Abstract
Heat stress cognate 70 (Hsc70) is a host protein associated with hepatitis B virus (HBV) replication. The goal of this study was to investigate whether Hsc70 could be an anti-HBV drug target. Our results showed that introducing Hsc70 increased HBV replication in HBV+ human hepatocytes (HepG2.2.15 cells). The coiled-coil region on Hsc70 (nucleotides 1533 to 1608; amino acids 511 to 536) was the key sequence for HBV replication. Knockdown of Hsc70 expression by RNA interference (RNAi) largely inhibited HBV replication with no cytotoxicity to the host. Using an Hsc70 mRNA screening assay, the natural compound oxymatrine (OMTR) was found to be a selective inhibitor for Hsc70 expression. Then, OMTR was used to investigate the potential of Hsc70 as an anti-HBV drug target. OMTR inhibited Hsc70 mRNA expression by 80% and HBV DNA replication by over 60% without causing cytotoxicity. The anti-HBV effect of OMTR appeared to be mediated by destabilizing Hsc70 mRNA. The half-life (T1/2) of Hsc70 mRNA decreased by 50% in OMTR-treated hepatocytes. The Hsc70 mRNA 3′-untranslated-region (UTR) sequence was the element responsible for OMTR's destabilization activity. OMTR suppressed HBV de novo synthesis at the reverse transcription stage from pregenomic RNA (pgRNA) to DNA and was active against either wild-type HBV or strains resistant to lamivudine, adefovir, and entecavir. Therefore, host Hsc70 could be a novel drug target against HBV, and OMTR appears to inhibit HBV replication by destabilizing Hsc70 mRNA. As the target is not a viral protein, OMTR is active for either wild-type HBV or strains resistant to reverse transcriptase (RT) inhibitors.
Antiviral chemotherapy can select for drug-resistant viral mutants (21). For chronic infections that need long-term chemotherapy, such as infection with hepatitis B virus (HBV), the challenge to clinical therapy is substantial (27, 31). Reverse transcriptase (RT) inhibitors, such as lamivudine, adefovir, entecavir, telbivudine, and tenofovir, are potent drugs for HBV infections, but their use in the clinical setting often selects for drug resistance (13, 14, 27, 31). The incidence of lamivudine resistance rises from 15 to 32% in the first year to 67 to 69% by the fifth year of treatment (7, 9, 28). Many drug-induced mutations in the HBV polymerase gene have been characterized. For instance, rtM204I/V/S (“rt” means “resistant”), rtL180M, rtL80V/I, and rtV173L are signature mutations for lamivudine; rtN236T and rtA181T/V are signature mutations for adefovir; rtS202G/I, rtI169T, rtS184G, and rtM250V are signature mutations for entecavir; rtM204I is a signature mutation for telbivudine; and rtA194T is a signature mutation for tenofovir (9, 27, 30, 31). The mutations in RT result from the intrinsic high variability due to the lack of an editing function of the enzyme (18, 20), and they alter the three-dimensional (3D) interaction between HBV polymerase and the drugs (27). This challenges the current anti-HBV strategy, which is directed at viral enzymes.
However, HBV strains rely heavily on host cell machinery to complete their life cycles. Indeed, a number of host proteins have been reported to be crucial for HBV replication (10, 17, 29). Our hypothesis is that (i) these cellular components might be drug targets to control the virus, and suppression of these cellular proteins might be able to inhibit HBV replication, and (ii) unlike those that target viral enzymes, drugs utilizing this mechanism would be active against either wild-type or drug-resistant HBV strains, because the virus is not the target of chemotherapy. However, inhibition of host proteins might be harmful to the host. The key to avoiding undesirable side effects is, first, to identify host targets that are crucial for viral replication but not essential or only conditionally required for cell survival and, second, to find compounds that selectively target these proteins.
Heat stress cognate 70 (Hsc70, or HspA8) is an ATP-binding protein of the heat stress protein 70 (Hsp70) family (16). It is the form of Hsp70 that is expressed in the absence of heat or other cell stress (1). This host protein was found to be required for the reverse transcription process in experiments using duck HBV DNA polymerase (2, 8). We anticipated that downregulation of this protein in the host would inhibit HBV replication in either wild-type or drug-resistant strains. Here, we report the results of testing this novel antiviral strategy that uses Hsc70 as a drug target to inhibit HBV.
MATERIALS AND METHODS
Compounds.
Oxymatrine (OMTR), lamivudine, adefovir, and entecavir with purity over 98.5% were from the National Institute for the Control of Pharmaceutical and Biological Products, State Federal Drug Administration (Beijing, China).
Cell lines.
Human HepG2 and Huh-7 liver cells were from the American Type Culture Collection (ATCC) (Frederick, MD). Human HepG2 hepatocytes transfected with the full genome of HBV (HepG2.2.15 cells) (19) were used for anti-HBV tests. The cells were cultivated in a basic minimal essential medium (MEM) (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS).
qRT-PCR and real-time PCR.
