Abstract
Protein complexes containing Prp19 play a central role during catalytic activation of the spliceosome, and Prp19 and its related proteins are major components of the spliceosome's catalytic core RNP. To learn more about the spatial organization of the human Prp19 (hPrp19)/CDC5L complex, which is comprised of hPrp19, CDC5L, PRL1, AD002, SPF27, CTNNBL1, and HSP73, we purified native hPrp19/CDC5L complexes from HeLa cells stably expressing FLAG-tagged AD002 or SPF27. Stoichiometric analyses indicated that, like Saccharomyces cerevisiae NTC (nineteen complex), the human Prp19/CDC5L complex contains four copies of hPrp19. Salt treatment identified a stable core comprised of CDC5L, hPrp19, PRL1, and SPF27. Protein-protein interaction studies revealed that SPF27 directly interacts with each component of the hPrp19/CDC5L complex core and also elucidated several additional, previously unknown interactions between hPrp19/CDC5L complex components. Limited proteolysis of the hPrp19/CDC5L complex revealed a protease-resistant complex comprised of SPF27, the C terminus of CDC5L, and the N termini of PRL1 and hPrp19. Under the electron microscope, purified hPrp19/CDC5L complexes exhibit an elongated, asymmetric shape with a maximum dimension of ∼20 nm. Our findings not only elucidate the molecular organization of the hPrp19/CDC5L complex but also provide insights into potential protein-protein interactions at the core of the catalytically active spliceosome.
Pre-mRNA splicing, the two consecutive transesterification reactions leading to intron removal and exon ligation, is catalyzed by the spliceosome, a highly dynamic, multiple-megadalton molecular machine (41). The major subunits of the spliceosome are the U1, U2, U4, U5, and U6 small nuclear ribonucleoprotein particles (snRNPs). Each snRNP consists of an RNA moeity, the snRNA, and a set of particle-specific proteins, plus seven Sm proteins (or Lsm proteins in the case of U6) that are found in all of the spliceosomal snRNPs. In addition, the spliceosome is comprised of numerous non-snRNP proteins, some of which are preassembled into stable heteromeric complexes.
Spliceosome assembly occurs in a stepwise and highly dynamic manner (41). Initially, the U1 snRNP binds the 5′ splice site, followed by the ATP-dependent recognition of the pre-mRNA's branch point sequence (BPS) by the U2 snRNP, forming the prespliceosome or A complex. The assembly of snRNPs on the pre-mRNA is completed by the addition of the U4/U6-U5 tri-snRNP, generating the precatalytic B complex, which is still catalytically inactive. In order to catalyze the first step of splicing, the spliceosome must undergo dramatic compositional and structural remodeling events, culminating in the destabilization of the U1 and U4 snRNPs and the formation of the catalytically activated spliceosome (B* complex). The first transesterification reaction then occurs, generating the C complex, which in turn catalyzes the second step of splicing. After catalysis, the postspliceosomal complex dissociates, releasing the mRNA and the U2, U5, and U6 snRNPs, which are recycled for subsequent rounds of splicing.
The spliceosome is a very protein-rich molecular machine. In addition to snRNP proteins, the spliceosome contains numerous non-snRNP proteins, many of which play essential roles during splicing (42). One such protein is Prp19, an evolutionary highly conserved splicing factor required for the activation of the spliceosome (8, 26). Both Saccharomyces cerevisiae and human Prp19 (hPrp19) proteins were also shown to exhibit E3 ubiquitin ligase activity in vitro (18, 28), but the significance of this activity for Prp19's function in splicing is currently not known. Prp19 is present in cells as part of a stable heteromeric complex both in yeast (i.e., NTC [nineteen complex]) (36) and in humans (i.e., the hPrp19/CDC5L complex) (2, 24). Mass spectrometry of immunoaffinity-purified human Prp19/CDC5L complexes showed that they consist of seven proteins: hPrp19, CDC5L, PRL1, AD002, SPF27, CTNNBL1 (β-catenin-like 1), and HSP73 (24). In total, at least 11 proteins were reported to be part of the yeast NTC (9, 26, 36). Homologs of four of the human Prp19/CDC5L complex components are also found in S. cerevisiae NTC, namely, Prp19p (hPrp19), Cef1p (CDC5L), Snt309p (SPF27), and Prp46p (PRL1). Whereas Prp19p, Cef1p, and Snt309p are stably bound in the yeast NTC, Prp46p dissociates from the complex upon gradient centrifugation, suggesting that it is only loosely associated with the complex (36). In contrast, proteins homologous to human CTNNBL1, HSP73, and AD002 are not found in the yeast NTC.
Yeast Prp19p was shown to form a tetramer in vivo and in vitro (29). Although the human Prp19 protein was shown to self-interact (17), it is presently unclear whether it also forms a tetramer. Indeed, little is currently known about the molecular architecture of the human Prp19/CDC5L complex. As most of its components are present in the salt-stable RNP core of the human C complex (see below), information about its organization should provide initial insights into potential protein-protein interactions in the core of the catalytically active spliceosome.
Comparative proteomic analyses of spliceosomal complexes A, B, and C indicated that the human Prp19/CDC5L complex interacts with the spliceosome prior to its catalytic activation but that its association is stabilized during the B complex-to-C complex transition (4, 12, 22, 24). A set of proteins (designated Prp19-related proteins) that interact with the human or yeast Prp19 complex and/or are present in the human 35S U5 snRNP also associate with the spliceosome at this stage (9, 22, 27). Immunodepletion/complementation studies with HeLa nuclear extract demonstrated that the hPrp19/CDC5L complex is required for pre-mRNA splicing prior to the first step of splicing (24). Like its human counterpart, the yeast NTC is required for pre-mRNA splicing. During catalytic activation, the NTC stabilizes the association of U5 and U6 with the spliceosome, thereby specifying interactions of the U5 and U6 snRNAs with the pre-mRNA (7, 8). Consistent with this, Prp19-related proteins and subunits of the hPrp19/CDC5L complex stably interact with the human U5 snRNP within the spliceosome and remain associated during both catalytic steps (22). Significantly, most hPrp19/CDC5L complex and related proteins, together with U5 proteins, remain associated with a 1 M NaCl-resistant RNP core of the C complex (6). This suggests that they are tightly incorporated components of the C complex and play an important role in maintaining its catalytically active RNA network. Finally, the hPrp19/CDC5L complex is released from the spliceosome as part of the 35S U5 snRNP, which is thought to be a disassembly intermediate of the postspliceosomal U2-, U5-, and U6-containing complex (22).
Here we have isolated preparative amounts of native human Prp19/CDC5L complexes by using HeLa cell lines expressing FLAG-tagged AD002 or SPF27. Subsequent analysis of the stoichiometry of the hPrp19/CDC5L complex indicated that it contains four copies of the hPrp19 protein and likely single copies of all other components. Treatment of the complex with the protease subtilisin or high concentrations of salt revealed a salt- and protease-resistant core composed of hPrp19, CDC5L, PRL1, and SPF27. A complex protein-protein interaction network involving these proteins, as well as the less stably associated AD002 and CTNNBL1 proteins, was also elucidated using a variety of methods. Finally, by performing negative-stain electron microscopy (EM) we determined the overall two-dimensional (2D) structure of the hPrp19/CDC5L complex. Together, our findings provide important insights into the molecular organization of the hPrp19/CDC5L complex and additionally pave the way for future functional and high-resolution structural analyses of this essential complex that is part of the spliceosome's catalytically active RNP core.
MATERIALS AND METHODS
Plasmids.
To generate FLAG/hemagglutinin (HA)-tagged AD002 and SPF27, cDNAs encoding these proteins were cloned into a modified pIRESneo plasmid (Clontech) containing an N-terminal FLAG/HA tag (25) using PCR-based techniques. A StrepII tag (40) was introduced into the pEU3-NII vector (32), generating pEU3-NII-StrepII. cDNAs encoding all seven hPrp19/CDC5L complex proteins were cloned into pEU3-NII-StrepII via PCR. Primers and cloning strategies are described in detail in the supplemental material.
In vitro splicing.
Nuclear extracts were prepared from HeLa cells according to the method described in reference 13. In vitro splicing reaction mixtures contained 5 nM 32P-labeled MINX pre-mRNA (43) and 40% (vol/vol) HeLa nuclear extract in buffer D (20 mM HEPES-KOH [pH 7.9], 100 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA [pH 8.0], 10% [vol/vol] glycerol, 0.5 mM dithioerythritol [DTE], 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) and were supplemented with 25 mM KCl, 3.6 mM MgCl2, 2 mM ATP, and 20 mM creatine phosphate. The reaction mixture was incubated at 30°C for 0 to 90 min. RNA was then isolated, separated on an 8.3 M urea-12% polyacrylamide gel, and visualized by autoradiography.
Purification of the human Prp19/CDC5L complex.
