Abstract
The nuclear receptor peroxisome proliferator activator receptor γ (PPARγ) is the target of antidiabetic thiazolidinedione drugs, which improve insulin resistance but have side effects that limit widespread use. PPARγ is required for adipocyte differentiation, but it is also expressed in other cell types, notably macrophages, where it influences atherosclerosis, insulin resistance, and inflammation. A central question is whether PPARγ binding in macrophages occurs at genomic locations the same as or different from those in adipocytes. Here, utilizing chromatin immunoprecipitation and high-throughput sequencing (ChIP-seq), we demonstrate that PPARγ cistromes in mouse adipocytes and macrophages are predominantly cell type specific. In thioglycolate-elicited macrophages, PPARγ colocalizes with the hematopoietic transcription factor PU.1 in areas of open chromatin and histone acetylation, near a distinct set of immune genes in addition to a number of metabolic genes shared with adipocytes. In adipocytes, the macrophage-unique binding regions are marked with repressive histone modifications, typically associated with local chromatin compaction and gene silencing. PPARγ, when introduced into preadipocytes, bound only to regions depleted of repressive histone modifications, where it increased DNA accessibility, enhanced histone acetylation, and induced gene expression. Thus, the cell specificity of PPARγ function is regulated by cell-specific transcription factors, chromatin accessibility, and histone marks. Our data support the existence of an epigenomic hierarchy in which PPARγ binding to cell-specific sites not marked by repressive marks opens chromatin and leads to local activation marks, including histone acetylation.
Peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear receptor that regulates essential aspects of adipocyte biology, including insulin sensitivity, lipogenesis, and survival, and is the target of anti-diabetic thiazolidinedione drugs (39, 69). Recent genome-wide studies of adipocytes (40, 53) have demonstrated that PPARγ localizes preferentially to lipid and carbohydrate metabolism genes, many of which are downregulated by PPARγ knockdown (62). It has also become apparent that in vivo PPARγ binding occurs predominantly as a heterodimer with retinoid X receptor (RXR) at direct repeats of the sequence AGGTCA separated by a single base pair, i.e., DR1 elements, as predicted by in vitro studies and a small number of previously known target genes (61). Furthermore, CCAAT/enhancer-binding proteins (C/EBPs) were found to colocalize with PPARγ at the majority of its binding sites and to have cooperative effects on target gene transcription (40).
The two isoforms of PPARγ, γ1 and γ2, are transcribed from alternative start sites and are most abundant in adipocytes, which require PPARγ for differentiation (39, 69), although other cell types express lower levels of the γ1 isoform (10, 72). Among these, macrophages have garnered much attention for their ability to affect metabolism in a number of tissues (54). Macrophages residing in lean fat maintain an anti-inflammatory environment and insulin sensitivity, whereas in obesity, proinflammatory macrophages infiltrate adipose tissue and exacerbate local inflammation and insulin resistance (43-45, 54). Interestingly, PPARγ is required for the beneficial effects of resident macrophages that are polarized toward an alternative phenotype (55), although it is not required for macrophage differentiation or phagocytic activity (30, 50). PPARγ deficiency in macrophages is associated with greater adipose tissue inflammation and increased susceptibility to diet-induced obesity, glucose intolerance, and insulin resistance (29, 55). Macrophage PPARγ also plays protective roles in atherosclerotic plaques, where it regulates oxidized low-density lipoprotein (oxLDL) uptake and reverse cholesterol transport (9, 59, 68). In addition, ligand-mediated activation of PPARγ has anti-inflammatory effects through a mechanism involving transrepression of NF-κB rather than direct PPARγ binding to canonical DR1 elements (56, 58). Taken together, such findings indicate that PPARγ in macrophages is functional, although the locations and mechanism of PPARγ recruitment in this cell type, including the presence of any colocalizing transcription factors, have not been addressed systematically.
Since PPARγ regulates various aspects of metabolism in macrophages and adipocytes, an important question is whether it occupies the same genomic locations in the two cell types. Alternatively, PPARγ may be recruited to cell-type-specific sites, allowing the regulation of specialized transcriptional pathways. Several recent studies have examined the occupancy of other DNA-binding factors on a genome-wide scale and found various degrees of overlap among cell types. General factors, such as RNA polymerase II and the insulator protein CTCF, vary little in their cistromes across cell types such that only a minority of their binding locations are cell type unique (2, 12). In the most extreme case, the coactivator p300 was found at only 1 to 3% of common enhancers across three embryonic mouse tissues (73). On the other hand, transcription factors such as FOXA1 and estrogen receptor have binding profiles with moderate overlap across cell types, i.e., 40% and 15%, respectively (17, 36, 46).
Here we determined the endogenous PPARγ cistrome in primary mouse macrophages. Macrophage PPARγ binds uniquely at genomic sites near immune genes, where it colocalizes with the hematopoietic transcription factor PU.1 in areas of open chromatin and histone acetylation. When expressed in preadipocytes, repressive histone marks exclude PPARγ from macrophage-unique sites, and PPARγ opens chromatin and increases histone acetylation at adipocyte sites. Thus, cell-specific PPARγ binding is characterized by the presence of cooperating transcription factors at sites of open chromatin and positive histone marks in the absence of repressive histone marks.
MATERIALS AND METHODS
Cell culture.
Macrophages were obtained from male C57BL/6 mice (Jackson Laboratory) via peritoneal lavage 3 days post-intraperitoneal injection with 1 ml of sterile 3% thioglycolate. Cells were harvested after 24 h of adherence purification in culture. 3T3-L1 preadipocytes were obtained from the American Type Culture Collection and differentiated from adipocytes as previously described (40). All cells were cultured in high-glucose Dulbecco modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum (U.S. Biotechnologies), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen).
Animals.
Six-week-old wild-type male C57BL/6 mice were obtained from Jackson Laboratory and maintained on a standard diet. All animal experiments were performed at the University of Pennsylvania according to protocols approved by the Institutional Animal Care and Use Committee.
Chromatin immunoprecipitation (ChIP).
ChIP was performed as described previously (40) using cross-linked 3T3-L1 nuclear lysates or macrophage whole-cell lysates. The following antibodies were used: anti-PPARγ (sc-7196 or 81b8; Santa Cruz or Cell Signaling Technologies, respectively), anti-RXR (sc-774; Santa Cruz), anti-H3K9Ace (ab4441; Abcam), anti-H3K9Me2 (ab1220; Abcam), anti-H3K27Me3 (07-449; Millipore), anti-PU.1 (sc-352x; Santa Cruz), anti-C/EBPα (sc-61; Santa Cruz), and anti-C/EBPβ (sc-150x; Santa Cruz). For the ChIP-chip experiments, the anti-H3K9Ace (06-942; Upstate) and anti-histone 3 (H3) (ab1791) antibodies were used and the ChIP was performed as previously described (65). For ChIP-quantitative PCR (ChIP-QPCR), enrichment was measured using Power SYBR green PCR master mix and the PRISM 7900 instrument (Applied Biosystems). Analysis was performed by the standard curve method and normalization to a nontarget control region. Primer sequences used for QPCR analysis can be found in Table S1 in the supplemental material. Primers for H3K3Ace ChIP-QPCR were located ∼500 bp away from the center of the PPARγ peak, which is generally depleted of nucleosomes; instead, the primers were localized to the nearest peak of acetylation.
Formaldehyde-assisted isolation of regulatory elements (FAIRE).
FAIRE was performed essentially as previously described (17, 23). Briefly, cells were cross-linked and processed as for ChIP. Sonication was performed for 5 min in addition to the time needed for ChIP. Samples were centrifuged, and supernatants were used for three consecutive phenol-chloroform-isoamyl alcohol extractions of the aqueous phase. After the final extraction, the FAIRE samples and inputs (obtained prior to the extractions) were incubated overnight at 65°C to reverse cross-linking, and DNA was isolated using phenol-chloroform-isoamyl alcohol. QPCR after FAIRE was performed using the same primers and protocol as for ChIP-QPCR.
ChIP-seq.
ChIP samples were amplified using the ChIP-Seq sample prep kit (Illumina) according to the manufacturer's instructions. For macrophages, cells from at least four animals were pooled per sequencing run to minimize the effects of biological variability. Two sequencing runs were performed for the macrophage PPARγ ChIP, and the data were concatenated; one sequencing run was performed for all other transcription factors and histone modifications. Sequence reads of 36 bp were obtained using the Solexa analysis pipeline and mapped to the mouse genome (mm8) using ELAND, allowing up to two mismatches. Only reads with a unique best match were included in further analysis. ChIP-seq data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/).
