteins were resolved by SDS-PAGE, followed by immunoblotting with the indicated antibodies. Histone H3 and actin were used as loading controls. Bands labeled with an asterisk are nonspecific proteins detected by anti-CENP-A antibody. (C) Growth curves of the cells indicated in panel B. The number of cells on day 8 after infection was set at 1. The data are means from two independent experiments. (D) SA-β-Gal staining performed on day 10 after the second infection. The proportions of SA-β-Gal-positive (n = 300) and SAHF-positive cells (n = 300) are shown. Scale bars, 100 μm. (E) Immunoblotting of TIG3 cells expressing vector control (Vec) or FLAG-CENP-A (first infection) in combination with vector control (Vec) or Ras (second infection). TCE were resolved by SDS-PAGE, followed by immunoblotting with the indicated antibodies. Actin was used as a loading control. (F) Growth curves of the cells indicated in panel E. The number of cells on day 9 after infection was set at 1. The data are means from two independent experiments.
TABLE 1.
shRNA target sequences and primers used for qPCR
| Primer | Sequence (5′-3′) |
|---|---|
| Control shRNA | CATTGCTATAGAGGCAGAT |
| CENP-A shRNA | AGGAGATCCGAAAGCTTCA |
| p53 shRNA | GACTCCAGTGGTAATCTAC |
| p16INK4a shRNA | GAGGAGGTGCGGGCGCTGC |
| p21CIP1 qPCR forward | GAGACTCTCAGGGTCGAAAACG |
| p21CIP1 qPCR reverse | GATTAGGGCTTCCTCTTGGAGAA |
| p16INK4a qPCR forward | CATAGATGCCGCGGAAGGT |
| p16INK4a qPCR reverse | AAGTTTCCCGAGGTTTCTCAGA |
| GAPDH qPCR forward | CAGGGCTGCTTTTAACTCTGGTA |
| GAPDH qPCR reverse | AATTTGCCATGGGTGGAATC |
Retrovirus production was performed as reported previously (34, 46). Briefly, Phoenix cells were transfected with retroviral vectors by using the calcium phosphate method (12). Viral supernatant was collected, filtered using a 0.45-μm-pore-size syringe filter, and supplemented with 8 μg of hexadimethrine bromide (Sigma-Aldrich)/ml. Infections with retrovirus were performed at 33.5°C for 12 h and were repeated three times to increase infection efficiency. Infected cells were purified and maintained using appropriate selection: 300 μg of G418/ml for TIG3 cells, 400 μg of G418/ml for HeLa cells, 2 μg of puromycin/ml for both cell lines, and 50 μg of hygromycin B/ml for both cell lines.
Growth curve.
To make growth curves, 4 × 104 cells in 2 ml of DMEM were plated on gridded glass coverslips (Matsunami Glass) in six-well plates. The number of cells in an area of 900 by 900 μm2 was counted in triplicate under a microscope.
Antibodies and protein analyses.
Total cell extracts were prepared from washed and pelleted cells by direct lysis in 2× sample buffer. Acid extraction, chromatin isolation, and preparation of soluble proteins in the nucleus and cytoplasm were performed as described previously with minor modifications (34, 37, 39, 46, 63). Total cell extracts (2 × 104 to 5 × 104 cells), chromatin-rich fractions (2 × 104 to 5 × 104 cells), acid extracts (30 μg), or soluble proteins (30 μg) were subjected to SDS-PAGE and analyzed by immunoblotting with the following primary antibodies: anti-CENP-A (07-574; Upstate Biotechnology), anti-CENP-B (07-735; Upstate Biotechnology), anti-hMis12 (A300-776A; Bethyl Laboratories), anti-heterochromatin protein 1β (HP1β) (MAB3448; Chemicon), anti-HP1γ (MAB3450; Chemicon), anti-histone H2B (07-371; Upstate Biotechnology), anti-histone macroH2A1 (07-219; Upstate Biotechnology), anti-histone H3 (ab1791; Abcam), anti-p16INK4a (NA29; Calbiochem), anti-p53 (OP43; Calbiochem), anti-p21CIP1 (sc-397; Santa Cruz Biotechnology), anti-cyclin A (sc-751; Santa Cruz Biotechnology), anti-cyclin B1 (sc-752; Santa Cruz Biotechnology), anti-CDC2 (610037; BD Transduction Laboratories), anti-pan-ras (OP41; Oncogene Research Products), anti-phospho-chk1 (serine 345, catalog no. 2341; Cell Signaling Technology), anti-phospho-chk2 (threonine 68, catalog no. 2661; Cell Signaling Technology), anti-chk1 (sc-8408; Santa Cruz Biotechnology), anti-chk2 (05-649; Upstate Biotechnology), anti-phospho-p53 (serine 15, catalog no. 9284; Cell Signaling Technology), and anti-actin (A4700; Sigma-Aldrich). Horseradish peroxidase-conjugated anti-mouse IgG (W4021; Promega) and anti-rabbit IgG (W4011; Promega) were used as the secondary antibodies.
For determination of the half-lives of CENP-A and cyclin A, cells treated with 50 μg of cycloheximide (Nacalai Tesque)/ml were collected at different time points, and total cell extracts were prepared for immunoblotting using antibodies to CENP-A (Upstate Biotechnology) and cyclin A (Santa Cruz Biotechnology). The half-life was estimated from the band density quantified by using ImageJ software (http://rsb.info.nih.gov/ij/).
Northern blotting analysis.
Total RNA was isolated by using TRIzol reagent (Invitrogen), and aliquots of 10 μg of total RNA were resolved by electrophoresis and transferred onto Hybond-N+ membranes (GE Healthcare Biosciences). DNA probes for CENP-A, H2B and GAPDH were labeled by the incorporation of [α-32P]dCTP using a random labeling kit (TaKaRa). The membranes were hybridized with the labeled probe at 65°C overnight in hybridization buffer (100 mM NaPO4 [pH 7.0], 5 mM EDTA, 1% SDS, 10% dextran sulfate, and 0.4 M NaCl). After washing the membranes, the signals were detected with a Typhoon 9410 imager (GE Healthcare Biosciences).
Quantification of mRNA levels by quantitative PCR (qPCR).
cDNA was synthesized from total RNA using a SuperScript first-strand synthesis system for RT-PCR (Invitrogen). Quantification of mRNA levels was carried out using the SYBR green I detection system on an ABI Prism 7300. PCR for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was used as an internal control. The relative abundances of p21CIP1 and p16INK4a mRNA were calculated after normalization using GAPDH mRNA. The primer sequences used are listed in Table 1.
BrdU incorporation assay and immunofluorescence.
Bromodeoxyuridine (BrdU) incorporation assay was performed as described previously (34, 46). Briefly, cells grown on coverslips were labeled with 10 μM BrdU for 1 h, fixed with 3% paraformaldehyde-2% glucose in phosphate-buffered saline (PBS), permeabilized with 0.2% Triton X-100 for 5 min, and pretreated with 5% goat serum in PBS with 0.1% Triton X-100 for 20 min. Cells were incubated with anti-BrdU antibody (catalog no. 555627; BD Pharmingen) containing 100 U of DNase (TaKaRa)/ml for 1 h, followed by Alexa Fluor 488-conjugated anti-mouse IgG (Molecular Probes) for 30 min. Subsequently, the cells were incubated with anti-phospho-specific histone H3 (serine 10) (06-570; Upstate Biotechnology) for 1 h, followed by Alexa Fluor 546-conjugated anti-rabbit IgG (Molecular Probes) for 30 min. DNA was stained with 100 ng of Hoechst 33342/ml for 5 min. Between each incubation step, cells were washed with PBS with 0.1% Triton X-100 (PBST). To determine the proportion of BrdU-positive or phospho-histone H3-positive cells, 300 cells were counted from each of the samples.
