Abstract
Mutations in mitochondrial tRNA genes are associated with a wide spectrum of human diseases. In particular, the tRNALeu(UUR) A3243G mutation causes mitochondrial encephalomyopathy, lactic acidosis, and stroke-like symptoms (MELAS) and 2% of cases of type 2 diabetes. The primary defect in this mutation was an inefficient aminoacylation of the tRNALeu(UUR). In the present study, we have investigated the molecular mechanism of the A3243G mutation and whether the overexpression of human mitochondrial leucyl-tRNA synthetase (LARS2) in the cytoplasmic hybrid (cybrid) cells carrying the A3243G mutation corrects the mitochondrial dysfunctions. Human LARS2 localizes exclusively to mitochondria, and LARS2 is expressed ubiquitously but most abundantly in tissues with high metabolic rates. We showed that the alteration of aminoacylation tRNALeu(UUR) caused by the A3243G mutation led to mitochondrial translational defects and thereby reduced the aminoacylated efficiencies of tRNALeu(UUR) as well as tRNAAla and tRNAMet. We demonstrated that the transfer of human mitochondrial leucyl-tRNA synthetase into the cybrid cells carrying the A3243G mutation improved the efficiency of aminoacylation and stability of mitochondrial tRNAs and then increased the rates of mitochondrial translation and respiration, consequently correcting the mitochondrial dysfunction. These findings provide new insights into the molecular mechanism of maternally inherited diseases and a step toward therapeutic interventions for these disorders.
Mutations in mitochondrial DNA (mtDNA) have been associated with a wide spectrum of clinical abnormalities, including neuromuscular disorders, heart failure, diabetes, and hearing and visual loss (2, 20, 28, 47). More than 50% of these mtDNA mutations are located in the 22 mitochondrial tRNA genes, including tRNALeu(UUR) (2). In particular, the A3243G mutation in the tRNALeu(UUR) gene causes mitochondrial encephalomyopathy, lactic acidosis, and stroke-like symptoms (MELAS) (14). This mtDNA mutation is also one of the most important causes of maternally inherited diabetes and deafness (35, 37, 45). The primary defect in this mutation was an inefficient aminoacylation of the tRNALeu(UUR) (1, 5, 9, 24, 38, 50). This mutation also affected the processing of the longer mitochondrial RNA precursors (22, 24, 41) and the base posttranscriptional modification of the tRNALeu(UUR) (19, 26, 50). In cytoplasmic hybrids (cybrids) harboring the nearly homoplasmic A3243G mutation, the level of aminoacylated tRNALeu(UUR) was reduced approximately 70% to 75% (5, 9, 12, 50). The deficient aminoacylation of tRNALeu(UUR) mainly contributed to a shortage of tRNALeu(UUR) (20), thereby causing the reduced rate of mitochondrial protein synthesis and respiration defects (5, 21, 24, 51).
The formation of aminoacylated mitochondrial tRNALeu(UUR) was catalyzed by the human mitochondrial leucyl-tRNA synthetase (LARS2) belonging to class I of aminoacyl-tRNA synthetases (3, 27, 43). This evolutionarily conservative tRNA synthetase, encoded by the nuclear gene LARS2 at chromosome 3p21.3, was composed of 903 amino acids with a mitochondrial signal sequence (3, 18, 27). In the yeast Saccharomyces cerevisiae, mitochondrial defects due to the yeast counterpart of the human tRNALeu(UUR) A3243G mutation were rescued by overexpression of the translation elongation factor EFTu (11) and mitochondrial leucyl-tRNA synthetase (6). Thus, it is anticipated that the overexpression of human LARS2 in the cybrid cells carrying the A3243G mutation would improve the efficiency of aminoacylation of tRNALeu(UUR), enhance the stability of tRNA, and then increase the rates of mitochondrial translation and respiration, consequently correcting the mitochondrial dysfunction. To test this hypothesis, stable transfectants were constructed by transferring a human LARS2 cDNA into a cybrid cell line carrying the nearly homoplasmic A3243G mutation and an isogenic control cybrid cell line harboring the homoplasmic wild-type version of tRNALeu(UUR). These stable transfectants were analyzed for the aminoacylation capacity of tRNAs, the stability of the tRNALeu(UUR), and the rates of mitochondrial translation and respiration as well as RNA processing. Furthermore, human LARS2 was further characterized by examining the gene expression in different tissues and subcellular locations.
MATERIALS AND METHODS
Cell lines and culture conditions.
143B.TK− is a human osteosarcoma-derived cell line (ATCC CRL 8303). The 43B cybrid cell line carrying the nearly homoplasmic A3243G mutation in the tRNALeu(UUR) gene and the isogenic HSI cybrid cell line carrying the homoplasmic wild-type version of the tRNALeu(UUR) gene were isolated by transfer mitochondria from myoblasts of the same MELAS subject carrying the heteroplasmic A3243G mutation into human mtDNA-less ρo206 cells (51). The 143B.TK− cells and cybrid cells were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS). The stable transfectants were grown in DMEM supplemented with 10% FBS and 15 ng/ml of phleomycin (Zeocin).
Isolation of human LARS2 cDNA.
To construct the plasmid pLARS2 containing the entire coding region of LARS2 cDNA, reverse transcription-PCR (RT-PCR) was performed by using Taq DNA polymerase (Promega) and total RNA isolated from 143B cells as template, with the primers 5′-ATGGCTTCTGTTTGGCAGAG (nucleotides [nt] 186 to 205) and 5′-TGTCTTTTCCCTTCCCCTG (nt 2980 to 2998) (GenBank accession no. NM_015340) (3). The predominant PCR product was purified by agarose gel electrophoresis and subsequently cloned into a PCR 2.1-Topo vector (Invitrogen). Nucleotide sequencing was done using a Dye Terminator cycle sequencing kit (Perkin-Elmer) and an ABI Prism 3700 genetic analyzer.
Northern blot analysis of LARS2 expression.
A 12-lane human multiple tissue RNA blot (Clontech) containing 2 μg poly(A)+ RNA/lane was used for this study. An 1,124-bp LARS2 cDNA fragment corresponding to the nucleotide positions 1309 to 2432 (3) was random prime labeled with [32P]dATP and hybridized with the RNA blots according to the manufacturer's instructions. Membranes were then washed to a final stringency of 0.1× SSC (0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS) at 65°C for 40 min. As an internal control, the human RNA blots were stripped and rehybridized with [32P]dATP-labeled human actin β subunit cDNA as a control.
Subcellular localization of human LARS2.
