Abstract
Monocytopoiesis involves the stepwise differentiation in the bone marrow (BM) of common myeloid precursors (CMPs) to monocytes. The basic helix-loop-helix transcription factor TAL1/SCL plays a critical role in other hematopoietic lineages, and while it had been reported to be expressed by BM-derived macrophages, its role in monocytopoiesis had not been elucidated. Using cell explant models of monocyte/macrophage (MM) differentiation, one originating with CMPs and the other from more committed precursors, we characterized the phenotypic and molecular consequences of inactivation of Tal1 expression ex vivo. While Tal1 knockout had minimal effects on cell survival and slightly accelerated terminal differentiation, it profoundly inhibited cell proliferation and decreased entry into and traversal of the G1 and S phases. In conjunction, steady-state levels of p16(Ink4a) mRNA were increased and those of Gata2 mRNA decreased. Chromatin immunoprecipitation analysis demonstrated the association of Tal1 and E47, one of its E protein DNA-binding partners, with an E box-GATA sequence element in intron 4 of the Gata2 gene and with three E boxes upstream of p16(Ink4a). Finally, wild-type Tal1, but not a DNA binding-defective mutant, rescued the proliferative defect in Tal1-null MM precursors. These results document the importance of this transcription factor in cell cycle progression and proliferation during monocytopoiesis and the requirement for direct DNA binding in these processes.
Mononuclear phagocytes serve as critical effectors of both innate and adaptive immunity and have important functions in the maintenance of tissue homeostasis. Macrophages derive from common myeloid precursors (CMPs) that differentiate in the bone marrow (BM). Monocytopoiesis is driven by specific growth factors that include interleukin-3 (IL-3) and macrophage colony-stimulating factor (M-CSF) and by transcription factors, including PU.1 and C/EBPα (10). This generates monoblasts, promonocytes, and finally monocytes, which are released into circulation (10, 12). While monocytes are capable of phagocytosis and cytokine secretion, their functional capabilities are enhanced with differentiation to macrophages in tissues.
The T-cell acute lymphoblastic leukemia 1 (TAL1) gene, also known as stem cell leukemia (SCL), encodes a class II basic helix-loop-helix (bHLH) transcription factor that regulates both embryonic and adult hematopoiesis (25, 28, 39, 42, 43, 45). In the adult, TAL1 has a recognized role in the differentiation of erythroid, megakaryocytic, and mast cell progenitors (1, 3, 13, 14, 31, 41, 54) and is expressed in vascular and lymphatic endothelial cells under both physiological and pathological conditions (49). Heterodimers of TAL1 and ubiquitous bHLH proteins known as E proteins regulate transcription by binding to specific regulatory regions in target genes in association with the zinc finger transcription factor Gata1 or Gata2. The sequence element recognized by those complexes includes an E box (CANNTG) that binds a bHLH heterodimer and a WGATAR motif recognized by a GATA protein (34, 35, 56, 58, 60). TAL1/E protein heterodimers can also bind E boxes to effect transcriptional repression (8, 11, 19, 20, 55). In murine erythroleukemia and T-cell acute lymphoblastic leukemia cell lines, TAL1 had been shown to interact with a number of nuclear corepressors, including mSin3A and HDAC1 (21, 23), Brg1 and HDAC2 (59), and Mtg16/Eto-2 and Mtgr1 (6, 44).
Although an important role for TAL1 has been demonstrated in other hematopoietic lineages, a function in mononuclear phagocyte production, while suggested by a number of observations, had not been established. First, TAL1 had been found to contribute to specific E box DNA-binding complexes in the M1 monocytic leukemia cell line (55). While studies with M1 cells and the human bipotent cell line TF-1 suggested that it was downregulated during monocytic differentiation (18, 50, 55), TAL1 mRNA or protein was detected in mouse peripheral blood mononuclear cells and BM macrophages (25) and zebrafish (62) and human (41) macrophages. In addition, higher and earlier expression of IL-6 receptor (Il6-r) and colony-stimulating factor 1 receptor (Csf1r) mRNA (our unpublished data) was observed in Tal1-transduced primary mouse BM monocyte/macrophage (MM) precursors and M1 cells induced to differentiate with mouse M-CSF (mM-CSF) and IL-6 (mIL-6), respectively. In contrast, knockdown of TAL1 in human cord blood CD34+ cells by short hairpin RNA reduced the numbers of both myeloid and erythroid cell progenitors (5). Finally, while deletion of Tal1 in adult hematopoietic stem cells (HSCs) did not affect granulocyte-macrophage or macrophage progenitor numbers (16, 30), 5-fluorouracil (5-FU) treatment of Tal1−/− mutant mice resulted in a transient but significant delay in the recovery of circulating monocytes (David Curtis, unpublished data).
In this study, we establish that Tal1 is expressed at all stages of monocytic differentiation from CMP to macrophage and report a previously unrecognized function of this hematopoietic transcription factor in cells of this lineage. Using an ex vivo method of Tal1 gene deletion, knockout of Tal1 in BM mononuclear cells was found to suppress cellular proliferation at a relatively late stage of monocytopoiesis and delay entrance into and progression through S phase. Finally, we identify two genomic targets of TAL1 in differentiating MM precursors whose altered expression likely contributed to the abnormal phenotype of knockout cells.
MATERIALS AND METHODS
Cell lines and cell culture reagents.
The mouse myeloid leukemia cell line M1, connective tissue cell line L-929, and retroviral packaging cell line BOSC23 were purchased from the American Type Culture Collection (Manassas, VA). M1 cells were maintained in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and 1% penicillin-streptomycin (Invitrogen) and induced to differentiate with 50 ng/ml mIL-6 (Stem Cell Technologies, Vancouver, Canada). L-929 cells were cultured in Eagle's minimum essential medium (Invitrogen)-15% FBS-2 mM l-glutamine (Invitrogen)-1 mM sodium pyruvate (Fisher Scientific, Pittsburgh, PA)-1% penicillin-streptomycin. The BOSC23 cell line was maintained in Dulbecco's minimum essential medium (DMEM; Invitrogen)-10% FBS-1% penicillin-streptomycin and GPT selection medium (Millipore, Billerica, MA).
For preparation of L-cell conditioned medium, 1.25 × 105 L-929 cells were seeded into 30 ml of medium. Medium was harvested 7 to 10 days later, centrifuged at 1,500 × g for 20 min, and then forced through a 40-μm filter to remove cellular debris. This conditioned medium served as a source of M-CSF in mouse MM progenitor cell cultures.
Plasmids and cDNAs.
The murine stem cell virus (MSCV)-based bicistronic vectors MSCV-IRES-GFP, MSCV-IRES-GFP-Tal1, and MSCV-IRES-GFP-Tal1T192P have been previously described (22). A Gata2 cDNA was obtained from J. Douglas Engel (University of Michigan, Ann Arbor), a Cre expression plasmid from Lishan Su (University of North Carolina, Chapel Hill), and the MSCV-IRES-YFP vector from Derek Persons (St. Jude Children's Research Hospital, Memphis, TN).
Mouse genotyping and breeding.
Three-week-old C57BL/6J mice were purchased from Jackson Labs (Bar Harbor, ME). Mice with a loxP-targeted Tal1 allele (SCLloxP/loxP) and SCLLacZ/WT mice, both in a C57BL/6 background, have been previously described (9, 16). Mice were housed in an AAALAC-accredited animal facility at Vanderbilt University Medical Center according to approved guidelines. SCLloxP/loxP and SCLLacZ/WT mice were intercrossed to generate SCLloxP/LacZ mice, with genotyping carried out by PCR analysis of DNA extracted from tail biopsy samples. The forward primer for the floxed and wild-type Tal1 alleles was TCCCAAGCCCAAAGATTTCCCCAATG, and that for the excised allele was GCAAGCTGGATGGATCAACATGGACCT. A common reverse primer with the sequence GCAAGCTGGATGGATCAACATGCACCT was used for all three alleles. For detection of the LacZ knock-in allele, the forward and reverse primer sequences were, respectively, GGATGGCGGGGCACACGAGGTAA and TGCCAGTTTGAGGGGACGACGACA.
Gene targeting.