For detection of centromere and chromatin proteins, cells were fixed with 3% paraformaldehyde in PBS at room temperature for 10 min, permeabilized with 0.2% Triton X-100 for 5 min, and pretreated with 3% bovine serum albumin in PBS for 20 min. Cells were incubated with appropriate primary antibodies: anti-CENP-A (Upstate Biotechnology or GTX13939; GeneTex), anti-CENP-B (Upstate Biotechnology), anti-HP1β (Chemicon), anti-HP1γ (Chemicon), and anti-phospho-histone H2A.X (serine 139, catalog no. 05-636; Upstate Biotechnology). Cells were then incubated with the appropriate Alexa Fluor 488- or Alexa Fluor 546-conjugated secondary antibodies (Molecular Probes). DNA was Hoechst stained in PBS for 5 min.
For observation, an Olympus IX81 microscope (Olympus) equipped with a ×40 objective lens (NA 1.30), a ×60 objective lens (NA 1.40), and a ×100 objective lens (NA 1.35), and a RETIGA Exi FAST cooled charge-coupled device camera (Roper Scientific) was used. Three-dimensional stacks of Z-sectioned images (0.3-μm intervals) were collected, deconvolved, and flattened using MetaMorph software (Molecular Devices). For measurement of the intensity of centromere protein levels and the area of CENP-B, nonprocessed three-dimensional stacks of Z-sectioned images, which had been collected on the same day with identical exposure, were directly flattened. The average pixel intensity per nucleus with nuclear background subtracted was measured from at least 300 cells using MetaMorph software. The pixel area of CENP-B signals, whose pixel intensity is over 1.5-fold of that of nuclear background, was measured in at least 300 cells using MetaMorph software. Statistical significances of quantification experiments were verified by using the Student t test.
Live cell analysis.
TIG3 cells expressing H2B-EGFP were cultured on 35-mm glass-bottom dishes (MatTek) in DMEM and observed using a Leica ASMDW live cell imaging system (Leica Microsystems) equipped with a ×63 objective lens (NA 1.30) and a temperature control unit. The cells were maintained at 37°C under a humidified atmosphere of 5% CO2. Three-dimensional stacks of Z-sectioned images (2-μm interval) were collected using a 30-ms exposure every 4 min. For deconvolution, the nonblind algorithm of ASMDW software was applied.
SA-β-Gal staining.
Senescence-associated β-galactosidase (SA-β-Gal) activity was detected as described previously (17, 34, 46). For observation, an Olympus IMT-2 microscope equipped with a ×20 objective lens (NA 0.40) and a camera (VB-6000; Keyence) was used.
RESULTS
Alteration of centromere proteins in senescent human cells.
To test the possibility that centromere proteins could influence the decision to enter the senescent state, we initially examined the levels of centromere-associated proteins in senescent human diploid fibroblasts (TIG3) by immunoblotting. Cellular senescence was induced by two independent mechanisms: extensive culture (replicative senescence) and retroviral delivery of oncogenic ras (ras-induced premature senescence) (25, 55). As the levels of these proteins in cell lysates is often influenced by the particular method used for protein extraction, we used three independent approaches to generate cell extracts: no-salt extraction for enrichment of chromatin-associated proteins (39), acid extraction for basic proteins (63), and total cell extraction.
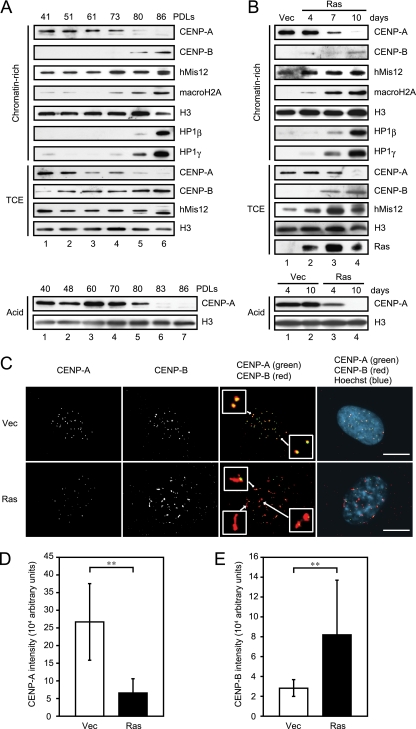
Of all of the candidate proteins examined, CENP-A exhibited the most striking change between proliferating and senescent cells; in both replicative and ras-induced senescent cells, it was reduced to less than 20% of the levels in proliferating cells (Fig. 1A and B). In contrast to CENP-A (the centromere-specific variant of histone H3), the levels of core histones, H2A, H2B, H3, and H4 remained unchanged between senescent and growing cells (Fig. 1A and B, data not shown). The levels of the other centromere proteins, CENP-B and hMis12, increased gradually, as the cells became senescent. Since these changes in the levels of the centromere proteins were reproducibly observed in both replicative and ras-induced senescent cells, we conclude that the reduction of CENP-A and the accumulation of CENP-B and hMis12, which may alter the centromere chromatin structure, represented physiologically significant phenomena associated with cellular senescence.
FIG. 1.
Alterations of centromere proteins in senescent human cells. (A and B) Immunoblotting of replicative (A) and ras-induced (B) senescent TIG3 cells. Chromatin-rich fractions, total cell extracts (TCE) (2 × 104 cells, upper panels), and acid extracts (30 μg, lower panels) were resolved by SDS-PAGE, followed by immunoblotting with the indicated antibodies. PDLs and the time points after infection are shown at the top. (C) Immunofluorescence of ras-induced senescent cells performed on day 10 after infection. Staining revealed CENP-A (green in overlay) and CENP-B (red in overlay). DNA was counterstained with Hoechst (blue in overlay). Insets represent enlarged images indicated by arrows. Scale bars, 10 μm. (D and E) Signal intensities of centromere-localizing CENP-A (D) and CENP-B (E) quantified in immunofluorescence performed on day 10 after infection. Graphs show the intensity of CENP-A in the control (n = 307) and the ras-induced senescent cells (n = 323) and that of CENP-B in the control (n = 329) and the ras-induced senescent cells (n = 313). The data are means ± the standard deviation (SD) (**, P < 0.01 [Student t test]).
In these experiments, we found that the levels of histone H2A variant, macro H2A, and the heterochromatin proteins, HP1β and HP1γ, also increased in the senescent TIG3 cells. These proteins have been reported to be incorporated into the specialized domains of facultative heterochromatin, designated senescence-associated heterochromatic foci (SAHF), in senescent human cells (44, 64). The number of cells showing elevated levels of SAHF increased with the onset of senescence; SAHF was detected in 4.3, 14.1, and 51.2% of TIG3 cells grown for 81, 83, and 86 PDLs, respectively. In the case of ras-induced premature senescence, SAHF was detected in 20.0 and 31.0% of the cells on days 7 and 10 after infection with ras, respectively. Although the increases in protein levels of macroH2A and HP1 have been suggested to promote the formation of SAHF in senescent human cells, our observations suggest that increased HP1 proteins may also promote alteration of the centromere structure, since HP1 proteins are essential components of the pericentric heterochromatin region.
Stimulating heterochromatin formation on centromeres in the senescent cells.
To confirm the association between alterations in the levels of CENP-A and CENP-B with the onset of senescence, we performed indirect immunofluorescence assays with antibodies against these centromere proteins (Fig. 1C). Consistent with the reduction of CENP-A levels detected by immunoblotting, the immunofluorescent signal of CENP-A was markedly weaker in ras-induced senescent cells than in the growing control cells, although CENP-A signals could still be detected on the centromeres in the senescent cells. In contrast, CENP-B signals, which were distributed over much larger areas around CENP-A, were markedly stronger in the senescent cells than in the control cells. Similar results were obtained upon processing replicative senescent cells, where p53 activation plays a predominant role in induction of senescence, for immunofluorescence microscopy (data not shown). We calculated the amounts of these proteins localized on the centromeres but not in the nucleoplasm from the intensity of the signals (see Materials and Methods) and found that the amount of centromere-localized CENP-A was reduced to 25% in the senescent cells compared to the proliferating cells, whereas the amount of CENP-B that associated with centromeres was increased ∼3-fold (Fig. 1D and E). In addition, the CENP-B signals in the senescent cells were distributed in six times larger areas than those in the growing cells. This result indicated that the centromere/kinetochore protein composition underwent significant changes upon senescence. We detected the reduction of CENP-A and the enrichment of CENP-B on centromeres prior to SAHF formation and were even able to observe it in SAHF-negative senescent cells. These observations suggested that alteration in the composition of molecules associating with centromeres is an early event that precedes SAHF formation in cellular senescence.