HeLa S3 cell lines stably expressing FLAG/HA-tagged human AD002 or SPF27 were generated as described previously (25). For affinity purification of human Prp19/CDC5L complexes, nuclear extract was prepared from HeLa cells expressing FLAG-AD002 or FLAG-SPF27. For the preparative isolation of the hPrp19/CDC5L complex, nuclear extract was mixed with an equal volume of G250 buffer (20 mM HEPES-KOH, pH 7.9, 1.5 mM MgCl2, 250 mM NaCl) containing 0.5 mM DTE and 0.5 mM PMSF and incubated overnight. The extract was cleared by centrifugation for 30 min at 125,171 × g in a 45 Ti rotor (Beckman Coulter), and the supernatant was loaded onto anti-FLAG-M2-agarose beads (Sigma). The beads were subsequently washed with G250 buffer containing 2.5% (vol/vol) glycerol, 0.5 mM DTE, and 0.5 mM PMSF, and bound material was eluted with G250 buffer containing 2.5% (vol/vol) glycerol, 1 mM dithiothreitol (DTT), and 0.2 mg/ml FLAG peptide (Sigma). Complexes were further purified via Superose 6 GL 10/300 gel filtration (GE Healthcare) in SEC100 buffer (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM DTT). Alternatively, they were subjected to 5-to-20% (vol/vol) glycerol gradient centrifugation (13 h at 164,326 × g in a Sorvall TH660 rotor) in G150 buffer (20 mM HEPES-KOH, pH 7.9, 1.5 mM MgCl2, 150 mM NaCl). Proteins present in each gradient or gel filtration fraction were analyzed on a 12% polyacrylamide-SDS gel and stained with Coomassie blue.
Iodoacetamide labeling and Sypro-Ruby staining.
For iodoacetamide modification, 50 μg of affinity-purified hPrp19/CDC5L complex was initially treated with 8 M urea and 10 mM β-mercaptoethanol for 90 min at 37°C. Subsequently, the pH was increased to 9.0 with 100 mM Tris-HCl buffer, and [14C]iodoacetamide (56 mCi/mmol; GE Healthcare) was added to a concentration of 50 mM. The reaction volume was incubated for 6 min at 37°C and stopped by adding β-mercaptoethanol (final concentration, 0.5 M). Aliquots of 12.5 μg, 4.2 μg, 1.4 μg, and 0.5 μg were separated by SDS gel electrophoresis, and the gel was stained with Sypro-Ruby as recommended by the manufacturer (Invitrogen). Fluorescence intensity and radioactivity of the proteins were monitored by fluorescence scanning and autoradiography, respectively, and quantified using the program Fuji Image Gauge v4.22.
Analytical ultracentrifugation.
Sedimentation velocity and equilibrium analysis were performed on an Optima XL-A analytical ultracentrifuge (Beckman-Coulter) by using an An60 Ti rotor and double-sector cells (path length, 12 mm). For sedimentation velocity experiments, 400-μl samples in phosphate-buffered saline (PBS; pH 8.0) were analyzed at 20°C and a rotor speed of 40,000 rpm. The evolution of the boundary was monitored by UV measurement at 280 nm. Buffer density, viscosity, and protein partial specific volumes were calculated using Sednterp software (version 1.09) based on the amino acid sequence and assuming no posttranslational modifications. Raw data were analyzed using the program Sedfit (version 11.8; see reference 34). For each analysis, 30 scans were continuously collected. The size distributions were calculated while floating the meniscus, frictional coefficient, and baseline but keeping the buffer parameters constant, with a confidence level of a P value of 0.68, a resolution (n) of 150 Svedberg units, and sedimentation coefficients between 2 and 40S. Sedimentation equilibrium data were obtained with 180-μl samples at rotor speeds of 3,500 and 5,500 rpm. The approach to equilibrium was monitored at 6-h intervals by using the program Sedfit. Final scans were recorded after no systematic deviation was observed between two subsequent scans. Species analysis was performed with the Sedphat software (33).
Salt and heparin dissociation of the hPrp19/CDC5L complex.
Twenty-five micrograms of affinity-purified hPrp19/CDC5L complex was incubated for 30 min on ice in the presence of 150 to 1,200 mM NaCl, 150 mM NaCl and 0.1 mg/ml heparin, or 150 mM NaCl and 0.4 M NaSCN. Subsequently, the treated complexes were loaded onto a 4-ml 5-to-20% (vol/vol) glycerol gradient containing 20 mM HEPES-KOH (pH 7.9), 1.5 mM MgCl2, and the corresponding concentrations of NaCl, heparin, and NaSCN and centrifuged at 4°C for 16 h at 164,326 × g in a TH660 rotor. Gradients were harvested from the top, and proteins in each fraction were analyzed on a 12% polyacrylamide-SDS gel and stained with Coomassie blue.
Sulfo-MBS cross-linking and antibodies.
Exchange of the buffer to PBS (pH 7.2) was performed with 50 μg of affinity-purified hPrp19/CDC5L complexes, using a preequilibrated Bio-Gel P6 column (Bio-Rad). The complex was divided into three equal aliquots and incubated for 30 min at 20°C with 0 μM, 1 μM, or 10 μM Sulfo-MBS (Pierce) in a final volume of 100 μl PBS (pH 7.2). The reaction was stopped by adding 12 μl of 0.5 M glycinamide-hydrochloride and 8 μl of 150 mM DTT and incubating for 30 min at 37°C. Proteins were precipitated with acetone, separated on a 10% polyacrylamide-SDS gel, and transferred to nitrocellulose membrane. Western blotting was performed using an ECL detection kit (GE Healthcare) with antibodies against each hPrp19/CDC5L complex protein. Antibodies against the following proteins were used in this study: 61K (23), CDC5L and AD002 (24), CTNNBL1 (kindly provided by Dierk Ingelfinger), hPrp19 (raised against residues 176 to 191), HSP73 (Santa Cruz), PRL1 (6), and SPF27 (raised against residues 37 to 50).
Far-Western overlay analysis and coimmunoprecipitation assays.
Capped mRNAs were synthesized in vitro by T7 runoff transcription of linearized pEU3-NII-StrepII-derived plasmids. Cell-free translation of mRNA transcribed in vitro was performed using wheat germ extract (Zoegene, Japan) in the presence of [35S]methionine or [14C]leucine (as required) essentially as described in reference 32. For far-Western analyses, affinity-purified hPrp19/CDC5L complex proteins were separated on a 10% polyacrylamide-SDS gel, transferred to nitrocellulose, and visualized by staining with Ponceau S. Far-Western overlays were performed with 35S-labeled proteins as described previously (1). For pulldown assays, 40 μl of in vitro-translated, unlabeled protein with a StrepII tag was incubated with 40 μl of translated 14C-labeled protein without a StrepII tag for 90 min at 0°C prior to the addition of 15 μl of Strep-Tactin Sepharose (IBA, Göttingen, Germany) and 200 μl of IPP150 buffer (20 mM HEPES-KOH [pH 7.9], 150 mM NaCl, 1.5 mM MgCl2, 0.05% [vol/vol] NP-40). The mixture was incubated with end-over-end rotation for 1 h. The Sepharose beads were washed four times with IPP150 buffer, and bound protein was recovered and analyzed by SDS-PAGE and autoradiography.
Limited proteolysis of hPrp19/CDC5L complexes.
Limited proteolysis was performed on ice with subtilisin (Sigma) by incubating 150 μg of affinity-purified hPrp19/CDC5L complex with 0.02 μg, 0.05 μg, 0.1 μg, 0.2 μg, or 0.4 μg subtilisin. The reaction was stopped by the addition of 1 μl of 0.1 M PMSF (in isopropanol) and analyzed by size exclusion chromatography on a SMART fast-performance liquid chromatography system using a Superose 6 PC 3.2/30 gel filtration column (GE Healthcare) in SEC100 buffer. Proteins from pairwise pooled fractions were separated on a 15% polyacrylamide-SDS gel, stained with Coomassie blue, and analyzed by mass spectrometry after digestion with trypsin.
Electron microscopy.
Negative staining was carried out by the double carbon film method (19). Particles were fixed using the GraFix procedure (20) on a 4-ml linear 5-to-20% (vol/vol) glycerol and 0-to-0.1% (vol/vol) glutaraldehyde gradient containing G150 buffer. Images were recorded on a CM 200 FEG electron microscope (Philips, Eindhoven, Netherlands) at a primary magnification of ×105,000 with a 4,096- by 4,096-pixel charge-coupled-device (CCD) camera (TemCam-F415; TVIPS, Gauting, Germany) with a Philips holder. Images were taken under low-dose conditions at an ∼1.5-μm defocus. Imagic-5 was used for image processing of ∼5,000 particles (39). Briefly, after a “reference-free” alignment procedure (14), images were subjected to multivariate statistical analysis (MSA) and classification (38). Based on the classification, ∼30 individual molecular images were averaged. The resulting class averages were used as reference images in subsequent rounds of alignment until the computed class averages were stable.
Mass spectrometry.
Proteins were separated by SDS gel electrophoresis, and Coomassie blue-stained protein bands were excised from the gel. Proteins were digested with trypsin as described in reference 35. Extracted peptides were separated on a C18 reversed-phase chromatography column and subsequently delivered to the mass spectrometer via liquid chromatography (LC)-online electrospray ionization-tandem mass spectrometry (ESI-MS-MS), using either a hybrid Linear Ion Trap-Orbitrap mass spectrometer (LTQ-Orbitrap XL; Thermo Scientific) or a high-resolution quadrupole time of flight analyzer (Micromass qToF II Ultima; Waters). Alternatively, dried droplets were prepared from LC peptide fractions (30) and analyzed by matrix-assisted laser desorption ionization-MS-MS (MALDI-MS-MS) with a tandem time of flight mass analyzer (4800 MALDI ToF/ToF analyzer; Applied Biosystems/MDS Sciex). Proteins were identified by searching fragment spectra of sequenced peptides against the NCBI nonredundant database by using Mascot as the search engine (31). Peptides whose N- or C-terminal end was generated by digestion with subtilisin, which preferentially cleaves large hydrophobic residues, were identified by manual inspection of the fragment spectra. In addition, the precise N terminus of the protease-resistant CDC5L fragment in the largest protease-resistant complex was determined by Edman sequencing.