BM cells from SCLloxP/LacZ mice were transduced with MSCV-GFP-Cre (where GFP is green fluorescent protein) or its parental vector using the methods described below. Cre expression resulted in excision of the single floxed Tal1 locus, generating SCLΔ/LacZ cells, which were effectively nullizygous, while compound heterozygotes (SCLloxP/LacZ genotype) that were transduced with the parental vector served as controls. GFP-expressing cells were isolated in a fluorescence-activated cell sorter and returned to culture for subsequent studies.
Fluorescence-activated cell sorting (FACS) and flow cytometry analysis.
FACS and standard flow cytometry analysis were carried out in FacsAria and BD FacsCanto II instruments, respectively, running FacsDiva software (Becton Dickinson, Franklin Lakes, NJ). Data analysis was performed using WinList software, and PKH26 dye dilution data were modeled with ModFit LT software (Verity Software House, Topsham, ME).
Isolation and culture of mouse BM CMPs.
CMPs were purified and cultured using previously published procedures (48), with minor modifications. Briefly, BM cells were isolated from 3- to 5-week-old C57BL/6J mice and depleted of lineage-committed cells using a Mouse Lineage Cell Depletion kit (Miltenyi Biotec, Auburn, CA) according to the manufacturer's recommendations. These lineage marker-negative cells were stained with Pacific Blue-conjugated anti-mouse CD117 (c-kit; Biolegend, San Diego, CA), phycoerythrin (PE)-conjugated anti-Ly6C (Miltenyi Biotec), and allophycocyanin (APC)-conjugated anti-CD31 (eBioscience, San Diego, CA) monoclonal antibodies (MAbs) or their isotype controls. The c-kithigh/CD-31high/Ly6Cneg population was then isolated by FACS and cultured in DMEM-15% FBS-0.5 μM β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO)-20 ng/ml recombinant mM-CSF-15 ng/ml IL-3-100 U/ml IL-1α (all from Stem Cell Technologies) to induce macrophage differentiation. Macrophages were activated by the addition of 150 U/ml gamma interferon (IFN-γ; BioLegend) and 100 ng/ml lipopolysaccharide (LPS; Sigma-Aldrich).
Retroviral transduction of terminally differentiating mouse BM MM precursor cells.
High-titer, ecotropic retrovirus-containing conditioned medium was collected 48 h after transfection of BOSC23 packaging cells (38) with MSCV-IRES-GFP or -YFP (yellow fluorescent protein) vectors. BM cells were isolated from 3- to 5-week-old mice and cultured for 24 h under the above-described conditions, and nonadherent cells were subjected to Pronase digestion (Roche, Branford, CT) and centrifugation through a step gradient of horse serum (HyClone, Logan, UT) (15, 47, 53). The resulting mononuclear cell preparations were mixed with conditioned medium containing viral particles and 4 μg/ml Polybrene (Sigma-Aldrich) and subjected to two rounds of centrifuge-assisted infection (9,000 × g for 1 h each at 37°C) at an interval of 3 to 4 h. Cells were returned to culture overnight and transduced a final time 12 h later. After another 24 h in culture, nonadherent cells were collected and analyzed for integration of the retroviral vector on the basis of GFP or YFP expression. GFP- and YFP-expressing cells were returned to culture in the presence of 20 ng/ml mM-CSF and 40% L-cell conditioned medium.
Quantitative real-time PCR analysis.
Total cellular RNA was extracted using RNeasy reagents (Qiagen, Valencia, CA), and genomic DNA was removed by the DNA-Free product (Ambion, Austin, TX). RNA was reverse transcribed using iScript (Bio-Rad, Hercules, CA), and specific transcripts were analyzed by real-time PCR using iQ SYBR green Supermix (Bio-Rad). Gene expression was normalized to ribosomal protein S16 (RPS16) RNA expression. The sequences of the primers used were as follows: Tal1 forward primer, ATAGCCTTAGCCAGCCGCTC; Tal1 reverse primer, GCCGCACTACTTTGGTGTGA; RPS16 forward primer, CACTGCAAACGGGGAAATGG; RPS16 reverse primer, CACCAGCAAATCGCTCCTTG; Gata2 forward primer, CCGACGAGGTGGATGTCTTC; Gata2 reverse primer, TGGGCTGTGCAACAAGTGTG; p16(Ink4a) forward primer, GGACATCAAGACATCGTGCGA; p16(Ink4a) reverse primer, ATTGGCCGCGAAGTTCCA.
Apoptosis analysis.
MM precursors cultured for the indicated times were stained with APC-conjugated annexin V (BD Biosciences, San Jose, CA) and the DNA-intercalating dye 7-aminoactinomycin D (7-AAD; Invitrogen). Cells were then analyzed by flow cytometry, with those excluding both stains scored as viable. Briefly, cells were suspended in 100 μl of binding buffer (10 mM HEPES buffer, pH 7.4, 140 mM sodium chloride, 2.5 mM calcium chloride) and incubated with 5 μl of APC-annexin V for 15 min at room temperature in the dark. After washing, cells were resuspended in 500 μl of binding buffer, stained with 2 μg of 7-AAD, and analyzed by flow cytometry.
Cell division analysis.
Cell division was analyzed with the membrane-intercalating red fluorescent dye PKH26 (PKH26 MINI kit; Sigma-Aldrich). Retrovirus-transduced cells were labeled with PKH26 according to the manufacturer's instructions, and cells positive for both PKH26 and GFP fluorescence were isolated and returned to culture. Cells were analyzed at the indicated times by flow cytometry, and the decrease in fluorescence intensity over time in culture was used to quantify cell division. This was modeled using ModFit LT software.
Cell differentiation analysis.
Differentiation of MM precursors was studied by flow cytometry analysis of cell surface antigens with PE-conjugated anti-Ly-6C (Miltenyi Biotec) and APC-conjugated anti-CD31 (eBioscience) MAbs and their isotype controls (48). Cell morphology was determined from Wright-Giemsa staining of cytospin preparations.
Cell cycle analysis.
Bromodeoxyuridine (BrdU) pulse-chase analysis was carried out using an APC BrdU Flow Kit (BD Biosciences) according to the manufacturer's recommendations. Cells were labeled with 10 μM BrdU for 45 min, washed with complete medium, and returned to culture in fresh medium. At specific times (0, 1, 2, and 4 h), labeled cells were washed with staining buffer (1× Dulbecco's phosphate-buffered saline, 3% FBS, 0.09% sodium azide), fixed in BD Cytofix/Cytoperm buffer, and processed for flow cytometry analysis. As described below, cells were also incubated with 10 μM BrdU for prolonged periods, including 12, 24, 36, and 48 h. These cells were processed for flow cytometry analysis identically to the others.
Chromatin immunoprecipitation (ChIP) analysis.
ChIP analysis was carried out using a commercial kit (Millipore) with certain modifications (59). Formaldehyde was added at a final concentration of 1% to 2 × 107 cells for 20 min at 37°C. For M1 cells, cross-linking was carried out with 1 × 108 cells cultured in the presence or absence of recombinant mIL-6 for 48 h. All subsequent steps were carried out according to the manufacturer's instructions. Antibodies used in immunoprecipitation of chromatin fragments included rabbit polyclonal anti-Tal1 (25), anti-E47 (sc-763X; Santa Cruz), and anti-E2A (Yae) (sc-416X; Santa Cruz), in addition to normal rabbit IgG (sc-2027; Santa Cruz). Immunoprecipitated DNA was analyzed by real-time PCR with iQ SYBR green Supermix (Bio-Rad), and occupancy was quantified for all antibodies and IgG and expressed as a percentage of the input. The sequences of the primers used were as follows: p16(Ink4a) −13 region forward, CCACTGGTCACACGACTG; p16(Ink4a) −13 region reverse, GCACGTCATGCACACG; p16(Ink4a) −2592 region forward, CCCTAGCCAAATCTGACAT; p16(Ink4a) −2592 region reverse, TCCAGGTAGTACATGCTTTCA; p16(Ink4a) −4638 region forward, CCTCTCCAGGGTAGTCATGG; p16(Ink4a) −4638 region reverse, GATGCAAGAATCACACAAAGG; p16(Ink4a) −5106 region forward, CACTTTGCTTATACTGGATGG; p16(Ink4a) −5106 region reverse, CCAGTGAAACCCTTGTATCTT; p16(Ink4a) −6932 region forward, GCTTTCACAGGACACAGAAGG; p16(Ink4a) −6932 region reverse, TTACTCTTTCCCACCATGACC; p16(Ink4a) 3′ UTR (untranslated region) forward, GGAGGGAGCAGAAGGAGG; p16(Ink4a) 3′ UTR reverse, ACAATCCAGCCATTATTCCC; Gata2 intron 4 forward, CCTGCCGGAGTTTCCTATCC; Gata2 intron 4 reverse, ACTGAGTCGAGGTGGCTCTGA; Gata2 3′ UTR forward, TCGATTCTGTGTGTGGTGGT; Gata2 3′ UTR reverse, ACTTCCGGTTAGGGTGCTCT.