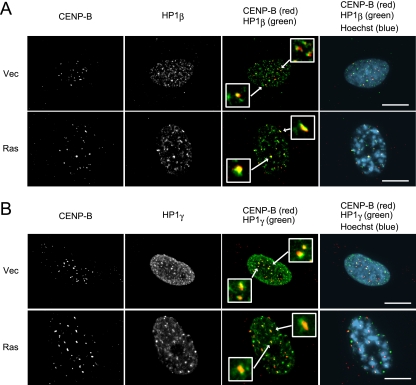
Since a recent study using human/mammalian artificial chromosomes demonstrated that CENP-B stimulates heterochromatin formation that can potentially lead to centromere inactivation (48), we speculated that the enhanced accumulation of CENP-B in senescent cells may promote heterochromatinization of the centromere. We therefore examined the localization of the HP1 proteins that constitute essential components of heterochromatin, alongside CENP-B by indirect immunofluorescence assays. Speckles of HP1β and HP1γ were distributed throughout the nuclei of proliferating cells (Fig. 2); some of these speckles were colocalized to centromeres with CENP-B, suggesting that HP1 proteins participated in pericentromeric heterochromatin formation in dividing cells. In ras-induced senescent cells, HP1 proteins were localized to centromeres as well as the condensed DNA foci (SAHF). The fluorescent signals of HP1 that colocalized with the diffuse CENP-B signals were much stronger than those in the control cells, indicating that the regions around centromeres that contained heterochromatin were greatly extended in senescent cells. Consistent with this observation, we detected enrichment of K9-trimethylated histone H3, which is another hallmark of heterochromatin, on centromeres in the senescent cells (data not shown). These results suggested that the enrichment of CENP-B at the centromeres of senescent cells may extend the region of heterochromatin formation at the centromere. We presumed that these changes in the centromere structure, i.e., the reduction of CENP-A and extended heterochromatinization, are key characteristics of the senescent cells.
FIG. 2.
Stimulating heterochromatin formation on centromeres in senescent cells. (A and B) Immunofluorescence of ras-induced senescent cells performed on day 10 after infection. Staining revealed CENP-B (red in overlay) and HP1β (green in overlay) (A) or HP1γ (green in overlay) (B). DNA was counterstained with Hoechst (blue in overlay). Insets represent enlarged images indicated by arrows. Scale bars, 10 μm.
The reduction of CENP-A is mediated by transcriptional and posttranslational control in the senescent cells.
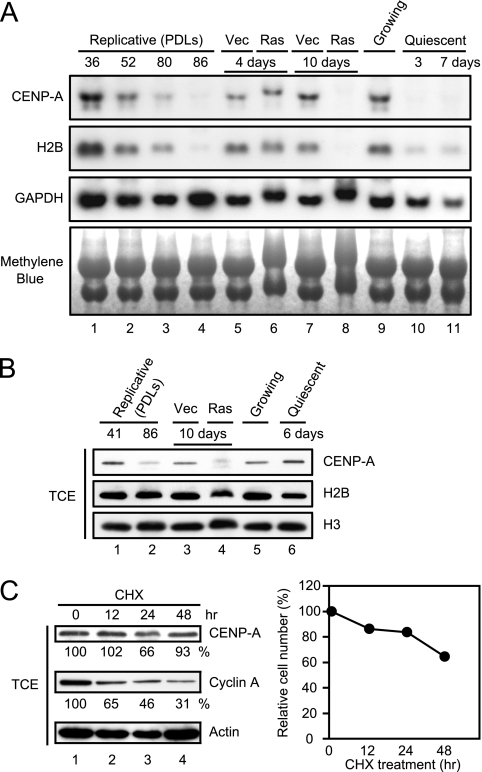
To investigate the cause of the reduction of CENP-A during cellular senescence, we performed Northern blotting analysis for CENP-A mRNA. It is assumed that CENP-A mRNA synthesis would be affected by cell proliferation ability, because its expression is regulated during the cell cycle and occurs in G2 phase in human cells (56). Therefore, we compared the expression of CENP-A mRNA in the senescent cells with that in serum-starved quiescent cells, which had also ceased to proliferate. As shown in Fig. 3A, the expression of CENP-A mRNA was gradually reduced in a time-dependent manner in the replicative and the ras-induced senescent cells. The reduction of CENP-A mRNA, however, was not specific to the senescent cells; quiescent cells also showed a marked decrease in CENP-A mRNA transcript levels. The expression of histone H2B mRNA was also reduced in both senescent and quiescent cells, indicating that the transcription of core histones as well as CENP-A ceased immediately when cells were arrested regardless of whether the arrest was promoted by senescence or quiescence.
FIG. 3.
Senescence-associated CENP-A reduction is mediated not only by transcriptional control but also by posttranslational proteolysis. (A) Northern blotting of the senescent and the quiescent TIG3 cells. The transcript levels of CENP-A, H2B, and GAPDH are shown. GAPDH and methylene blue staining were used as loading controls. PDLs and the time points after infection or treatment are indicated at the top. (B) Immunoblotting of the senescent and the quiescent cells. TCE (2 × 104 cells) were resolved by SDS-PAGE, followed by immunoblotting with the indicated antibodies. (C) The protein levels of CENP-A and cyclin A were determined in TCE prepared from cells treated with 50 μg of cycloheximide/ml for the indicated time points (left panel). The integrated density at 0 h was set at 100%. The half-lives of CENP-A and cyclin A were estimated by the band density as described in Materials and Methods. Growth curve of the drug-treated cells (right panel). The number of cells at 0 h was set at 100%.
In contrast to the level of CENP-A transcript, which was reduced in both senescent and quiescent cells, the level of CENP-A protein was substantially different between senescent and quiescent cells. CENP-A levels were markedly reduced in senescent cells, while quiescent cells retained similar levels of CENP-A to their actively growing counterparts (Fig. 3B). The levels of core histones in both senescent and quiescent cells were similar to those found in actively growing cells. The half-life of CENP-A protein was roughly estimated as more than 10 days in proliferating cells treated with protein synthesis inhibitor, cycloheximide, which immediately inhibited cell proliferation (Fig. 3C). In the same preparation, the half-life of cyclin A was estimated as 1.2 days, indicating that CENP-A is exceptionally stable protein (Fig. 3C, left panel). This unusual stability of CENP-A protein suggests that reduction of CENP-A in senescent cells is not solely caused by transcriptional repression. We, therefore, conclude that both transcriptional downregulation and posttranslational proteolysis are involved in the senescence-associated reduction of CENP-A protein.
Primary human fibroblasts depleted of CENP-A exhibit senescencelike phenotypes.