ChIP-seq peak calling.
All transcription factor binding data were analyzed with GLITR (71) using default parameters and an iteration number of 100. GLITR first filtered each data set such that each start coordinate was represented once. Then, a pseudo-ChIP data set that had the same number of tags as each filtered data set was generated. Pseudo-ChIP sequence tags were sampled from the input sequence tag data available at http://web.me.com/kaestnerlab1/GLITR/, and the remaining input sequence tags in this set were used as background for the GLITR sampling procedure. The following statistical cutoffs were used to generate the lists of high-confidence binding intervals for transcription factors: false-discovery rate (FDR), ≤0.5% except for PU.1 (FDR, ≤0.1%); fold change over input, ≥5; number of reads at the peak summit, ≥9. These criteria and filtering out of regions enriched in inputs were applied to minimize nonspecific effects, e.g., from PCR bias or local chromatin structure. H3K9Ace enrichment was analyzed using the significance tester for accumulation of reads (STAR) (http://www.cbil.upenn.edu/STAR; our unpublished data). Briefly, an algorithm was developed to determine regions with a locally significant accumulation of reads while controlling for false positives. For each sample, a set of permutation controls was generated by randomizing the reads via a uniform distribution across the genome. For an error rate of alpha, a cutoff for peak height was chosen that gives a proportion of the (average) signal over the permuted controls to the signal in the ChIP sample of no more than alpha. This gives a cutoff for significant peak height that controls the proportion of false positives in the set of all calls, otherwise known as the FDR. Prior to the comparison with the permutation controls, regions previously identified as common input bias were filtered out. Calls were made using a window length of 500 bp with a displacement of 50 bp as appropriate for histone modifications and an FDR of <5%.
Region overlap was generated in the UCSC Genome Browser (http://genome.ucsc.edu) such that sites were considered overlapping if there was at least 1 bp in common between the regions. To generate the “adipocyte” genome-wide data set for PPARγ, the union of the following data sets was generated in the UCSC Genome Browser: ChIP-chip at FDR < 5% (40), ChIP-seq at FDR < 0.1% (53) converted to mm8 coordinates, and ChIP-seq generated in this study and analyzed with GLITR as described above.
Mapping of PPARγ binding to genes differentially expressed in PPARγ-deficient macrophages.
For each gene in the mouse genome (mm8 assembly), the nearest ChIP region was identified using the Gene Centered Annotation module of CEAS (63). The list of all genes whose nearest macrophage PPARγ ChIP peak was located within 100 kb of the transcription start site (TSS) was compared to the lists of genes differentially expressed or unchanged in PPARγ-deficient macrophages (29). Fisher's exact test was used to determine whether the fraction of downregulated/upregulated genes with PPARγ binding within 100 kb was significantly different from the fraction of unchanged genes that also contained PPARγ-binding regions within the same distance.
ChIP-seq average profiles.
Average signal profiles were generated with the Sitepro module of CEAS (63) using a bed file of centers of PPARγ-binding regions and a WIG file of ChIP-seq signals. Briefly, for each PPARγ-binding peak identified by GLITR, the mm8 coordinates of the center 1 bp were obtained and used to generate a new bed file for a specific category, e.g., macrophage unique or adipocyte unique. A similar approach was taken for 600 control regions of similar genomic distribution generated in CisGenome (32). MACS software (74) was used to generate WIG files of ChIP-seq signals in macrophages and adipocytes for H3K9Ace, PU.1, and C/EBPβ binding. For each PPARγ binding or control region, H3K9Ace was examined 5 kb upstream and downstream from the center 1 bp in 100-bp segments. Next, all 10-kb regions were aligned at the center 1 bp and the average signal per 100-bp segment was computed for the category. For transcription factors, a 20-bp sliding window and 1-kb range were used. Where applicable, standard error was computed across all members of a category of regions, and an unpaired t test was used to evaluate statistical significance.
Enriched motif analysis.
The enrichment of motifs within the PPARγ ChIP-seq data set was calculated based on a test for positional bias relative to the center of the ChIP-enriched regions analogous to what has been described previously for FOXA1 genome-wide binding (46). DNA binding motifs were discovered de novo using MDscan (42) and assessed for significance using the same positional bias statistic.
Nearest-gene analysis.
For each macrophage PPARγ-binding region, the nearest gene was determined using CisGenome (32). Only genes with a binding region within 100 kb of the TSS were used for gene ontology (GO) analysis in PANTHER (http://www.pantherdb.org/) (66, 67) to examine the biological processes in which nearest genes are involved; the entire mouse genome (NCBI, Mus musculus genes) was used as a reference list. Bonferroni correction for multiple testing was used.
ChIP-chip.
The custom “PPARγ binding site” array used for H3 and H3K9Ace hybridization in adipocytes has been described elsewhere (40). Briefly, 740 distal PPARγ-binding regions, i.e., located ≥10 kb away from the TSS of the nearest gene, were selected and centered within 10 kb of genomic sequence. The nonrepetitive chromosomal sequence of these 10-kb regions (February 2006 Mouse Genome Assembly [mm8]) was obtained from the UCSC Genome Browser (http://genome.ucsc.edu) and tiled with overlapping 60-mer oligonucleotides (Agilent). Hybridization and data analysis were performed as previously described (40, 65). To generate the average H3K9Ac and H3 profiles, the 740 regions of 10 kb in length were aligned at the center where PPARγ binding occurs, followed by calculation of the average signal for each location. A 500-bp sliding window was used to smooth the profiles.
RNA isolation and quantitative PCR (QPCR).
RNA isolation and QPCR analysis were performed as previously described (40) using primers listed in Table S1 in the supplemental material, Power SYBR green PCR master mix (Applied Biosystems), and the PRISM 7900 instrument (Applied Biosystems). Analysis was performed using the standard curve method and normalization of all genes of interest to the housekeeping control Pabpc1.
Immunoblotting.
Immunoblotting was performed as described previously (40). The primary antibodies used for immunoblotting were as follows: anti-PPARγ (81b8; Cell Signaling Technologies), anti-PU.1 (sc-352x; Santa Cruz), anti-C/EBPβ (sc-150x; Santa Cruz), anti-HDAC2 (sc-7899; Santa Cruz), and anti-Ran (610341; BD Biosciences).
Retroviral transduction of 3T3-L1 preadipocytes.
The pMSCV-PPARγ2 and pMSCV-Empty vectors were provided by Bob Roeder (Rockefeller University, NY) (21). Retroviral supernatants were generated by transfecting packaging BOSC23 cells (57) at >90% confluence with retroviral vectors using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Retroviral supernatants were harvested at 48 and 72 h. Infection of 3T3-L1 preadipocytes was performed at ∼50% confluence by adding retroviral supernatant and Polybrene to a final concentration of 1 μg/ml. After 24 h, selection with puromycin (1 μg/ml) was initiated. Transduced cells were harvested 1 day after they reached confluence.
RESULTS
Cell-type-specific binding of PPARγ in macrophages and adipocytes.
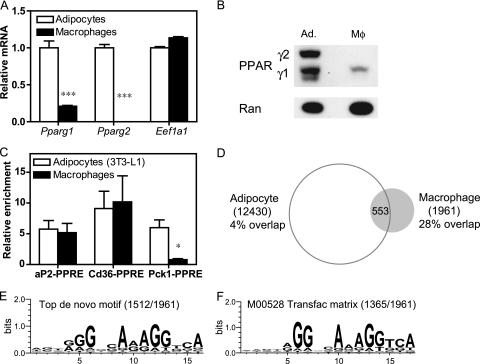
Mouse 3T3-L1 adipocytes and thioglycolate-elicited mouse macrophages were chosen for comparison because both are frequently employed for in vitro studies of PPARγ function (29, 39, 56). As expected from earlier studies showing that PPARγ1 is expressed at highest levels in adipose tissue and that PPARγ2 is adipocyte specific (10, 72), PPARγ1 mRNA and protein levels were substantially lower in macrophages than in differentiated 3T3-L1 adipocytes, and PPARγ2 was detected only in the adipocytes (Fig. 1A and B). Nevertheless, ChIP revealed measurable and reproducible recruitment of endogenous PPARγ at the PPARγ response elements (PPREs) of the Cd36 and Fabp4/aP2 genes in macrophages as well as adipocytes, while the PPRE at Pck1 was occupied only in the adipocytes (Fig. 1C). This result indicates that ChIP for endogenous PPARγ is feasible and robust in macrophages.
FIG. 1.