RNA was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA), and intracellular DNA was extracted using a QIAamp DNA Mini Kit (Qiagen, Valencia, CA) following the instructions from the venders. Quantitative real-time PCR was performed using the SYBR green technique in the Bio-Rad iQ5 Multicolor Real-Time Detection System (Bio-Rad, Hercules, CA). Hsc70, Hsp90, and HspA4 mRNA assays were done with a SuperScript III Platinum SYBR Green One-Step quantitative real-time RT-PCR (qRT-PCR) kit (Invitrogen), and the HBV DNA assay was done with a Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen). Primers were designed with Primer 5.0 (Premier Co., Canada) and tested for specificity using the BLASTN program. The Hsc70, Hsp90, HspA4, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNAs were detected using human primers (Hsc70 primers, F, 5′-TGCTGCTGCTATTGCTTACG-3′, and R, 5′-TCAATAGTGAGGATTGACACATCA-3′; Hsp90 primers, F, 5′-AGAGCCTACGTTCCTGCACT-3′, and R, 5′-GACCATTCTTCTAGAGCATTCAGG-3′; HspA4 primers, F, 5′-GGAGGAACCACATGTTGAAGA-3′, and R, 5′-TGGATCCAGCTTGAGAGGTC-3′; GAPDH primers, F, 5′-ACCCACTCCTCCACCTTTG-3′, and R, 5′-CTGTAGCCAAATTCGTTGTCAT-3′). HBV DNA and GAPDH DNA detection used human HBV DNA primers (F, 5′-GGCTTTCGGAAAATTCCTATG-3′, and R, 5′-AGCCCTACGAACCACTGAAC-3′) and GAPDH DNA primers (F, 5′-GACTAACCCTGCGCTCCTG-3′, and R, 5′-CATCACCCGGAGGAGAAAT-3′), respectively. The data were analyzed using Bio-Rad iQ5 software. The copy number of the study genes in the untreated cells was defined as 1 (the actual range was between 105 and 107 copies), and the numbers of gene copies in the treated cells were plotted relative to that value.
Introduction of Hsc70.
HepG2.2.15 cells were seeded in a 6-well plate at 5 × 105 per well. The cells were cotransfected with a tetracycline-controlled vector, pTet-On Advanced, as well as vector pTRE-Tight (Clontech, Mountain View, CA). The rtTA-Advanced (rtTA2s-M2) in pTet-On Advanced controls the expression of the Hsc70 gene inserted at the multiple-cloning site (MCS) of pTRE-Tight. rtTA-Advanced binds TRE-Tight and activates transcription in the presence of either doxycycline (Dox) or tetracycline. The vector transfection was carried out for 24 h at 37°C. Transfection efficiency was monitored using the Block-iT Alexa Fluor red fluorescent oligonucleotide (Invitrogen). About 85 to 90% of the cells were transfected with the vector in the experiments. The cells were then treated with Dox (1 μg/ml) or solvent (control) for 24 h, followed by measurement of Hsc70 mRNA and HBV DNA. The experiments were repeated at least 3 times, with 2 or 3 replicates in each experiment.
Plasmid construction and deletion analysis of Hsc70.
The Hsc70 open reading frame (ORF) was amplified from HepG2 cellular cDNA and inserted into the BamHI and SalI sites of the expression vector pcDNA3.1/Neo (+). A series of deletion mutants of Hsc70 were created by overlap extension PCR. The PCR products were digested with BamHI and SalI and cloned into pcDNA3.1/Neo (+). PCR was run on the PCR system DNA engine (PTC-200) (Bio-Rad, Hercules, CA). The specific trans-intronic primers used for the PCR were as follows: Hsc70 (product, 1,938 bp), F, 5′-GTGGATCCATGTCCAAGGGACCT-3′, and R, 5′-GCTCTAGAGGCTTAATCAACCTCTT-3′; Hsc70/del-CTD (product, 1,620 bp), F, 5′-GTGGATCCATGTCCAAGGGACCT-3′, and RCTD, 5′-GGTCGACTAACTTGGATGACACCTTGTCCC-3′; Hsc70/del-CC (product, 1,857 bp), F, 5′-GTGGATCCATGTCCAAGGGACCT-3′, RCC, 5′-CAAACGGCCCTTGTCATTAGTG-3′, FCC, 5′-ACTAATGACAAGGGCCGTTTGTCATCCAAGAATTCACTTGAGTCC-3′, and R, 5′-GCTCTAGAGGCTTAATCAACCTCTT-3′; and Hsc70/del-NLS (product, 1,875 bp), F, 5′-GTGGATCCATGTCCAAGGGACCT-3′, RNLS, 5′-AGGAACACCTCGGGGTGCAGGA-3′, FNLS, 5′-CTGCACCCCGAGGTGTTCCTAAGAGTACGGGAAAAGAGAACAAGATTAC-3′, and R, 5′-GCTCTAGAGGCTTAATCAACCTCTT-3′.
HepG2.2.15 cells (5 × 105/well) were seeded in 6-well microplates (Corning, NY) and grown for 24 h in basic MEM (Gibco) with 10% fetal bovine serum (HyClone, Logan, UT). The cells were then transfected with Hsc70 wild-type or deletion mutant construct (2 μg plasmid/well) using Fugene HD Transfection Reagent (Roche, Mannheim, Germany). Forty-eight hours later, the Hsc70 mRNA and HBV DNA in the transfected HepG2.2.15 cells were examined by real-time RT-PCR or real-time PCR.
Specific RNA interference for human Hsc70 in hepatocytes.
HepG2.2.15 cells were plated in a 6-well plate at a density of 2 × 105 per well in growth medium (Opti-MEM; 10% FBS; antibiotic free; Invitrogen). Hsc70 gene-specific small interfering RNA (siRNA) (5′-ACUGAUGUCCUUCUUAUGCUUGCGC-3′ or 5′-AAACUUGCCAAGCAGGUUGUUAUCC-3′; Invitrogen) was transfected into the cells using Lipofectamine RNAiMax (Invitrogen) with a nonspecific control siRNA as a reference. Twenty-four and 48 h after transfection, the cells were harvested for RNA and DNA extraction. Total RNA was analyzed for Hsc70 mRNA expression using real-time RT-PCR; HBV DNA was examined using real-time PCR.
Hsc70 mRNA half-life assay.