The coding region of LARS2 cDNA lacking its natural stop codon was obtained by PCR using pLARS2 cDNA as the template. Primers 5′-CTCGAGATGGCTTCTGTTTGGCAGAG (nt 186 to 205) and 5′-GGA CCT TTG CAC CAG GAA GTT GAT GA (nt 2870 to 2891) were used for the PCR amplification. PCR products were digested with EcoRI and cloned into pBluescript II KS(+) (Promega). After sequence determination, the inserts were subcloned into pEGFP-N1 (Clontech) to generate the pEGFP-N1-LARS2. Resultant constructs were transfected into 143B cells using the SuperFect transfection reagent (Qiagen, Inc.) according to the manufacturer's protocol. Immunofluorescence analysis was performed as detailed elsewhere (29, 30, 49).
Construction of stable transfectants.
The insert of pLARS2 was subcloned into pcDNA3.1/Zeo(−) (Invitrogen). The resultant constructs or vector only were transfected into 43B and HSI cell lines using the SuperFect transfection reagent (Qiagen, Inc.) according to the manufacturer's protocol. The stable transfectants were isolated by culturing cells in DMEM supplemented with 15 ng/ml of phleomycin and 10% FBS for 4 weeks. The resultant clones were examined for the expression of LARS2 by Northern blot analysis. Briefly, equal amounts (5 μg) of total RNA extracted from cybrids and transfectants were fractionated by electrophoresis through a 1.8% agarose-formaldehyde gel, transferred onto a positively charged membrane (Roche), and initially hybridized with a LARS2 RNA probe, as described above. RNA blots were then stripped and rehybridized with a human actin β subunit cDNA (29). Probes were synthesized on the corresponding restriction enzyme-linearized plasmid using a digoxigenin (DIG) RNA labeling kit (Roche).
Mitochondrial DNA analysis.
Total DNA samples were isolated from the cultured cybrid and transfectant cell lines and from 143B.TK− cells by using the Puregene DNA isolation kit (Gentra Systems). The presence and amount of the A3243G mutation were examined as detailed elsewhere (34, 51). The quantification of DNA copy number was performed by slot blot hybridization, as detailed elsewhere (15).
Mitochondrial tRNA analysis.
Total mitochondrial RNA preparations were obtained by using the Totally RNA kit (Ambion) from mitochondria isolated from cybrid cell lines (∼4.0 × 107 cells), as previously described (25). Two micrograms of total mitochondrial RNA was electrophoresed through a 10% polyacrylamide-7 M urea gel in Tris-borate-EDTA (TBE) buffer (after the sample was heated at 65°C for 10 min) and then electroblotted onto a positively charged nylon membrane (Roche) for the hybridization analysis with specific oligodeoxynucleotide probes. For the detection of tRNALeu(UUR), tRNALeu(CUN), tRNALys, tRNAMet, tRNASer(UCN), and tRNAAla, the following nonradioactive DIG-labeled oligodeoxynucleotides specific for each RNA were used: tRNALeu(UUR), tRNALeu(CUN), tRNALys, tRNAMet, tRNASer(UCN), tRNAAla, and 5S RNA as detailed elsewhere (16, 17, 31, 33). DIG-labeled oligodeoxynucleotides were generated by using the DIG oligonucleotide tailing kit (Roche). The hybridization and quantification of density in each band were carried out as detailed elsewhere (16, 17).
Mitochondrial tRNA aminoacylation analysis.
Total mitochondrial RNAs were isolated under acid conditions. Two micrograms of total mitochondrial RNAs was electrophoresed at 4°C through an acid (pH 5.2) 10% polyacrylamide-7 M urea gel to separate the charged and uncharged tRNA as detailed elsewhere (10, 48). The gels were then electroblotted onto a positively charged nylon membrane (Roche) for the hybridization analysis with oligodeoxynucleotide probes as described above. Quantification of density in each band was performed as detailed previously (48).
Mitochondrial mRNA analysis.
Equal amounts (5 μg) of total mitochondrial RNA were fractionated by electrophoresis through a 1.8% agarose-formaldehyde gel, transferred onto a positively charged membrane (Roche), and hybridized with the RNA probes specific for ND1, ND4, ND6, 12S rRNA, and 16 rRNA, respectively (31).
Analysis of mitochondrial protein synthesis.
Pulse-labeling of the cell lines for 30 min with [35S]methionine-[35S]cysteine in methionine-free DMEM in the presence of emetine, electrophoretic analysis of the translation products, and quantification of radioactivity in the whole electrophoretic patterns or in individual well-resolved bands were carried out as detailed previously (4, 15, 16).
O2 consumption measurements.
Rates of O2 consumption in intact cells were determined with a YSI 5300 oxygraph (Yellow Springs Instruments) on samples of 5 × 106 cells in 1.5 ml of special DMEM lacking glucose and supplemented with 10% dialyzed FBS (23).
RESULTS
Expression analysis and subcellular location of human LARS2.
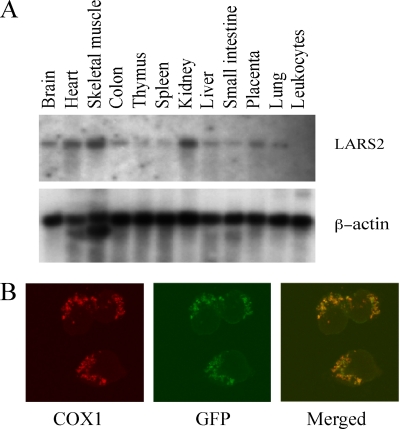
To investigate the tissue-specific expression, a LARS2 cDNA probe was hybridized with RNA blots of multiple human tissues. Figure 1A illustrates that LARS2 was ubiquitously expressed but that there were significant variations in the steady-state LARS2 mRNA levels among tissues. Compared to a high-level expression of LARS2 in the tissues with a high metabolic rate such as skeletal muscle, heart, and kidney, the expression levels of LARS2 in small intestine, spleen, thymus, and leukocytes were very low, while LARS2 was expressed moderately in the brain, colon, placenta, liver, spleen, and lung.
FIG. 1.
Subcellular localization and gene expression analysis of human LARS2. (A) Multiple-tissue Northern blot analysis of LARS2 expression. A human 12-lane multiple-tissue blot (Clontech) containing 2 μg of poly(A)+ RNA per lane was hybridized with a 32P-labeled fragment containing a human 1,124-bp LARS2 cDNA probe according to the manufacturer's protocol. The blot was then stripped and rehybridized with 32P-labeled human β-actin probe as a control. (B) Subcellular localization of human LARS2 in 143B cells. Cells were transiently transfected with a LARS2 cDNA fused with GFP or pEGFP. The fusion protein was visualized by indirect immunofluorescence using antibodies to mouse COX1 and to GFP. A merged image is shown on the right.