PPARγ binding in macrophages compared to adipocytes. (A) Gene expression analysis by QPCR of the Pparg1 and Pparg2 isoforms in thioglycolate-elicited macrophages and differentiated 3T3-L1 adipocytes. Eukaryotic translation elongation factor 1 alpha 1 (Eef1a1) was used as a negative control. Data were normalized to the housekeeping gene Pabpc1 and are presented as mean ± standard error (n = 3 to 4). ***, P < 0.001. (B) Immunoblot analysis demonstrating PPARγ abundance in macrophages (Mφ) and adipocytes (Ad.), using Ran as a loading control. The migration of the PPARγ1 and PPARγ2 isoforms is indicated. (C) ChIP-QPCR analysis of PPARγ enrichment at the known PPARγ response elements (PPRE) near the aP2, Cd36, and Pck1 genes. Data were normalized to a nontarget genomic site and are presented as mean ± standard error (n = 3 to 4). *, P < 0.05, unpaired t test. (D) Venn diagram representing the overlap of macrophage and adipocyte PPARγ cistromes, i.e., those having at least 1 bp in common. The adipocyte data set is the union of a ChIP-seq experiment performed in this study and the previously published data sets using ChIP-seq (53) and ChIP-chip (40). (E and F) Highest scoring sequence motifs from de novo motif analysis (E) and known motif analysis (F). Indicated in parentheses is the number of macrophage PPARγ-binding regions that contain the motif. x axes, residue number within motif.
ChIP-seq for PPARγ in macrophages was performed, and peak calling using GLITR (71) identified 1,961 high-confidence binding regions. The locations of these peaks were then compared with two previously published genome-wide PPARγ data sets in 3T3-L1 adipocytes (40, 53) and results of a ChIP-seq experiment performed here in parallel with the macrophage sequencing, which had a high degree of overlap with the previous studies (see Fig. S1 in the supplemental material). Although ∼28% of the macrophage regions (553/1,961) were common to adipocytes and macrophages, the majority (1,408/1,961) did not overlap with any of the three adipocyte data sets described above and were considered macrophage unique (Fig. 1D). A de novo motif search within the PPARγ peaks in macrophages revealed a matrix that strongly resembles the known PPARγ binding element, i.e., a direct repeat of the sequence AGGTCA separated by a single base pair (DR1 element), which was present in 77% (1,512/1,961) of the regions (Fig. 1E). The PPARγ-binding regions were also queried for known transcription factor motifs from the TRANSFAC database (48), demonstrating that several known matrices for the DR1 element are the most highly enriched known motifs; for example, the M00528 matrix for PPAR-RXR was found in 70% (1,365/1,961) of the regions (Fig. 1F). In addition, RXR occupancy was detectable by ChIP-QPCR at PPARγ regions in macrophages (see Fig. S2 in the supplemental material). Taken together, these findings indicate that PPARγ in macrophages binds to DR1 sequences as a heterodimer with RXR in a manner similar to that described for adipocytes (31, 40, 53).
PPARγ colocalizes with the hematopoietic factor PU.1 and with C/EBPβ in a macrophage-specific manner.
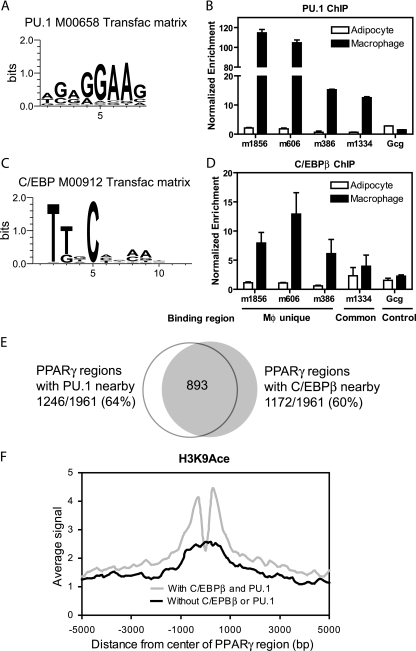
In adipocytes, the majority of PPARγ binding occurs in tandem with C/EBP transcription factors (40). To determine whether PPARγ binds coordinately with other transcription factors in macrophages, its binding regions were queried for enrichment of known transcription factor binding motifs. The second most highly enriched motif after the DR1 element was for Ets factors (Fig. 2A; see Table S2 in the supplemental material). PU.1 is the most critical Ets factor in macrophages (13, 15, 19, 64), and its occupancy near several PPARγ-binding sites was confirmed by ChIP-QPCR (Fig. 2B). PU.1 is not expressed in adipocytes (see Fig. S3), and thus the lack of PU.1 ChIP enrichment in adipocytes is expected. Also enriched were C/EBP motifs (Fig. 2C; also see Table S2). Indeed, C/EBPβ, which is also known as NF-IL-6 based on its ability to activate cytokine gene expression in macrophages (1), also localized to the same PPARγ-binding regions as PU.1 (Fig. 2D). Interestingly, although C/EBPβ is expressed at similar levels in adipocytes and macrophages, it does not bind at macrophage-unique PPARγ regions in adipocytes (Fig. 2D).
FIG. 2.
PPARγ colocalizes in a macrophage-specific manner with C/EBPβ and PU.1. (A) Example of a PU.1/Ets TRANSFAC matrix enriched in the PPARγ-binding regions in macrophages. (B) ChIP-QPCR analysis of PU.1 enrichment in adipocytes and macrophages at several PPARγ-binding regions predicted to have PU.1 colocalization. Data were normalized to a nontarget genomic site and are presented as mean ± standard deviation (n = 2). (C) Example of a C/EBP TRANSFAC matrix enriched in the PPARγ-binding regions in macrophages. (D) ChIP-QPCR analysis of C/EBPβ enrichment as for panel B. Indicated are the regions that are bound by PPARγ only in macrophages (Mφ unique), a region bound by PPARγ in both cell types (Common), and a negative-control site at the glucagon TSS (Gcg, Control). (E) Venn diagram representing the overlap between the sets of macrophage PPARγ-binding regions with PU.1 and C/EBPβ, i.e., those having at least 1 bp in common. (F) Average profiles of H3K9Ace ChIP-seq signals around macrophage PPARγ-binding regions that have colocalization with both PU.1 and C/EBPβ (n = 776) or neither factor (n = 516). All profiles are centered at the middle 1 bp of the binding regions. The average signal represents the average number of reads across a category of binding regions per 100-bp interval.
To better characterize the overlap of PU.1 and C/EBPβ binding with that of PPARγ, we determined their macrophage cistromes. Peak calling analysis of ChIP-seq data in thioglycolate-elicited macrophages identified 46,356 high-confidence binding regions for PU.1 and 24,421 for C/EBPβ (see Table S3 in the supplemental material). Transcription factor motif analysis identified C/EBP motifs in the PU.1 regions and Ets motifs in the C/EBP data in addition to the factors' cognate motifs (see Table S3). As predicted by this analysis, the majority of C/EBPβ-binding regions (average length of 300 bp) overlapped by ≥1 bp with PU.1 ChIP-seq regions (average length of 270 bp) (see Fig. S4 in the supplemental material). Since the actual binding sites for PU.1 and C/EBP are ∼10 bp long (Fig. 2A and C), the overlap between a 300-bp C/EBP ChIP-seq region and a 270-bp PU.1 region does not imply that the actual binding sites overlap each other but most likely indicates that the two factors bind near one another (i.e., within ∼570 bp at most). Remarkably, a similar intersection analysis revealed that 64% (1,246/1,961) of PPARγ-binding regions overlapped with PU.1-binding regions and 60% (1,172/1,961) with C/EBPβ regions, while 45% (893/1,961) overlapped with both factors (Fig. 2E). Of note, only 2% (45/1,961) of the PPARγ regions had any overlap with 46,350 control genomic regions with similar lengths and genomic distributions (P < 0.001 versus 60% and 64% overlap; Fisher's exact test). Moreover, the level of H3K9Ace was higher around PPARγ-binding sites with PU.1 and C/EBβ colocalization (Fig. 2F). Taken together, these data suggest that PU.1 and C/EBPβ are functionally cooperative with PPARγ at macrophage enhancers.
Differential enrichment of PU.1 and C/EBPβ at PPARγ-binding regions correlates with biological functions of nearby genes.