Western blot analysis.
Whole-cell lysates of MM precursors were prepared at days 1, 4, 6, and 8 of culture. Western blot analysis was performed according to previously described procedures (21) with antibodies to Tal1 (sc-12982; Santa Cruz) and β-actin (ab 8226; Abcam, Cambridge, MA).
RESULTS
Tal1 is expressed continuously during murine monocytopoiesis.
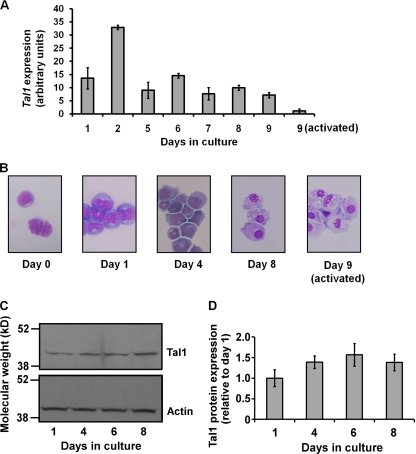
To characterize Tal1 expression in monocyte progenitors, we made use of a previously developed method for isolating CMPs from mouse BM and inducing their differentiation (48). Using quantitative reverse transcription (RT)-PCR analysis, Tal1 mRNA was detected at all stages from the common myeloid progenitor to the postmitotic macrophage (Fig. 1A). Although Tal1 message fluctuated in abundance, it was detectable throughout, as well as in LPS- and IFN-γ-activated macrophages, albeit at lower levels than in unactivated cells (Fig. 1A). Macrophage differentiation was verified by morphological analysis of Wright-Giemsa-stained cytospin preparations (Fig. 1B), quantitative RT-PCR analysis of well-established macrophage gene markers (data not shown) (48), and flow cytometry analysis of the cell surface antigens CD31 and Ly6C. As previously described (48), CMPs (CD31+ Ly6C−) differentiated sequentially to CD31+ Ly6C+, CD31− Ly6C+, and CD31− Ly6C− cells (data not shown).
FIG. 1.
Tal1 mRNA expression during in vitro differentiation of CMPs to macrophages and following LPS/IFN-γ activation. (A) Total RNAs prepared from purified precursors and differentiated cells at the indicated times in culture were converted to cDNA for real-time PCR analysis employing SYBR green fluorescence. Each bar represents the mean ± the standard deviation from three independent PCRs as normalized to RPS16 mRNA expression. Results are representative of three independent experiments. (B) Morphology of differentiating cells from Wright-Giemsa stains of cytospin preparations. (C) Western blot analysis of Tal1 expression in cultures of terminally differentiating mouse BM MM precursors. Results are representative of two independent analyses. (D) Graph of normalized Tal1 expression from Western blot analysis.
Expression studies were also carried out with MM precursors explanted at a later stage of differentiation. Using methods developed by Stanley and colleagues (15, 47, 53), terminal differentiation of mouse BM monoblasts and promonocytes was induced in vitro with recombinant murine M-CSF. Western blot analysis of cellular extracts prepared after 1, 4, 6, and 8 days of culture revealed only modest changes in Tal1 expression over this period (Fig. 1C and D).
Loss of Tal1 impairs proliferation of mouse MM precursors.
For subsequent studies, monoblasts and promonocytes were isolated from mouse BM using established procedures (15, 47, 53) and Tal1 expression was manipulated in them in culture. In the absence of a suitable promoter to direct Cre expression to monocytic progenitors in vivo, a strategy was employed to delete the gene in MM precursors isolated from mice with a floxed Tal1 allele. To that end, BM mononuclear cells from SCLloxP/LacZ mice were transduced with a retrovirus, MSCV-GFP-Cre, expressing Cre recombinase to render them nullizygous for a functional Tal1 gene and these cells were then induced to differentiate in the presence of recombinant M-CSF (15, 53). Semiquantitative PCR analysis of genomic DNA from day 8 Tal1−/− cells revealed that the loxP-flanked allele was almost totally deleted by retroviral delivery of Cre, although low levels of the floxed allele were detected at later times in culture, likely due to a competitive advantage of the SCLloxP/LacZ cells over SCLΔ/LacZ cells (data not shown). In any case, RT-PCR analysis showed essentially complete abolition of Tal1 expression in gene-targeted cells (Fig. 2A).
FIG. 2.
Effect of increased and decreased Tal1 expression on proliferation of MM precursors in vitro. Retrovirus-transduced, GFP-expressing cells were sorted and cultured under identical conditions at equivalent initial cell concentrations. (A) Real-time PCR analysis of Tal1 mRNA abundance in MM precursor cells isolated from SCLloxP/LacZ mice and transduced with the parental MSCV-GFP vector (SCLloxP/LacZ) or the MSCV-GFP-Cre vector (SCLΔ/LacZ). Total RNA was prepared at the indicated times. Each bar represents the mean ± the standard deviation from three independent PCRs as normalized to RPS16 mRNA expression. Results are representative of six independent experiments. (B to D) Number of cells per plate as a function of time in culture for SCLloxP/LacZ and SCLΔ/LacZ MM precursors (B), wild-type MM precursors transduced with MSCV-GFP or MSCV-GFP-Cre (C), and wild-type MM precursors transduced with MSCV-GFP-Tal1 or MSCV-GFP-Cre, SCLloxP/WT MM precursors transduced with MSCV-GFP-Cre, and SCLloxP/LacZ MM precursors transduced with MSCV-GFP-Cre (D). Each data point represents the mean ± the standard deviation from triplicate determinations, with similar trends noted in four independent experiments. (E) PKH26 staining in cells of the indicated genotypes. GFP+ PKH26+ cells were isolated by dual-color FACS and cultured under identical conditions at equivalent starting cell concentrations. Day 1 cells were modeled to represent generation I, and loss of PKH26 fluorescence intensity due to cell division, measured at days 6 and 8, was fitted to the model shown. The data depicted are representative of three independent experiments. Gen, generation.
Cell counts at timed intervals in culture revealed a severe proliferative defect in Tal1 knockout cells compared to vector-transduced cells (Fig. 2B). While the number of cells in cultures infected with the control vector increased approximately threefold, Cre-transduced cells showed little or no change. Toxic effects of Cre expression and retroviral infection (data not shown) were ruled out by transduction of wild-type cells with Cre DNA and the empty vector, respectively, with both populations found to proliferate at rates similar to those of nontransduced cells (Fig. 2C). In contrast, Tal1-overexpressing cells had a higher proliferative capacity than wild-type cells (Fig. 2D). Finally, heterozygous knockout cells accumulated in slightly lower numbers than wild-type cells, although this was not apparent until late in culture (days 6 and 8) (Fig. 2D). Thus, loss of Tal1 in differentiating MM precursors significantly impaired cellular proliferation, with Tal1 gene expression correlating closely with proliferative potential.