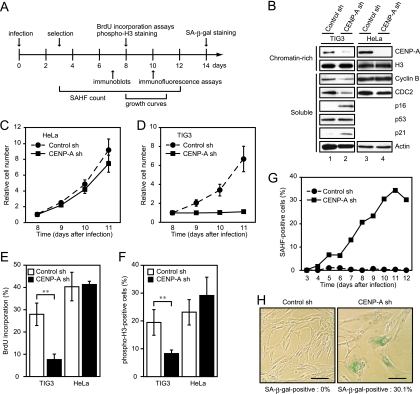
Although CENP-A was shown to be reduced in senescent cells, the question remained as to whether CENP-A reduction was a cause, or a consequence, of cellular senescence. To address this question, we examined the effects of CENP-A depletion in human primary cells with functional p53 and Rb. The experiments are shown schematically in Fig. 4A. We used TIG3 cells expressing ecotropic receptors that enable the efficient retrovirus-mediated transduction of exogenous DNAs. Ecotropic receptor-expressing HeLa cells were also generated for comparison. CENP-A shRNA effectively reduced the protein level to below 10% of the mock-depleted controls (Fig. 4B). Transduction of CENP-A shRNA did not alter the levels of core histones, highlighting the specificity of the CENP-A shRNA. Consistent with the previous report, HeLa cells with CENP-A depleted continued to proliferate, although these cells showed mild growth retardation (Fig. 4C). In marked contrast to HeLa cells, the depletion of CENP-A immediately blocked proliferation of TIG3 cells (Fig. 4D). A BrdU incorporation assay (BrdU is a marker of DNA synthesis) demonstrated that CENP-A depletion in TIG3 cells reduced the proportion of cells transiting S phase of the cell cycle (Fig. 4E). The proportion of M-phase cells was also reduced in the CENP-A-depleted TIG3 cells, as determined by immunostaining for S10-phosphorylated histone H3 (a marker of late G2 and mitotic cells) (Fig. 4F); although 8% of CENP-A-depleted TIG3 cells still harbored phosphorylated histone H3, the majority of these cells appeared to be in G2 phase, as determined from their nuclear morphology (data not shown). A similar reduction in the proportions of S- and M-phase cells was not observed in HeLa cells depleted of CENP-A. Consistent with these observations, immunoblotting analysis demonstrated that CENP-A depletion in TIG3 cells led to a reduction in the levels of cyclin B and CDC2, which are essential for cell cycle progression, and an elevation of the levels of the CDK inhibitors, p16INK4a and p21CIP1 (Fig. 4B). These changes in cell cycle regulators were not found in HeLa cells depleted of CENP-A. Taken together, these results suggested that CENP-A depletion in human primary cells led to an immediate cell cycle arrest, presumably in G1 and G2 phases of the cell cycle.
FIG. 4.
CENP-A depletion causes senescencelike proliferation arrest. (A) Experimental design. (B) Immunoblotting of CENP-A-depleted TIG3 and HeLa cells performed on day 7 after infection. Chromatin-rich fractions (2 × 104 TIG3 cells or 5 × 104 HeLa cells) and soluble proteins (30 μg) were resolved by SDS-PAGE, followed by immunoblotting with the indicated antibodies. Histone H3 and actin were used as loading controls. (C and D) Growth curves of HeLa (C) and TIG3 (D) cells with CENP-A depleted. The number of cells on day 8 after infection was set at 1. The data are means ± the SD from three (C) and four (D) independent experiments. (E and F) BrdU incorporation assay (E) combined with immunostaining with anti-phospho-specific histone H3 antibody (F) performed on day 8 after infection. The data are means ± the SD from three independent experiments (**, P < 0.01 [Student t test]). (G) SAHF formation in TIG3 cells with CENP-A depleted. Fixed cells were stained with Hoechst to visualize DNA. The proportion of SAHF-positive cells (n = 300) is shown. (H) SA-β-Gal staining performed on day 14 after infection. The proportion of SA-β-Gal-positive cells (n = 300) is shown. Scale bars, 100 μm.
The elevation of p16INK4a and p21CIP1 in TIG3 cells depleted of CENP-A is highly reminiscent of cells that have been promoted to senesce by a variety of means. To determine whether TIG3 cells depleted of CENP-A exhibited other phenotypes that have been associated with senescence, we examined SAHF formation in these cells by Hoechst staining. The frequency of SAHF-positive cells depleted of CENP-A increased gradually and reached 30% 10 days after infection (Fig. 4G); this frequency is comparable with that achieved in ras-induced senescent TIG3 cells. TIG3 cells depleted of CENP-A also exhibited the increase in SA-β-Gal activity that is the cytological marker of senescent cells (Fig. 4H). This acquisition of multiple markers of the senescent state led us to the conclusion that CENP-A deletion caused senescencelike proliferation arrest in primary human cells.
To confirm that the growth arrest phenotypes of CENP-A shRNA-treated TIG3 cells were caused by depletion of CENP-A protein, we tested whether these phenotypes were suppressed by the overexpression of CENP-A. To do this, an shRNA insensitive CENP-A cDNA, in which the target site of shRNA was mutated (Fig. 5A), was retrovirally transduced into TIG3 cells and CENP-A shRNA was subsequently expressed in these cells. The cells expressing triple FLAG-tagged shRNA-insensitive CENP-A maintained the high level of exogenous CENP-A protein even after CENP-A shRNA expression, whereas endogenous CENP-A protein was depleted in these cells (Fig. 5B). Noticeably accumulation of p16INK4a induced by CENP-A shRNA expression was substantially suppressed in these cells. Furthermore, the other senescencelike phenotypes caused by CENP-A shRNA, such as growth arrest, SAHF formation, and an increased SA-β-Gal activity, were at least partially suppressed by the expression of FLAG-CENP-A (Fig. 5C and D). We conclude that the senescencelike phenotypes observed in CENP-A shRNA-treated cells are indeed caused by CENP-A depletion. It would be noteworthy that the ectopic expression of CENP-A did not significantly delay ras-induced or replicative senescence (Fig. 5E and F, data not shown). CENP-A reduction therefore does not appear to be a single cause to induce cellular senescence.
CENP-A depletion stimulates heterochromatin formation on centromeres.
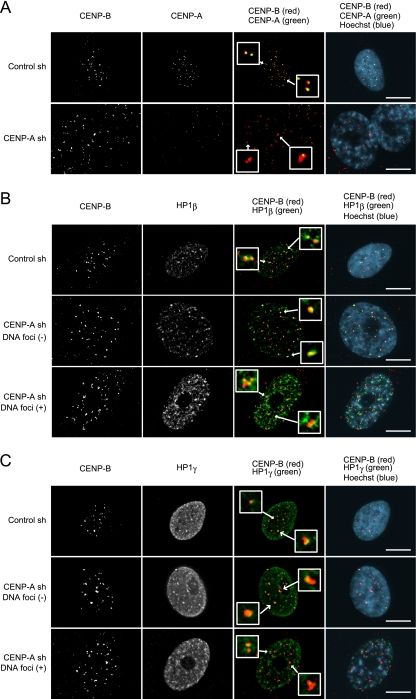
As shown in Fig. 1C and 2, ras-induced senescent cells exhibited extension of the heterochromatinization of the centromere. To examine whether the senescence induced by CENP-A depletion also caused extension of centromeric heterochromatin, the localization of CENP-B and HP1 proteins was determined by immunofluorescence assays in TIG3 cells depleted of CENP-A. CENP-B was diffusely distributed around a faint signal of CENP-A in these cells regardless of whether the cells had formed DNA foci (SAHF) or not, and HP1β and HP1γ proteins were enriched on centromeres alongside CENP-B (Fig. 6). The altered distributions of CENP-B and HP1 proteins were similar to those observed in ras-induced senescent cells (Fig. 1C and 2). Furthermore, immunoblotting analysis demonstrated that these proteins accumulated in TIG3 cells depleted of CENP-A to a similar degree compared to that observed in replicative and ras-induced senescent cells (data not shown). These results indicated that CENP-A depletion in primary cells mirrors ras-induced and replicative senescence by altering the degree to which heterochromatin forms at centromeres as well as altering the protein composition in these centromeres.
FIG. 6.
Stimulating heterochromatin formation on centromeres in CENP-A-depleted TIG3 cells. (A to C) Immunofluorescence of TIG3 cells depleted of CENP-A performed on day 10 after infection. Staining revealed CENP-B (red in overlay) and CENP-A (green in overlay) (A), HP1β (green in overlay) (B), or HP1γ (green in overlay) (C). DNA was counterstained with Hoechst (blue in overlay). In panels B and C, DNA focus-negative (middle panels) and DNA focus-positive cells depleted of CENP-A (lower panels) are shown. Insets represent enlarged images indicated by arrows. Scale bars, 10 μm.
Senescencelike phenotypes induced by CENP-A depletion require p53 but not p16INK4a-Rb.