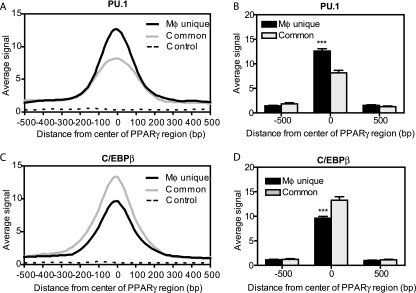
PU.1 is macrophage specific, whereas C/EBPβ is expressed in both macrophages and adipocytes. We therefore tested for differential recruitment of these factors at distinct classes of PPARγ-binding regions. Indeed, the enrichment of PU.1 in macrophages was much greater at macrophage-unique PPARγ-binding sites than at sites common to macrophages and adipocytes, whether assessed across the region as a running average (Fig. 3A) or at the center of the PPARγ-binding regions (Fig. 3B). In contrast, the enrichment of C/EBPβ was much higher at common regions (Fig. 3C and D).
FIG. 3.
Differential PU.1 and C/EBP enrichment at macrophage-unique versus common PPARγ-binding regions. (A) Average profile of PU.1 ChIP-seq signals in macrophages around PPARγ-binding regions that are either unique to macrophages (Mφ unique, n = 1,408) or shared with adipocytes (common, n = 553), as well as at control genomic regions (control, n = 600). The average signal represents the average number of reads per 20-bp segment over a 1-kb distance, centered on the middle 1 bp of the PPARγ-binding regions. (B) Comparison of average PU.1 signals between Mφ unique (n = 1,408) and Common (n = 553) regions at the middle 1 bp of the PPARγ-binding regions and 500 bp downstream and upstream. (C) Average profile of C/EBPβ ChIP-seq signals in macrophages around PPARγ-binding regions as for panel A. (D) Comparison of average C/EBPβ signal between Mφ unique and common regions at the middle 1 bp of the PPARγ-binding regions and 500 bp downstream and upstream as for panel C. ***, P < 0.001, unpaired t test.
A potential explanation for these findings may be that common and macrophage-unique regions target different types of genes. To explore this possibility, each PPARγ region in the macrophage-unique or common category was annotated with the nearest RefSeq gene within a 100-kb distance of the TSS. GO analysis of the genes near common PPARγ-binding regions revealed strong enrichment of biological processes related to lipid metabolism (Table 1), similar to what has been reported for PPARγ binding in adipocytes (40, 53). On the other hand, genes located near macrophage-unique PPARγ-binding sites are highly enriched for immunity and defense as well as cytokine/chemokine-mediated signaling (Table 2). This is in accordance with the well-established role of PU.1 as a master regulator of immune functions in macrophages (15, 19, 64). Thus, in addition to its functions in lipid metabolism, PPARγ may play macrophage-specific roles, such as modulating immune responses with the aid of the hematopoietic factor PU.1. This finding is consistent with recent studies implicating PPARγ in the function of alternative macrophages (5, 20, 55) and bone marrow-derived dendritic cells (34, 51).
TABLE 1.
Biological functions of genes located near common PPARγ-binding regionsa
| GO biological processb | P valuec |
|---|---|
| Lipid, fatty acid, and steroid metabolism | 4.80E−9 |
| Fatty acid metabolism | 1.56E−4 |
| Carbohydrate metabolism | 2.26E−3 |
| Apoptosis | 3.05E−3 |
| Fatty acid beta-oxidation | 5.43E−3 |
In this analysis, only binding regions whose nearest gene was within 100 kb of the TSS were considered.
Summary of gene ontology (GO) categories of the nearest genes to regions that are bound by PPARγ in both adipocytes and macrophages.
P value reflects Bonferroni correction for multiple testing.
TABLE 2.
Biological functions of genes located near macrophage-unique PPARγ-binding regionsa
| GO biological processb | P value |
|---|---|
| Protein modification | 8.33E−10 |
| Cytokine- and chemokine-mediated immunity | 6.69E−9 |
| Immunity and defense | 1.28E−8 |
| Cytokine- and chemokine-mediated signaling | 4.37E−8 |
| Signal transduction | 7.33E−8 |
Analysis was performed as for Table 1.
Summary of gene ontology (GO) categories of the nearest genes to macrophage-unique PPARγ-binding regions.
Macrophage PPARγ binding is related to gene activation and histone acetylation.
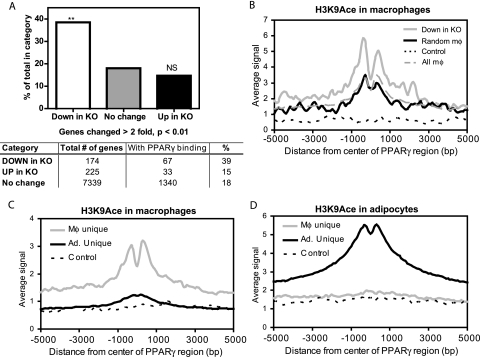
To assess the functional significance of PPARγ binding in macrophages, the distribution of its binding regions was examined relative to the location of differentially expressed genes in PPARγ-deficient macrophages. The generation and expression profiling of these cells have been described elsewhere (29). Of the 174 genes that are significantly decreased in PPARγ-deficient macrophages compared to wild-type controls, 67 (39%) have at least one PPARγ-binding region located within 100 kb of the transcription start site (TSS). This is statistically higher than the levels for genes that were upregulated or unchanged in the PPARγ-deficient macrophages, 15% and 18%, respectively (Fig. 4A). Gene ontology (GO) analysis of the 67 downregulated genes with macrophage PPARγ binding nearby showed that they were enriched for lipid metabolism and immunity as well as apoptosis (see Table S4 in the supplemental material).
FIG. 4.
PPARγ binding correlates with PPARγ-dependent gene expression and histone acetylation. (A) Percentage of genes that have a PPARγ binding site within 100 kb of the TSS among the genes that are downregulated (down in KO), upregulated (up in KO), or unchanged (no change) in PPARγ-deficient macrophages (KO) compared to wild-type controls, described previously (29). **, P < 0.01 and NS (not significant) versus “no change,” Fisher's exact test. (B) Average H3K9Ace profiles for the 96 macrophage PPARγ-binding regions located within 100 kb of downregulated genes (down in KO), 96 macrophage regions selected at random from the entire set of macrophage PPARγ-binding regions (random mφ), 96 negative-control regions (control), and the entire set of 1,961 macrophage PPARγ-binding regions (all mφ). Distances from the centers of the binding regions are shown in base pairs (bp). (C and D) Genome-wide H3K9Ace in macrophages (C) and in adipocytes (D). Average profiles of H3K9Ace ChIP-seq signal around three categories of PPARγ-binding regions: macrophage (Mφ) unique (n = 1,408), adipocyte (Ad.) unique (n = 11,892), and genomic regions where no binding was detected for PPARγ in either cell type (control) (n = 500). All profiles are centered at the middle 1 bp of the binding regions. The average signal represents the average number of reads across a category of binding regions per 100-bp interval.
To confirm that PPARγ binding at these locations is functional, acetylated lysine 9 of histone 3 (H3K9Ace) was quantified around the 96 PPARγ regions located within 100 kb of significantly downregulated genes. Histone acetylation, including H3K9Ace, is highly enriched at enhancer regions (27, 28) and was recently employed to assess the functionality of genome-wide PPARγ binding in adipocytes (40). Indeed, the average H3K9Ace signal is significantly higher around PPARγ regions near the downregulated genes than in a similar number of randomly selected macrophage PPARγ regions, all of the PPARγ regions, or controls without PPARγ binding (Fig. 4B). In addition, 77% (74/96) of the regions near downregulated genes fall within areas of significantly enriched H3K9Ace compared to the rest of the genome, while only 58% (1,143/1,962) of all macrophage regions have this characteristic. Thus, PPARγ binding at the locations identified by this approach is associated with high levels of histone acetylation, which generally has activating effects on transcription (35). Moreover, examination of histone acetylation on a genome-wide scale in macrophages revealed that H3K9Ace is markedly enriched at macrophage-unique PPARγ-binding regions but only minimally at adipocyte-unique regions (Fig. 4C). Reciprocally, in adipocytes only the adipocyte-specific PPARγ-binding regions demonstrate substantially increased histone acetylation (Fig. 4D).
Cell-type-specific DNA accessibility correlates with PPARγ binding.