Dye dilution analysis of transduced cells using the fluorescent membrane-intercalating dye PKH26 showed both an absolute reduction and a delay in cell division in SCLΔ/LacZ cells compared to SCLloxP/LacZ cells (Fig. 2E). With >98% of the day 1 cells of both genotypes assigned to the first generation (Fig. 2E, i and iv), a model in which three (or more) generations of cells appeared over the 6- to 8-day culture period best fit the data. This would correspond to a fourfold [or 2(3 − 1)] increase in cell number over the first generation, slightly higher than what was observed for control SCLloxP/LacZ cells. After 6 days, 34.8% of the SCLΔ/LacZ cells were still first generation, 46.8% were second generation, and only 17.8% could be classified by dye content as third generation (Fig. 2E, v). Over the same time period, 13.8% of the control SCLloxP/LacZ cells were first generation, 21.1% were second generation, and 62.8% were third generation (Fig. 2E, ii). While slightly less than half of the SCLΔ/LacZ cells (48.9%) were second generation after 8 days, most of the SCLloxP/LacZ cells were third generation, with some fourth generation cells (6.5%) also represented (Fig. 2E, iii and vi). Similar results were obtained when mean fluorescence intensities were compared (data not shown).
Tal1 knockout MM precursors show reduced viability and accelerated differentiation.
To determine whether the reduced accumulation of SCLΔ/LacZ cells resulted from decreased viability, apoptosis was analyzed using annexin V and 7-AAD staining, with nonapoptotic cells characterized by the ability to exclude both stains (Fig. 3A). The percentage of viable cells in the SCLΔ/LacZ group was very slightly decreased relative to that of SCLloxP/LacZ cells at days 4 and 6 but was not different at days 1 and 8 of culture. These results show that Tal1 gene loss had minimal effects on cell survival and that apoptosis could not account for the marked reduction in cell number observed.
FIG. 3.
Viability and differentiation of SCLloxP/LacZ and SCLΔ/LacZ MM precursor cells in culture. (A) Viability of cells was determined by flow cytometry analysis. Viable cells were characterized by absent APC-annexin V binding and 7-AAD staining. Each bar represents the mean percentage ± the standard deviation of cells negative for both markers (from triplicate plates). (B) Flow cytometry analysis of APC-CD31 (y axis) and PE-Ly6C (x axis) staining of SCLloxP/LacZ and SCLΔ/LacZ cells at days 1, 2, and 4 of culture. A representative profile from three independent experiments is shown.
Cellular differentiation was then assessed by flow cytometry analysis of cell surface antigens CD31 and Ly6C (Fig. 3B). A higher percent of the most mature CD31− Ly6C− cells was noted in the SCLΔ/LacZ population (59.2% at day 2 and 91.2% at day 4) than in the SCLloxP/LacZ population (52.2% at day 2 and 82.6% at day 4) at early times in culture. In contrast, less differentiated CD31− Ly6C+ cells made up a smaller proportion of SCLΔ/LacZ cultures (26.9% at day 2 and 8.5% at day 4) than of SCLloxP/LacZ cultures (36.1% at day 2 and 16.4% at day 4). Expression of another widely used macrophage marker, F4/80, did not reliably discriminate between different stages of monocytopoiesis in our studies, with approximately 90% of the cells in both groups becoming F4/80 positive after only 24 h in culture (data not shown). In sum, these data indicate that SCLΔ/LacZ cells lacking a functional Tal1 gene differentiated slightly more rapidly than SCLloxP/LacZ cells.
SCLΔ/LacZ cells are defective in cell cycle progression.
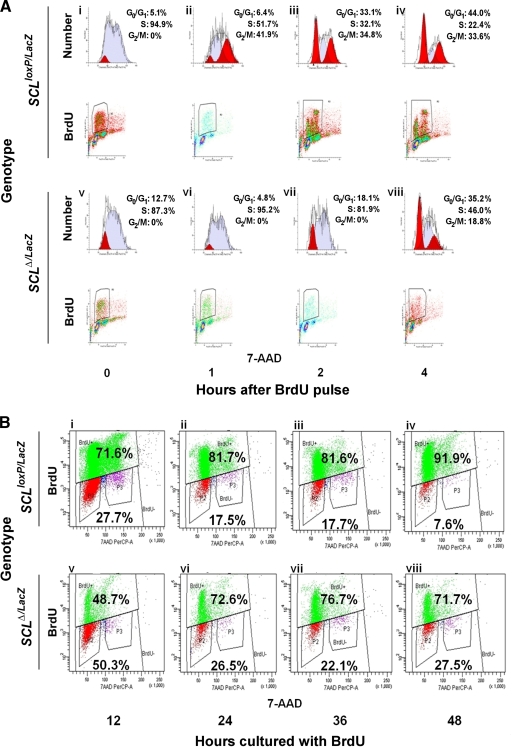
To better characterize the proliferative defect in Tal1 knockout cells, a BrdU pulse-chase analysis was carried out. To that end, GFP-expressing cells were cultured for 3 days, labeled with BrdU for 45 min, and followed through at least one cell cycle by flow cytometry (Fig. 4A). Analyzed immediately after pulsing, the majority of the BrdU-labeled cells from both the SCLloxP/LacZ and SCLΔ/LacZ groups were in S phase (94.9% and 87.3%, respectively), with smaller percentages in early S phase or late G0/G1 (5.1% and 12.7%, respectively; shaded dark in the 0-h population, Fig. 4A, i and v). At later times, SCLloxP/LacZ cells progressed rapidly to G2/M and then to G0/G1, with a concomitant decrease in the percentage in S phase (Fig. 4A, ii, iii, and iv). In contrast, while some SCLΔ/LacZ cells advanced to late S phase 1 h after pulsing, as evidenced by the two peaks of labeling (Fig. 4A, vi), these cultures had a larger proportion of cells in G0/G1 after 2 and 4 h (18.1% and 35.2%, respectively), lacked a distinct G2/M population (0% and 18.8%, respectively), and contained a higher percentage of cells still in S phase (81.9% and 46.0%) than did the control cells (Fig. 4A, vii and viii). These results provide evidence of slowed cell cycle progression in Tal1 knockout cells, with delayed entry into and traversal of S phase.
FIG. 4.
Cell cycle analysis of SCLloxP/LacZ and SCLΔ/LacZ MM precursor cells. (A) Flow cytometry analysis of BrdU incorporation versus DNA content (7-AAD staining) for SCLloxP/LacZ and SCLΔ/LacZ cells at 0, 1, 2, and 4 h after pulsing. G0/G1, S, and G2/M populations were modeled computationally. A representative profile from three independent experiments is shown. (B) Flow cytometry analysis of SCLloxP/LacZ and SCLΔ/LacZ cells cultured with BrdU for 12, 24, 36, and 48 h. DNA content (x axis) is plotted versus BrdU incorporation (y axis). Values inside the upper and lower quadrants denote the percentages of BrdU-positive and -negative cells, respectively.
As only cells able to incorporate the BrdU label can be analyzed by the pulse-chase approach, MM precursors were also cultured with BrdU for longer times (12, 24, 36, and 48 h) before analysis by flow cytometry (Fig. 4B). As could have been predicted, the SCLΔ/LacZ population contained a higher proportion (27.5% versus 7.6% for SCLloxP/LacZ cells, Fig. 4B, iv and viii) of cells that never incorporated BrdU during a 48-h labeling period. Of note, the number of BrdU-positive SCLΔ/LacZ cells decreased slightly over time in culture (Fig. 4B, vii and viii), consistent with their increased tendency toward apoptosis (Fig. 3A). Finally, and most directly, double staining with Hoechst 33342 and pyronin Y showed a threefold higher percentage of cells in G0 in SCLΔ/LacZ than in SCLloxP/LacZ cultures (data not shown). Together, these studies suggest a role for TAL1 in cell cycle progression in MM precursors that can largely account for the proliferative defect associated with homozygous Tal1 gene loss.
Tal1 knockout MM precursor cells contain lower Gata2 and higher p16(Ink4a) mRNA levels.