HepG2.2.15 cells were treated or not with 0.4 mg/ml (equal to 1.51 mM) of OMTR for 24 h. Then, 5 μg/ml of actinomycin D (A. G. Scientific, San Diego, CA) was added to block transcription. Total cellular RNAs were harvested at different time points after actinomycin D treatment to analyze the Hsc70 mRNA level by real-time RT-PCR. The Hsc70 mRNA levels were normalized to that of GAPDH, and the remaining levels were plotted against time to estimate the half-life (T1/2) of Hsc70 mRNA.
Hsc70 protein expression detection.
The HepG2.2.15 cells treated with OMTR (0.4 mg/ml; 1.51 mM) were collected. Aliquots of the cells were taken and lysed in RIPA lysis buffer containing phenylmethylsulfonyl fluoride (PMSF) and protease inhibitor cocktail (Santa Cruz Biotechnology). Equal amounts of lysate were subjected to SDS-polyacrylamide gel electrophoresis and then blotted onto polyvinylidene difluoride (PVDF) membranes (Millipore) using a Semi-Dry Electrophoretic Transfer System (Bio-Rad, CA). Then, the membranes were blocked with 5% nonfat milk in TBST buffer (20 mmol/liter Tris-HCl, 137 mmol/liter NaCl, 0.05% Tween 20) at 37°C for 1 h and probed with mouse monoclonal antibodies against Hsc70 (Santa Cruz Biotechnology, Santa Cruz, CA), followed by incubation with goat anti-mouse antibody labeled with horseradish peroxidase. The signals were detected by enhanced chemiluminescence (ECL) (Santa Cruz Biotechnology, Santa Cruz, CA). Hsc70 protein expression was also examined by flow cytometry using rabbit antibody against Hsc70 and Cy5-labeled mouse anti-rabbit monoclonal antibody (Abcam Biotechnology).
Hsc70 mRNA 3′ UTR.
The luciferase (Luc) reporter plasmid pLuc was constructed by insertion of the luciferase cDNA region into the HindIII and XbaI sites of the pcDNA3.0 plasmid (Invitrogen). With Hsc70 mRNA as the template, the Hsc70 5′-untranslated-region (UTR) and 3′-UTR sequences were reverse transcribed and amplified by PCR using HindIII-tailed and XbaI-tailed primers, respectively. We cut the 5′-UTR or 3′-UTR cDNA fragment with HindIII and XbaI and then separately inserted the fragment into the HindIII or XbaI site of pLuc. All constructs were sequenced for confirmation, and the clones were further propagated to obtain plasmid DNA. The HepG2.2.15 cells were transfected with the plasmid pLuc, pLuc-3′UTR, or pLuc-5′UTR for 24 h. The transfected cells were divided into 2 dishes for drug treatment or as an untreated control. After overnight incubation, the cells were treated with OMTR or dilution buffer (solvent control). Total RNA was extracted 12 h later to measure the luciferase mRNA using a quantitative real-time RT-PCR assay.
Anti-HBV effect of OMTR in cell culture.
HepG2.2.15 cells were treated with OMTR at 0.4 mg/ml (1.51 mM) for 12, 24, or 36 h. Cells were harvested, and total RNA was isolated for measurement of the mRNA levels of Hsc70 and HBV; the intracellular DNA (including genomic DNA and HBV DNA) was isolated as well for HBV DNA determination. Real-time RT-PCRs were done as described above for the quantitative measurement of HBV mRNA and Hsc70 mRNA, and real-time PCR was done for HBV DNA. Host HspA4 or Hsp90 mRNA was also measured as a reference in some experiments. For the detection of viral de novo synthesis, the treated cells were washed, lysed, and centrifuged at 14,000 rpm for the isolation of intracellular HBV replicative intermediates by the method described previously (6). The newly synthesized HBV DNA replicative intermediates were quantified by real-time PCR. The dose-dependent effect was shown as described previously (23).
Anti-HBV activity of OMTR in drug-resistant HBV strains was also tested. In these experiments, 105 Huh-7 cells were plated in 24-well plates. After overnight incubation, the cells were transiently transfected with pcDNA3.1 containing the wild-type HBV strain or the Lamr (rtL180M plus rtM204V), Advr (rtA181V plus rtN236T), or Etvr (rtL180M plus rtM204V plus rtS202G) strain for 24 h. The supernatant was discarded, and the cells were cultivated with fresh medium containing 0.4 mg/ml (1.51 mM) OMTR. HBV DNA replicative intermediates were quantified after 24 h of incubation.
Long-term treatment.
HepG2.2.15 cells were exposed to OMTR (30 μg/ml; 113 μM) or left untreated. The cells were washed with phosphate-buffered saline (PBS) every 3 days and then fed with fresh culture medium containing the same concentration of the study drug. The cell passage was taken every 6 or 7 days, followed by cultivation as described above. The treatment continued for 8 months. Intracellular HBV DNA and the numbers of surviving cells were determined every 30 days, using quantitative real-time PCR and trypan blue staining, respectively.
Statistical analysis.
After validation of the test for homogeneity of variance, the results were examined with Student's t test. For all of the figures, the bars (means and standard deviations [SD]) represent the average results across experiments, with 2 or 3 replicates within each experiment. To ensure differences, a P value of <0.01 was considered statistically significant. The statistics were done with SPSS11.0 software.
RESULTS
Host Hsc70 is a supportive factor for HBV replication.