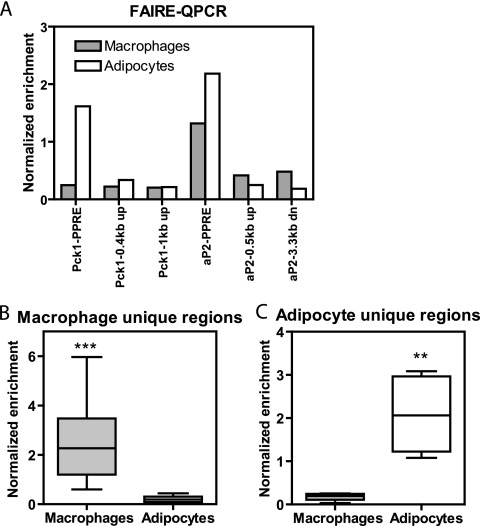
H3K9Ace was increased at PPARγ-binding locations, yet the H3K9Ace signal was relatively decreased at the exact center of the regions (Fig. 4B to D), suggesting local nucleosomal depletion where PPARγ binds. This phenomenon has been described at active promoters and enhancers (4, 17, 24, 28, 33, 41). Indeed, examination of 740 PPARγ-binding regions in adipocytes by ChIP-chip also demonstrated that the center of PPARγ-binding regions colocalizes with a decrease in histone 3 enrichment (see Fig. S5 in the supplemental material). Therefore, chromatin accessibility was assessed in macrophages and adipocytes by formaldehyde-assisted isolation of regulatory elements (FAIRE). This assay has been validated as an alternative to DNase I hypersensitivity assays (23, 24) and was used recently to show DNA accessibility at active FOXA1 enhancers (17). Indeed, a high FAIRE-QPCR signal was noted at the aP2 PPRE in both macrophages and adipocytes, at Pck1 only in adipocytes, and not at distal regions not bound by PPARγ (Fig. 5A). When FAIRE-QPCR was employed to compare DNA accessibility at a number of macrophage-unique PPARγ regions (see Fig. S6A in the supplemental material), an increased FAIRE signal was found in macrophages but not in adipocytes (Fig. 5B). Conversely, FAIRE was increased in adipocytes only when assessed at sites uniquely bound by PPARγ in this cell type (Fig. 5C; see Fig. S6B in the supplemental material).
FIG. 5.
PPARγ correlates with DNA accessibility in a cell-type-specific manner. (A) FAIRE-QPCR at aP2-PPRE and Pck1-PPRE and two downstream (dn) and/or upstream (up) regions for each PPRE. Enrichment in macrophages and adipocytes was normalized to a control genomic region. (B and C) Box-whisker plots of FAIRE-QPCR in macrophages and adipocytes at macrophage-unique regions (n = 13) and adipocyte (Ad.) unique regions (n = 6). Enrichment was normalized as for panel A and measured in two to four biological replicates, **, P < 0.01; ***, P < 0.001, paired t test.
Macrophage-unique PPARγ-binding sites are marked by repressive histone methylation in adipocytes.
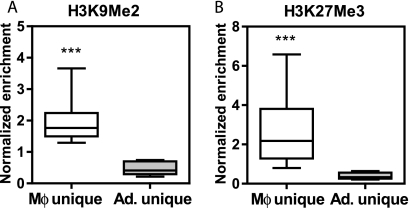
Dimethyl lysine 9 of histone 3 (H3K9Me2) and trimethyl lysine 27 of histone 3 (H3K27Me3) correlate negatively with gene transcription (3) and characterize heterochromatin (70). In adipocytes, H3K9Me2 was significantly enriched at macrophage-unique PPARγ-binding regions compared to the adipocyte-unique sites (Fig. 6A). Similar findings pertain to H3K27Me3, which is greatly enhanced at macrophage-unique sites in adipocytes (Fig. 6B). These data strongly suggest that the cell specificity of PPARγ binding may be restricted by the presence of heterochromatin in the nonpermissive cell types.
FIG. 6.
Evidence of chromatin silencing in adipocytes at macrophage-unique PPARγ-binding regions. (A) H3K9Me2. (B) H3K27Me3. Box-whisker plots of ChIP-QPCR enrichment of the repressive chromatin marks H3K9Me2 and H3K27Me3 in adipocytes at macrophage (Mφ) unique (n = 13) and adipocyte (Ad.) unique (n = 9) regions. Enrichment at target sites was normalized to a control genomic region and was measured in three biological replicates for each cell type. *, P < 0.05; ***, P < 0.001, unpaired t test.
PPARγ binding induces open chromatin and histone acetylation at sites lacking repressive histone marks.
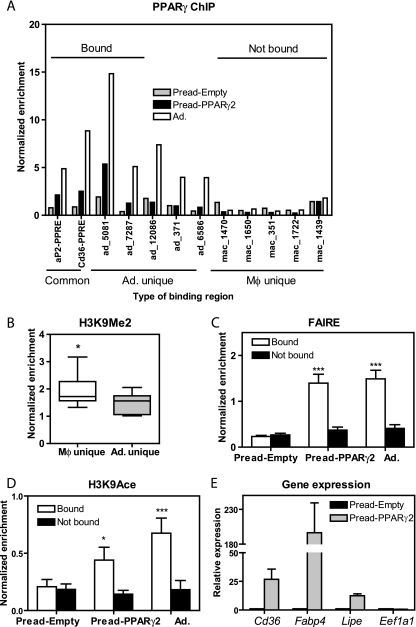
The data thus far demonstrate that cell-type-specific PPARγ binding correlates positively with open chromatin and histone acetylation and negatively with repressive marks like histone H3K9Me2 and H3K27Me3, but they do not address causality. To test whether PPARγ binding could induce changes in the chromatin landscape, PPARγ2 was ectopically expressed in preadipocytes (see Fig. S7A in the supplemental material), which otherwise have low levels of PPARγ1 and no measurable PPARγ2 (see Fig. S7B in the supplemental material). ChIP-QPCR for PPARγ at several target regions demonstrated recruitment at a subset of adipocyte-binding sites, consistent with previous studies of ectopic PPARγ expression in fibroblast-like cells (6). However, PPARγ binding was not observed at macrophage-unique sites (Fig. 7A). Remarkably, the macrophage-unique PPARγ binding sites were marked in preadipocytes by the repressive H3K9Me2 modification, whereas this mark was much less pronounced at adipocyte-unique PPARγ binding sites (Fig. 7B). FAIRE assessment of chromatin accessibility in the preadipocytes transduced with empty virus (Pread-Empty) was similar at the macrophage and adipocyte PPARγ binding sites (Fig. 7C). However, PPARγ expression increased the preadipocyte FAIRE signal at the sites bound by PPARγ, whereas the FAIRE signal did not increase at the nonbound macrophage-unique sites (Fig. 7C). Moreover, H3K9Ace was also markedly increased at sites bound by PPARγ in the preadipocytes (Fig. 7D). Open chromatin and histone acetylation are characteristics of functional enhancers, and, consistent with this, ectopic PPARγ increased expression of several target genes located near sites that are bound by PPARγ in the retrovirally transduced cells and adipocytes (Fig. 7E). Taken together, these findings indicate that PPARγ binding causes changes in chromatin organization and local histone acetylation; however, this occurred only at a subset of sites that were not previously marked by repressive histone modifications.
FIG. 7.
PPARγ binding induces open chromatin and histone acetylation. (A) ChIP-QPCR at three types of PPARγ-binding regions: common (n = 2), adipocyte (Ad.) unique (n = 5), and macrophage (Mφ) unique (n = 5). Enrichment was normalized as in Fig. 1C and measured in mature adipocytes (Ad.) and in preadipocytes transduced with pMSCV-Empty virus (Pread-Empty) or pMSCV-PPARγ2 (Pread-PPARγ2). “Bound” refers to the adipocyte regions that were occupied by PPARγ in Pread-PPARγ2 (n = 4); “not bound” refers to five macrophage regions that show no recruitment in Pread-PPARγ2 and Ad. (B) H3K9Me2 in preadipocytes, analyzed as in Fig. 6A. (C) FAIRE-QPCR at the four regions bound by PPARγ in Pread-PPARγ2 (“bound,” in panel A) and compared to the five macrophage-unique regions (“not bound” in panel A). The same comparison was carried out for Pread-Empty and Ad.; *** P < 0.001, two-way analysis of variance (ANOVA) (bound versus not bound). (D) H3K9Ace ChIP-QPCR at the same bound (n = 4) and not bound (n = 5) PPARγ regions in preadipocytes transduced with pMSCV-PPARγ2 (Pread-PPARγ2) or pMSCV-Empty retrovirus (Pread-Empty), and mature adipocytes (Ad.); *, P < 0.05, ***, P < 0.001, two-way ANOVA (bound versus not bound). (E) QPCR analysis of PPARγ target gene expression in Pread-PPARγ2 and Pread-Empty. The genes that were examined are Cd36, aP2/Fabp4, Lipe/HSL, and eukaryotic translation elongation factor 1 alpha 1 (Eef1a1) as a negative control. Data were normalized to the housekeeping gene Pabpc1 and are presented as mean ± standard deviation (n = 2).