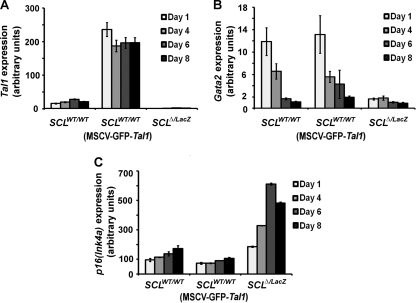
Tal1 gene loss would be expected to impact the transcription of its target genes, especially direct targets like those coregulated with E proteins. Accordingly, real-time PCR analysis was used to evaluate expression of genes known to have a role in MM differentiation, including putative TAL1 targets, with comparisons made among SCLWT/WT, Tal1-overexpressing, and SCLΔ/LacZ cells. The same bicistronic retroviral vector used to deliver Cre to MM precursors for inactivation of the floxed Tal1 allele was also employed in enforcing Tal1 expression (Fig. 5A). These studies revealed that Gata2 mRNA was reduced seven- and fourfold, respectively, at days 1 and 4 in SCLΔ/LacZ relative to SCLWT/WT cells (Fig. 5B). Although the level of Gata2 expression decreases physiologically with differentiation of MM precursors (Fig. 5B and reference 48), Tal1-null (SCLΔ/LacZ) cells showed even lower Gata2 expression than wild-type controls at days 6 and 8. In contrast, enforced Tal1 expression did not increase the abundance of Gata2 mRNA at any time point examined (Fig. 5B).
FIG. 5.
Tal1 (A), Gata2 (B), and p16(Ink4a) (C) gene expression analysis in wild-type MM precursor cells transduced with Cre or Tal1 cDNA and SCLloxP/LacZ MM precursor cells transduced with Cre cDNA (SCLΔ/LacZ cells). Cells were grown in culture for the indicated times after sorting, and transcript abundance was measured by real-time RT-PCR analysis. PCRs were done in triplicate, and results are expressed in arbitrary units relative to RPS16 mRNA expression. Expression profile shown is representative of three independent experiments.
Because of the delay in cell cycle progression observed with Tal1 loss (Fig. 4A and B) and the previously published evidence of their regulation by E proteins, expression of the genes for several cyclin-dependent kinase (CDK) inhibitors was analyzed. p16(Ink4a), which was shown to be a target of repression by Tal1 (17, 33, 37), was upregulated approximately three- to fourfold in SCLΔ/LacZ cells and reduced, albeit to a lesser extent, in Tal1-overexpressing cells (Fig. 5C). In contrast, expression of p21(Cip1), another putative target of Tal1 and E proteins (29, 37, 40), was only slightly increased in SCLΔ/LacZ cells compared to SCLWT/WT cells (data not shown). While elevation of p16(Ink4a) and p21(Cip1) could relate to the cell cycle delay in Tal1 knockout cells, especially given the phase of the cell cycle affected, the only slight reduction in these mRNAs cannot explain the increased proliferation by Tal1-overexpressing cells (Fig. 2D). Finally, expression of Csf1r, the receptor for M-CSF and itself a marker of macrophage differentiation, was slightly elevated in SCLΔ/LacZ cells (data not shown) and could, potentially, have contributed to their accelerated differentiation (Fig. 3B).
Gata2 intron 4 and p16(Ink4a) upstream regions are occupied by a Tal1-E protein complex in MM precursors.
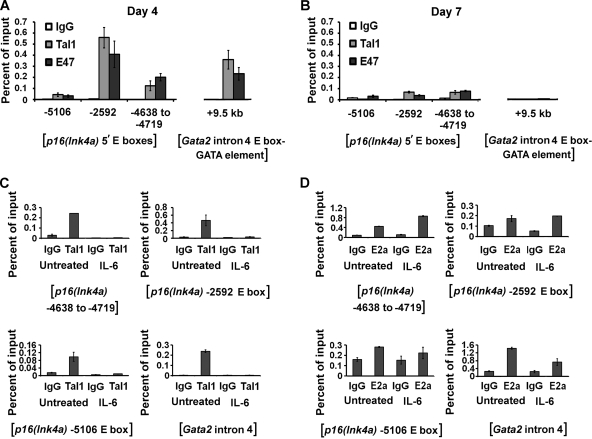
The above-described studies revealed that knockout, and to a lesser extent overexpression, of Tal1 affected the expression of Gata2 and p16(Ink4a). Previous studies defined an enhancer in intron 4 of the Gata2 gene (kb +9.5) with a consensus E box-GATA DNA-binding motif that is occupied by Tal1, E proteins, and GATA proteins in endothelial and fetal liver cells (27, 57). Moreover, this DNA sequence element appeared to be essential for the proper expression of Gata2 in endothelial cells (27). To ascertain whether Tal1 occupies this region in MM precursors, a ChIP analysis was carried out on sonicated chromatin from day 4 and day 7 wild-type (SCLWT/WT) cells with polyclonal antibodies to Tal1 and E47. The amount of DNA immunoprecipitated was then quantified in a real-time PCR analysis using region-specific primers and expressed as a percentage of the input (Fig. 6A and B).
FIG. 6.
ChIP analysis of Tal1 and E47 occupancy of the p16(Ink4a) and Gata2 genes in wild-type MM precursors cultured for 4 (A) or 7 (B) days. DNA derived from chromatin fragments immunoprecipitated with antibodies to Tal1 or E47 or normal rabbit IgG was quantified by real-time PCR analysis using region-specific primers. Each bar represents the mean abundance ± the standard deviation of each fragment relative to the input as determined from triplicate PCR measurements. ChIP analysis of Tal1 (C) and E2a (D) occupancy of the p16(Ink4a) and Gata2 genes in M1 cells cultured with or without IL-6 for 48 h. DNA derived from chromatin fragments immunoprecipitated with antibodies to Tal1 or E2a or normal rabbit IgG was quantified by real-time PCR analysis using region-specific primers. Each bar represents the mean abundance ± the standard deviation of each fragment relative to the input as determined from triplicate PCR measurements.
Antibodies to Tal1 and E47, but not normal rabbit IgG, were able to precipitate this intronic fragment from day 4 cells (Fig. 6A) but not day 7 cells (Fig. 6B), concordant with the decline in Gata2 expression over this same time period. Primer sequences designed for the Gata2 3′ UTR did not support any amplification (data not shown), indicating specific occupancy of this intronic region in MM precursor cells.
Scanning a 12-kb region upstream of the p16(Ink4a) translational start site identified five preferred E box sequences (CATCTG or CAGATG) for TAL1/E protein binding (bases −13, −2592, −4638, −5106, and −6932). PCR primers complementary to each of these regions and to the gene's 3′ UTR were then designed, and a quantitative ChIP analysis was carried out. Three of these E boxes, at −2592, −4638, and −5106, were shown by this approach to be occupied by Tal1 and E47 in day 4 cells (Fig. 6A). In contrast, no binding was detected at −13, −6932, or the 3′ UTR at any time point (data not shown), and another, noncanonical E box in the promoter region (−437) likewise was not occupied (data not shown). Finally, Tal1 and, unexpectedly, E47 occupancy of the −2592 and −4638 elements was very low and essentially absent at the −5106 E box at day 7 (Fig. 6B), despite the increase in p16(Ink4a) expression with differentiation (Fig. 5C). This suggests that E proteins other than E47 are involved or that other mechanisms for p16(Ink4a) gene transactivation operate at these later times.
p16(Ink4a) expression increases with IL-6-induced differentiation (2) of the murine myeloid leukemia cell line M1 in parallel with a decrease in Tal1 expression and Tal1/E protein DNA-binding activity (55). As this phenocopies the results of Tal1 gene knockout, a ChIP analysis was also carried out on chromatin from untreated and IL-6-treated M1 cells. As predicted from its drastic decline in expression (55), Tal1 occupancy was detected only in uninduced cells (Fig. 6C). In contrast, E2A association with these same E box elements was detected in both IL-6-treated and untreated cells, indicating that its E12 and/or E47 protein products acted to augment the expression of this gene unopposed by TAL1 when these cells were induced to differentiate (Fig. 6D).
Tal1 function in MM precursor cell proliferation requires direct DNA binding.