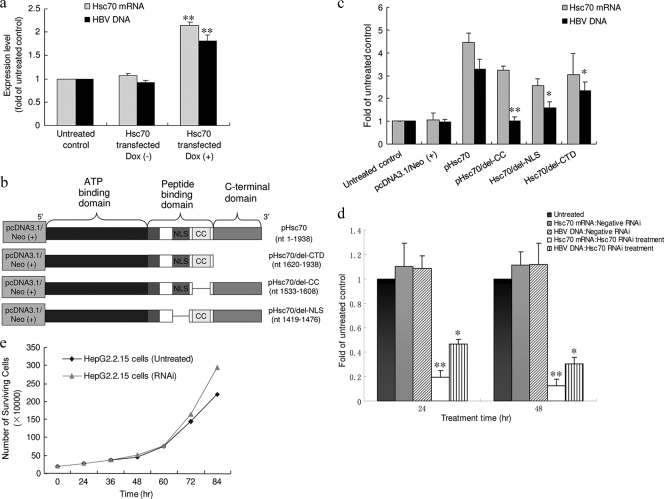
To introduce Hsc70, HepG2.2.15 HBV+ human hepatocytes were transfected with a tetracycline-controlled Hsc70 expression vector system in which Hsc70 transcription could be triggered by either Dox or tetracycline. In this study, Dox was used. As Dox was added to the culture medium, the transcription of the inserted Hsc70 gene at the MCS was subsequently turned on. The intracellular Hsc70 mRNA and HBV DNA were measured 24 h after Dox treatment. As shown in Fig. 1a, turning on the expression of external Hsc70 with Dox increased intracellular Hsc70 mRNA in the HepG2.2.15 cells, in parallel with which there was a 1.8-fold elevation of the intracellular HBV DNA. The results support previous observations for Hsc70 (2, 8, 17). The experiment was repeated more than 3 times, and HBV DNA was significantly higher in the cells with external Hsc70 than in those without addition of Hsc70 (P < 0.001).
FIG. 1.
Host Hsc70 expression regulates HBV DNA replication. (a) HepG2.2.15 cells were cotransfected with the two vectors mentioned in Materials and Methods; 24 h later, Dox (1 μg/ml) was add to the culture media. The amounts of Hsc70 mRNA and HBV DNA in the untreated HepG2.2.15 cells was defined as 1, and the amounts of Hsc70 or HBV DNA in the cells with [Dox (+)] or without [Dox (−)] Dox were plotted relative to that value. *, P < 0.001 relative to Dox (−). The experiment was performed 4 times. Shown are means and SD. (b) Diagram of the wild type and deletion mutants of the Hsc70 gene, which were used for the construction of pHsc70, pHsc70/del-CTD, pHsc70/del-CC, and pHsc70/del-NLS with pcDNA3.1/Neo (+). (c) HepG2.2.15 cells were transfected with pHsc70, pHsc70/del-NLS, pHsc70/del-CC, or pHsc70/del-CTD for 48 h. The mRNA expression of Hsc70, Hsc70/del-NLS, Hsc70/del-CC, and Hsc70/del-CTD was analyzed by real-time RT-PCR, with parallel measurement of HBV DNA. The experiment was performed 3 times. Shown are means and SD. *, P < 0.01, and **, P < 0.001 versus the pHsc70 group. (d) HepG2.2.15 cells were transfected with Hsc70 siRNA (816 to 840 bp) or negative RNAi (reference control) for 24 or 48 h, respectively. Total cellular RNA, as well as intracellular DNA, was isolated to measure Hsc70 mRNA and HBV DNA. The amount of Hsc70 mRNA or HBV DNA in the treated cells was normalized to that of the untreated control. The experiment was performed more than 6 times. Shown are means and SD. *, P < 0.01, and **, P < 0.001 versus untreated controls. (e) Growth curves for HepG2.2.15 cells with or without Hsc70 RNAi. The numbers of surviving cells were plotted against the time of treatment.
Human Hsc70 protein contains an ATP-binding domain (ABD) (nucleotides [nt] 1 to 1146; amino acids [aa] 1 to 382), a peptide-binding domain (PBD) (nt 1155 to 1629; aa 385 to 543), and a C-terminal domain (CTD) (nt 1620 to 1938; aa 540 to 646) (15, 22). Within the PBD, there are a nuclear localization signal (NLS) region (nt 1419 to 1476; aa 473 to 492) and a coiled-coil (CC) region (nt 1533 to 1608; aa 511 to 536) (15, 22). To determine which element in the Hsc70 gene is involved in supporting HBV replication, wild-type Hsc70, as well as three Hsc70 gene mutants with deletion of the NLS, CC, or CTD were created (Fig. 1b) and individually inserted into the BamHI and SalI sites of pcDNA3.1/Neo (+). We then transfected wild-type Hsc70 (pHsc70) and Hsc70 mutants (pHsc70/del-NLS, pHsc70/del-CC, and pHsc70/del-CTD) into the HepG2.2.15 cells. Expression of Hsc70 or its mutants, as well as HBV DNA, in the cells was examined 48 h later. The results are shown in Fig. 1c. Transfection with either the wild-type or mutant Hsc70 increased their mRNA expression in the HepG2.2.15 cells. While the Hsc70 mutant with deletion of the CTD or NLS caused partial loss of support for HBV replication compared to wild-type Hsc70, deletion of the CC completely abolished the activity of Hsc70 in assistance to HBV. This result supports the observation shown in Fig. 1a and indicates that the CC sequence in the PBD region encodes a key domain for HBV replication. Also, it is possible that CC region deletion alters the conformation of the remaining section of Hsc70 and thus abolishes the function.
Specific siRNAs were used to silence the endogenous Hsc70 gene. Treatment with specific Hsc70 siRNA (816 to 840 bp) reduced Hsc70 mRNA expression (Fig. 1d), but without a negative effect on HepG2.2.15 cell growth; the doubling time was between 20 and 36 h for both lines (Fig. 1e). This suggested that reduction of Hsc70 did not damage the cells. Importantly, treatment of the HepG2.2.15 cells with siRNA significantly reduced the intracellular HBV DNA load (Fig. 1d). Hsc70 RNA interference (RNAi) with another siRNA sequence showed a similar effect (data not shown). We assumed that compounds with downregulatory activity on Hsc70 expression might be anti-HBV agents.