DISCUSSION
A critical step toward understanding the cell-type-specific functions of PPARγ is elucidating its recruitment across the genome in various cell types. In this study, PPARγ binding was compared in mouse macrophages and adipocytes, both of which play important roles in regulating metabolism (54, 69). PPARγ cistromes in adipocytes and macrophages are largely distinct, although a small subset of the binding locations is shared between cell types. The ability of PPARγ to occupy cell-type-unique locations is associated with chromatin remodeling and active chromatin marks such as H3K9Ace, while access to binding sites of a different cell type may be prevented through chromatin silencing. In macrophages, PPARγ colocalizes with C/EBPβ as well as the hematopoietic factor PU.1, which may explain the ability of PPARγ to regulate not only a subset of the metabolic genes it binds in adipocytes but also a unique set of immune genes.
PPARγ binding in macrophages occurs at DR1 elements through heterodimerization with RXR, as previously described for adipocytes (31, 40, 53). It is interesting that, although macrophages express only PPARγ1 and their overall level of PPARγ protein is much lower than that of adipocytes, the ChIP enrichment levels at regions common to both cell types are similar. A notable difference, however, is that there are fewer regions occupied by PPARγ in macrophages than in any of the three adipocyte data sets discussed here. Thus, the higher protein levels in adipocytes and/or the expression of the γ2 isoform may allow PPARγ to bind and activate a larger number of genes in adipocytes, where it is required for differentiation, mature function, and survival (69). In contrast, macrophages do not require PPARγ for differentiation, phagocytic activity, or proinflammatory cytokine production (50) but employ PPARγ in more specialized functions, such as maintenance of the alternative macrophage phenotype (5, 20, 55), cholesterol uptake and efflux in atherosclerotic plaques (49), antigen cross-presentation to T lymphocytes (34), and dendritic cell immunogenicity (51).
One of the major differences at PPARγ-binding regions in macrophages compared to adipocytes is the nearby presence of the hematopoietic transcription factor PU.1. It belongs to the Ets family of transcription factors and is most highly expressed in myeloid cells (13, 19), from which mature macrophages are derived. PU.1 is absolutely required for the development of monocytes (19, 64) and can reprogram lymphocytes and fibroblasts into monocytes when expressed exogenously together with C/EBP (18, 37, 52). In fact, the genome-wide binding studies performed here demonstrate that PU.1 colocalizes extensively with C/EBPβ across the genome. Importantly, the motif search performed on the PPARγ-binding regions did not reveal enrichment for NF-κB sites, although motifs for other transcription factors associated with immunity and inflammation were identified, including AP-1 and STAT, suggesting that PPARγ may also cooperate with those factors in addition to PU.1 and C/EBP. Thus, through colocalization with potent regulators of immune function in macrophages, PPARγ may be able to access and regulate a number of immune genes in addition to a subset of the metabolic genes it binds in adipocytes (40).
Further support for the notion that PPARγ can regulate both immune and metabolic genes in macrophages is gained from the distribution of PPARγ-binding regions relative to differentially expressed genes in PPARγ-deficient macrophages (29). A higher percentage of the downregulated genes have PPARγ-binding regions than the upregulated genes (39% compared to 15%), consistent with findings in adipocytes following PPARγ knockdown (62). This suggests that in both cell types direct PPARγ binding has activating rather than repressive effects on target gene transcription.
To begin to address the mechanisms driving cell-type-specific binding of PPARγ in macrophages and adipocytes, several chromatin features were examined in the vicinity of PPARγ sites. In the absence of PPARγ occupancy, DNA accessibility is low at PPARγ-binding sites, suggesting that the DNA may be sequestered in nucleosomes (60). In contrast, occupied PPARγ sites are characterized by a high degree of DNA accessibility, which indicates that the chromatin organization has been altered and histone octamers have been released or repositioned (26, 60). This type of chromatin remodeling may occur prior to or following PPARγ binding. Although the former possibility cannot be excluded, PPARγ was able to induce chromatin remodeling at adipocyte regions when retrovirally expressed in undifferentiated preadipocytes. PPARγ can interact with SWI/SNF ATP-dependent remodeling complexes (8, 14), and, therefore, the increased accessibility may be a consequence of PPARγ-dependent recruitment of chromatin remodelers. However, it remains unclear which, if any, remodeling complexes are responsible for this activity and how PPARγ recruits them to target sites. Nevertheless, a similar induction of DNA accessibility has been described for other factors, including FOXA1 (16, 17), glucocorticoid receptor (33), and others (4).
In addition to altering DNA accessibility, PPARγ was also found to increase H3K9Ace at sites to which it bound in retrovirally transduced preadipocytes. Acetylation of histone tails, including H3K9 acetylation, is mediated by histone acetyltransferases such as p300/CBP and Gcn5 (35, 47), some of which have been shown to interact with and coactivate PPARγ (11, 22, 69). Notably, histone acetylation occurs at enhancers and promoters (27, 28) and is predominantly associated with transcriptional activation (35, 38, 41). It should be noted that in addition to coactivators, PPARγ can also associate with corepressors such as NCoR and SMRT, which in turn recruit histone HDAC3 (25, 69). Therefore, it is also possible that the high level of acetylation observed at occupied PPARγ binding sites in macrophages and adipocytes reflects decreased recruitment of corepressors.
Although PPARγ was able to induce both DNA accessibility and H3K9Ace in PPARγ-transduced preadipocytes, it did so selectively: only at a subset of adipocyte regions tested and not at sites that are unique to macrophages. This finding was correlated with the presence of repressive H3K9Me2 chromatin marks at macrophage-unique PPARγ-binding regions in preadipocytes. The presence of these modifications, along with the concurrent hypoacetylation and lack of DNA accessibility, strongly suggests chromatin silencing. H3K9Me2/3 and H3K27Me2/3 are associated with transcriptional repression across the human genome (3). They can occur in constitutive heterochromatin, i.e., gene-poor repetitive domains at centromeres and telomeres, as well as in facultative heterochromatin, which contains genes that can undergo silencing or activation depending on the cellular context (7, 70). Given the developmental divergence between macrophages and adipocytes, it is possible that the observed silencing effects may be established early during the differentiation programs of these cell types and may span large domains, including subsets of PPARγ-binding regions and the genes they regulate. Indeed, the fibroblast-like preadipocytes are committed to adipocyte differentiation and, consistent with this, our data implicate H3K9Me2 as a mark that restricts PPARγ binding to sites bound in the mature adipocyte.
The findings presented here demonstrate that cell-type-specific PPARγ binding can facilitate the formation of cell-type-specific enhancers that target unique sets of genes related to the specification of a cell type. The ability of PPARγ to access binding sites is limited and may be defined by repressive mechanisms like chromatin silencing and active mechanisms such as cell-type-specific colocalization with factors like PU.1 and C/EBPβ. Moreover, our data support the existence of an epigenomic hierarchy in which PPARγ binding to cell-specific sites not marked by repressive marks, including H3K9Me2, opens chromatin and leads to local activation marks, including H3K9 acetylation. A more comprehensive understanding of the mechanisms by which PPARγ regulates transcription in cell-type-specific ways may be helpful in designing pharmacological agents that can target individual cell types or distinct pathways in order to optimize therapeutic results and minimize side effects.
Supplementary Material
Acknowledgments
We thank Clifford A. Meyer, Josiah Altschuler, Hyunjin Shin, Tao Liu, and X. Shirley Liu (Dana-Farber Cancer Institute) for enriched motif tools and for sharing CEAS prior to its publication (63). We also thank Ana Cristancho and Michael Schupp for insightful discussions and reagents and Jonathan Schug (University of Pennsylvania), Mathieu Lupien (Dartmouth), Christian J. Stoeckert, Jr. (University of Pennsylvania), and Klaus H. Kaestner (University of Pennsylvania) for their helpful comments.
This work was supported by NIH R01 DK49780 (to M.A.L.), the George S. Cox Medical Research Institute, and the Picower Foundation.