To determine the cellular localization of human LARS2, pEGFP-N1-LARS2 expressing the LARS2-green fluorescent protein (GFP) fusion protein was transfected into the 143B cell line. Figure 1B shows that the immunofluorescence pattern of transfected 143B cells was double labeled with a monoclonal antibody specific for the GFP and mouse monoclonal antibody to COXI, a subunit of cytochrome c oxidase (COX) complex in the mitochondrial inner membrane. A typical mitochondrial staining pattern was observed with both antibodies, and superimposition of two panels showed complete overlap of the two patterns, demonstrating that human LARS2 localizes exclusively to mitochondria. This suggested that this protein, like other nucleus-encoding mitochondrial proteins such as MTO1, GTPBP3, and TRMU (29, 30, 49), was synthesized on cytoplasmic ribosomes and was imported into and then functioned in the mitochondrion.
Construction of stable transfectants expressing the human LARS2.
A 2.8-kb human LARS2 cDNA expressed in a pcDNA3.1 vector or the vector only was transfected into the 43B cybrid cell line carrying the A3243G mutation and the isogenic HSI cell line with the wild-type version of tRNALeu(UUR) (5). The HSI cell line carrying the homoplasmic wild-type version of the tRNALeu(UUR) gene was isogenic with its respective mutant 43B cell line, as the wild-type and mutated mtDNA were derived from the same heteroplasmic patient cells (5, 51). These stable transfectants were isolated by culturing cells in DMEM supplemented with 15 ng/ml of phleomycin and 10% FBS for 4 weeks. The expression level of the LARS2 cDNA in resultant stable transfectants was examined by a Northern blot analysis, as shown in Fig. 2A. Here, the levels of exogenous LARS2 mRNA (2.8 kb) harboring only the coding region were more than 5-fold higher than those of endogenous LARS2 mRNA (4.2 kb) representing full-length mRNA in transfectants 43B-LARS2 and HSI-LARS2. To test if the transfer of LARS2 may change the proportion of mutated mtDNA molecules and mtDNA copy number in transfectant lines carrying the A3243G mutation, the presence and degree of the A3243G mutation in transfectants were examined by methods described elsewhere (34, 51). As shown in Fig. 2B, the presence and degree of the A3243G mutation in the transfectants 43B-LARS2, expressing the LARS2 cDNA, and 43B-V, carrying the vector only, were comparable with those of the parental cybrid cell line 43B carrying the A3243G mutation. Furthermore, an analysis of the mtDNA copy numbers of these 43B- and HSI-derived transfectants expressing the LARS2 cDNA and HSI-V carrying the vector as well as parental cybrids failed to reveal any significant difference in the mtDNA/nuclear ribosomal DNA (rDNA) ratios between these transfectants and parental cybrids. These four transfectants (43B-LARS2, 43B-V, HSI-LARS2, and HSI-V) were then used for further characterization.
FIG. 2.
(A) Construction of stable transfectants. Expression analysis of human LARS2 in the transfectants. Equal amounts (5 μg) of total mitochondrial RNA from mutant cell lines and control cell lines were electrophoresed through a 1.8% agarose-formaldehyde gel, transferred onto a positively charged membrane, and hybridized first with a DIG-labeled LARS2-specific RNA probe. After the blot was stripped, it was hybridized with a human β-actin probe as a control. 43B is a cybrid cell line carrying a nearly homoplasmic A3243G mutation, the HSI cell line is an isogenic wild-type cybrid line derived from the same individuals as those for 43B-V and HSI-V are transfectant lines with vector only, and 43B-LARS2 and HSI-LARS2 are transfectant lines expressing the human LARS2. The 4.2-kb band and 2.8-kb bands correspond to the length of endogenous and exogenous LARS2 mRNA, respectively. (B) Quantification of the A3243G mutation in the tRNALeu(UUR) gene in transfectants and parental cybrids. PCR products around the A3243G mutation were digested with ApaI and analyzed by electrophoresis in a 6% polyacrylamide gel stained with ethidium bromide. The A3243G mutation creates the site for restriction enzyme ApaI (14). Transfectants and their parental cybrid cell lines are indicated.
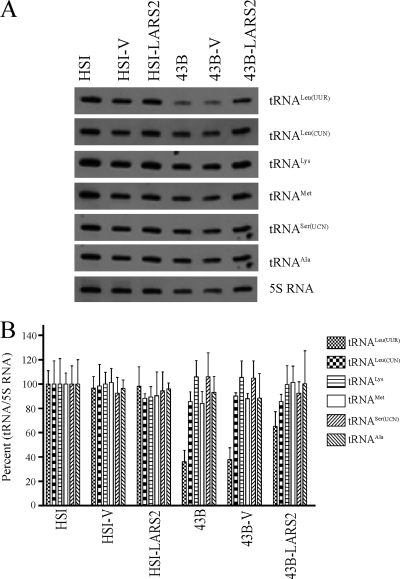
Aminoacylation capacity of mitochondrial tRNAs.
We first investigated the effects of the transfer of human LARS2 on the aminoacylation deficiency of tRNALeu(UUR) in transfectants 43B-LARS2 and 43B-V and their parental mutant cybrid cell line 43B as well as transfectants HSI-LARS2 and HSI-V and their parental wild-type cybrid cell line HSI in vivo. Total mitochondrial RNA was isolated from cell lines under acidic conditions to preserve the aminoacyl-tRNA linkage (46). The aminoacylated tRNAs were separated from nonaminoacylated tRNA species on acidic denaturing polyacrylamide-urea gels and then electroblotted and hybridized with specific probes for tRNALeu(UUR) as well as tRNALeu(CUN), tRNALys, tRNAMet, tRNASer(UCN), and tRNAAla, respectively. To further distinguish nonaminoacylated tRNA from aminoacylated tRNA, samples of mitochondrial tRNAs were deacylated by being heated for 10 min at 60°C at pH 8.3 and then run in parallel (10). Notably, the efficiencies of aminoacylated tRNAMet and tRNAAla, in addition to those of tRNALeu(UUR), as shown in Fig. 3, were markedly reduced in mutant 43B cells, relative to wild-type HSI cells. In particular, the proportions of aminoacylated tRNAs in the 43B cells were 24.9%, 61.8%, 66.3%, 38.7%, 71.2%, and 56.1% in the tRNALeu(UUR), tRNALeu(CUN), tRNALys, tRNAMet, tRNASer(UCN), and tRNAAla, respectively, while 64.1% of tRNALeu(UUR), 66% of tRNALeu(CUN), 67.4% of tRNALys, 73.1% of tRNAMet, 66.1% of tRNASer(UCN), and 73% of tRNAAla were aminoacylated in the isogenic HSI cells. In contrast, the aminoacylated levels of these six tRNAs in the 43B-V cells and HSI-V cells carrying only the vector as well as HSI-LARS2 cells expressing the LARS2 cDNA only were comparable with those of 43B cells and HSI cells, respectively. Strikingly, the expression of LARS2 in the 43B cybrids led to an ∼100% increase in the efficiency of aminoacylation of tRNALeu(UUR) (from 24.9% to 50.5%), but the level was still below that of controls (64.1%). Furthermore, the efficiencies of aminoacylation of tRNALeu(CUN), tRNALys, tRNAMet, tRNASer(UCN), and tRNAAla in the 43B-LARS2 cell line expressing LARS2 were 106.6%, 111.6%, 133.33%, 98.5%, and 118% of those in parental 43B cell line, respectively. These suggested that the deficiencies of aminoacylation of tRNAs were partially restored by the overexpression of human LARS2 in the 43B cells.