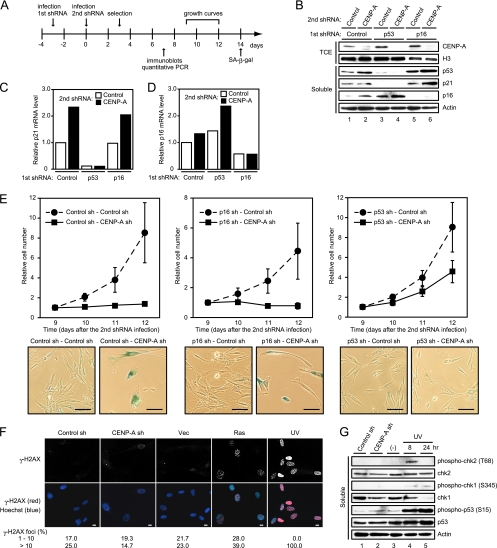
As mentioned above, CENP-A depletion induced senescencelike phenotypes in primary human cells, but not in HeLa cells lacking both functional p53 and p16INK4a-Rb pathways for proliferation control. This observation suggested that these pathways may play crucial roles in the induction of senescencelike phenotypes triggered by CENP-A depletion. To test this possibility, the effects of CENP-A depletion were analyzed in TIG3 cells from which either p53 or p16INK4a had been depleted (Fig. 7A). The efficient depletion of p53 or p16INK4a was confirmed by immunoblotting (Fig. 7B) and quantitative RT-PCR (Fig. 7C and D). Depletion of CENP-A in mock-depleted and p16INK4a-depleted TIG3 cells induced p21CIP1 expression (Fig. 7C) and strongly inhibited cell proliferation (Fig. 7E, left upper and middle upper panels), indicating that p16INK4a was dispensable for senescencelike proliferation arrest caused by CENP-A depletion. On the other hand, p53 appeared to be essential for this proliferation arrest; CENP-A depletion in p53-depleted TIG3 cells did not stop proliferating (Fig. 7E, right upper panel), even though the expression of p16INK4a was elevated (Fig. 7B and D). Consistent with the absence of growth arrest, cells depleted of both p53 and CENP-A harbored cyclin B and hyperphosphorylated Rb at levels comparable to those seen in cells depleted of p53 (data not shown). As expected, depletion of p53 suppressed SAHF formation (data not shown) and induction of SA-β-Gal activity in TIG3 cells depleted of CENP-A (Fig. 7E, lower panels). These results indicated that the senescencelike phenotypes induced by CENP-A depletion require the p53 pathway but not the p16INK4a-Rb pathway.
FIG. 7.
Senescencelike phenotypes induced by CENP-A depletion require p53. (A) Experimental design. (B) Immunoblotting of TIG3 cells expressing control, p53, or p16INK4a shRNA (first shRNA) in combination with control or CENP-A shRNA (second shRNA). TCE and soluble proteins were resolved by SDS-PAGE, followed by immunoblotting with the indicated antibodies. Histone H3 and actin were used as loading controls. (C and D) Quantitative RT-PCR of the cells indicated in panel B. The relative abundances of p21CIP1 mRNA (C) and p16INK4a mRNA (D) are shown after normalization using GAPDH mRNA. (E) Growth curves of TIG3 cells expressing control (left upper), p16INK4a (middle upper), or p53 shRNA (right upper) in combination with control or CENP-A shRNA. The number of cells on day 9 after the second shRNA infection was set at 1. The data are means ± the SD from three independent experiments. SA-β-Gal staining was performed on day 14 after the second shRNA infection (lower panels). Scale bars, 100 μm. (F) Immunofluorescence of CENP-A-depleted and ras-induced senescent TIG3 cells performed on day 10 after infection. Staining revealed γ-H2AX (red in overlay). DNA was counterstained with Hoechst (blue in overlay). As a positive control, cells were treated with UV. The numbers of cells (n = 300) containing 1 to 10 and >10 foci are shown. Scale bars, 10 μm. (G) Immunoblotting of CENP-A-depleted and UV-treated cells. Soluble proteins were resolved by SDS-PAGE, followed by immunoblotting with the indicated antibodies. Actin was used as a loading control.
Previous studies clearly demonstrated that p53 activation in oncogene-induced senescence is due to activation of the DNA damage response (DDR) (6, 16, 35). To test whether DDR plays a crucial role in p53 activation in senescencelike cells depleted of CENP-A, we examined the presence of DNA damage foci by immunofluorescence assay using antibody against phosphorylated histone H2A.X (γ-H2AX). As shown in Fig. 7F, γ-H2AX foci were clearly increased by the expression of oncogenic Ras or UV treatment, whereas these foci were not induced by CENP-A depletion. Moreover, phosphorylated chk2 on threonine 68 and phosphorylated chk1 on serine 345, both of which are associated with DDR, were not detectable by immunoblotting in cells depleted of CENP-A, and phosphorylated p53 on serine 15 was, if any, only slightly increased in these cells (Fig. 7G). Based on the results, CENP-A depletion does not appear causally linked to DDR. Taking these observations together, we conclude that CENP-A depletion triggers unconventional type of senescence that does not require the activation of DNA damage signaling.
TIG3 cells depleted of both p53 and CENP-A exhibit mitotic defects.
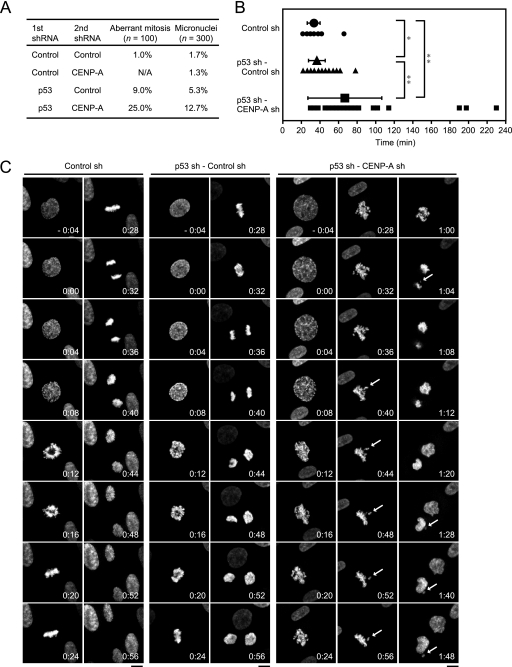
Since inactivation of p53 suppressed the CENP-A depletion-associated senescence, we next analyzed the consequences of this suppression. We focused upon mitotic phenotypes because CENP-A depletion in HeLa cells leads to defects in chromosome segregation. We examined 100 postmetaphase mitotic cells in each sample by Hoechst staining to measure the frequency of aberrant mitotic profiles, such as lagging chromosomes and chromosome bridges. The proportions of cells showing mitotic abnormalities were 1, 9, and 25% in the cells coexpressing shRNAs against control and control, p53 and control, and p53 and CENP-A, respectively (Fig. 8A). Mitotic cells were seldom found in the cells coexpressing shRNAs against control and CENP-A. Micronuclei in interphase or senescent cells (n = 300) were observed in 1.7, 1.3, 5.3, and 12.7% of the cells coexpressing shRNAs directed against control and control, control and CENP-A, p53 and control, and p53 and CENP-A, respectively. These results indicated that inactivation of p53 in cells depleted of CENP-A increased the frequency of errors in chromosomes segregation during mitosis.
FIG. 8.