Footnotes
Published ahead of print on 22 February 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akira, S., H. Isshiki, T. Sugita, O. Tanabe, S. Kinoshita, Y. Nishio, T. Nakajima, T. Hirano, and T. Kishimoto. 1990. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 9:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrera, L. O., Z. Li, A. D. Smith, K. C. Arden, W. K. Cavenee, M. Q. Zhang, R. D. Green, and B. Ren. 2008. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 18:46-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823-837. [DOI] [PubMed] [Google Scholar]
- 4.Biddie, S. C., S. John, and G. L. Hager. 2010. Genome-wide mechanisms of nuclear receptor action. Trends Endocrinol. Metab. 21:3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouhlel, M. A., B. Derudas, E. Rigamonti, R. Dievart, J. Brozek, S. Haulon, C. Zawadzki, B. Jude, G. Torpier, N. Marx, B. Staels, and G. Chinetti-Gbaguidi. 2007. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 6:137-143. [DOI] [PubMed] [Google Scholar]
- 6.Bugge, A., L. Grontved, M. M. Aagaard, R. Borup, and S. Mandrup. 2009. The PPARgamma2 A/B-domain plays a gene-specific role in transactivation and cofactor recruitment. Mol. Endocrinol. 23:794-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campos, E. I., and D. Reinberg. 2009. Histones: annotating chromatin. Annu. Rev. Genet. 43:559-599. [DOI] [PubMed] [Google Scholar]
- 8.Caramel, J., S. Medjkane, F. Quignon, and O. Delattre. 2008. The requirement for SNF5/INI1 in adipocyte differentiation highlights new features of malignant rhabdoid tumors. Oncogene 27:2035-2044. [DOI] [PubMed] [Google Scholar]
- 9.Chawla, A., W. A. Boisvert, C. H. Lee, B. A. Laffitte, Y. Barak, S. B. Joseph, D. Liao, L. Nagy, P. A. Edwards, L. K. Curtiss, R. M. Evans, and P. Tontonoz. 2001. A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell 7:161-171. [DOI] [PubMed] [Google Scholar]
- 10.Chawla, A., E. J. Schwarz, D. D. Dimaculangan, and M. A. Lazar. 1994. Peroxisome proliferator-activated receptor (PPAR) gamma: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology 135:798-800. [DOI] [PubMed] [Google Scholar]
- 11.Chen, S., B. A. Johnson, Y. Li, S. Aster, B. McKeever, R. Mosley, D. E. Moller, and G. Zhou. 2000. Both coactivator LXXLL motif-dependent and -independent interactions are required for peroxisome proliferator-activated receptor gamma (PPARgamma) function. J. Biol. Chem. 275:3733-3736. [DOI] [PubMed] [Google Scholar]
- 12.Cuddapah, S., R. Jothi, D. E. Schones, T. Y. Roh, K. Cui, and K. Zhao. 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 19:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahl, R., and M. C. Simon. 2003. The importance of PU.1 concentration in hematopoietic lineage commitment and maturation. Blood Cells Mol. Dis. 31:229-233. [DOI] [PubMed] [Google Scholar]
- 14.Debril, M. B., L. Gelman, E. Fayard, J. S. Annicotte, S. Rocchi, and J. Auwerx. 2004. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J. Biol. Chem. 279:16677-16686. [DOI] [PubMed] [Google Scholar]
- 15.DeKoter, R. P., M. B. Kamath, and I. B. Houston. 2007. Analysis of concentration-dependent functions of PU.1 in hematopoiesis using mouse models. Blood Cells Mol. Dis. 39:316-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eeckhoute, J., J. S. Carroll, T. R. Geistlinger, M. I. Torres-Arzayus, and M. Brown. 2006. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 20:2513-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eeckhoute, J., M. Lupien, C. A. Meyer, M. P. Verzi, R. A. Shivdasani, X. S. Liu, and M. Brown. 2009. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res. 19:372-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng, R., S. C. Desbordes, H. Xie, E. S. Tillo, F. Pixley, E. R. Stanley, and T. Graf. 2008. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc. Natl. Acad. Sci. U. S. A. 105:6057-6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman, A. D. 2007. Transcriptional control of granulocyte and monocyte development. Oncogene 26:6816-6828. [DOI] [PubMed] [Google Scholar]
- 20.Gallardo-Soler, A., C. Gomez-Nieto, M. L. Campo, C. Marathe, P. Tontonoz, A. Castrillo, and I. Corraliza. 2008. Arginase I induction by modified lipoproteins in macrophages: a peroxisome proliferator-activated receptor-gamma/delta-mediated effect that links lipid metabolism and immunity. Mol. Endocrinol. 22:1394-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge, K., M. Guermah, C. X. Yuan, M. Ito, A. E. Wallberg, B. M. Spiegelman, and R. G. Roeder. 2002. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature 417:563-567. [DOI] [PubMed] [Google Scholar]
- 22.Gelman, L., G. Zhou, L. Fajas, E. Raspe, J. C. Fruchart, and J. Auwerx. 1999. p300 interacts with the N- and C-terminal part of PPARgamma2 in a ligand-independent and -dependent manner, respectively. J. Biol. Chem. 274:7681-7688. [DOI] [PubMed] [Google Scholar]
- 23.Giresi, P. G., J. Kim, R. M. McDaniell, V. R. Iyer, and J. D. Lieb. 2007. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 17:877-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giresi, P. G., and J. D. Lieb. 2009. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (formaldehyde assisted isolation of regulatory elements). Methods 48:233-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan, H. P., T. Ishizuka, P. C. Chui, M. Lehrke, and M. A. Lazar. 2005. Corepressors selectively control the transcriptional activity of PPARgamma in adipocytes. Genes Dev. 19:453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hager, G. L., J. G. McNally, and T. Misteli. 2009. Transcription dynamics. Mol. Cell 35:741-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heintzman, N. D., G. C. Hon, R. D. Hawkins, P. Kheradpour, A. Stark, L. F. Harp, Z. Ye, L. K. Lee, R. K. Stuart, C. W. Ching, K. A. Ching, J. E. Antosiewicz-Bourget, H. Liu, X. Zhang, R. D. Green, V. V. Lobanenkov, R. Stewart, J. A. Thomson, G. E. Crawford, M. Kellis, and B. Ren. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459:108-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heintzman, N. D., R. K. Stuart, G. Hon, Y. Fu, C. W. Ching, R. D. Hawkins, L. O. Barrera, S. Van Calcar, C. Qu, K. A. Ching, W. Wang, Z. Weng, R. D. Green, G. E. Crawford, and B. Ren. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39:311-318. [DOI] [PubMed] [Google Scholar]
- 29.Hevener, A. L., J. M. Olefsky, D. Reichart, M. T. Nguyen, G. Bandyopadyhay, H. Y. Leung, M. J. Watt, C. Benner, M. A. Febbraio, A. K. Nguyen, B. Folian, S. Subramaniam, F. J. Gonzalez, C. K. Glass, and M. Ricote. 2007. Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J. Clin. Invest. 117:1658-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, J. T., J. S. Welch, M. Ricote, C. J. Binder, T. M. Willson, C. Kelly, J. L. Witztum, C. D. Funk, D. Conrad, and C. K. Glass. 1999. Interleukin-4-dependent production of PPAR-gamma ligands in macrophages by 12/15-lipoxygenase. Nature 400:378-382. [DOI] [PubMed] [Google Scholar]
- 31.IJpenberg, A. I., E. Jeannin, W. Wahli, and B. Desvergne. 1997. Polarity and specific sequence requirements of peroxisome proliferator-activated receptor (PPAR)/retinoid X receptor heterodimer binding to DNA. A functional analysis of the malic enzyme gene PPAR response element. J. Biol. Chem. 272:20108-20117. [DOI] [PubMed] [Google Scholar]
- 32.Ji, H., H. Jiang, W. Ma, D. S. Johnson, R. M. Myers, and W. H. Wong. 2008. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 26:1293-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.John, S., P. J. Sabo, T. A. Johnson, M. H. Sung, S. C. Biddie, S. L. Lightman, T. C. Voss, S. R. Davis, P. S. Meltzer, J. A. Stamatoyannopoulos, and G. L. Hager. 2008. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell 29:611-624. [DOI] [PubMed] [Google Scholar]
- 34.Klotz, L., S. Hucke, D. Thimm, S. Classen, A. Gaarz, J. Schultze, F. Edenhofer, C. Kurts, T. Klockgether, A. Limmer, P. Knolle, and S. Burgdorf. 2009. Increased antigen cross-presentation but impaired cross-priming after activation of peroxisome proliferator-activated receptor gamma is mediated by up-regulation of B7H1. J. Immunol. 183:129-136. [DOI] [PubMed] [Google Scholar]
- 35.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128:693-705. [DOI] [PubMed] [Google Scholar]