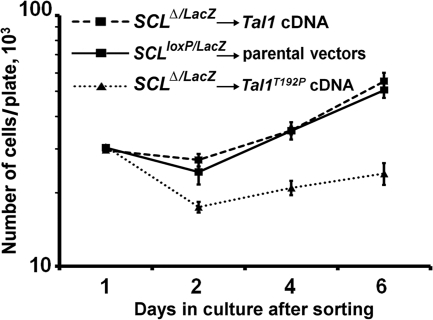
TAL1 has been shown to regulate transcription through direct binding to DNA and as a non-DNA-binding cofactor (34, 56, 58). To determine whether the proliferative defect in SCLΔ/LacZ cells could be rescued by a wild-type Tal1 cDNA, SCLloxP/LacZ MM precursors were infected with MSCV-GFP-Cre and MSCV-YFP-Tal1, while control cells were transduced with the corresponding parental vectors. Equal numbers of GFP- and YFP-expressing cells were then sorted, and cell numbers were determined at specific times. These studies showed that the defective proliferation of Tal1 knockout cells could be completely rescued by the Tal1 cDNA, with the growth curve of Tal1-transduced SCLΔ/LacZ cells virtually identical to that of control cells (Fig. 7). Since Tal1 deletion resulted in the downregulation of Gata2 expression, a Gata2 cDNA was also tested in rescue experiments. However, cells transduced with the Gata2 cDNA were nonviable after only 1 day in culture, demonstrating an adverse effect of Gata2 overexpression in a Tal1-null background (data not shown). Finally, to investigate whether DNA-binding activity is required for TAL1 function in MM proliferation, SCLΔ/LacZ cells were transduced with a cDNA for a well-characterized DNA binding-defective mutant form of Tal1, Tal1T192P (21). Significantly, cells transduced with this protein behaved identically to SCLΔ/LacZ cells and showed no recovery of proliferative capacity (Fig. 7). This result indicates that, similar to its actions in erythroid model systems (26), Tal1 DNA-binding activity is required for outgrowth of murine MM precursors in explant cultures.
FIG. 7.
Rescue analysis in Tal1 knockout cells. Cell counts in MM precursors from SCLloxP/LacZ mice simultaneously transduced with MSCV-GFP-Cre and either a Tal1 or a Tal1T192P cDNA in the retroviral vector MSCV-IRES-YFP. GFP- and YFP-expressing cells were isolated by dual-color FACS. The Tal1T192P cDNA encodes a DNA binding-defective protein. Parental vector-transduced cells were used as controls, and the total number of cells per plate for each day is plotted. Each data point represents the mean ± the standard deviation from three plates of cells.
DISCUSSION
Previous studies have established the importance of TAL1 in erythroid, megakaryocytic, and mast cell differentiation. Although derived from the same progenitor cell as these, the CMP, there is conflicting evidence for TAL1 function in MM production. First, work carried out with the TF-1 (18) and M1 (55) cell lines showed that TAL1 expression declined with macrophage differentiation and that enforced expression of this transcription factor inhibited this process (51), although monocytic differentiation of TF-1 cells was assessed with the CD45 and FcγRII (CD32) markers, neither of which is macrophage specific. Regardless, TAL1 expression has been detected in primary mouse macrophages by several groups and in mouse peripheral blood mononuclear cells and BM macrophages (25) and in zebrafish (62) and human (41) macrophages. In addition, the murine Tal1 cDNA was originally cloned from a BM macrophage cDNA library (4).
We investigated here whether the abundance of Tal1 mRNA was altered during in the differentiation of CMPs to monocytes and whether Tal1 knockout affected the survival, proliferation, or differentiation of mouse MM precursors. While deletion of Tal1 in HSCs in IFN-inducible MxI-Cre mice did not change the granulocyte-macrophage progenitor number significantly (16), loss of Tal1 in cells also nullizygous for the related Lyl1 gene did reduce myeloid colony-forming activity, raising the possibility of genetic redundancy (46). Deletion of the Tal1 gene ex vivo, as done in these studies, may also have brought out an abnormal phenotype, either because of the acuteness of gene loss or because growth in culture simulates stress monocytopoiesis. In support of the latter notion, SCLΔ/LacZ mice showed a small but significant delay in the recovery of circulating monocytes after 5-FU treatment (Curtis, unpublished).
The complete loss of Tal1 caused proliferative and cell cycle defects in macrophage precursors, and wild-type and heterozygous knockout cells also differed in their growth rates. This was most apparent at later times in culture (Fig. 2D), however, suggesting that more differentiated cells such as monoblasts or promonocytes are more sensitive to Tal1 gene dosage. In contrast, overexpression of Tal1 in MM progenitors resulted in a substantial increase in cell number (Fig. 2D), similar to what was reported for the 32D and HL-60 cell lines (7). As enforcement of Tal1 expression in mouse MM precursors was associated with Il6-r and Csf1r upregulation (data not shown), the increased proliferation of these cells could reflect a heightened sensitivity to cytokines.
The genes for several CDK inhibitors, including p16(Ink4a) and p21(Cip1), have been shown to be positively regulated by E proteins, while Tal1/E protein heterodimers were suggested to repress their transcription (8, 17, 19, 33, 37, 40). Further, restoration of E2A activity in the Jurkat cell line, in which Tal1-E2A function is low, profoundly depressed growth and increased apoptosis (36). In Tal1 knockout MM precursor cells, abrogation of Tal1 inhibition of E protein action was likely a major contributor to both the upregulation of p16(Ink4a) and p21(Cip1) expression and the impairment of cell cycle progression and proliferation observed. The gradual increase in p16(Ink4a) mRNA abundance (Fig. 5C) in the face of nearly invariant Tal1 expression during late-stage monocytopoiesis (Fig. 1A and C) suggests that while Tal1 may be required for initial suppression of p16(Ink4a) transcription, it is less important for maintenance of gene repression. This is also supported by the decreased Tal1/E protein occupancy of E boxes upstream of the p16(Ink4a) gene at day 7 of culture compared to day 4 (Fig. 6A and B). The further upregulation of p16(Ink4a) expression in Tal1−/− Lyl1−/− CMPs (Curtis, unpublished) suggests that there is, in addition, genetic redundancy in these bHLH proteins, at least for this target gene. In contrast, p16(Ink4a) mRNA abundance was only slightly decreased in Tal1-overexpressing cells (Fig. 5C), making it unlikely to account for their increased proliferative rate (Fig. 2D). Regardless of the timing of its actions, the direct association of Tal1 with three preferred E box sites in the p16(Ink4a) upstream region in both primary MM precursors and the M1 cell line provides the best evidence that this gene is a target of this transcription factor in cells.
While p16(Ink4a) transcription was inhibited by Tal1 in differentiating cells of this lineage, the Gata2 gene appeared to be transactivated by Tal1. A region in intron 4 of Gata2 was shown previously to have strong enhancer activity in endothelial and mouse fetal liver cells (27, 57), and the present data are compatible with its also being active in mononuclear phagocytes. Indeed, the slight acceleration of differentiation in Tal1 knockout cells could be explained by their reduced levels of Gata2, as a similar finding was reported for Gata2-deficient juvenile myelomonocytic leukemia cells (61) and brown adipocytes (32, 52). Furthermore, Gata2 knockdown reduced G1-S transition while increasing myeloid gene expression in G1ME cells (24), and the further decrease in Gata2 expression in SCLΔ/LacZ cells could also have contributed to their abnormal cell cycle progression. In contrast to p16(Ink4a) transcription, Gata2 transcription was not affected by Tal1 overexpression and was reduced only with Tal1 nullizygosity, reflecting a difference in the quantitative requirement for Tal1 in genes activated and repressed by this transcription factor.
Finally, the proliferative defect in SCLΔ/LacZ cells was fully rescued by the reintroduction of a Tal1 cDNA, which confirms that this phenotype is attributable to Tal1 loss and is not an indirect effect of retroviral transduction or Cre expression. The failure of a DNA binding-defective mutant to rescue proliferation in these cells, in contrast, shows that DNA-binding activity is as important for Tal1 function in this lineage as in erythroid progenitors (26).
In summary, these studies demonstrate that the bHLH transcription factor Tal1 is expressed throughout monocytopoiesis and that loss of Tal1 impairs the proliferation of late-stage MM precursors. This was associated with decreased exit from G0, delayed entrance into S from G1, and slowed traversal of S phase, resulting in part from the altered expression of a direct Tal1 target, the CDK inhibitor gene p16(Ink4a).
Acknowledgments
We thank J. Douglas Engel for providing a Gata2 cDNA, Lishan Su for the Cre plasmid, Derek Persons for the MSCV-IRES-YFP expression vector, Margaret Goodell and George Souroullas for helpful discussions, and Angel Lee for technical advice. We also thank Catherine Alford for her assistance with flow cytometry analysis.