Cosuppression of p53 and CENP-A causes aberrant mitosis. (A) The cells coexpressing shRNAs against control and control, control and CENP-A, p53 and control, or p53 and CENP-A were fixed and stained with Hoechst on day 15 after the second shRNA infection. The proportions of aberrant mitosis, including lagging chromosomes and chromosome bridges in mitotic cells (n = 100) and micronuclei in the interphase and/or the senescent cells (n = 300), are shown. Aberrant mitosis in the cells coexpressing shRNAs against control and CENP-A could not be assessed because we found few of these cells undergoing mitosis. (B) The duration of mitosis in H2B-EGFP-expressing TIG3 cells transduced with shRNA(s) against control, p53 and control, or p53 and CENP-A. Live cell time-lapse analysis was performed from days 7 to 10 after infection. The duration of mitosis, defined as the interval between apparent chromosome condensation and onset of decondensation of chromosomes, was measured from the cells transduced with shRNA(s) against control (n = 52, •), p53 and control (n = 112, ▴), or p53 and CENP-A (n = 54, ▪). The data are means ± the SD (**, P < 0.01; *, P < 0.05 [Student t test]). The small black dots are data points. (C) Live cell time-lapse fluorescence of the cells in panel B at the indicated time points (h:min). The time point of apparent chromosome condensation was set at 0:00. White arrows indicate misaligned and lagging chromosomes, and micronuclei. Scale bars, 10 μm.
To investigate the details of mitotic abnormalities in TIG3 cells with cosuppression of both p53 and CENP-A, we monitored mitotic progression using enhanced green fluorescent protein (EGFP)-tagged histone H2B in living TIG3 cells. We measured the duration of mitosis defined as the interval between apparent chromosome condensation and onset of decondensation of chromosomes. The control cells averaged 33 ± 6 min (n = 52) in mitosis (Fig. 8B). The cells depleted of p53 averaged 36 ± 9 min (n = 112), which was slightly longer than that of the control cells. In contrast, cells from which both p53 and CENP-A had been depleted averaged 66 ± 39 min (n = 54), indicating a significant extension of the duration of mitosis. This extension of mitosis was mainly due to prolongation of metaphase. In these cells, chromosomes tended to remain randomly distributed much longer than in the control cells and took ∼60 min to congress at the spindle equator (Fig. 8C). In contrast, the chromosomes finished aligning at the spindle equator of p53-depleted cells within ∼30 min after the chromosome condensation. In addition, some of the cells with cosuppression of p53 and CENP-A entered anaphase without proper chromosome alignment and as a consequence produced lagging chromosomes and chromosome bridges. Micronuclei were often formed in the subsequent interphase of such cells. These results indicated that suppression of CENP-A depletion-associated senescence by inactivation of p53 resulted in mitotic defects, which consequently led to the production of aneuploid cells. Therefore, p53 may rescue primary cells with CENP-A depleted from mitotic defects by inducing cellular senescence.
DISCUSSION
In this study, we found an unexpected physiological link between cellular senescence and centromere dysfunction in primary human cells. Our observations indicated that the level of CENP-A protein was markedly reduced in senescent human cells, and that forced reduction of CENP-A caused premature senescence in primary human fibroblasts. The premature senescence that was induced by the forced reduction of CENP-A levels was p53 dependent. In the absence of p53, CENP-A reduction failed to block cell division and led to marked elevation in the levels of chromosome missegregation and subsequently the propagation of cells with chromosomal abnormalities. These findings suggest that, in primary human cells, certain types of stress that damage the centromere structure and/or function induce p53-dependent senescence, which suppresses the production of potentially hazardous cells.
CENP-A is downregulated in senescent human cells.
We showed that CENP-A mRNA expression was reduced in both replicative and ras-induced senescent human cells. A previous study indicated a reduction in the levels of CENP-A transcripts in senescent IMR90 human fibroblasts (43). Similarly, the levels of CENP-A mRNA were reduced in dermal fibroblasts derived from elderly human subjects and from those with Hutchinson-Gilford progeria syndrome, a rare genetic disorder characterized by accelerated aging (33). Therefore, the reduction of CENP-A mRNA levels appears to be a common feature of cellular senescence and individual aging. However, this reduction is not specific to senescence; we noted that a marked reduction of CENP-A mRNA was also observed in the quiescent cells that had exited from the cell cycle and contained only a small G2 population. Since CENP-A transcription is regulated by the cell cycle (56), it is unlikely that the reduction of CENP-A transcript level is specific to cellular senescence, even though a reduction in the level of CENP-A transcript shows a strong association with the reduced proliferation potential of senescent cells.
In contrast, CENP-A protein levels were only reduced in the senescent cells and not in the quiescent cells. This result suggests that the reduction of CENP-A protein in the senescent cells is mediated in part by a posttranslational mechanism. Although the molecular mechanism of CENP-A reduction remains to be clarified, our preliminary results implicated that CENP-A protein is ubiquitinated in senescent cells (data not shown). A previous study demonstrated that cullin-4A, human ring finger protein 2 (ring 1B) and hypothetical protein FLJ23109, which have been reported or assumed to possess ubiquitin ligase activity, are coimmunoprecipitated with anti-CENP-A antibody from HeLa interphase nuclear extract (45). Taking these together, we speculate that CENP-A is degraded via the ubiquitin-proteasome-dependent pathway in these cells. It is noteworthy that CENP-A also undergoes destruction when human cells are infected with herpes simplex virus type 1 (32). Therefore, CENP-A destruction may be a part of the cellular response against certain types of genotoxic stress.
Interestingly, linker histone H1 protein level is decreased in senescent human WI38 cells, presumably because of posttranslational regulation (21). A reduction in the mitotic exit network kinase, WARTS/LATS1, was also reported in senescent human cells, and was attenuated by the addition of MG132 (58); in senescent cells, reduction of this kinase may inhibit proliferation by blocking cytokinesis. These results imply the presence of a senescence-associated proteolysis pathway in primary human cells. Were such a pathway to exist, it would help to maintain metabolism and biosynthesis in senescent cells by recycling proteins that are no longer required for nondividing cells. It is also possible that such the proteolysis ensures irreversible growth arrest by destruction of proteins essential for proliferation.
CENP-A reduction enhances centromeric heterochromatin formation.
We demonstrated here that CENP-B and HP1 proteins accumulated considerably on the centromeres of senescent human cells. Since enhanced accumulation of these proteins was observed in not only senescent TIG3 cells produced by retroviral delivery of oncogenic ras or extensive culturing but also in cells transduced with CENP-A shRNA, this accumulation is triggered, at least in part, by the reduction of CENP-A protein level. Consistent with these observation, it was reported that unusual diffuse CENP-B signals were detected by immunofluorescence assays in Cenpa-null mutant mouse embryos (27). Recent studies have demonstrated the remarkable role of CENP-B in heterochromatin formation in the centromere: CENP-B enhances histone H3K9 trimethylation and DNA methylation in chromosomally integrated alphoid DNA, thereby stimulating heterochromatin formation (48); in fission yeast, the disruption of CENP-B homologs, Abp1 and Cbh1, causes a reduction of Swi6, a homolog of HP1, at the centromeric chromatin and a decrease in heterochromatin-specific modifications of histone H3 (41). Furthermore, stimulating the formation of a heterochromatin state by forced binding of silencers or targeted nucleation of HP1 to the centromere results in the inactivation of a functional kinetochore (42). Based on these observations, we speculate that, during senescence, primary human cells alter their centromere states from a functional centromere, which is required for faithful segregation of chromosomes, to an inactivated centromere, which is likely to contribute to establishment of the senescent state. The loss of CENP-A and the extended heterochromatinization mediated by CENP-B and HP1 proteins on the centromere in senescent cells are assumed to promote centromere inactivation. Further qualitative and quantitative studies are needed to understand the structural and the functional changes of centromere in the senescence process.
Is the reduction of CENP-A a cause or a consequence of cellular senescence?
We found that CENP-A depletion by shRNA induced senescencelike phenotypes in the primary cells and that CENP-A appeared to be actively degraded in the senescent cells. These observations raise a question as to whether CENP-A reduction is a cause or an effect of cellular senescence. One possible explanation is that CENP-A is degraded in senescent cells, simply because CENP-A is dispensable in these cells. Although we do not have a clear answer to this question at this point, we favor the hypothesis that there may be a positive-feedback circuit between CENP-A degradation and induction of cellular senescence. We postulate that normal human cells possess a mechanism for monitoring centromere/kinetochore integrity, which activates the p53-dependent senescence pathway in response to centromere/kinetochore defects, such as insufficient incorporation of CENP-A at the centromere. Once the cells enter the senescent state, accelerated CENP-A proteolysis may destroy the kinetochore structure completely and so ensure a permanent block to cell proliferation.