- 36.Krum, S. A., G. A. Miranda-Carboni, M. Lupien, J. Eeckhoute, J. S. Carroll, and M. Brown. 2008. Unique ERalpha cistromes control cell type-specific gene regulation. Mol. Endocrinol. 22:2393-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laiosa, C. V., M. Stadtfeld, H. Xie, L. de Andres-Aguayo, and T. Graf. 2006. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity 25:731-744. [DOI] [PubMed] [Google Scholar]
- 38.Lee, K. K., and J. L. Workman. 2007. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8:284-295. [DOI] [PubMed] [Google Scholar]
- 39.Lefterova, M. I., and M. A. Lazar. 2009. New developments in adipogenesis. Trends Endocrinol. Metab. 20:107-114. [DOI] [PubMed] [Google Scholar]
- 40.Lefterova, M. I., Y. Zhang, D. J. Steger, M. Schupp, J. Schug, A. Cristancho, D. Feng, D. Zhuo, C. J. Stoeckert, Jr., X. S. Liu, and M. A. Lazar. 2008. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 22:2941-2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128:707-719. [DOI] [PubMed] [Google Scholar]
- 42.Liu, X. S., D. L. Brutlag, and J. S. Liu. 2002. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 20:835-839. [DOI] [PubMed] [Google Scholar]
- 43.Lumeng, C. N., J. L. Bodzin, and A. R. Saltiel. 2007. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 117:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lumeng, C. N., S. M. Deyoung, J. L. Bodzin, and A. R. Saltiel. 2007. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes 56:16-23. [DOI] [PubMed] [Google Scholar]
- 45.Lumeng, C. N., S. M. Deyoung, and A. R. Saltiel. 2007. Macrophages block insulin action in adipocytes by altering expression of signaling and glucose transport proteins. Am. J. Physiol. Endocrinol. Metab. 292:E166-E174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lupien, M., J. Eeckhoute, C. A. Meyer, Q. Wang, Y. Zhang, W. Li, J. S. Carroll, X. S. Liu, and M. Brown. 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132:958-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marmorstein, R., and R. C. Trievel. 2009. Histone modifying enzymes: structures, mechanisms, and specificities. Biochim. Biophys. Acta 1789:58-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matys, V., E. Fricke, R. Geffers, E. Gossling, M. Haubrock, R. Hehl, K. Hornischer, D. Karas, A. E. Kel, O. V. Kel-Margoulis, D. U. Kloos, S. Land, B. Lewicki-Potapov, H. Michael, R. Munch, I. Reuter, S. Rotert, H. Saxel, M. Scheer, S. Thiele, and E. Wingender. 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31:374-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore, K. J., M. L. Fitzgerald, and M. W. Freeman. 2001. Peroxisome proliferator-activated receptors in macrophage biology: friend or foe? Curr. Opin. Lipidol. 12:519-527. [DOI] [PubMed] [Google Scholar]
- 50.Moore, K. J., E. D. Rosen, M. L. Fitzgerald, F. Randow, L. P. Andersson, D. Altshuler, D. S. Milstone, R. M. Mortensen, B. M. Spiegelman, and M. W. Freeman. 2001. The role of PPAR-gamma in macrophage differentiation and cholesterol uptake. Nat. Med. 7:41-47. [DOI] [PubMed] [Google Scholar]
- 51.Nencioni, A., F. Grunebach, A. Zobywlaski, C. Denzlinger, W. Brugger, and P. Brossart. 2002. Dendritic cell immunogenicity is regulated by peroxisome proliferator-activated receptor gamma. J. Immunol. 169:1228-1235. [DOI] [PubMed] [Google Scholar]
- 52.Nerlov, C., and T. Graf. 1998. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 12:2403-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nielsen, R., T. A. Pedersen, D. Hagenbeek, P. Moulos, R. Siersbaek, E. Megens, S. Denissov, M. Borgesen, K. J. Francoijs, S. Mandrup, and H. G. Stunnenberg. 2008. Genome-wide profiling of PPARγ:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 22:2953-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Odegaard, J. I., and A. Chawla. 2008. Mechanisms of macrophage activation in obesity-induced insulin resistance. Nat. Clin. Pract. Endocrinol. Metab. 4:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Odegaard, J. I., R. R. Ricardo-Gonzalez, M. H. Goforth, C. R. Morel, V. Subramanian, L. Mukundan, A. R. Eagle, D. Vats, F. Brombacher, A. W. Ferrante, and A. Chawla. 2007. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature 447:1116-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pascual, G., A. L. Fong, S. Ogawa, A. Gamliel, A. C. Li, V. Perissi, D. W. Rose, T. M. Willson, M. G. Rosenfeld, and C. K. Glass. 2005. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature 437:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U. S. A. 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ricote, M., and C. K. Glass. 2007. PPARs and molecular mechanisms of transrepression. Biochim. Biophys. Acta 1771:926-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ricote, M., J. Huang, L. Fajas, A. Li, J. Welch, J. Najib, J. L. Witztum, J. Auwerx, W. Palinski, and C. K. Glass. 1998. Expression of the peroxisome proliferator-activated receptor gamma (PPARgamma) in human atherosclerosis and regulation in macrophages by colony stimulating factors and oxidized low density lipoprotein. Proc. Natl. Acad. Sci. U. S. A. 95:7614-7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schnitzler, G. R. 2008. Control of nucleosome positions by DNA sequence and remodeling machines. Cell Biochem. Biophys. 51:67-80. [DOI] [PubMed] [Google Scholar]
- 61.Schoonjans, K., B. Staels, and J. Auwerx. 1996. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J. Lipid Res. 37:907-925. [PubMed] [Google Scholar]
- 62.Schupp, M., A. G. Cristancho, M. I. Lefterova, E. A. Hanniman, E. R. Briggs, D. J. Steger, M. Qatanani, J. C. Curtin, J. Schug, S. A. Ochsner, N. J. McKenna, and M. A. Lazar. 2009. Re-expression of GATA2 cooperates with peroxisome proliferator-activated receptor-gamma depletion to revert the adipocyte phenotype. J. Biol. Chem. 284:9458-9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shin, H., T. Liu, A. K. Manrai, and X. S. Liu. 2009. CEAS: cis-regulatory element annotation system. Bioinformatics 25:2605-2606. [DOI] [PubMed] [Google Scholar]
- 64.Simon, M. C., M. Olson, E. Scott, A. Hack, G. Su, and H. Singh. 1996. Terminal myeloid gene expression and differentiation requires the transcription factor PU.1. Curr. Top. Microbiol. Immunol. 211:113-119. [DOI] [PubMed] [Google Scholar]
- 65.Steger, D. J., M. I. Lefterova, L. Ying, A. J. Stonestrom, M. Schupp, D. Zhuo, A. L. Vakoc, J.-E. Kim, J. Chen, M. A. Lazar, G. A. Blobel, and C. R. Vakoc. 2008. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 28:2825-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thomas, P. D., M. J. Campbell, A. Kejariwal, H. Mi, B. Karlak, R. Daverman, K. Diemer, A. Muruganujan, and A. Narechania. 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13:2129-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas, P. D., A. Kejariwal, N. Guo, H. Mi, M. J. Campbell, A. Muruganujan, and B. Lazareva-Ulitsky. 2006. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 34:W645-W650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tontonoz, P., L. Nagy, J. G. Alvarez, V. A. Thomazy, and R. M. Evans. 1998. PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell 93:241-252. [DOI] [PubMed] [Google Scholar]
- 69.Tontonoz, P., and B. M. Spiegelman. 2008. Fat and beyond: the diverse biology of PPARgamma. Annu. Rev. Biochem. 77:289-312. [DOI] [PubMed] [Google Scholar]
- 70.Trojer, P., and D. Reinberg. 2007. Facultative heterochromatin: is there a distinctive molecular signature? Mol. Cell 28:1-13. [DOI] [PubMed] [Google Scholar]
- 71.Tuteja, G., P. White, J. Schug, and K. H. Kaestner. 2009. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 37:e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vidal-Puig, A., M. Jimenez-Linan, B. B. Lowell, A. Hamann, E. Hu, B. Spiegelman, J. S. Flier, and D. E. Moller. 1996. Regulation of PPAR gamma gene expression by nutrition and obesity in rodents. J. Clin. Invest. 97:2553-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Visel, A., M. J. Blow, Z. Li, T. Zhang, J. A. Akiyama, A. Holt, I. Plajzer-Frick, M. Shoukry, C. Wright, F. Chen, V. Afzal, B. Ren, E. M. Rubin, and L. A. Pennacchio. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457:854-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, Y., T. Liu, C. A. Meyer, J. Eeckhoute, D. S. Johnson, B. E. Bernstein, C. Nussbaum, R. M. Myers, M. Brown, W. Li, and X. S. Liu. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.