RESULTS
Isolation of human Prp19/CDC5L complexes.
In previous studies, hPrp19-containing complexes were isolated from HeLa nuclear extracts by anti-CDC5L immunoaffinity chromatography (2, 24). However, only limited amounts could be obtained using this method, thereby preventing an in-depth analysis of the structure and molecular organization of the human Prp19/CDC5L complex. In order to purify preparative amounts of this complex, we established HeLa S3 cell lines stably expressing N-terminally FLAG/HA-tagged AD002 and SPF27, both integral components of the hPrp19/CDC5L complex. Western blotting confirmed the presence of the tagged proteins in nuclear extract prepared from these cell lines (Fig. 1A). Nuclear extract from both stable cell lines exhibited the same splicing activity as the wild-type extract (Fig. 1B). Furthermore, the tagged proteins were incorporated into affinity-purified spliceosomes to the same extent as the wild-type proteins (data not shown). Thus, the presence of a FLAG/HA tag does not alter the function of either protein and appears to have little or no effect on the structure of the hPrp19/CDC5L complex.
FIG. 1.
Characterization of HeLa cell lines stably expressing FLAG/HA-tagged AD002 and SPF27. (A) Nuclear extract was prepared from FLAG/HA-AD002, FLAG/HA-SPF27, or control cell lines as indicated above. Proteins were recovered from increasing amounts of nuclear extract (as indicated above each lane) and separated by SDS-PAGE. Proteins were transferred to nitrocellulose and further analyzed by Western blotting with anti-FLAG (lanes 1 to 12, middle panels), anti-AD002 (lanes 1 to 6, upper panel), and anti-SPF27 antibodies (lanes 7 to 12, upper panel). For a loading control, all membranes were also probed with anti-61K antibodies (lanes 1 to 12, bottom panel). The position of the endogenous and the FLAG/HA-tagged protein is indicated on the right of each panel. Note that in the middle panel, the anti-FLAG-SPF27 signal does not increase when more than 2.5 μl extract is analyzed, as the amount of anti-FLAG SPF27 present is saturating for the anti-FLAG antibody. (B) The presence of a FLAG/HA tag does not affect pre-mRNA splicing. Splicing was performed with 32P-labeled MINX pre-mRNA in HeLa nuclear extract prepared from the control cell line (lanes 1 to 6), FLAG/HA-AD002 cell line (lanes 7 to 12), or FLAG/HA-SPF27 (lanes 13 to 18) for 0 to 90 min as indicated. RNA was recovered and separated on an 8.3 M urea-10% polyacrylamide gel. The 32P-labeled pre-mRNA and splicing intermediates or products (indicated schematically on the left) were detected by autoradiography.
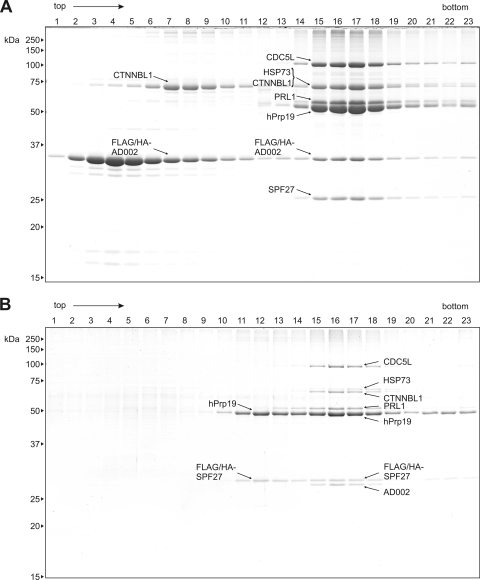
Immunoaffinity selection was performed with anti-FLAG agarose and nuclear extract prepared from both of these cell lines. Complexes were eluted under native conditions with FLAG peptide and further purified by glycerol gradient centrifugation. Proteins were isolated from fractions across the gradient and analyzed by SDS-PAGE (Fig. 2A and B). In both cases, a multicomponent complex peaking in fractions 15 to 18 was observed. This affinity-purified complex was comprised of hPrp19, CDC5L, HSP73, CTNNBL1 (β-catenin-like 1), PRL1, AD002, and SPF27, as determined by mass spectrometry, confirming it is the hPrp19/CDC5L complex. Interestingly, a fraction of FLAG/HA-AD002 and CTNNBL1 (Fig. 2A, lanes 7 to 9), as well as FLAG/HA-SPF27 and hPrp19 (Fig. 2B, lanes 11 to 13), cosedimented, indicating that these proteins can interact with one another.
FIG. 2.
Affinity purification of the hPrp19/CDC5L complex. Complexes present in nuclear extract prepared from HeLa cell lines expressing FLAG/HA-tagged AD002 or SPF27 were immunoaffinity selected using anti-FLAG-M2-agarose and subsequently eluted under native conditions with FLAG peptide. Affinity-purified complexes were then fractionated on a linear 5-to-20% glycerol gradient. Proteins present in each gradient fraction after affinity purification from the FLAG/HA-AD002 cell line (A) and the FLAG/HA-SPF27 cell line (B) were analyzed by SDS-PAGE and stained with Coomassie blue. The molecular masses of marker proteins are indicated on the left. Proteins were identified by mass spectrometry and are indicated by arrows as follows: CDC5L (gi 11067747), HSP73 (gi 5729877), CTNNBL1 (gi 18644734), PRL1 (gi 4505895), hPrp19 (gi 7657381), AD002 (gi 6523797), and SPF27 (gi 5031653).
Analysis of the stoichiometry of the native hPrp19/CDC5L complex reveals hPrp19 tetramerization.
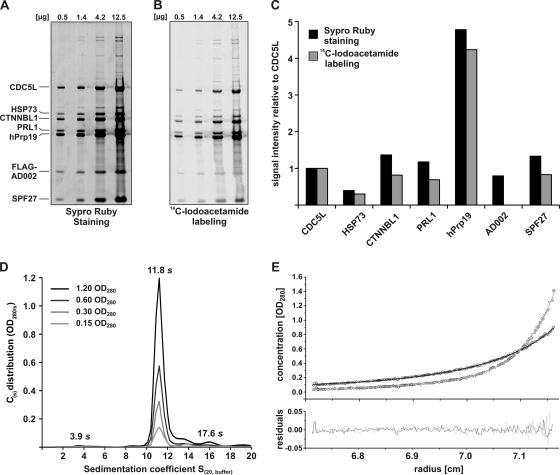
We next determined the stoichiometry of the human Prp19/CDC5L complex. For this purpose, we stained SDS-PAGE-separated hPrp19/CDC5L complex proteins with Sypro-Ruby (Fig. 3A) or we labeled denatured proteins with [14C]iodoacetamide prior to SDS-PAGE (Fig. 3B). Band intensities were quantified and normalized against the CDC5L signal (Fig. 3C). As the signal obtained via iodoacetamide labeling of AD002 was not linear over the concentration range analyzed (Fig. 3B), its relative amount could not be quantified using this method. Both methods revealed that approximately four times more hPrp19 is present relative to the CDC5L protein. Except for HSP73, which seems to be underrepresented, all other components appear to be present in equimolar amounts.
FIG. 3.
The native human Prp19/CDC5L complex contains four copies of hPrp19. (A) Determination of the protein stoichiometry by fluorescence staining. Four different amounts of hPrp19/CDC5L complex proteins (as indicated above each lane) were separated by SDS gel electrophoresis and stained with Sypro-Ruby. The fluorescence was detected by fluorescence imaging (FLA-7000; Fujifilm). (B) Determination of the protein stoichiometry by radioactive labeling. Denatured proteins (0.5 to 12.5 μg, as indicated) were labeled with [14C]iodoacetamide, separated by SDS-PAGE and detected by autoradiography. (C) Band intensities obtained after Sypro-Ruby staining and [14C]iodoacetamide labeling were quantified using Fuji Image Gauge v4.22 and normalized against the CDC5L signal. (D) Continuous size distribution [C(S)] analysis of the hPrp19/CDC5L complex. Four different concentrations of the complex were analyzed (optical densities at 280 nm [OD280] of 1.2, 0.6, 0.3, and 0.15) as indicated in the figure. The calculated C(S) is plotted against the sedimentation coefficient (S). The S values in the panel are corrected according to standard conditions [S(20,w)]. (E) Multispeed sedimentation equilibrium analysis of the hPrp19/CDC5L complex at two different rotor speeds (gray, 5,500 rpm; black, 3,500 rpm). The OD280 values along the cell were recorded and are shown in the upper panel. The lower panel shows the residuals between the fit (straight line in the upper panel) and the raw data (dots in the upper panel).