Oxymatrine downregulates Hsc70 expression with an inhibitory effect on HBV replication.
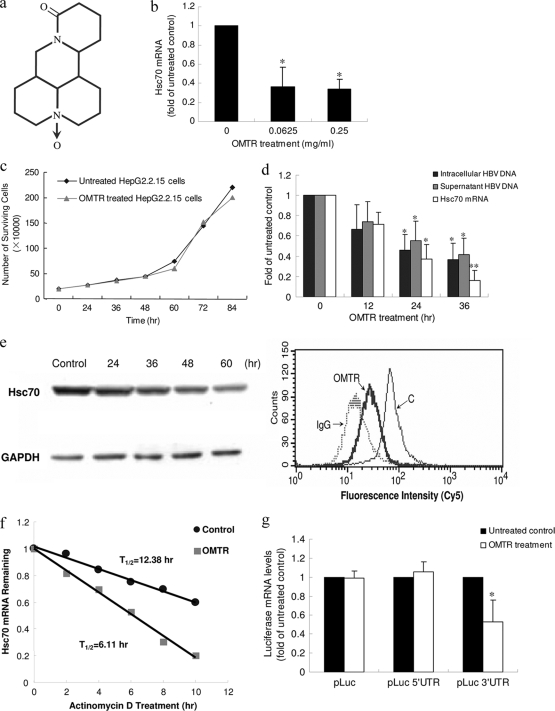
A rational screening for Hsc70 inhibitors was then conducted using an Hsc70 mRNA expression assay as a model. The screening was initiated with natural or semisynthesized natural compounds in our chemical library. Over 70 candidate compounds with potential activity against viruses were selected. The structures of the compounds were characterized, and the compounds showed no activity against retroviral RT. The cells were treated with the compounds for 24 h, and Hsc70 mRNA expression was determined. Among the compounds tested, OMTR (molecular weight [MW], 264.31) (Fig. 2a), an alkaloid extracted from the kushen plant (Sophora flavescens Ait) (11), showed significant activity in downregulating Hsc70 expression in human HepG2 liver cells (Fig. 2b). The results were of great interest to us, because the compound has been used to treat viral hepatitis in China since the 1990s (3, 4, 12, 24). However, its anti-HBV mechanism remains unclear. We used OMTR as a chemical probe to investigate the potential of Hsc70 as an anti-HBV drug target.
FIG. 2.
Downregulation of Hsc70 expression by OMTR inhibits HBV replication. (a) Chemical structure of OMTR. (b) OMTR downregulated Hsc70 mRNA expression in HBV-free HepG2 cells. The cells were treated with OMTR for 24 h at a concentration of 0.0625 or 0.25 mg/ml (equal to 0.23 and 0.94 mM, respectively). The experiment was performed more than 5 times. Shown are means and SD. *, P < 0.01 versus untreated control. (c) Treatment of HepG2.2.15 cells with OMTR at 0.4 mg/ml (1.51 mM) for 36 h caused no change in cell growth. The experiment was repeated twice. (d) HepG2.2.15 cells treated with OMTR (0.4 mg/ml; 1.51 mM) showed a time-dependent reduction of Hsc70 mRNA and HBV DNA. Shown are means and SD. *, P < 0.01, and **, P < 0.001 versus untreated control. (e) Expression of Hsc70 protein in cells decreased in Western blot and flow cytometry assays (24-h treatment; IgG, background; C, untreated control). The experiments were performed 5 times for each assay. (f) HepG2.2.15 cells were untreated or treated with OMTR (0.4 mg/ml; 1.51 mM) for 24 h. Then, actinomycin D was added to the cells for different intervals, and Hsc70 mRNA was analyzed. Normalized Hsc70 mRNA was plotted as the percent remaining. Decay curves were plotted versus time. The experiment was repeated twice. (g) OMTR destabilized Hsc70 mRNA through a 3′-UTR mediated mechanism. Plasmids pLuc, pLuc-3′UTR, and pLuc-5′UTR were transiently transfected into HepG2.2.15 cells, followed by OMTR treatment. The effects of OMTR on the wild-type or chimeric pLuc transcripts were determined by measuring Luc mRNA by quantitative real-time RT-PCR assay. The experiment was performed three times. Shown are means and SD. *, P < 0.01 versus untreated control.
OMTR treatment showed no toxicity in HepG2.2.15 cells at the study doses (Fig. 2c), whereas it significantly downregulated the expression of Hsc70 mRNA (Fig. 2d). The effect was confirmed by Western blotting at the protein level, showing a subsequent reduction of Hsc70 protein in the OMTR-treated cells (Fig. 2e). A flow-cytometric assay exhibited the same results (Fig. 2e). The downregulatory effect was reversible, and the intracellular Hsc70 mRNA rebounded after 72 h of exposure to a single dose of OMTR (see Fig. S1 in the supplemental material). In parallel with the downregulation of Hsc70, OMTR reduced intracellular, as well as supernatant, HBV DNA by over 60% (Fig. 2d).