FIG. 3.
In vivo aminoacylation assays. (A) Equal amounts (2 μg) of total mitochondrial RNA purified from six cell lines (the same as in Fig. 2A) under acid conditions were electrophoresed at 4°C through an acid (pH 5.2) 10% polyacrylamide-7 M urea gel, electroblotted, and hybridized with a DIG-labeled oligonucleotide probe specific for the mitochondrial tRNALeu(UUR). The blots were then stripped and rehybridized with DIG-labeled tRNALeu(UUR), tRNALeu(CUN), tRNALys, tRNAMet, tRNASer(UCN), and tRNAAla, respectively. Samples were deacylated (DA) by being heated for 10 min at 60°C at pH 8.3. 43B is a cybrid cell line carrying a nearly homplasmic A3243G mutation, the HSI cell line is an isogenic wild-type cybrid line derived from the same individuals as those for 43B-V and HSI-V are transfectant lines with vector only, and 43B-LARS2 and HSI-LARS2 are transfectant lines expressing the human LARS2. (B) In vivo aminoacylated proportions of tRNALeu(UUR), tRNALeu(CUN), tRNALys, tRNAMet, tRNASer(UCN), and tRNAAla in the mutant and controls. The calculations were based on three independent determinations. The error bars indicate two standard errors of the means.
The steady-state level of mitochondrial tRNA.
To evaluate if the overexpression of LARS2 increased the stability of tRNALeu(UUR) in transfectants carrying the A3243G mutation, we determined the steady-state level of the tRNALeu(UUR) by isolating total mitochondrial RNAs from cell lines, separating them on a 10% polyacrylamide-7 M urea gel, and electroblotting and hybridizing with a nonradioactive DIG-labeled oligodeoxynucleotide probe specific for tRNALeu(UUR). After the blots were stripped, the DIG-labeled oligodeoxynucleotide probes, including tRNALeu(CUN), tRNALys, and tRNAMet as representatives of the whole H-strand transcription unit and tRNASer(UCN) and tRNAAla derived from the L-strand transcription unit (16) as well as a nucleus-encoded mitochondrial 5S RNA (36), were hybridized with the same blots for normalization purposes. For comparison, the average level of each tRNA in control or mutant cell lines was normalized to the average levels in the same cell line for reference 5S RNA. As shown in Fig. 4, the average steady-state levels of tRNALeu(UUR) in the 43B, 43B-V, 43-LARS2, HSI-V, and HSI-LARS2 cell lines were 36%, 38%, 65%, 97%, and 98% of the isogenic HSI cybrid cell line after normalization to 5S RNA, respectively. However, the average steady-state levels of tRNASer(UCN), tRNAMet, tRNALeu(CUN), and tRNALys and tRNAAla in 43B, 43B-V, 43-LARS2, HSI-V, and HSI-LARS2 cell lines were comparable with those of the wild-type HSI cybrid cell line after normalization to 5S RNA.
FIG. 4.
Assays for the steady-state levels of mitochondrial tRNALeu(CUN). (A) Northern blot analysis of mitochondrial tRNA. Equal amounts (2 μg) of total mitochondrial RNA samples from the various cell lines were electrophoresed through a denaturing polyacrylamide gel, electroblotted, and hybridized with DIG-labeled oligonucleotide probes specific for the tRNALeu(UUR), tRNALeu(CUN), tRNALys, tRNAMet, tRNASer(UCN), and tRNAAla as well as 5S RNA, respectively. (B) Quantification of the mitochondrial tRNA levels. Average relative tRNALeu(UUR), tRNALeu(CUN), tRNALys, tRNAMet, tRNASer(UCN), and tRNAAla contents per cell were normalized to the average content per cell of 5S RNA in control cell lines and in mutant cell lines, respectively. The values for the latter are expressed as percentages of the average values for the control cell lines. The calculations were based on three independent determinations of tRNALeu(UUR) content in each cell line and three determinations of the content of each reference RNA marker in each cell line. The error bars indicate two standard errors of the means.
Mitochondrial RNA processing.
We then tested if the transfer of human LARS2 restores the defects in mitochondrial RNA processing in the 43B cell line carrying the A3243G mutation. For this purpose, RNA transfer hybridization experiments were performed with total mitochondrial RNA from transfectants and parental cybrids using the nonradioactive DIG-labeled ND1 RNA probe (16, 30). After the blots were stripped, a set of DIG-labeled RNA probes (ND4, ND6, 12S rRNA, and 16S rRNA) were rehybridized with the same blots for normalization purposes. As shown in Fig. 5, RNA19 comprising the 12S rRNA plus tRNAVal plus 16S rRNA plus tRNALeu(UUR) plus ND1 was accumulated in the 43B cell line, confirming the defects in mitochondrial RNA processing caused by the A3243G mutation (22, 24, 41). Interestingly, the reduction in the levels of RNA19, ND1, ND4, and ND6 mRNA was observed in the 43B-LARS2 transfectant, while the levels of 16S rRNA plus tRNALue(UUR) plus ND1 as well as 12S rRNA appeared to be unchanged in this transfectant compared with those of the parental 43B cell line.
FIG. 5.
Expression analysis of mitochondrial RNA. Equal amounts (5 μg) of total mitochondrial RNA from mutant cell lines and control cell lines were electrophoresed through a 1.8% agarose-formaldehyde gel, transferred onto a positively charged membrane, and hybridized first with a DIG-labeled ND1-specific RNA probe. After the blot was stripped, it was hybridized with DIG-labeled RNA probes ND4, ND6, 12S rRNA, and 16S rRNA, respectively. RNA19 consists of 12S rRNA plus tRNAVal plus 16S rRNA plus tRNALeu(UUR) plus ND1 (22, 24).