To confirm these results, we also performed sedimentation velocity and equilibrium analytical ultracentrifugation (Fig. 3D and E). Analysis of the velocity data at four different concentrations showed that 84% of the hPrp19/CDC5L complex exhibited a weight-averaged sedimentation coefficient at 20°C in water [S(20,w)] of 11.8 (Fig. 3D). Furthermore, two additional species were identified that sediment at 3.9S (2.1%) and 17.6S (5%). However, over the range of analyzed concentrations, no concentration-dependent associations were observable. Based on a single-species model, the apparent molecular mass of the 11.8S complex is approximately 600 kDa. Interestingly, these analyses also revealed that the hPrp19/CDC5L complex has a frictional ratio value of 2.1, indicating that the particle's shape is elongated. To analyze the molecular mass of the hPrp19/CDC5L complex in more detail, we additionally performed multispeed sedimentation equilibrium analyses at two different rotor speeds (Fig. 3E). Using the information from the continuous size distribution [C(S)] analysis, a molecular mass of 608 kDa ± 16.4 kDa for the 11.8S species was obtained using the Sedphat program (33). This is consistent with the presence of four copies of hPrp19 and single copies of the other proteins. Thus, within the hPrp19/CDC5L complex, hPrp19 is present as a tetramer.
hPrp19, CDC5L, PRL1, and SPF27 form a salt-stable core of the hPrp19/CDC5L complex.
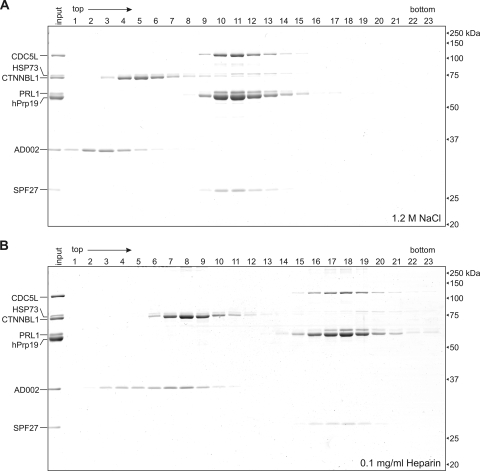
To analyze the molecular organization of the hPrp19/CDC5L complex in more detail, we treated FLAG-AD002 purified complexes with buffer containing increasing concentrations of NaCl or containing 0.1 mg/ml heparin. Subsequently, we performed glycerol gradient centrifugation under the same conditions. As shown in Fig. 4, hPrp19, CDC5L, PRL1, and SPF27 comigrate after treatment of the hPrp19/CDC5L complex with 1.2 M NaCl (Fig. 4A, lanes 10 to 13) or 0.1 mg/ml heparin (Fig. 4B, lanes 17 to 19), indicating that they form a stable subcomplex. The intense staining of the hPrp19 band, together with the fact that no free hPrp19 is observed at the top of the gradient, indicates that the hPrp19 tetramer is present in this stable subcomplex. In contrast, AD002, CTNNBL1, and HSP73 are found in the top fractions of the gradients, demonstrating that they have dissociated and are thus more loosely associated components. Similar results were obtained when complexes were treated with a 0.4 M concentration of the chaotropic salt NaSCN (see Fig. S2A in the supplemental material). However, a portion of AD002 and CTNNBL1 still comigrated after treatment with heparin (Fig. 4B, lane 8) or 300 mM NaCl (Fig. S2B), indicating that they interact. However, at higher NaCl concentrations and at 0.4 M NaSCN, the interaction between the two proteins was completely disrupted (i.e., they no longer comigrated during gradient centrifugation) (Fig. 4A; see Fig. S2A in the supplemental material). We also note that CTNNBL1 and HSP73 appear to comigrate after dissociation from the hPrp19/CDC5L complex, suggesting that they interact with one another. However, as CTNNBL1 and HSP73 have nearly identical molecular masses and we cannot distinguish between ∼75 kDa (single proteins) and 150 kDa (dimers) in this region of the gradient, and as they also do not appear to be present in equimolar amounts, it is likely that CTNNBL1 and HSP73 dissociate as individual proteins. Taken together, these data demonstrate that the stable core of the human Prp19/CDC5L complex is comprised of hPrp19, CDC5L, PRL1, and SPF27.
FIG. 4.
CDC5L, PRL1, hPrp19, and SPF27 form a salt-stable subcomplex. Purified hPrp19/CDC5L complexes were incubated with 1.2 M NaCl (A) or 0.1 mg/ml heparin (B) for 30 min on ice. Subsequently, the complexes were fractionated on a linear 5-to-20% glycerol gradient containing the same salt or heparin concentration. Proteins across the gradient were analyzed by SDS-PAGE and stained with Coomassie blue. The positions of the proteins are indicated on the left, and the molecular masses of marker proteins are indicated on the right.
Cross-linking elucidates protein-protein interactions within the native hPrp19/CDC5L complex.
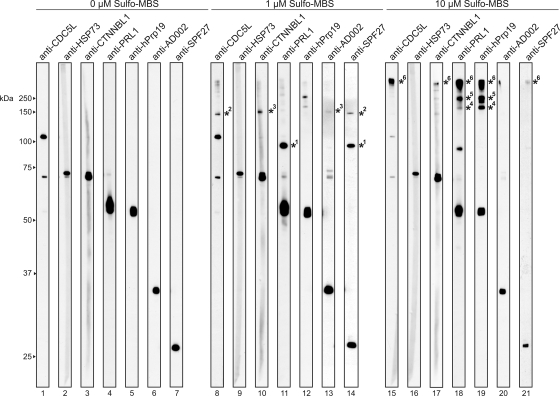
To further analyze interactions among the hPrp19/CDC5L complex proteins, we performed protein-protein cross-linking with native complexes and the heterobifunctional agent Sulfo-MBS. After SDS-PAGE, cross-links were identified by Western blotting with antibodies specific for each hPrp19/CDC5L subunit (Fig. 5, lanes 1 to 7). At 1 μM Sulfo-MBS, a strong cross-link between the PRL1 and SPF27 proteins was detected (Fig. 5, lanes 11 and 14). Furthermore, cross-links were observed between CDC5L and SPF27 (Fig. 5, lanes 8 and 14) and between CTNNBL1 and AD002 (Fig. 5, lanes 10 and 13). These data demonstrate physical interactions between each of the cross-linked protein pairs within the hPrp19/CDC5L complex. At 10 μM Sulfo-MBS, several high-molecular-mass cross-links were observed. One cross-link appeared to contain solely hPrp19 (Fig. 5, lane 19), although a weak signal at this position was also observed with anti-PRL1 antibodies (Fig. 5, lane 18). Based on its size (∼200 kDa), it could represent the hPrp19 tetramer or alternatively three copies of hPrp19 plus PRL1, as both proteins exhibit molecular masses of ∼50 kDa. An additional band at 250 kDa was recognized by antibodies against hPrp19 and PRL1, suggesting an interaction of the hPrp19 tetramer with the PRL1 protein (Fig. 5, lanes 18 and 19). Finally, a high-molecular-mass cross-linked complex was detected with antibodies against CDC5L, PRL1, and hPrp19 (Fig. 5, lanes 15, 18, and 19) and to a lesser extent by antibodies against CTNNBL1 and SPF27 (Fig. 5, lanes 17 and 21). Although only a faint signal can be seen with the anti-SPF27 antibodies at the position of this cross-link, the overall signal for SPF27 is generally lower (Fig. 5, lane 21). Therefore, it is likely that this cross-linked complex corresponds to the hPrp19/CDC5L core complex alone or together with CTNNBL1. Taken together, these data confirm physical links between PRL1-SPF27, CDC5L-SPF27, CTNNBL1-AD002, and hPrp19-PRL1.
FIG. 5.
Identification of protein-protein interactions in the hPrp19/CDC5L complex via cross-linking. Purified hPrp19/CDC5L complexes were incubated with 0 μM (lanes 1 to 7), 1 μM (lanes 8 to 14), or 10 μM (lanes 15 to 21) Sulfo-MBS. Cross-linked complexes were analyzed by Western blotting with the antibodies indicated above each lane. Cross-linked proteins are indicated by an asterisk with a number, where proteins in the same cross-linked complex are indicated by the same number next to a given asterisk.
Analysis of protein-protein interactions with in vitro-translated proteins.