To investigate the mode of action of OMTR on Hsc70 downregulation, the T1/2 of Hsc70 mRNA was determined in the OMTR-treated HepG2.2.15 cells. The results showed that OMTR treatment reduced the T1/2 of Hsc70 mRNA by 50% in the HepG2.2.15 cells (Fig. 2f), indicating a destabilization effect of OMTR on Hsc70 mRNA. Then, we designed and performed the following experiment to determine whether the 3′-UTR or 5′-UTR sequence of Hsc70 mRNA is involved in the destabilization effect of OMTR. Luc cDNA was used as a reporter and inserted into pcDNA3.0, forming the pLuc plasmid; the Hsc70 3′-UTR or Hsc70 5′-UTR sequence was inserted into the cytomegalovirus promoter-driven pLuc at the 3′ or 5′ end of the Luc coding sequence, respectively. The wild-type (pLuc) or chimeric (pLuc-3′UTR or pLuc-5′UTR) plasmid was transfected into the HepG2.2.15 cells. After 24 h of incubation, the transfected cells were treated or not with OMTR for 12 h. Then, we lysed the cells for total RNA, and the expression of Luc mRNA was determined using a real-time RT-PCR assay. As shown in Fig. 2g, OMTR decreased the Luc mRNA by about 50% in the pLuc-3′UTR-transfected cells, but not in those transfected with pLuc-5′UTR; OMTR did not affect the expression of the wild-type Luc mRNA (with pLuc as a control). The results indicate that OMTR decreased the mRNA stability of the heterologous Luc-Hsc70 transcript and that destabilization is mediated through a regulatory sequence(s) in the Hsc70 3′-UTR region. The pathway that is responsible for this action needs further investigation.
Selectivity of OMTR.
To investigate the selectivity of OMTR, the expression of HspA4 and Hsp90 mRNA in the OMTR-treated HepG2.2.15 cells was examined, as HspA4 is in the same Hsc70 family and Hsp90 is the most abundant cellular heat stress protein. As shown in Fig. S2a in the supplemental material, OMTR at 0.4 mg/ml (1.51 mM) largely inhibited Hsc70 expression but showed no effect on that of HspA4 and Hsp90 in the cells; HBV DNA decreased in parallel. Furthermore, well-known anti-HBV drugs were also tested to learn their effects on Hsc70 expression. Lamivudine and adefovir at their anti-HBV 50% infectious concentrations (IC50) showed almost no effect on the expression of Hsc70 mRNA (see Fig. S2b in the supplemental material), indicating the unique effect of OMTR on Hsc70.
Anti-HBV activity of OMTR in vitro.
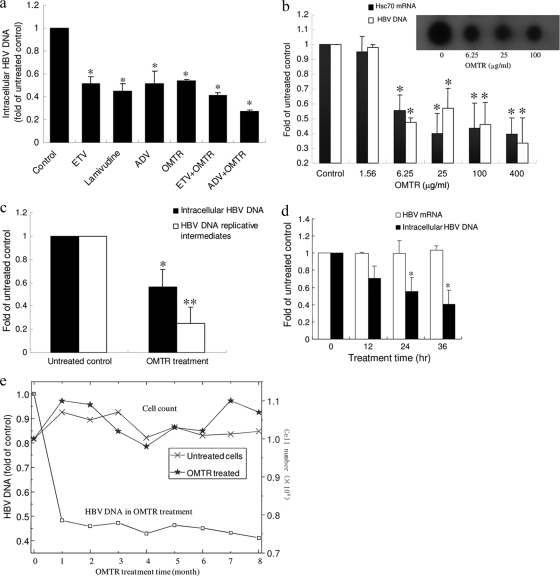
Treatment of HepG2.2.15 cells with OMTR for 36 h decreased intracellular HBV DNA by 62% and supernatant HBV DNA by 60% (Fig. 2d). Here, we tried to compare the efficacy of OMTR with those of known anti-HBV drugs, alone or in combination; therefore, a suboptimal dose of OMTR (30 μg/ml; 113 μM) was used. Although OMTR reached an anti-HBV efficacy similar to those of lamivudine, adefovir, and entecavir, the concentration of OMTR was still higher than those of the RT inhibitors, indicating that the activity of OMTR was not as potent as those of the known drugs (Fig. 3a), probably due to its indirect effect on HBV. A combination of OMTR with adefovir or entecavir caused an additional reduction of the intracellular HBV DNA (Fig. 3a). In the anti-HBV tests using HepG2.2.15 cells, we observed a dose-dependent reduction of HBV DNA, as well as an activity plateau of OMTR between 6.25 μg/ml (23.6 μM) and 0.4 mg/ml (1.51 mM), with an inhibition rate in the 43 to 65% range (Fig. 3b). The inset in Fig. 3b shows a dot blot confirmation. There could be two explanations for the activity plateau. First, it suggests that OMTR might suppress de novo synthesis of HBV but that it has little effect on the HBV genomic DNA integrated into the host cell genome in HepG2.2.15 cells; these HBV DNA copies constitute the base of the plateau. Second, the plateau also reflects the indirect mechanism of OMTR on HBV inhibition. The 50% effective concentration (EC50) of OMTR was 0.031 mg/ml (0.116 mM), and the CC50 (cytotoxic concentration that kills 50% of cells) in the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay was 3.12 mg/ml (11.8 mM), resulting in a therapeutic ratio of over 100.
FIG. 3.