Mitochondrial protein synthesis.
To assess if the overexpression of LARS2 enhanced the rate of overall mitochondrial protein synthesis in the transfectant carrying the A3243G mutation, samples of cultures of transfectants as well as their parental cybrid cell lines were labeled with short [35S]methionine pulses in the presence of emetine to inhibit the cytosolic protein synthesis (4). As shown in Fig. 6A, the patterns of the mtDNA-encoded polypeptides of parental cybrid cell lines and their transfectants were qualitatively identical, in terms of electrophoretic mobility of the various polypeptides, to those of the isogenic control cybrid cell line HSI. However, the 43B cells exhibited a decrease in the total rate of labeling of mitochondrial translation products, relative to the control cybrid. In contrast, the overexpression of LARS2 apparently increased the total rate of labeling of mitochondrial translation products in the 43B-LARS2 transfectant line. Figure 6B illustrates a quantification of the results of three labeling experiments and three electrophoretic runs, which was carried out by Image-Quant program analysis of appropriate exposures of the fluorograms and normalization to the data obtained for the HSI sample included in each gel. The overall rates of labeling of the mitochondrial translation products in the 43B cell line and 43B-V and 43B-LARS2 transfectants were 32.3%, 31.1%, and 48.3%, respectively, of the mean value measured in the control HSI cell line. This translated to a 51% increase of the rate of mitochondrial translation in 43B-LARS2 cells, compared to the parental cybrid cell line 43B. In contrast, the rates of mitochondrial translation of HSI-LARS2 and HSI-V transfectants were comparable with that of the parental HSI cell line.
FIG. 6.
Analysis of rates of mitochondrial protein labeling. (A) Electrophoretic patterns of the mitochondrial translation products of different cell lines labeled for 30 min with [35S]methionine in the presence of 100 μg of emetine per ml. Samples containing equal amounts of protein (20 μg) were run in SDS-polyacrylamide gradient gels. COI, COII, and COIII, subunits I, II, and III of cytochrome c oxidase, respectively; ND1, ND2, ND3, ND4, ND4L, ND5, and ND6, subunits 1, 2, 3, 4, 4L, 5, and 6 of the respiratory-chain NADH dehydrogenase, respectively; A6 and A8, subunits 6 and 8 of the H+-ATPase, respectively; CYTB, apocytochrome b. (B) Quantification of the rates of labeling of the mitochondrial translation products. The rates of mitochondrial protein labeling in three mutant cell lines and three control cell lines, determined as described elsewhere (16, 17), are expressed as percentages of the value for HSI in each gel, with error bars indicating standard errors of the means. Three independent labeling experiments and three electrophoretic analyses of each labeled preparation were carried out on each cell line.
Analysis of respiration.
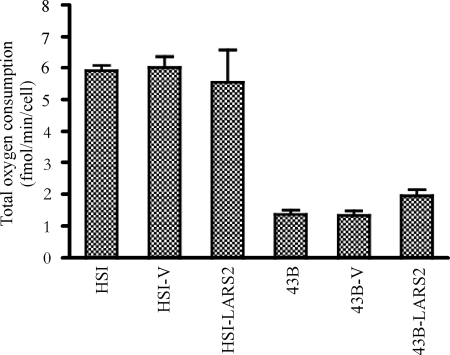
To examine if the overexpression of LARS2 rescued the respiratory deficiencies caused by the A3243G mutation, the endogenous respiration rates of transfectants and their parental cybrid cell lines were measured by determining the O2 consumption rate in intact cells, as described previously (23). As can be seen in Fig. 7, the rate of total O2 consumption in 43B cell lines was 24%, relative to the mean value measured in the control HSI cell line. However, the rate of total O2 consumption in the 43B-LARS2 transfectant cell line expressing LARS2 was 142% of that of the parental 43B cell line, while the rate of total oxygen consumption of HSI-LARS2 was 92% of that of the parental HSI cell line. However, rates for 43B-V and HSI-V transfectants were comparable with those of parental 43B and HSI cell lines. These data demonstrated that the increase of the rate of mitochondrial translation products by overexpression of LARS2 indeed enhanced the rate of respiration in the transfectant 43B-LARS2 carrying the A3243G mutation.
FIG. 7.
Respiration assay. Average rates of total O2 consumption per cell were measured in different cell lines. Four to eight determinations were carried out for each cell line, with error bars indicating standard errors of the means.
DISCUSSION
In the present study, we have further investigated the molecular pathogenesis of the MELAS-associated tRNALeu(UUR) A3243G mutation and whether the overexpression of human LARS2 in the cybrid cells carrying the A3243G mutation corrects the mitochondrial dysfunctions. The A3243G mutation changes the A to G at conventional position 14 (A14) of the mitochondrial tRNALeu(UUR) (13) and thus alters the structure and function of tRNALeu(UUR). In particular, the tRNALeu(UUR) carrying the A3243G mutation was less charged to a lesser extent by the leucyl-tRNA synthetase, thereby altering aminoacylation. Alteration in aminoacylation of tRNALeu(UUR) may result in stalling of ribosomes at leucine codons, mistranslation of UUR codons, or translational shifting (1, 21, 42). This was evidenced by the disproportionate decrease of translation in some polypeptides (42). The idea that an inefficient aminoacylation of tRNALeu(UUR) is the primary defect in MELAS was further supported by the recent isolation of a suppressor mutation in the anticodon of the mitochondrial tRNALeu(CUN) gene in an A3243G lung carcinoma cell line (8, 9). Furthermore, the A3243G mutation disturbed the processing of the tRNALeu(UUR) precursor (22, 24, 41) as well as the posttranscriptional modification of uridine at the first position of the anticodon in the tRNALeu(UUR) (19, 26, 50). The mutant tRNALeu(UUR) may be metabolically less stable and subject to turnover, thereby lowering the steady-state level of tRNALeu(UUR). As a result, a failure in tRNA metabolism is responsible for defective mitochondrial protein synthesis and an impaired oxidative phosphorylation. In this investigation, it is interesting that the lowered efficiencies of aminoacylated mitochondrial tRNAMet and tRNAAla were also observed in the cybrid cell line carrying the A3243G mutation. It is likely that the reduced ratio of charged to uncharged tRNALeu(UUR) may mediate mitochondrial tRNA metabolism, thereby reducing the aminoacylated efficiencies of other mitochondrial tRNAs. Indeed, the mutation in TRMU, encoding a highly conserved 5-methylaminomethyl-2-thiouridylate-methyltransferase responsible for the biosynthesis of 5-taurinomethyl-2-thiouridine (τm5s2U) of mitochondrial tRNALys, tRNAGlu, and tRNAGln in the wobble position, not only lowered the steady-state level of tRNALys, tRNAGlu, and tRNAGln but also reduced the steady-state level of other tRNAs such as tRNALeu(UUR), tRNASer(UCN), tRNAMet, and tRNAHis (17).