This work was supported by grant R01 HL049118 from the National Institutes of Health and a Merit Review Award from the Department of Veterans Affairs (both to S.J.B.).
Footnotes
Published ahead of print on 1 March 2010.
REFERENCES
- 1.Akashi, K., D. Traver, T. Miyamoto, and I. L. Weissman. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404:193-197. [DOI] [PubMed] [Google Scholar]
- 2.Amanullah, A., B. Hoffman, and D. A. Liebermann. 2000. Deregulated E2F-1 blocks terminal differentiation and loss of leukemogenicity of M1 myeloblastic leukemia cells without abrogating induction of p15INK4B and p16INK4A. Blood 96:475-482. [PubMed] [Google Scholar]
- 3.Begley, C. G., P. D. Aplan, S. M. Denning, B. F. Haynes, T. A. Waldmann, and I. R. Kirsch. 1989. The gene SCL is expressed during early hematopoiesis and encodes a differentiation-related DNA-binding motif. Proc. Natl. Acad. Sci. U. S. A. 86:10128-10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begley, C. G., J. Visvader, A. R. Green, P. D. Aplan, D. Metcalf, I. R. Kirsch, and N. M. Gough. 1991. Molecular cloning and chromosomal localization of the murine homolog of the human helix-loop-helix gene SCL. Proc. Natl. Acad. Sci. U. S. A. 88:869-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunet de la Grange, P., F. Armstrong, V. Duval, M. C. Rouyez, N. Goardon, P. H. Romeo, and F. Pflumio. 2006. Low SCL/TAL1 expression reveals its major role in adult hematopoietic myeloid progenitors and stem cells. Blood 108:2998-3004. [DOI] [PubMed] [Google Scholar]
- 6.Cai, Y., Z. Xu, J. Xie, A.-J. L. Ham, M. J. Koury, S. W. Hiebert, and S. J. Brandt. 2009. ETO2/MTG16 and MTGR1 are heteromeric corepressors of the TAL1/SCL transcription factor in murine erythroid progenitors. Biochem. Biophys. Res. Commun. 390:295-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Condorelli, G. L., A. Tocci, R. Botta, F. Facchiano, U. Testa, L. Vitelli, M. Valtieri, C. M. Croce, and C. Peschle. 1997. Ectopic TAL-1/SCL expression in phenotypically normal or leukemic myeloid precursors: proliferative and antiapoptotic effects coupled with a differentiation blockade. Mol. Cell. Biol. 17:2954-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doyle, K., Y. Zhang, R. Baer, and M. Bina. 1994. Distinguishable patterns of protein-DNA interactions involving complexes of basic helix-loop-helix proteins. J. Biol. Chem. 269:12099-12105. [PubMed] [Google Scholar]
- 9.Elefanty, A. G., C. G. Begley, D. Metcalf, L. Barnett, F. Köntgen, and L. Robb. 1998. Characterization of hematopoietic progenitor cells that express the transcription factor SCL, using a lacZ “knock-in” strategy. Proc. Natl. Acad. Sci. U. S. A. 95:11897-11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman, A. D. 2007. Transcriptional control of granulocyte and monocyte development. Oncogene 26:6816-6828. [DOI] [PubMed] [Google Scholar]
- 11.Goldfarb, A. N., and K. Lewandowska. 1995. Inhibition of cellular differentiation by the SCL/tal oncoprotein: transcriptional repression by an Id-like mechanism. Blood 85:465-471. [PubMed] [Google Scholar]
- 12.Gordon, S., and P. R. Taylor. 2005. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5:953-964. [DOI] [PubMed] [Google Scholar]
- 13.Green, A. R., T. Lints, J. Visvader, R. Harvey, and C. G. Begley. 1992. SCL is coexpressed with GATA-1 in hemopoietic cells but is also expressed in developing brain. Oncogene 7:653-660. [PubMed] [Google Scholar]
- 14.Green, A. R., E. Salvaris, and C. G. Begley. 1991. Erythroid expression of the ‘helix-loop-helix’ gene, SCL. Oncogene 6:475-479. [PubMed] [Google Scholar]
- 15.Guilbert, L. J., and E. R. Stanley. 1986. The interaction of 125I-colony-stimulating factor-1 with bone marrow-derived macrophages. J. Biol. Chem. 261:4024-4032. [PubMed] [Google Scholar]
- 16.Hall, M. A., D. J. Curtis, D. Metcalf, A. G. Elefanty, K. Sourris, L. Robb, J. R. Göthert, S. M. Jane, and C. G. Begley. 2003. The critical regulator of embryonic hematopoiesis, SCL, is vital in the adult for megakaryopoiesis, erythropoiesis, and lineage choice in CFU-S12. Proc. Natl. Acad. Sci. U. S. A. 100:992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansson, A., C. Manetopoulos, J. I. Jonsson, and H. Axelson. 2003. The basic helix-loop-helix transcription factor TAL1/SCL inhibits the expression of the p16INK4A and pTα genes. Biochem. Biophys. Res. Commun. 312:1073-1081. [DOI] [PubMed] [Google Scholar]
- 18.Hoang, T., E. Paradis, G. Brady, F. Billia, K. Nakahara, N. N. Iscove, and I. R. Kirsch. 1996. Opposing effects of the basic helix-loop-helix transcription factor SCL on erythroid and monocytic differentiation. Blood 87:102-111. [PubMed] [Google Scholar]
- 19.Hofmann, T. J., and M. D. Cole. 1996. The TAL1/Scl basic helix-loop-helix protein blocks myogenic differentiation and E-box dependent transactivation. Oncogene 13:617-624. [PubMed] [Google Scholar]
- 20.Hsu, H. L., I. Wadman, J. T. Tsan, and R. Baer. 1994. Positive and negative transcriptional control by the TAL1 helix-loop-helix protein. Proc. Natl. Acad. Sci. U. S. A. 91:5947-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, S., and S. J. Brandt. 2000. mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol. Cell. Biol. 20:2248-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, S., Y. Qiu, Y. Shi, Z. Xu, and S. J. Brandt. 2000. P/CAF-mediated acetylation regulates the function of the basic helix-loop-helix transcription factor TAL1/SCL. EMBO J. 19:6792-6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, S., Y. Qiu, R. W. Stein, and S. J. Brandt. 1999. p300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein. Oncogene 18:4958-4967. [DOI] [PubMed] [Google Scholar]
- 24.Huang, Z., L. C. Dore, Z. Li, S. H. Orkin, G. Feng, S. Lin, and J. D. Crispino. 2009. GATA-2 reinforces megakaryocyte development in the absence of GATA-1. Mol. Cell. Biol. 29:5168-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kallianpur, A. R., J. E. Jordan, and S. J. Brandt. 1994. The SCL/TAL-1 gene is expressed in progenitors of both the hematopoietic and vascular systems during embryogenesis. Blood 83:1200-1208. [PubMed] [Google Scholar]
- 26.Kassouf, M. T., H. Chagraoui, P. Vyas, and C. Porcher. 2008. Differential use of SCL/TAL-1 DNA-binding domain in developmental hematopoiesis. Blood 112:1056-1067. [DOI] [PubMed] [Google Scholar]
- 27.Khandekar, M., W. Brandt, Y. Zhou, S. Dagenais, T. W. Glover, N. Suzuki, R. Shimizu, M. Yamamoto, K. C. Lim, and J. D. Engel. 2007. A Gata2 intronic enhancer confers its pan-endothelia-specific regulation. Development 134:1703-1712. [DOI] [PubMed] [Google Scholar]
- 28.Labastie, M. C., F. Cortes, P. H. Romeo, C. Dulac, and B. Peault. 1998. Molecular identity of hematopoietic precursor cells emerging in the human embryo. Blood 92:3624-3635. [PubMed] [Google Scholar]
- 29.Liu, Y., M. Encinas, J. X. Comella, M. Aldea, and C. Gallego. 2004. Basic helix-loop-helix proteins bind to TrkB and p21Cip1 promoters linking differentiation and cell cycle arrest in neuroblastoma cells. Mol. Cell. Biol. 24:2662-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikkola, H. K., J. Klintman, H. Yang, H. Hock, T. M. Schlaeger, Y. Fujiwara, and S. H. Orkin. 2003. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature 421:547-551. [DOI] [PubMed] [Google Scholar]