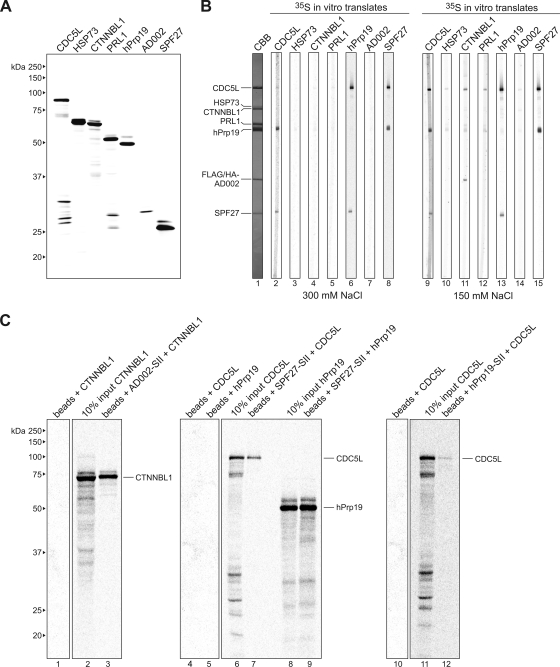
To elucidate additional protein-protein interactions in the hPrp19/CDC5L complex, we performed far-Western analyses. Thus, hPrp19/CDC5L complex proteins were separated by SDS-PAGE, transferred to nitrocellulose (Fig. 6B, lane 1), and incubated with in vitro-translated, 35S-labeled proteins (Fig. 6A). Consistent with the aforementioned results, at 300 mM salt, 35S-labeled SPF27 interacted with CDC5L and hPrp19 (Fig. 6B, lane 8). Reciprocally, 35S-labeled CDC5L and hPrp19 interacted with SPF27 (Fig. 6B, lanes 2 and 6). Furthermore, 35S-labeled CDC5L also interacted with Prp19 and vice versa (Fig. 6B, lanes 2 and 6). In addition to detecting these interactions, we also detected interactions between CTNNBL1 and AD002 and also between CTNNBL1 and CDC5L, which were observed only at 150 mM salt (Fig. 6B, lane 11). Although indirect interactions cannot be excluded when binding studies are performed with in vitro-translated proteins (which contain numerous other proteins), these data suggest a direct link between AD002-CTNNBL1 and a protein of the hPrp19/CDC5L complex core. Similar to previously published results (3), we also detected an interaction between PRL1 and CDC5L at 150 mM but not at 300 mM NaCl (Fig. 6B, lanes 5 and 12) and an interaction between CDC5L and itself (Fig. 6B, lanes 2 and 9).
FIG. 6.
Far-Western and coimmunoprecipitation analyses reveal additional interactions between components of the hPrp19/CDC5L complex. (A) SDS-PAGE analysis of in vitro-translated 35S-labeled proteins (as indicated above each lane). Proteins were visualized by autoradiography. The molecular masses of marker proteins are indicated on the left. (B) Far-Western overlay analysis of protein-protein interactions between individual hPrp19/CDC5L complex proteins. SDS-PAGE-separated hPrp19/CDC5L complex proteins were blotted onto nitrocellulose strips and visualized by staining with Coomassie blue (CBB; lane 1) or were incubated with 35S-labeled, in vitro-translated hPrp19/CDC5L complex proteins in buffer containing 300 mM (lanes 2 to 8) or 150 mM (lanes 9 to 15) NaCl and visualized by autoradiography (lanes 2 to 15). hPrp19/CDC5L complex proteins are indicated on the left. (C) Coimmunoprecipitation confirms interactions between SPF27 and both CDC5L and hPrp19, between CTNNBL1 and AD002, and between CDC5L and hPrp19. In vitro-translated 14C-labeled CTNNBL1, CDC5L, and hPrp19, as indicated above each lane, were incubated alone (lanes 1, 4, 5, and 10) or with StrepII (SII)-tagged AD002 (lane 3), SPF27 (lanes 7 and 9), or hPrp19 (lane 12), and pulldowns were subsequently performed with Strep-Tactin Sepharose beads. Coprecipitated proteins or 10% of the input (lanes 2, 6, 8, and 11) were analyzed by SDS-PAGE and visualized by autoradiography. The presence of the StrepII-tagged protein was confirmed by Coomassie blue staining (data not shown). The identities of the coprecipitated proteins are indicated on the right.
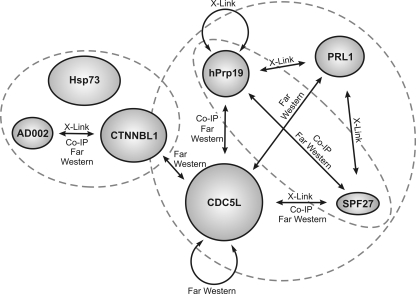
Interaction partners in the hPrp19/CDC5L complex were also analyzed by in vitro pulldown assays. For this purpose, AD002, SPF27, and hPrp19 containing a StrepII tag were translated in vitro and incubated with in vitro-translated 14C-labeled CDC5L, hPrp19, or CTNNBL1. Proteins containing the StrepII tag were then precipitated with StrepTactin Sepharose, and coprecipitating proteins were analyzed by SDS-PAGE. As shown in Fig. 6C, we could confirm interactions between AD002 and CTNNBL1 (lane 3), SPF27 and CDC5L (lane 7), SPF27 and hPrp19 (lane 9), and CDC5L and hPrp19 (lane 12). No interaction with the StrepTactin Sepharose alone was observed with radiolabeled CDC5L, hPrp19, or CTNNBL1 (lanes 1, 4, 5, and 10). A summary of interactions within the hPrp19/CDC5L complex is shown in Fig. 7. Taken together, these data indicate a strong interaction network among proteins of the hPRP19/CDC5L complex, in particular among proteins comprising its highly stable core.
FIG. 7.
Schematic overview of the protein-protein interactions within the hPrp19/CDC5L complex. Interactions identified in this study are indicated by an arrow together with the methods demonstrating a given interaction. The dashed circles enclose proteins that were found in stable subcomplexes. Co-IP, coimmunoprecipitation.
Limited proteolysis of the hPrp19/CDC5L complex delineates protein interaction domains.
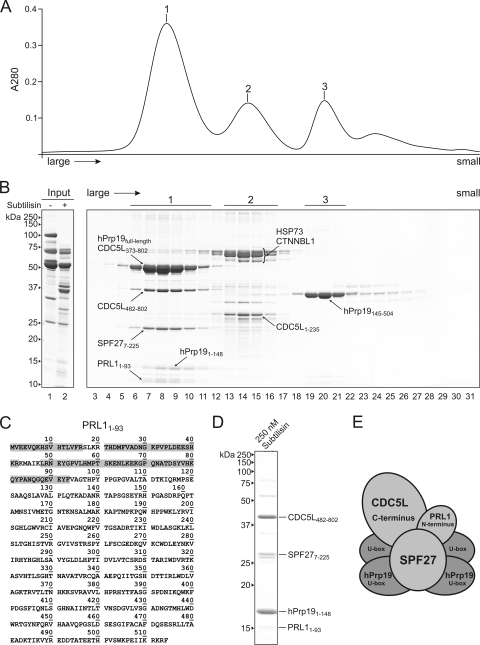
To further probe the structural organization of the native hPrp19/CDC5L complex, we performed limited proteolysis. Purified complexes were incubated with increasing amounts of the protease subtilisin (see Fig. S4 in the supplemental material), and the proteolysed components were then fractionated via gel filtration and analyzed by SDS-PAGE (Fig. 8A and B). A large protease-resistant complex and two smaller ones could be identified after treatment with 30 nM subtilisin (Fig. 8A and B, peaks 1 to 3). Exhaustive LC-offline MALDI-tandem mass spectrometry was performed after digestion with trypsin to delineate regions of each protein present in this complex (see Fig. S5 in the supplemental material and Fig. 8C). Nearly full-length SPF27 (i.e., lacking only the most N-terminal six amino acids) and full-length hPrp19, together with an N-terminal domain of hPrp19 that contains a U-box domain (amino acids [aa] 1 to 148), were identified in the largest complex. An overview of predicted domains in the hPrp19/CDC5L complex proteins is shown in Fig. S3 in the supplemental material. Two fragments of the CDC5L protein, both consisting of the C-terminal half of the protein, aa 373 to 802 and 482 to 802, were also detected; the precise N terminus of the latter could be confirmed by Edman sequencing. Furthermore, a fragment consisting of aa 1 to 93 of the PRL1 protein could be identified. Thus, consistent with our salt dissociation data, full-length SPF27 and fragments of hPrp19, CDC5L, and PRL1 form a protease-resistant core complex.
FIG. 8.
Limited protease digestion delineates interaction domains of hPrp19/CDC5L complex proteins. Purified hPrp19/CDC5L complexes were incubated with 30 nM protease subtilisin, and the proteolysed fragments were fractionated by size via gel filtration. (A) Three protease-resistant complexes could be separated by gel filtration. The absorption at 280 nm of proteins eluting from the gel filtration column was plotted against time. (B) SDS-PAGE analysis of the undigested hPrp19/CDC5L complex (lane 1) or the protease-treated complex prior to gel filtration (lane 2) or after gel filtration fractionation (lanes 3 to 31). Exhaustive LC-offline tandem mass spectrometry was performed to identify protein fragments in the protease-resistant complexes. Proteins and their protease-resistant amino acids delineated by MS are indicated. The molecular masses (kDa) of marker proteins are indicated on the left. (C) Mass spectrometric analysis of the protease-resistant fragment of PRL1. Peptides identified via MALDI-MS-MS are indicated with a gray box. (D) Composition of the largest protease-resistant complex observed after incubation with 250 nM subtilisin. (E) Schematic model of interaction domains in the protease-resistant core of the hPrp19/CDC5L complex.
The second protease-resistant complex contained full-length HSP73 and CTNNBL1 (and slightly truncated versions thereof) (Fig. 8A and B). A 150-kDa marker protein also peaked at this position in a separate gradient (data not shown), indicating that HSP73 and CTNNBL1 are most likely present in this peak in the form of a complex. Interestingly, we could also identify an N-terminal fragment of CDC5L (aa 1 to 235) comigrating with these proteins in the same peak. This is consistent with our far-Western blot analyses, in which CDC5L interacted with CTNNBL1, indicating that the AD002-CTNNBL1 dimer indeed binds the hPrp19/CDC5L complex core via the N terminus of CDC5L. The third peak contained solely a 35-kDa fragment of the hPrp19 protein (Fig. 8B). Mass spectrometric analysis demonstrated that it is the C-terminal part of the protein (aa 145 to 504), containing the WD40 domain.