In the present investigation, we have shown that the overexpression of human LARS2 in the cybrid cells carrying the A3243G mutation corrected the mitochondrial dysfunctions. In particular, the overexpression of LARS2 in the 43B cybrid cell line carrying the A3243G mutation raised the percentage of aminoacylated tRNALeu(UUR) from 24.9% to 50.5%. Despite ∼100% increasing efficiency of charged tRNALeu(UUR), the partial function of tRNALeu(UUR) in the 43B cybrid cell line would cause the level of aminoacylated tRNALeu(UUR) in 43B cell lines expressing LARS2 to be below that of control cell line HSI. The fact that overexpression of LARS2 did not elevate the level of aminoacylated tRNALeu(UUR) in the wild-type cybrid cell line HSI indicated that 60% of aminoacylated tRNALeu(UUR) in cells appeared to be the maximum threshold level to maintain the normal function. On the other hand, the improvement of aminoacylation of tRNALeu(UUR) likely modulated the uncharged/charged ratios of other mitochondrial tRNAs, thereby enhancing the efficiency of aminoacylation of other tRNAs, including tRNAMet and tRNAAla. Furthermore, the high-level expression of LARS2 increased the steady-state level of tRNALeu(UUR) in the 43B cells from 36.1% to 65.2% of the level in wild-type cybrid cells. The overexpression of LARS2 may stabilize metabolically unstably mutant tRNALeu(UUR) by increasing the proportion of aminoacylated tRNA in the cells, as in the cases of overexpression of yeast LARS2 in the mitochondrial tRNALeu mutant cells (6) and human VARS2L in a cybrid cell line carrying the tRNAVal C1624T mutation (40). In addition, the modified human FARS2 displayed significantly improved aminoacylation efficiency for the mitochondrial tRNAPhe G611A mutation (32). Alternatively, an increasing LARS2 expression in 43B cells may facilitate the correct folding and stabilization of the hypomodified tRNALeu(UUR) (39). Moreover, an improvement of aminoacylation of tRNALeu(UUR) by high levels of LARS2 appeared to lead to more efficient processing of the longer RNA precursors (22, 24), evidenced by the fact that a reduction in the RNA19 was observed in the 43B-LARS2 cell line compared with the parental 43B cell line.
In the present study, we have shown that increasing expression of LARS2 in the mutant 43B cybrid cell line raised 50% of rates of mitochondrial translation in the mutant 43B cybrid cell line but not in the wild-type HSI cybrid cell line. The improved translation by overexpression of LARS2 could be the result of more efficient charging of tRNALeu(UUR) and more efficient processing of the longer RNA precursors in mutant cells. The restoration of mitochondrial translation by overexpression of LARS2 appeared to be more efficient than that by overexpression of EFTu and EFG2 in cybrid cells carrying the A3243G mutation (42). In fact, EFTu mediates the entry of the aminoacylated tRNA into the A site of the ribosome and then promotes the addition of amino acids to the polypeptide chain. Thus, the overexpression of EFTu may enhance the overall mitochondrial translational efficiency, while overexpression of LARS2 improves the efficiency of aminoacylation of tRNALeu(UUR) and then increases the efficiency and fidelity of mitochondrial translation. Subsequently, the increasing expression of LARS2 in the mutant 43B cell line improved the respiration capacity even though it still was far short of the respiration rate in the wild-type HSI cybrid cell line, as in the case of overexpression of PGC-1α/β in the cybrid cell line carrying the A3243G mutation (44). Indeed, the degree of increase in the rate of oxygen consumption in 43B-LARS2 cells correlated well with the increase in the rate of mitochondrial protein synthesis, suggesting that the improvement of mitochondrial translation was responsible for the increasing oxidative phosphorylation. However, this result was not in good agreement with the observation that the full recovery of respiration chain function occurred by means of overexpression of LARS2 in a mutant cybrid cell line carrying the A3243G mutation (39). This discrepancy may be attributed to different nuclear backgrounds, as in the case of cell lines carrying other mtDNA mutations (7, 15). In conclusion, we demonstrated that the overexpression of human mitochondrial leucyl-tRNA synthetase corrected the mitochondrial dysfunction caused by the MELAS-associated tRNALeu(UUR) A3243G mutation. These findings will provide new insights into the pathophysiology of maternally inherited diseases and a step toward therapeutic interventions for these disorders.
Acknowledgments
This paper is dedicated to the memory of Giuseppe Attardi, a pioneer in mitochondrial genetics and biomedicine.
We are grateful to Anne Chomyn (California Institute of Technology) for cell lines and Linda Spremulli (University of North Carolina) for the human LARS2 cDNA plasmid.
This work was supported by Public Health Service grants RO1NS44015 from the National Institute of Neurological Disorders and Stroke and RO1DC05230 and RO1DC07696 from the National Institute on Deafness and Other Communication Disorders to M.X.G.
Footnotes
Published ahead of print on 1 March 2010.