Anti-HBV activity of OMTR in vitro. (a) OMTR was used to treat HepG2.2.15 cells alone or in combination with known anti-HBV drugs. The treatment was continued for 72 h, followed by HBV DNA measurement. ETV, entecavir (0.6 μg/ml; 2.16 μM); ADV, adefovir (3 μg/ml; 5.9 μM); lamivudine (3 μg/ml; 13 μM); OMTR (30 μg/ml; 113 μM). The experiment was repeated twice. Shown are means and SD. *, P < 0.01 versus untreated control. (b) HepG2.2.15 cells were treated with OMTR at the indicated concentrations, which were equal to 5.9, 23.6, 94.5, and 378 μM and 1.51 mM. After 72 h of incubation, Hsc70 mRNA and HBV DNA were examined. The amounts of Hsc70 mRNA and HBV DNA in the untreated cells were defined as 1, and the amounts in the cells treated with OMTR were plotted relative to those values. (Inset) Dot blot detection of the HBV DNA. Shown are means and SD. *, P < 0.01 versus untreated control. (c) The anti-HBV effect of OMTR on HBV de novo synthesis was examined in HepG2.2.15 cells. After 24 h of treatment with OMTR (0.4 mg/ml; 1.51 mM), cellular genomic DNA, as well as HBV DNA replicative intermediates, was isolated and measured by real-time PCR. The experiment was performed 3 times. Shown are means and SD. *, P < 0.01, and **, P < 0.001 versus untreated control. (d) HepG2.2.15 cells were treated with OMTR (0.4 mg/ml; 1.51 mM) for 12, 24, or 36 h, followed by measurement of the HBV DNA and HBV mRNA. The amount of DNA or RNA extracted from the treated cells was standardized to the mean level of the untreated cells, defined as 1.0. The experiment was performed 3 times. Shown are means and SD. *, P < 0.01 versus untreated control. (e) HepG2.2.15 cells were continuously exposed to a suboptimal dose of OMTR (30 μg/ml; 113 μM) for 8 months. The HBV DNA level and cell count were determined every 30 days. The HBV DNA in the untreated flasks at each time point was defined as 1, and the amount of HBV DNA of the treated cells at that time point was plotted relative to that value. The result for each time point represents an average of 2 flasks.
For the first explanation, we then examined OMTR's activity on HBV replicative intermediates in HepG2.2.15 cells. After treatment with OMTR for 24 h, the intracellular HBV DNA replicative intermediates, as well as HBV DNA, were examined. OMTR reduced the replicative intermediates by 75%, higher than its inhibition of genomic HBV DNA (54%) (Fig. 3c), supporting the assumption mentioned above. In addition to HBV DNA, HBV mRNA was also examined to investigate which step in the HBV life cycle was affected by OMTR. As shown in Fig. 3d, although the intracellular HBV DNA decreased with OMTR treatment, the HBV mRNA remained stable. It seems that the reverse transcription from pregenomic RNA (pgRNA) to HBV DNA is interrupted by OMTR. The unused HBV pgRNA might degrade through cellular mechanisms. Furthermore, other HBV life cycle events that involve reverse transcription (such as packaging) might also be interrupted by OMTR.
We then tested the effect of OMTR in a long-term treatment regimen. A suboptimal dose of OMTR (30 μg/ml; 113 μM) was used in the treatment to learn the possible mutation-related viral-load rebound, as well as cytotoxicity. HepG2.2.15 cells were treated for 8 months. HBV DNA declined in the first 2 months and remained low for at least 7 months (Fig. 3e). HBV DNA rebound was not observed. At each time point, the cells were counted and compared to the untreated control. As shown in Fig. 3e, the number of surviving cells in the OMTR group was similar to that in the untreated group, indicating good safety of OMTR in long-term therapy.
OMTR's activity against drug-resistant HBV strains.
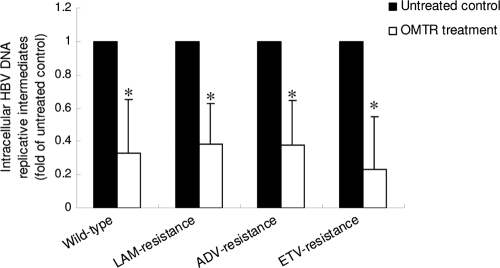
The above-mentioned results suggested that downregulation of host Hsc70 expression might be the main anti-HBV mechanism of OMTR. If so, OMTR should be effective against HBV that is resistant to RT inhibitors. Therefore, we measured OMTR's activity against the HBV DNA replicative intermediates of the drug-resistant HBV strains. In this experiment, Huh-7 cells transiently transfected with lamivudine-resistant (rtL180M plus rtM204V), adefovir-resistant (rtA181V plus rtN236T), or entecavir-resistant (rtL180M plus rtM204V plus rtS202G) HBV strains were used. After treatment with OMTR for 24 h, the intracellular HBV DNA replicative intermediates were isolated and examined. As shown in Fig. 4, treatment with OMTR for 24 h suppressed viral replication by 62 to 78% in all of the HBV strains. The inhibition efficacy of OMTR in the three drug-resistant HBV strains was similar to that in the wild-type HBV, indicating the effectiveness of OMTR against drug-resistant HBV.
FIG. 4.
OMTR was effective against drug-resistant HBV. Huh-7 cells were transfected with wild-type HBV or HBV strains resistant to lamivudine (LAM), adefovir (ADV), or entecavir (ETV). The cells were then treated with OMTR (0.4 mg/ml; 1.51 mM) for 24 h, followed by measurement of the HBV DNA replicative intermediates by the method described in Materials and Methods. The experiment was performed twice. Shown are means and SD. *, P < 0.01 versus untreated control.
DISCUSSION
Although natural anti-HBV products have been found in China and used with chronic hepatitis B (CHB) patients (5, 26), the mechanisms of action often remain unclear. This gives us opportunities to explore novel anti-HBV targets and mechanisms. Drugs that have identified chemical structures, confirmed anti-HBV effects in patients, and negative effects on viral enzymes (for instance, RT) are of particular interest to us. This has led us to search for a host-based antiviral strategy, the goal of which is to discover drugs that are able to create an intracellular environment in which viral proliferation is not supported and the chemotherapeutic target is not on the viruses. Antiviral agents working through this mechanism inhibit the replication of not only wild-type HBV, but also drug-resistant mutants. Clinical studies have shown that injection of OMTR (0.4 g/day for 6 months) in CHB patients caused over 40% of the patients to become negative for HBV DNA (12), and viral-load measurement showed that OMTR injection (0.4 g/day for 3 months) decreased the HBV viral load by about 2 log units on average (4). The anti-HBV effect of OMTR in patients basically agrees with that described here in HepG2.2.15 cells, but with higher efficacy, because the HBV viral-load test in the clinical setting measures the de novo synthesized viruses in blood. The T1/2 of OMTR in human blood was about 2.4 h (25), and the minimal blood concentration requirement for its effect on HBV has not been reported, to our knowledge. Our clinical studies showed that oral administration of OMTR (0.4 g/day for 12 months) in the treatment of patients with drug-resistant CHB caused about a 1.5-log-unit reduction of HBV DNA on average, similar to its efficacy in the naïve CHB cohort (R. Xue, Y.-P. Wang, W. Zhao, Z.-X. Zhou, S. Ning, and J.-D. Jiang, unpublished observations). Antiviral drugs working through host mechanisms could be at least part of the answer to the grand challenge of drug resistance in viruses. In addition, combinations of these agents with RT inhibitors might provide new options to achieve increased efficacy against HBV.