When subtilisin was increased to 250 nM, only the N-terminal region of hPrp19 (aa 1 to 148), containing the U-box domain, was present in the largest protease-resistant complex, together with an N-terminal fragment of PRL1 (aa 1 to 93), a C-terminal fragment of CDC5L (aa 482 to 802), and SPF27 lacking only six amino acids at its N terminus (Fig. 8D; see Fig. S5 in the supplemental material). Notably, in vitro-translated SPF27 on its own is completely digested by 250 nM subtilisin (data not shown). Thus, SPF27 is highly resistant to protease digestion within the hPrp19/CDC5L complex, apparently due to its interaction with all other core components. The staining intensity of the hPrp19 N-terminal fragment indicates that multiple copies of it are present. Indeed, the amino acids known to mediate hPrp19 self-interaction (i.e., aa 56 to 74) (17) are still present. Taken together, these data suggest that the N-terminal fragment of the hPrp19 protein is present as a tetramer in the protease-resistant core of the hPrp19/CDC5L complex together with SPF27, the N terminus of PRL1, and the C terminus of CDC5L (Fig. 8E).
Elucidation of the 2D structure of the hPrp19/CDC5L complex via electron microscopy.
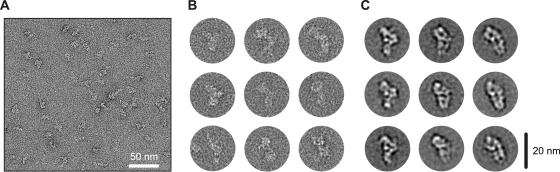
To determine the structure of native hPrp19/CDC5L complexes, affinity-purified particles were subjected to GraFix (20), negatively stained with uranyl formate, and analyzed by electron microscopy. The raw images (Fig. 9A and B) reveal a homogeneous, well-defined population of particles similar in size and shape. Two-dimensional class averages of the hPrp19/CDC5L complex were obtained by averaging individual images after “reference-free” image alignment, MSA, and classification (Fig. 9C). The most frequently observed class averages reveal an elongated, asymmetric shape with a maximum dimension of ∼20 nm and a width of ∼9 nm. This is consistent with our sedimentation velocity data, which indicated that the hPrp19/CDC5L complex has an elongated shape. Additionally, some class averages appear triangular and branched with a slightly reduced maximal length; these likely represent different views of the hPrp19/CDC5L complex, as opposed to structural heterogeneity in the complex.
FIG. 9.
Electron microscopy of affinity-purified human Prp19/CDC5L complexes. (A) Electron micrograph of negatively stained hPrp19/CDC5L complexes. Images were taken at a primary magnification of ×105,000 under low-dose conditions. (B) A representative selection of individual raw images. (C) A gallery of class averages of native hPrp19/CDC5L complexes obtained by averaging individual raw images.
DISCUSSION
In this study, we affinity purified the human Prp19/CDC5L complex from HeLa cell lines stably expressing FLAG-AD002 or FLAG-SPF27 and determined its molecular architecture and EM structure. Salt treatment revealed a stable core complex composed of CDC5L, PRL1, hPrp19, and SPF27. Consequently, we identified the protein-protein interaction network among the core components of the hPrp19/CDC5L complex. In addition, protease treatment coupled with MS allowed us to delineate those domains of the core proteins involved in protein-protein interactions. CTNNBL1, HSP73, and an N-terminal fragment of CDC5L cofractionated after protease treatment, and an interaction between AD002 and CTNNBL1 was identified by copurification and cross-linking. Association of these less stably bound subunits appears to be mediated by contacts between CTNNBL1 and CDC5L. Taken together, our data provide the first insights into the molecular organization and structure of the human Prp19/CDC5L complex. As the core components of the hPrp19/CDC5L complex are also part of the stable RNP core of the spliceosomal C complex, our data also potentially provide information about the organization of the spliceosome's catalytic RNP core.
Purification of the human Prp19/CDC5L complex.
The hPrp19/CDC5L complex was isolated from HeLa cells stably expressing FLAG-AD002 or FLAG-SPF27. Importantly, extracts prepared from either cell line exhibited the same splicing activity as the wild-type extract (Fig. 1). Furthermore, FLAG-tagged proteins were incorporated into spliceosomes to the same extent as their nontagged endogenous counterparts. Thus, insertion of a tag at the N terminus of AD002 or SPF27 does not appear to alter its activity or that of the hPrp19/CDC5L complex, indicating that it is also structurally intact. Indeed, MS analysis of affinity-purified hPrp19/CDC5L complexes after glycerol gradient centrifugation revealed a protein composition identical to that previously described for immunoaffinity-purified complexes (Fig. 2) (24). Interestingly, dimers consisting of AD002-CTNNBL1 or SPF27-hPrp19 were coisolated together with the hPrp19/CDC5L complexes from nuclear extract prepared from the FLAG-AD002 or FLAG-SPF27 stable cell line, respectively. This was the first indication that these proteins interact with one another; indeed, AD002-CTNNBL1 and SPF27-hPrp19 interactions within the hPrp19/CDC5L complex were confirmed via cross-linking, far-Western, and/or coimmunoprecipitation assays.
The stoichiometry of the hPrp19/CDC5L complex.
Quantification of the hPrp19/CDC5L complex stoichiometry by iodoacetamide labeling and Sypro-Ruby staining, along with analytical ultracentrifugation and cross-linking, indicated that in the human Prp19/CDC5L complex, like in the yeast NTC (29), there are four copies of hPrp19 (Fig. 3 and 4). HSP73 appears to be underrepresented; thus, it may be very loosely associated and partially lost during purification. All other proteins appeared to be present in single copies. These results are generally consistent with quantitative mass spectrometry analyses of the hPrp19/CDC5L complex (employing labeled internal standard peptides for absolute quantification), except that CDC5L was found in 1 or 2 copies depending on the conditions used for protein denaturation prior to protease digestion and subsequent absolute quantification by MS (32a). Interestingly, CDC5L can interact with itself, at least when it is present as an individual protein (Fig. 6; see also reference 16). In the yeast NTC, Cef1, the yeast homolog of human CDC5L, appears to be present as a single copy (29). Given that only one copy of CDC5L is present in our hPrp19/CDC5L complexes, CDC5L dimerization and CDC5L association with the hPrp19 complex would appear to be mutually exclusive. As CDC5L appears to have multiple roles in the cell (5, 21), dimerization may be important for one or more of CDC5L's additional functions. The presence of a single copy of AD002 and SPF27 in the native hPrp19/CDC5L complex is also supported by our affinity purification results (Fig. 2). That is, only tagged AD002 or tagged SPF27, but not their endogenous, untagged counterparts, was present in purified complexes, indicating that the hPrp19/CDC5L complex contains only one copy of each of these proteins.
Identification of a highly stable core of the hPrp19/CDC5L complex.
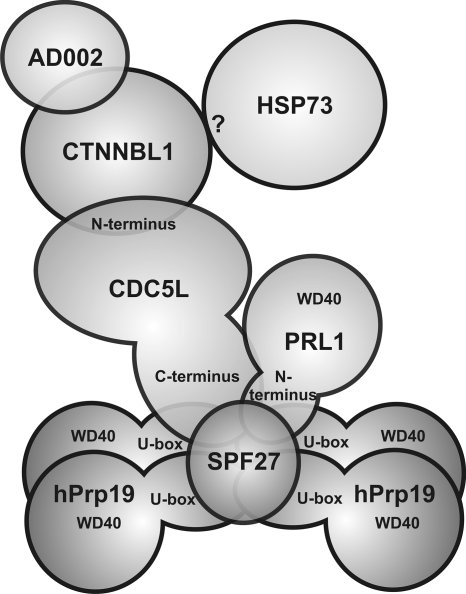
A subcomplex of the hPrp19/CDC5L complex, consisting of CDC5L, PRL1, hPrp19, and SPF27, was stable in the presence of 1.2 M NaCl, 0.4 M NaSCN, and 0.1 mg/ml heparin (Fig. 4; see Fig. S2 in the supplemental material), and also fragments of these proteins were found together in a protease-resistant complex. The existence of an extensive interaction network between the stable core components of the hPrp19/CDC5L complex (CDC5L, PRL1, hPrp19, and SPF27) was confirmed by several independent approaches, including cross-linking, far-Western blotting, and in vitro pulldown analysis. Prior to our analyses, the only known molecular interaction involving proteins of the human Prp19/CDC5L complex core was that between the C terminus of CDC5L and the WD40 domain of PRL1 (3). In addition to observing this known interaction, we observed several novel interactions (i.e., hPrp19-CDC5L, hPrp19-PRL1, SPF27-hPrp19, SPF27-PRL1, and SPF27-CDC5L) by a variety of methods. A model of the human Prp19/CDC5L complex which incorporates all of the information presented here is shown in Fig. 10.
FIG. 10.
Model of the molecular organization of the hPrp19/CDC5L complex.