REFERENCES
- 1.Börner, G. V., M. Zeviani, V. Tiranti, F. Carrara, S. Hoffmann, K. D. Gerbitz, H. Lochmüller, D. Pongratz, T. Klopstock, A. Melberg, E. Holme, and S. Pääbo. 2000. Decreased aminoacylation of mutant tRNAs in MELAS but not in MERRF patients. Hum. Mol. Genet. 9:467-475. [DOI] [PubMed] [Google Scholar]
- 2.Brandon, M. C., M. T. Lott, K. C. Nguyen, S. Spolim, S. B. Navathe, P. Baldi, and D. C. Wallace. 2005. MITOMAP: a human mitochondrial genome database—2004 update. Nucleic Acids Res. 33:D611-D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullard, J. M., Y. C. Cai, and L. L. Spremulli. 2000. Expression and characterization of the human mitochondrial leucyl-tRNA synthetase. Biochim. Biophys. Acta 1490:245-258. [DOI] [PubMed] [Google Scholar]
- 4.Chomyn, A. 1996. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 264:197-211. [DOI] [PubMed] [Google Scholar]
- 5.Chomyn, A., J. A. Enriquez, V. Micol, P. Fernandez-Silva, and G. Attardi. 2000. The mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episode syndrome-associated human mitochondrial tRNALeu(UUR) mutation causes aminoacylation deficiency and concomitant reduced association of mRNA with ribosomes. J. Biol. Chem. 275:19198-19209. [DOI] [PubMed] [Google Scholar]
- 6.De Luca, C., C. Besagni, L. Frontali, M. Bolotin-Fukuhara, and S. Francisci. 2006. Mutations in yeast mt tRNAs: specific and general suppression by nuclear encoded tRNA interactors. Gene 377:169-176. [DOI] [PubMed] [Google Scholar]
- 7.Deng, J. H., Y. Li, J. S. Park, J. Wu, P. Hu, J. Lechleiter, and Y. Bai. 2006. Nuclear suppression of mitochondrial defects in cells without the ND6 subunit. Mol. Cell. Biol. 26:1077-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunbar, D. R., P. A. Moonie, M. Zeviani, and I. J. Holt. 1996. Complex I deficiency is associated with 3243G:C mitochondrial DNA in osteosarcoma cell cybrids. Hum. Mol. Genet. 5:123-129. [DOI] [PubMed] [Google Scholar]
- 9.El Meziane, A., S. K. Lehtinen, N. Hance, L. G. Nijtmans, D. Dunbar, I. J. Holt, and H. T. Jacobs. 1998. A tRNA suppressor mutation in human mitochondria. Nat. Genet. 18:350-353. [DOI] [PubMed] [Google Scholar]
- 10.Enríquez, J. A., and G. Attardi. 1996. Analysis of aminoacylation of human mitochondrial tRNAs. Methods Enzymol. 264:183-196. [DOI] [PubMed] [Google Scholar]
- 11.Feuermann, M., S. Francisci, T. Rinaldi, C. De Luca, H. Rohou, L. Frontali, and M. Bolotin-Fukuhara. 2003. The yeast counterparts of human ‘MELAS’ mutations cause mitochondrial dysfunction that can be rescued by overexpression of the mitochondrial translation factor EF-Tu. EMBO Rep. 4:53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flierl, A., H. Reichmann, and P. Seibel. 1997. Pathophysiology of the MELAS 3243 transition mutation. J. Biol. Chem. 272:27189-27196. [DOI] [PubMed] [Google Scholar]
- 13.Florentz, C., B. Sohm, P. Tryoen-Toth, J. Putz, and M. Sissler. 2003. Human mitochondrial tRNAs in health and disease. Cell. Mol. Life Sci. 60:1356-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goto, Y., I. Nonaka, and S. Horai. 1990. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348:651-653. [DOI] [PubMed] [Google Scholar]
- 15.Guan, M. X., N. Fischel-Ghodsian, and G. Attardi. 1996. Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum. Mol. Genet. 5:963-971. [DOI] [PubMed] [Google Scholar]
- 16.Guan, M. X., J. A. Enriquez, N. Fischel-Ghodsian, R. S. Puranam, C. P. Lin, M. A. Maw, and G. Attardi. 1998. The deafness-associated mitochondrial DNA mutation at position 7445, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase subunit ND6 gene expression. Mol. Cell. Biol. 18:5868-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan, M. X., Q. Yan, X. Li, Y. Bykhovskaya, J. Gallo-Teran, P. Hajek, N. Umeda, H. Zhao, G. Garrido, E. Mengesha, T. Suzuki, I. del Castillo, J. L. Peters, R. Li, Y. Qian, X. Wang, E. Ballana, M. Shohat, J. Lu, X. Estivill, K. Watanabe, and N. Fischel-Ghodsian. 2006. Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am. J. Hum. Genet. 79:291-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartl, F. U., and W. Neupert. 1990. Protein sorting to mitochondria: evolutionary conservations of folding and assembly. Science 247:930-938. [DOI] [PubMed] [Google Scholar]
- 19.Helm, M., C. Florentz, A. Chomyn, and G. Attardi. 1999. Search for differences in post-transcriptional modification patterns of mitochondrial DNA-encoded wild-type and mutant human tRNALys and tRNALeu(UUR). Nucleic Acids Res. 27:756-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs, H. 2003. Disorders of mitochondrial protein synthesis. Hum. Mol. Genet. 12:R293-R301. [DOI] [PubMed] [Google Scholar]
- 21.Janssen, G. M., P. J. Hensbergen, F. J. van Bussel, C. I. Balog, J. A. Maassen, A. M. Deelder, and A. K. Raap. 2007. The A3243G tRNALeu(UUR) mutation induces mitochondrial dysfunction and variable disease expression without dominant negative acting translational defects in complex IV subunits at UUR codons. Hum. Mol. Genet. 16:2472-2481. [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann, P., Y. Koga, S. Shanske, M. Hirano, S. DiMauro, M. P. King, and E. A. Schon. 1996. Mitochondrial DNA and RNA processing in MELAS. Ann. Neurol. 40:172-180. [DOI] [PubMed] [Google Scholar]
- 23.King, M. P., and G. Attardi. 1989. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science 246:500-503. [DOI] [PubMed] [Google Scholar]
- 24.King, M. P., Y. Koga, M. Davidson, and E. A. Schon. 1992. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNALeu(UUR) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Mol. Cell. Biol. 12:480-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King, M. P., and G. Attardi. 1993. Post-transcriptional regulation of the steady-state levels of mitochondrial tRNAs in HeLa cells. J. Biol. Chem. 268:10228-10237. [PubMed] [Google Scholar]
- 26.Kirino, Y., T. Yasukawa, S. Ohta, S. Akira, K. Ishihara, K. Watanabe, and T. Suzuki. 2004. Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease Proc. Natl. Acad. Sci. U. S. A. 101:15070-15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiss, H., D. Kedra, Y. Yang, M. Kost-Alimova, C. Kiss, K. P. O'Brian, I. Fransson, G. Klein, S. Imreh, and J. P. Dumanski. 1999. A novel gene containing LIM domains (LIMD1) is located within the common eliminated region 1 (C3CER1) in 3p21.3. Hum. Genet. 105:552-559. [DOI] [PubMed] [Google Scholar]