Cellular senescence as a self-defense mechanism against centromere dysfunction.
CENP-A depletion-associated senescence, which is dependent on the integrity of the p53 tumor suppressor protein, appears to act as a fail-safe mechanism that prevents the cells from executing defective mitoses. Interestingly, aberrant accumulation of CENP-A and CENP-H, which was presumed to lead to massive aneuploid formation, was reported in human colorectal cancer cells (2, 60, 61). Failure in CENP-A reduction mechanism may thus cause missegregation and tumorigenesis. It should be noted that widely conserved SAC, which induces metaphase arrest in response to defective spindle-kinetochore interaction, also prevents the cells from the fatal consequences of mitotic progression when kinetochore structure/function is defective (40, 52). Although the relationship between CENP-A depletion and SAC in primary human cells remains to be established, it is possible that SAC generates a signal that activates the p53 pathway. Alternatively, SAC and the pathways involved in cellular senescence may redundantly protect the cells from defective mitosis. It has been reported that SAC signaling cannot properly respond to defects in a subset of centromere proteins, such as CENP-A and Mis6/CENP-I (31, 51, 54). Furthermore, the studies of BubR1-insufficient and Bub3/Rae1-haploinsufficient mice and Bub1-depleted primary human cells indicated that the defects in these checkpoint proteins cause senescence and aging (3-5, 22). We have analyzed the effects of reduction of several centromere/kinetochore proteins and found that depletion of hMis12, as well as BubR1, also caused senescencelike proliferation arrest in primary human cells (K. Maehara et al., unpublished data). Based on these observations and the results of the present study, primary cells appear to induce cellular senescence in response to fatal centromere/kinetochore dysfunction in circumstances under which some of key centromere proteins and/or the SAC are not functioning properly. Under these conditions, senescence seems to not only prevent the cells from producing abnormal chromosomes but also to protect the organism from the potentially hazardous consequences of proliferation of cells harboring chromosomal abnormalities that arose as a consequence of defective mitosis.
Acknowledgments
We thank the members of our laboratory, K. Ishii and Y. Ogiyama (Osaka University) for their support and discussion and H. Kondoh (Kyoto University), H. Kimura (Osaka University), and I. Hagan (Paterson Institute for Cancer Research, University of Manchester) for critical reading of the manuscript. We thank N. Jones (Paterson Institute for Cancer Research, University of Manchester), G. P. Nolan (Stanford University), R. Agami (the Netherlands Cancer Institute), and M. Ikeno (School of Medicine, Keio University) for materials and K. Yoda (Nagoya University) for technical advice regarding the measurement of signal intensity.
This study was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas (to K.T.); a Grant-in-Aid for Young Scientists (B) (to S.S.) from the Ministry of Education, Culture, Sports, Science, and Technology; a Grant-in-Aid for Scientific Research (C) (to K.M.) from the Japan Society for the Promotion of Science; and the Ichiro Kanehara Foundation (to K.M.).
Footnotes
Published ahead of print on 16 February 2010.
REFERENCES
- 1.Alcorta, D. A., Y. Xiong, D. Phelps, G. Hannon, D. Beach, and J. C. Barrett. 1996. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 93:13742-13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amato, A., T. Schillaci, L. Lentini, and A. Di Leonardo. 2009. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol. Cancer 8:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, D. J., K. B. Jeganathan, J. D. Cameron, M. Thompson, S. Juneja, A. Kopecka, R. Kumar, R. B. Jenkins, P. C. de Groen, P. Roche, and J. M. van Deursen. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36:744-749. [DOI] [PubMed] [Google Scholar]
- 4.Baker, D. J., K. B. Jeganathan, L. Malureanu, C. Perez-Terzic, A. Terzic, and J. M. van Deursen. 2006. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J. Cell Biol. 172:529-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, D. J., C. Perez-Terzic, F. Jin, K. Pitel, N. J. Niederländer, K. Jeganathan, S. Yamada, S. Reyes, L. Rowe, H. J. Hiddinga, N. L. Eberhardt, A. Terzic, and J. M. van Deursen. 2008. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat. Cell Biol. 10:825-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartkova, J., N. Rezaei, M. Liontos, P. Karakaidos, D. Kletsas, N. Issaeva, L. V. Vassiliou, E. Kolettas, K. Niforou, V. C. Zoumpourlis, M. Takaoka, H. Nakagawa, F. Tort, K. Fugger, F. Johansson, M. Sehested, C. L. Andersen, L. Dyrskjot, T. Ørntoft, J. Lukas, C. Kittas, T. Helleday, T. D. Halazonetis, J. Bartek, and V. G. Gorgoulis. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444:633-637. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Porath, I., and R. A. Weinberg. 2005. The signals and pathways activating cellular senescence. Int. J. Biochem. Cell Biol. 37:961-976. [DOI] [PubMed] [Google Scholar]
- 8.Blower, M. D., and G. H. Karpen. 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3:730-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243-247. [DOI] [PubMed] [Google Scholar]
- 10.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 11.Buchwitz, B. J., K. Ahmad, L. L. Moore, M. B. Roth, and S. Henikoff. 1999. A histone-H3-like protein in C. elegans. Nature 401:547-548. [DOI] [PubMed] [Google Scholar]
- 12.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407-421. [DOI] [PubMed] [Google Scholar]
- 14.Collado, M., M. A. Blasco, and M. Serrano. 2007. Cellular senescence in cancer and aging. Cell 130:223-233. [DOI] [PubMed] [Google Scholar]
- 15.Deng, Y., S. S. Chan, and S. Chang. 2008. Telomere dysfunction and tumour suppression: the senescence connection. Nat. Rev. Cancer 8:450-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Micco, R., M. Fumagalli, A. Cicalese, S. Piccinin, P. Gasparini, C. Luise, C. Schurra, M. Garré, P. G. Nuciforo, A. Bensimon, R. Maestro, P. G. Pelicci, and F. d'Adda di Fagagna. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444:638-642. [DOI] [PubMed] [Google Scholar]
- 17.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, M. Peacocke, and J. Campisi. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U. S. A. 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Earnshaw, W. C., and N. Rothfield. 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91:313-321. [DOI] [PubMed] [Google Scholar]
- 19.Earnshaw, W. C., K. F. Sullivan, P. S. Machlin, C. A. Cooke, D. A. Kaiser, T. D. Pollard, N. F. Rothfield, and D. W. Cleveland. 1987. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 104:817-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foltz, D. R., L. E. T. Jansen, B. E. Black, A. O. Bailey, J. R. Yates III, and D. W. Cleveland. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458-469. [DOI] [PubMed] [Google Scholar]
- 21.Funayama, R., M. Saito, H. Tanobe, and F. Ishikawa. 2006. Loss of linker histone H1 in cellular senescence. J. Cell Biol. 175:869-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gjoerup, O. V., J. Wu, D. Chandler-Militello, G. L. Williams, J. Zhao, B. Schaffhausen, P. S. Jat, and T. M. Roberts. 2007. Surveillance mechanism linking Bub1 loss to the p53 pathway. Proc. Natl. Acad. Sci. U. S. A. 104:8334-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goshima, G., T. Kiyomitsu, K. Yoda, and M. Yanagida. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160:25-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara, E., R. Smith, D. Parry, H. Tahara, S. Stone, and G. Peters. 1996. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol. Cell. Biol. 16:859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayflick, L., and P. S. Moorhead. 1961. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25:585-621. [DOI] [PubMed] [Google Scholar]