- 31.Mouthon, M. A., O. Bernard, M. T. Mitjavila, P. H. Romeo, W. Vainchenker, and D. Mathieu-Mahul. 1993. Expression of tal-1 and GATA-binding proteins during human hematopoiesis. Blood 81:647-655. [PubMed] [Google Scholar]
- 32.Okitsu, Y., S. Takahashi, N. Minegishi, J. Kameoka, M. Kaku, M. Yamamoto, T. Sasaki, and H. Harigae. 2007. Regulation of adipocyte differentiation of bone marrow stromal cells by transcription factor GATA-2. Biochem. Biophys. Res. Commun. 364:383-387. [DOI] [PubMed] [Google Scholar]
- 33.O'Neil, J., J. Shank, N. Cusson, C. Murre, and M. Kelliher. 2004. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell 5:587-596. [DOI] [PubMed] [Google Scholar]
- 34.Ono, Y., N. Fukuhara, and O. Yoshie. 1998. TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3. Mol. Cell. Biol. 18:6939-6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono, Y., N. Fukuhara, and O. Yoshie. 1997. Transcriptional activity of TAL1 in T cell acute lymphoblastic leukemia (T-ALL) requires RBTN1 or -2 and induces TALLA1, a highly specific tumor marker of T-ALL. J. Biol. Chem. 272:4576-4581. [DOI] [PubMed] [Google Scholar]
- 36.Park, S. T., G. P. Nolan, and X.-H. Sun. 1999. Growth inhibition and apoptosis due to restoration of E2A activity in T cell acute lymphoblastic leukemia cells. J. Exp. Med. 189:501-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, S. T., and X.-H. Sun. 1998. The Tal1 oncoprotein inhibits E47-mediated transcription. Mechanism of inhibition. J. Biol. Chem. 273:7030-7037. [DOI] [PubMed] [Google Scholar]
- 38.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U. S. A. 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porcher, C., W. Swat, K. Rockwell, Y. Fujiwara, F. W. Alt, and S. H. Orkin. 1996. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 86:47-57. [DOI] [PubMed] [Google Scholar]
- 40.Prabhu, S., A. Ignatova, S. T. Park, and X.-H. Sun. 1997. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol. Cell. Biol. 17:5888-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pulford, K., N. Lecointe, K. Leroy-Viard, M. Jones, D. Mathieu-Mahul, and D. Y. Mason. 1995. Expression of TAL-1 proteins in human tissues. Blood 85:675-684. [PubMed] [Google Scholar]
- 42.Robb, L., N. J. Elwood, A. G. Elefanty, F. Köntgen, R. Li, L. D. Barnett, and C. G. Begley. 1996. The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J. 15:4123-4129. [PMC free article] [PubMed] [Google Scholar]
- 43.Robb, L., I. Lyons, R. Li, L. Hartley, F. Köntgen, R. P. Harvey, D. Metcalf, and C. G. Begley. 1995. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc. Natl. Acad. Sci. U. S. A. 92:7075-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuh, A. H., A. J. Tipping, A. J. Clark, I. Hamlett, B. Guyot, F. J. Iborra, P. Rodriguez, J. Strouboulis, T. Enver, P. Vyas, and C. Porcher. 2005. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Mol. Cell. Biol. 25:10235-10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shivdasani, R. A., E. L. Mayer, and S. H. Orkin. 1995. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373:432-434. [DOI] [PubMed] [Google Scholar]
- 46.Souroullas, G. P., J. M. Salmon, F. Sablitzky, D. J. Curtis, and M. A. Goodell. 2009. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell 4:180-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanley, E. R. 1997. Murine bone marrow-derived macrophages, p. 301-304. In J. W. Pollard and John M. Walker (ed.), Methods in molecular biology, vol. 75. Basic cell culture protocols, 2nd ed. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 48.Tagoh, H., R. Himes, D. Clarke, P. J. Leenen, A. D. Riggs, D. Hume, and C. Bonifer. 2002. Transcription factor complex formation and chromatin fine structure alterations at the murine c-fms (CSF-1 receptor) locus during maturation of myeloid precursor cells. Genes Dev. 16:1721-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang, T., Y. Shi, S. R. Opalenik, D. M. Brantley-Sieders, J. Chen, J. M. Davidson, and S. J. Brandt. 2006. Expression of the TAL1/SCL transcription factor in physiological and pathological vascular processes. J. Pathol. 210:121-129. [DOI] [PubMed] [Google Scholar]
- 50.Tanigawa, T., N. Elwood, D. Metcalf, D. Cary, E. DeLuca, N. A. Nicola, and C. G. Begley. 1993. The SCL gene product is regulated by and differentially regulates cytokine responses during myeloid leukemic cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 90:7864-7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanigawa, T., N. Nicola, G. A. McArthur, A. Strasser, and C. G. Begley. 1995. Differential regulation of macrophage differentiation in response to leukemia inhibitory factor/oncostatin-M/interleukin-6: the effect of enforced expression of the SCL transcription factor. Blood 85:379-390. [PubMed] [Google Scholar]
- 52.Tsai, J., Q. Tong, G. Tan, A. N. Chang, S. H. Orkin, and G. S. Hotamisligil. 2005. The transcription factor GATA2 regulates differentiation of brown adipocytes. EMBO Rep. 6:879-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tushinski, R. J., I. T. Oliver, L. J. Guilbert, P. W. Tynan, J. R. Warner, and E. R. Stanley. 1982. Survival of mononuclear phagocytes depends on a lineage-specific growth factor that the differentiated cells selectively destroy. Cell 28:71-81. [DOI] [PubMed] [Google Scholar]
- 54.Visvader, J., C. G. Begley, and J. M. Adams. 1991. Differential expression of the LYL, SCL and E2A helix-loop-helix genes within the hemopoietic system. Oncogene 6:187-194. [PubMed] [Google Scholar]
- 55.Voronova, A. F., and F. Lee. 1994. The E2A and tal-1 helix-loop-helix proteins associate in vivo and are modulated by Id proteins during interleukin 6-induced myeloid differentiation. Proc. Natl. Acad. Sci. U. S. A. 91:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wadman, I. A., H. Osada, G. G. Grütz, A. D. Agulnick, H. Westphal, A. Forster, and T. H. Rabbitts. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wozniak, R. J., M. E. Boyer, J. A. Grass, Y. Lee, and E. H. Bresnick. 2007. Context-dependent GATA factor function: combinatorial requirements for transcriptional control in hematopoietic and endothelial cells. J. Biol. Chem. 282:14665-14674. [DOI] [PubMed] [Google Scholar]
- 58.Xu, Z., S. Huang, L. S. Chang, A. D. Agulnick, and S. J. Brandt. 2003. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol. Cell. Biol. 23:7585-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu, Z., X. Meng, Y. Cai, M. J. Koury, and S. J. Brandt. 2006. Recruitment of the SWI/SNF protein Brg1 by a multiprotein complex effects transcriptional repression in murine erythroid progenitors. Biochem. J. 399:297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu, Z., X. Meng, Y. Cai, H. Liang, L. Nagarajan, and S. J. Brandt. 2007. Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins. Genes Dev. 21:942-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, Z., T. Kondo, C. S. Voorhorst, S. C. Nabinger, L. Ndong, F. Yin, E. M. Chan, M. Yu, O. Würstlin, C. P. Kratz, C. M. Niemeyer, C. Flotho, E. Hashino, and R. J. Chan. 2009. Increased c-Jun and reduced GATA2 expression promotes aberrant monocytic differentiation induced by activating PTPN11 mutants. Mol. Cell. Biol. 29:4376-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, X. Y., and A. R. Rodaway. 2007. SCL-GFP transgenic zebrafish: in vivo imaging of blood and endothelial development and identification of the initial site of definitive hematopoiesis. Dev. Biol. 307:179-194. [DOI] [PubMed] [Google Scholar]