- 28.Larsson, N. G., and D. A. Clayton. 1995. Molecular genetic aspects of human mitochondrial disorders. Annu. Rev. Genet. 29:151-178. [DOI] [PubMed] [Google Scholar]
- 29.Li, X., and M.-X. Guan. 2002. A human mitochondrial GTP binding protein related to tRNA modification may modulate phenotypic expression of the deafness-associated mitochondrial 12S rRNA mutation. Mol. Cell. Biol. 22:7701-7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, X., R. Li, X. Lin, and M.-X. Guan. 2002. Isolation and characterization of the putative nuclear modifier gene MTO1 involved in the pathogenesis of deafness-associated mitochondrial 12 S rRNA A1555G mutation. J. Biol. Chem. 277:27256-27264. [DOI] [PubMed] [Google Scholar]
- 31.Li, X., N. Fischel-Ghodsian, F. Schwartz, Q. Yan, R. A. Friedman, and M. X. Guan. 2004. Biochemical characterization of the mitochondrial tRNASer(UCN) T7511C mutation associated with nonsyndromic deafness. Nucleic Acids Res. 32:867-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ling, J., H. Roy, D. Qin, M. A. Rubio, J. D. Alfonzo, K. Fredrick, and M. Ibba. 2007. Pathogenic mechanism of a human mitochondrial tRNAPhe mutation associated with myoclonic epilepsy with ragged red fibers syndrome. Proc. Natl. Acad. Sci. U. S. A. 104:15299-15304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, Y., R. Li, Z. Li, X. J. Wang, L. Yang, S. Wang, and M. X. Guan. 2009. Mitochondrial transfer RNAMet 4435A>G mutation is associated with maternally inherited hypertension in a Chinese pedigree. Hypertension 53:1083-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, J., D. Wang, R. Li, W. Li, J. Ji, J. Zhao, W. Ye, L. Yang, Y. Qian, Y. Zhu, and M. X. Guan. 2006. Maternally transmitted diabetes mellitus associated with the mitochondrial tRNALeu(UUR) A3243G mutation in a four-generation Han Chinese family. Biochem. Biophy. Res. Commun. 348:115-119. [DOI] [PubMed] [Google Scholar]
- 35.Maassen, J. A., L. M. 't Hart, E. van Essen, R. J. Heine, G. Nijpels, R. S. Jahangir Tafrechi, A. K. Raap, G. M. C. Janssen, and H. H. Lemkes. 2004. Mitochondrial diabetes: molecular mechanisms and clinical presentation. Diabetes 53(Suppl. 1):S103-S109. [DOI] [PubMed] [Google Scholar]
- 36.Magalhaes, P. J., A. L. Andreu, and E. A. Schon. 1998. Evidence for the presence of 5S rRNA in mammalian mitochondria. Mol. Biol. Cell 9:2375-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohkubo, K., A. Yamano, M. Nagashima, Y. Mori, K. Anzai, Y. Akehi, R. Nomiyama, T. Asano, A. Urae, and J. Ono. 2001. Mitochondrial gene mutations in the tRNALeu(UUR) region and diabetes: prevalence and clinical phenotypes in Japan. Clin. Chem. 47:1641-1648. [PubMed] [Google Scholar]
- 38.Park, H., E. Davidson, and M. P. King. 2003. The pathogenic A3243G mutation in human mitochondrial tRNALeu(UUR) decreases the efficiency of aminoacylation. Biochemistry 42:958-964. [DOI] [PubMed] [Google Scholar]
- 39.Park, H., E. Davidson, and M. P. King. 2008. Overexpressed mitochondrial leucyl-tRNA synthetase suppresses the A3243G mutation in the mitochondrial tRNALeu(UUR) gene. RNA 14:2407-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rorbach, J., A. A. Yusoff, H. Tuppen, D. P. Abg-Kamaludin, Z. M. Chrzanowska-Lightowlers, R. W. Taylor, D. M. Turnbull, R. McFarland, and R. N. Lightowlers. 2008. Overexpression of human mitochondrial valyl tRNA synthetase can partially restore levels of cognate mt-tRNAVal carrying the pathogenic C25U mutation. Nucleic Acids Res. 36:3065-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossmanith, W., and R. Karwan. 1998. Impairment of tRNA processing by point mutations in mitochondrial tRNALeu(UUR) associated with mitochondrial diseases. FEBS Lett. 433:269-274. [DOI] [PubMed] [Google Scholar]
- 42.Sasarman, F., H. Antonicka, and E. A. Shoubridge. 2008. The A3243G tRNALeu(UUR) MELAS mutation causes amino acid misincorporation and a combined respiratory chain assembly defect partially suppressed by overexpression of EFTu and EFG2. Hum. Mol. Genet. 17:3697-3707. [DOI] [PubMed] [Google Scholar]
- 43.Sohm, B., M. Sissler, H. Park, M. P. King, and C. Florentz. 2004. Recognition of human mitochondrial tRNALeu(UUR) by its cognate leucyl-tRNA synthetase. J. Mol. Biol. 339:17-29. [DOI] [PubMed] [Google Scholar]
- 44.Srivastava, S., F. Diaz, L. Iommarini, K. Aure, A. Lombes, and C. T. Moraes. 2009. PGC-1alpha/beta induced expression partially compensates for respiratory chain defects in cells from patients with mitochondrial disorders. Hum. Mol. Genet. 18:1805-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Ouweland, J. M. W., H. H. P. J. Lemkes, W. Ruitenbeek, L. A. Sandkuijl, M. F. de Vijlder, P. A. A. Struijvenberg, J. J. P. van de Kamp, and J. A. Maassen. 1992. Mutation in mitochondrial tRNALeu(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat. Genet. 1:368-371. [DOI] [PubMed] [Google Scholar]
- 46.Varshney, U., C. P. Lee, and U. L. RajBhandary. 1991. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 1266:24712-24718. [PubMed] [Google Scholar]
- 47.Wallace, D. C. 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39:359-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, X., J. Lu, Y. Zhu, A. Yang, L. Yang, R. Li, B. Chen, Y. Qian, X. Tang, J. Wang, X. Zhang, and M. X. Guan. 2008. Mitochondrial tRNAThr G15927A mutation may modulate the phenotypic manifestation of ototoxic 12S rRNA A1555G mutation in four Chinese families. Pharmacogenet. Genom. 18:1059-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan, Q., X. Li, G. Faye, and M. X. Guan. 2005. Mutations in MTO2 related to tRNA modification impair mitochondrial gene expression and protein synthesis in the presence of a paromomycin resistance mutation in mitochondrial 15 S rRNA. J. Biol. Chem. 280:29151-29157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasukawa, T., T. Suzuki, T. Suzuki, T. Ueda, S. Ohta, and K. Watanabe. 2000. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAsLeu(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J. Biol. Chem. 275:4251-4257. [DOI] [PubMed] [Google Scholar]
- 51.Yoneda, M., A. Chomyn, A. Martinuzzi, O. Hurko, and G. Attardi. 1992. Marked replicative advantage of human mtDNA carrying a point mutation that causes the MELAS encephalomyelopathy. Proc. Natl. Acad. Sci. U. S. A. 89:11164-11168. [DOI] [PMC free article] [PubMed] [Google Scholar]