- 26.Hollstein, M., D. Sidransky, B. Vogelstein, and C. C. Harris. 1991. p53 mutations in human cancers. Science 253:49-53. [DOI] [PubMed] [Google Scholar]
- 27.Howman, E. V., K. J. Fowler, A. J. Newson, S. Redward, A. C. MacDonald, P. Kalitsis, and K. H. A. Choo. 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. U. S. A. 97:1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hudson, D. F., K. J. Fowler, E. Earle, R. Saffery, P. Kalitsis, H. Trowell, J. Hill, N. G. Wreford, D. M. de Kretser, M. R. Cancilla, E. Howman, L. Hii, S. M. Cutts, D. V. Irvine, and K. H. A. Choo. 1998. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 141:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izuta, H., M. Ikeno, N. Suzuki, T. Tomonaga, N. Nozaki, C. Obuse, Y. Kisu, N. Goshima, F. Nomura, N. Nomura, and K. Yoda. 2006. Comprehensive analysis of the ICEN (interphase centromere complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 11:673-684. [DOI] [PubMed] [Google Scholar]
- 30.Kapoor, M., R. Montes de Oca Luna, G. Liu, G. Lozano, C. Cummings, M. Mancini, I. Ouspenski, B. R. Brinkley, and G. S. May. 1998. The cenpB gene is not essential in mice. Chromosoma 107:570-576. [DOI] [PubMed] [Google Scholar]
- 31.Liu, S. T., J. C. Hittle, S. A. Jablonski, M. S. Campbell, K. Yoda, and T. J. Yen. 2003. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 5:341-345. [DOI] [PubMed] [Google Scholar]
- 32.Lomonte, P., K. F. Sullivan, and R. D. Everett. 2001. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 276:5829-5835. [DOI] [PubMed] [Google Scholar]
- 33.Ly, D. H., D. J. Lockhart, R. A. Lerner, and P. G. Schultz. 2000. Mitotic misregulation and human aging. Science 287:2486-2492. [DOI] [PubMed] [Google Scholar]
- 34.Maehara, K., K. Yamakoshi, N. Ohtani, Y. Kubo, A. Takahashi, S. Arase, N. Jones, and E. Hara. 2005. Reduction of total E2F/DP activity induces senescence-like cell cycle arrest in cancer cells lacking functional pRB and p53. J. Cell Biol. 168:553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mallette, F. A., M. F. Gaumont-Leclerc, and G. Ferbeyre. 2007. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 21:43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masumoto, H., H. Masukata, Y. Muro, N. Nozaki, and T. Okazaki. 1989. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsushime, H., D. E. Quelle, S. A. Shurtleff, M. Shibuya, C. J. Sherr, and J. Y. Kato. 1994. D-type cyclin-dependent kinase activity in mammalian cells. Mol. Cell. Biol. 14:2066-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland, and M. M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94:607-613. [DOI] [PubMed] [Google Scholar]
- 39.Méndez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musacchio, A., and E. D. Salmon. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell. Biol. 8:379-393. [DOI] [PubMed] [Google Scholar]
- 41.Nakagawa, H., J. K. Lee, J. Hurwitz, R. C. Allshire, J. Nakayama, S. I. Grewal, K. Tanaka, and Y. Murakami. 2002. Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 16:1766-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakano, M., S. Cardinale, V. N. Noskov, R. Gassmann, P. Vagnarelli, S. Kandels-Lewis, V. Larionov, W. C. Earnshaw, and H. Masumoto. 2008. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell 14:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narita, M., M. Narita, V. Krizhanovsky, S. Nuñez, A. Chicas, S. A. Hearn, M. P. Myers, and S. W. Lowe. 2006. A novel role for high-mobility group A proteins in cellular senescence and heterochromatin formation. Cell 126:503-514. [DOI] [PubMed] [Google Scholar]
- 44.Narita, M., S. Nuñez, E. Heard, M. Narita, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113:703-716. [DOI] [PubMed] [Google Scholar]
- 45.Obuse, C., H. Yang, N. Nozaki, S. Goto, T. Okazaki, and K. Yoda. 2004. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells 9:105-120. [DOI] [PubMed] [Google Scholar]
- 46.Ohtani, N., P. Brennan, S. Gaubatz, E. Sanij, P. Hertzog, E. Wolvetang, J. Ghysdael, M. Rowe, and E. Hara. 2003. Epstein-Barr virus LMP1 blocks p16INK4a-RB pathway by promoting nuclear export of E2F4/5. J. Cell Biol. 162:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okada, M., I. M. Cheeseman, T. Hori, K. Okawa, I. X. McLeod, J. R. Yates III, A. Desai, and T. Fukagawa. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446-457. [DOI] [PubMed] [Google Scholar]
- 48.Okada, T., J. Ohzeki, M. Nakano, K. Yoda, W. R. Brinkley, V. Larionov, and H. Masumoto. 2007. CENP-B controls centromere formation depending on the chromatin context. Cell 131:1287-1300. [DOI] [PubMed] [Google Scholar]
- 49.Palmer, D. K., K. O'Day, M. H. Wener, B. S. Andrews, and R. L. Margolis. 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez-Castro, A. V., F. L. Shamanski, J. J. Meneses, T. L. Lovato, K. G. Vogel, R. K. Moyzis, and R. Pedersen. 1998. Centromeric protein B null mice are viable with no apparent abnormalities. Dev. Biol. 201:135-143. [DOI] [PubMed] [Google Scholar]
- 51.Régnier, V., P. Vagnarelli, T. Fukagawa, T. Zerjal, E. Burns, D. Trouche, W. Earnshaw, and W. Brown. 2005. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 25:3967-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rieder, C. L., and H. Maiato. 2004. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell 7:637-651. [DOI] [PubMed] [Google Scholar]
- 53.Ruas, M., and G. Peters. 1998. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim. Biophys. Acta 1378:F115-177. [DOI] [PubMed] [Google Scholar]
- 54.Saitoh, S., K. Ishii, Y. Kobayashi, and K. Takahashi. 2005. Spindle checkpoint signaling requires the Mis6 kinetochore subcomplex, which interacts with Mad2 and mitotic spindles. Mol. Biol. Cell 16:3666-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 56.Shelby, R. D., O. Vafa, and K. F. Sullivan. 1997. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136:501-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stoler, S., K. C. Keith, K. E. Curnick, and M. Fitzgerald-Hayes. 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9:573-586. [DOI] [PubMed] [Google Scholar]
- 58.Takahashi, A., N. Ohtani, K. Yamakoshi, S. Iida, H. Tahara, K. Nakayama, K. I. Nakayama, T. Ide, H. Saya, and E. Hara. 2006. Mitogenic signalling and the p16INK4a-Rb pathway cooperate to enforce irreversible cellular senescence. Nat. Cell Biol. 8:1291-1297. [DOI] [PubMed] [Google Scholar]
- 59.Takahashi, K., E. S. Chen, and M. Yanagida. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288:2215-2219. [DOI] [PubMed] [Google Scholar]
- 60.Tomonaga, T., K. Matsushita, M. Ishibashi, M. Nezu, H. Shimada, T. Ochiai, K. Yoda, and F. Nomura. 2005. Centromere protein H is up-regulated in primary human colorectal cancer and its overexpression induces aneuploidy. Cancer Res. 65:4683-4689. [DOI] [PubMed] [Google Scholar]
- 61.Tomonaga, T., K. Matsushita, S. Yamaguchi, T. Oohashi, H. Shimada, T. Ochiai, K. Yoda, and F. Nomura. 2003. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 63:3511-3516. [PubMed] [Google Scholar]
- 62.Voorhoeve, P. M., and R. Agami. 2003. The tumor-suppressive functions of the human INK4A locus. Cancer Cell 4:311-319. [DOI] [PubMed] [Google Scholar]
- 63.Zeitlin, S. G., C. M. Barber, C. D. Allis, and K. F. Sullivan. 2001. Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J. Cell Sci. 114:653-661. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, R., M. V. Poustovoitov, X. Ye, H. A. Santos, W. Chen, S. M. Daganzo, J. P. Erzberger, I. G. Serebriiskii, A. A. Canutescu, R. L. Dunbrack, J. R. Pehrson, J. M. Berger, P. D. Kaufman, and P. D. Adams. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8:19-30. [DOI] [PubMed] [Google Scholar]