Interestingly, some but not all of the aforementioned intermolecular interactions are also observed between the yeast homologs of CDC5L, hPrp19, PRL1, and SPF27, which are all components of the yeast NTC (9, 36). For example, Cef1p (the yeast homolog of human CDC5L) interacts with both Prp19 and Prp46p (the yeast homolog of human PRL1), but it does not interact with Snt309 (the yeast homolog of SPF27). Snt309p interacts with the Prp19 tetramer and Prp46 interacts with Cef1 (9, 26), but neither of these proteins interacts with the yeast homologs of any of the other core components of the human Prp19/CDC5 complex. Thus, protein-protein interactions involving these common proteins appear to differ somewhat between yeast and humans, and the overall architecture of the NTC core, at least in part, may also differ. Indeed, the yeast NTC contains 6 additional proteins not found in the human complex (Syf1, Syf2, Syf3, Isy1, NTC20, and Cwc2), some of which also contact Prp19, Cef1, or Prp46 (26), and at present it is not clear which of the yeast NTC proteins comprise its salt-stable core. However, Prp46 is readily lost from the NTC upon gradient centrifugation (26, 36), and thus it is not a highly stable NTC component (in contrast to its human homolog, PRL1). Taken together, these results point to clear differences in the organization of the stable core of human and yeast Prp19 complexes.
Although four proteins are shared by the NTC and hPrp19/CDC5L complex, a completely different set of additional proteins is recruited to each complex. This likely is a consequence of the presence of additional or different domains in the human proteins. Whereas the length and domain structure of Prp19 are highly conserved between yeast and humans, this is not the case for CDC5L and SPF27. For example, human CDC5L is 803 amino acids long, in contrast to Cef1, which is only 590 amino acids long. While the N-terminal halves of these proteins share over 40% homology and contain two SANT (c-Myb) domains, the C-terminal halves are much less conserved, with CDC5L containing a second coiled-coil domain at the far end of its C terminus that is lacking in Cef1 (see Fig. S3 in the supplemental material). Likewise, SPF27 and Snt309 share very limited homology (24%), and SPF27 contains a coiled-coil domain that is not found in Snt309 (see Fig. S3; also data not shown). These differences also likely contribute to the dissimilarities in SPF27 interactions and Snt309 interactions (or CDC5L interactions and Cef1 interactions) and the other shared proteins of both complexes.
AD002, CTNNBL1, and HSP73 are less stably associated with the hPrp19/CDC5L complex.
AD002, CTNNBL1, and HSP73 dissociate from the core complex already at moderate salt concentrations (e.g., 300 mM NaCl). In contrast to the components of the stable hPrp19/CDC5L complex core, homologs of human CTNNBL1, AD002, and HSP73 either do not exist in yeast (CTNNBL1) or are not found (AD002 and HSP73) in the yeast NTC but are nonetheless present in the yeast spliceosome (9, 15, 36). AD002 and CTNNBL1 interact with one another (Fig. 2 and 4 to 6), and CTNNBL1 also interacts with CDC5L. Furthermore, CTNNBL1 is found together with an N-terminal fragment of CDC5L in a protease-resistant complex (Fig. 8). Thus, CTNNBL1 and its interaction partner, AD002, are likely linked to the hPrp19/CDC5L complex core, at least in part, via the N terminus of CDC5L (Fig. 10). No direct interaction involving HSP73 could be observed via cross-linking, far-Western, or coimmunoprecipitation assays. Nonetheless, HSP73 appears to migrate in a complex together with CTNNBL and the N terminus of CDC5L after protease digestion, suggesting that it associates with the hPrp19/CDC5L complex via interacting with one or more of these proteins (Fig. 10).
Elucidation of domains of the hPrp19/CDC5L proteins involved in protein-protein interactions.
Limited protease digestion, followed by gel filtration and MS analyses, revealed a protease-resistant complex of the human Prp19/CDC5L complex that is composed of nearly full-length SPF27, the N-terminal U-box-containing domain of hPrp19, an N-terminal fragment of PRL1, and a C-terminal fragment of CDC5L. Interestingly, the U-box domains and ∼80 amino acids downstream are highly homologous in humans and yeast Prp19 proteins, and these regions were shown to be sufficient for tetramerization of yeast Prp19 (29), with the U-box domain alone capable of forming a dimer in vitro (28). This supports the conclusion that the N-terminal fragment of hPrp19 is indeed present in the form of a tetramer in this protease-resistant complex (Fig. 8E and 10). The C terminus of CDC5L could contact both hPrp19 and SPF27 in this complex. In contrast, as the C terminus of CDC5L was previously shown to bind the C-terminal WD40 domain of PRL1 (and this region is lacking in the protease-resistant complex), it likely does not contact PRL1. Indeed, the C-terminal WD40 domain of PRL1 is completely digested after treatment with low concentrations (30 nM) of subtilisin (see Fig. S5 in the supplemental material). Amino acids 1 to 93 of PRL1, which contain no known motifs (see Fig. S3), potentially contact the N-terminal region of hPrp19 and/or SPF27. Finally, SPF27 potentially contacts all other components of the protease-resistant core. Indeed, in contrast to free SPF27, it remains nearly intact after protease treatment, suggesting that it is either well protected against protease digestion (and thus potentially buried within the protease-resistant complex) or that it becomes highly structured (and thus more resistant to protease digestion) upon integration into the hPRP19/CDC5L complex. This is further consistent with the idea that it potentially plays an important role as a scaffold protein (together with Prp19) and/or in maintaining the integrity of the hPrp19/CDC5L complex. That is, in contrast to yeast, in which Snt309 seems to interact solely with the Prp19 tetramer (11) and is only required for yeast cell growth at elevated temperatures (10), the human SPF27 protein might play a more central role, interacting with all other core components, apparently via those regions identified by MS in the protease-resistant complex.
The human Prp19/CDC5L complex has an asymmetric and elongated morphology.
EM of the human Prp19/CDC5L complex revealed an elongated, asymmetric shape with a maximum dimension of ∼20 nm and a width of ∼9 nm. This agrees well with our analytical ultracentrifugation data, which revealed a frictional ratio of 2.1 for the complex, indicating that it is indeed elongated. Previous studies showed that recombinant Prp19 from S. cerevisiae forms a tetramer, and EM analyses indicated that this tetrameric complex is an elongated particle with a side length of 40 nm (29). The reason for this apparent difference in size between the yeast Prp19 tetramer and the entire human Prp19/CDC5L complex is not clear. Prp19 is also present as a tetramer in the human Prp19/CDC5L complex, and the two proteins have similar molecular masses and overall domain structures. Thus, the presence of much smaller and/or fewer copies of Prp19 in the human complex cannot explain this difference. It is possible that the human complex is very compact and that the hPrp19 tetramer is highly condensed due to its interaction with other components. Furthermore, in contrast to the yeast Prp19 tetramer, the human Prp19/CDC5L complex was prepared under GraFix conditions, which was shown to stabilize individual macromolecules and thereby provide reliable and high-resolution structures of macromolecular complexes by EM (20). The presence of a cross-linking reagent thus might fix the human complex in its native but more compact conformation.
The asymmetric shape of the hPrp19/CDC5L complex, together with the presence of a hPrp19 tetramer but apparently single copies of the other components of the complex, suggests an asymmetric assembly pathway. That is, although four copies of Prp19 are present, they are not recognized by the same interaction partner multiple times. This suggests that Prp19 tetramerization leads to the formation of a unique interaction interface which recruits one or more of the other core components of the hPrp19/CDC5L complex. As a dimer consisting of SPF27 and what appears to be tetrameric Prp19 (based on its migration behavior and the staining intensity of the Prp19 band) was present in nuclear extract prepared from cells expressing FLAG-tagged SPF27 (Fig. 2), it is tempting to speculate that SPF27 initially binds the hPrp19 tetramer and that this interaction in turn would promote the interaction of CDC5L and PRL1. Indeed, the association of Cef1 (the yeast homolog of human CDC5L) with the yeast NTC requires Prp19 oligomerization (36), and Snt309 (the yeast homolog of human SPF27) is required to maintain the Prp19-Cef1 interaction, at least at elevated temperatures (11). However, as we showed that CDC5L and PRL1 interact with hPrp19 on their own in vitro, it is also conceivable that they bind independently of SPF27 and even potentially prior to hPrp19 tetramerization. Finally, the AD002-CTNNBL1 dimer alone or possibly together with HSP73 likely binds after formation of the stable hPrp19/CDC5L core. However, future in vitro reconstitution studies with purified recombinant proteins are needed to shed more light on the assembly pathway of the hPrp19/CDC5L complex.
Insight into a major building block of the RNP core of the step 1 spliceosome.
Taken together, this work provides a detailed view of the architecture of the human Prp19/CDC5L complex and provides a framework for future high-resolution structural analyses. Furthermore, the elucidation of the molecular organization of the hPrp19/CDC5L complex provides the first insight into the possible architecture of a major building block of the RNP core of the catalytically active spliceosome. That is, all proteins of the highly stable core of the hPrp19/CDC5L complex, in addition to AD002, plus ∼30 other proteins, are present in the 1 M salt-stable RNP core of the human spliceosomal C complex (6), and they are thus thought to help maintain its catalytically active RNA network. In addition, with the exception of CTNNBL1, all proteins of the hPrp19/CDC5L complex are also present in the 35S U5 snRNP, a presumed dissociation product of the postspliceosomal intron-containing complex (22). Based on its exceptional stability, it is highly likely that the protein interaction network found in hPrp19/CDC5L complex core is also conserved in the core of the step 1 spliceosome.
Supplementary Material
Acknowledgments
We thank Thomas Conrad, Peter Kempkes, Hossein Kohansal, and Monika Raabe for excellent technical assistance. We are grateful to Gunter Meister (Max Planck Institute for Biochemistry) and Dierk Ingelfinger (German Cancer Research Center) for providing the pIRESneo-FLAG/HA plasmid and the anti-CTNNBL1 antibody, respectively. We thank Holger Stark and Berthold Kastner for help with the EM analyses.
This work was supported by grants from the EU (EURASNET contract no. 518238; 6th framework) to R.L. and H.U., and the Ernst Jung Stiftung to R.L. M.G. was supported by the Stiftung Stipendien-Fonds des Verbandes der Chemischen Industrie, and E.W. was supported by the Studienstiftung des Deutschen Volkes.
Footnotes
Published ahead of print on 22 February 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Achsel, T., K. Ahrens, H. Brahms, and R. Lührmann. 1998. The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 18:6756-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajuh, P., B. Kuster, K. Panov, J. C. Zomerdijk, M. Mann, and A. I. Lamond. 2000. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J. 19:6569-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajuh, P., J. Sleeman, J. Chusainow, and A. I. Lamond. 2001. A direct interaction between the carboxyl-terminal region of CDC5L and the WD40 domain of PLRG1 is essential for pre-mRNA splicing. J. Biol. Chem. 276:42370-42381. [DOI] [PubMed] [Google Scholar]
- 4.Behzadnia, N., M. M. Golas, K. Hartmuth, B. Sander, B. Kastner, J. Deckert, P. Dube, C. L. Will, H. Urlaub, H. Stark, and R. Lührmann. 2007. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 26:1737-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein, H. S., and S. R. Coughlin. 1997. Pombe Cdc5-related protein. A putative human transcription factor implicated in mitogen-activated signaling. J. Biol. Chem. 272:5833-5837. [DOI] [PubMed] [Google Scholar]
- 6.Bessonov, S., M. Anokhina, C. L. Will, H. Urlaub, and R. Lührmann. 2008. Isolation of an active step I spliceosome and composition of its RNP core. Nature 452:846-850. [DOI] [PubMed] [Google Scholar]
- 7.Chan, S. P., and S. C. Cheng. 2005. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J. Biol. Chem. 280:31190-31199. [DOI] [PubMed] [Google Scholar]
- 8.Chan, S. P., D. I. Kao, W. Y. Tsai, and S. C. Cheng. 2003. The Prp19p-associated complex in spliceosome activation. Science 302:279-282. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. H., W. C. Yu, T. Y. Tsao, L. Y. Wang, H. R. Chen, J. Y. Lin, W. Y. Tsai, and S. C. Cheng. 2002. Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res. 30:1029-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, H. R., S. P. Jan, T. Y. Tsao, Y. J. Sheu, J. Banroques, and S. C. Cheng. 1998. Snt309p, a component of the Prp19p-associated complex that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19p. Mol. Cell. Biol. 18:2196-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, H. R., T. Y. Tsao, C. H. Chen, W. Y. Tsai, L. S. Her, M. M. Hsu, and S. C. Cheng. 1999. Snt309p modulates interactions of Prp19p with its associated components to stabilize the Prp19p-associated complex essential for pre-mRNA splicing. Proc. Natl. Acad. Sci. U. S. A. 96:5406-5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deckert, J., K. Hartmuth, D. Boehringer, N. Behzadnia, C. L. Will, B. Kastner, H. Stark, H. Urlaub, and R. Lührmann. 2006. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26:5528-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dube, P., P. Tavares, R. Lurz, and M. van Heel. 1993. The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J. 12:1303-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabrizio, P., J. Dannenberg, P. Dube, B. Kastner, H. Stark, H. Urlaub, and R. Lührmann. 2009. The evolutionary conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell 36:593-608. [DOI] [PubMed] [Google Scholar]
- 16.Gräub, R., H. Lancero, A. Pedersen, M. Chu, K. Padmanabhan, X. Q. Xu, P. Spitz, R. Chalkley, A. L. Burlingame, D. Stokoe, and H. S. Bernstein. 2008. Cell cycle-dependent phosphorylation of human CDC5 regulates RNA processing. Cell Cycle 7:1795-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grillari, J., P. Ajuh, G. Stadler, M. Loscher, R. Voglauer, W. Ernst, J. Chusainow, F. Eisenhaber, M. Pokar, K. Fortschegger, M. Grey, A. I. Lamond, and H. Katinger. 2005. SNEV is an evolutionarily conserved splicing factor whose oligomerization is necessary for spliceosome assembly. Nucleic Acids Res. 33:6868-6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatakeyama, S., M. Yada, M. Matsumoto, N. Ishida, and K. I. Nakayama. 2001. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276:33111-33120. [DOI] [PubMed] [Google Scholar]
- 19.Kastner, B. 1998. Purification and electron microscopy of spliceosomal snRNPs, p. 95-140. In J. Schenkel (ed.), RNP particles, splicing and autoimmune diseases. Springer lab manual. Springer, Heidelberg, Germany.
- 20.Kastner, B., N. Fischer, M. M. Golas, B. Sander, P. Dube, D. Boehringer, K. Hartmuth, J. Deckert, F. Hauer, E. Wolf, H. Uchtenhagen, H. Urlaub, F. Herzog, J. M. Peters, D. Poerschke, R. Lührmann, and H. Stark. 2008. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5:53-55. [DOI] [PubMed] [Google Scholar]
- 21.Lei, X. H., X. Shen, X. Q. Xu, and H. S. Bernstein. 2000. Human Cdc5, a regulator of mitotic entry, can act as a site-specific DNA binding protein. J. Cell Sci. 113:4523-4531. [DOI] [PubMed] [Google Scholar]
- 22.Makarov, E. M., O. V. Makarova, H. Urlaub, M. Gentzel, C. L. Will, M. Wilm, and R. Lührmann. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205-2208. [DOI] [PubMed] [Google Scholar]
- 23.Makarova, O. V., E. M. Makarov, S. Liu, H. P. Vornlocher, and R. Lührmann. 2002. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 21:1148-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarova, O. V., E. M. Makarov, H. Urlaub, C. L. Will, M. Gentzel, M. Wilm, and R. Lührmann. 2004. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 23:2381-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malik, S., and R. G. Roeder. 2003. Isolation and functional characterization of the TRAP/mediator complex. Methods Enzymol. 364:257-284. [DOI] [PubMed] [Google Scholar]
- 26.Ohi, M. D., and K. L. Gould. 2002. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 8:798-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohi, M. D., A. J. Link, L. Ren, J. L. Jennings, W. H. McDonald, and K. L. Gould. 2002. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 22:2011-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohi, M. D., C. W. Vander Kooi, J. A. Rosenberg, W. J. Chazin, and K. L. Gould. 2003. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 10:250-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohi, M. D., C. W. Vander Kooi, J. A. Rosenberg, L. Ren, J. P. Hirsch, W. J. Chazin, T. Walz, and K. L. Gould. 2005. Structural and functional analysis of essential pre-mRNA splicing factor Prp19p. Mol. Cell. Biol. 25:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ons, S., F. Richter, H. Urlaub, and R. R. Pomar. 2009. The neuropeptidome of Rhodnius prolixus brain. Proteomics 9:788-792. [DOI] [PubMed] [Google Scholar]
- 31.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 32.Sawasaki, T., T. Ogasawara, R. Morishita, and Y. Endo. 2002. A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl. Acad. Sci. U. S. A. 99:14652-14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Schmidt, C., C. Lenz, M. Grote, R. Lührmann, and H. Urlaub. Determination of protein stoichiometry within protein complexes using absolute quantification (AQUA) and multiple reaction monitoring (MRM). Anal. Chem., in press. [DOI] [PubMed]
- 33.Schuck, P. 2003. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 320:104-124. [DOI] [PubMed] [Google Scholar]
- 34.Schuck, P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78:1606-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 36.Tarn, W. Y., C. H. Hsu, K. T. Huang, H. R. Chen, H. Y. Kao, K. R. Lee, and S. C. Cheng. 1994. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 13:2421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reference deleted.
- 38.van Heel, M. 1989. Classification of very large electron microscopical image data sets. Optik 82:114-126. [Google Scholar]
- 39.van Heel, M., G. Harauz, E. V. Orlova, R. Schmidt, and M. Schatz. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17-24. [DOI] [PubMed] [Google Scholar]
- 40.Voss, S., and A. Skerra. 1997. Mutagenesis of a flexible loop in streptavidin leads to higher affinity for the Strep-tag II peptide and improved performance in recombinant protein purification. Protein Eng. 10:975-982. [DOI] [PubMed] [Google Scholar]
- 41.Wahl, M. C., C. L. Will, and R. Lührmann. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136:701-718. [DOI] [PubMed] [Google Scholar]
- 42.Will, C. L., and R. Lührmann. 2006. Spliceosome structure and function, p. 369-400. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Zillmann, M., M. L. Zapp, and S. M. Berget. 1988. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol. Cell. Biol. 8:814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.