Targeting viral enzymes is an effective approach leading to potent drugs; however, newly discovered antiviral agents that act at different binding sites on enzymes or different viral proteins generate new drug-resistant mutations. It has become crucial to identify novel drug targets for the control of drug resistance. As knockdown of Hsc70 did not interfere with cell replication, we believe that host Hsc70, required for HBV replication, is a novel target for the inhibition of wild-type, as well as drug-resistant, HBV, and the compound OMTR appears to inhibit HBV replication through downregulation of host Hsc70 expression.
Supplementary Material
Acknowledgments
This work was supported by the Grand Challenges in Global Health Initiatives of the Bill and Melinda Gates Foundation and NIH (GC10-577; J.-D. Jiang) and the 973 Program, as well as the 11th 5-Year Program Key Drug R&D of the Ministry of Science and Technology, People's Republic of China (J.-D. Jiang).
We are grateful for help from Dong-Ping Xi's laboratory in the Beijing 302 Hospital for support in the experiment using drug-resistant HBV strains.
Footnotes
Published ahead of print on 22 February 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Arispe, N., M. Doh, and A. De Maio. 2002. Lipid interaction differentiates the constitutive and stress-induced heat shock proteins Hsc70 and Hsp70. Cell Stress Chaperones 7:330-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, J., and M. Nassal. 2003. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J. Biol. Chem. 278:36128-36138. [DOI] [PubMed] [Google Scholar]
- 3.Cai, X., G. J. Wang, Y. Qu, C. H. Fan, R. Q. Zhang, and W. S. Xu. 1997. Clinical efficacy of oxymatrine in the treatment of chronic hepatitis B. Acad. J. Sec. Mil. Med. Univ. 18:47-49. [Google Scholar]
- 4.Chen, Y. X., B. Y. Mao, and J. H. Jiang. 2002. Relationship between serum load of HBV-DNA and therapeutic effect of oxymatrine in patients with chronic hepatitis B. Zhongguo Zhong Xi Yi Jie He Za Zhi 22:335-336. [PubMed] [Google Scholar]
- 5.Chen, Z. X., S. J. Zhang, S. X. Lao, H. T. Hu, C. Y. Zhang, S. H. Guan, and Y. L. Gu. 2005. He Jie Tang in the treatment of chronic hepatitis B patients. World J. Gastroenterol. 11:6638-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delaney, W. E. IV, and H. C. Isom. 1998. Hepatitis B virus replication in human HepG2 cells mediated by hepatitis B virus recombinant baculovirus. Hepatology 28:1134-1146. [DOI] [PubMed] [Google Scholar]
- 7.Doo, E., and T. J. Liang. 2001. Molecular anatomy and pathophysiologic implications of drug resistance in hepatitis B virus infection. Gastroenterology 120:1000-1008. [DOI] [PubMed] [Google Scholar]
- 8.Hu, J., D. Flores, D. Toft, X. Wang, and D. Nguyen. 2004. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J. Virol. 78:13122-13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai, C. L., V. Ratziu, M. F. Yuan, and T. Poynard. 2003. Viral hepatitis B. Lancet 362:2089-2094. [DOI] [PubMed] [Google Scholar]
- 10.Lambert, C., and R. Prange. 2003. Chaperone action in the posttranslational topological reorientation of the hepatitis B virus large envelope protein: implications for translocational regulation. Proc. Natl. Acad. Sci. U. S. A. 100:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling, J. Y., G. Y. Zhang, Z. J. Cui, and C. K. Zhang. 2007. Supercritical fluid extraction of quinolizidine alkaloids from Sophora flavescens Ait and purification by high-speed counter-current chromatography. J. Chromatogr. A 1145:123-127. [DOI] [PubMed] [Google Scholar]
- 12.Lu, L. G., M. D. Zeng, Y. M. Mao, J. Q. Li, M. B. Wan, C. Z. Li, C. W. Chen, Q. C. Fu, J. Y. Wang, W. M. She, X. Cai, J. Ye, X. Q. Zhou, H. Wang, S. M. Wu, M. F. Tang, J. S. Zhu, W. X. Chen, and H. Q. Zhang. 2003. Oxymatrine therapy for chronic hepatitis B: a randomized double-blind and placebo-controlled multi-center trial. World J. Gastroenterol. 9:2480-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcellin, P., T. T. Chang, S. G. Lim, M. J. Tong, W. Sievert, M. L. Shiffman, L. Jeffers, Z. Goodman, M. S. Wulfsohn, S. Xiong, J. Fry, C. L. Brosgart, and the Adefovir Dipivoxil 437 Study Group. 2003. Adefovir dipivoxil for the treatment of hepatitis B e antigen-positive chronic hepatitis B. N. Engl. J. Med. 348:808-816. [DOI] [PubMed] [Google Scholar]
- 14.Mohanty, S. R., S. S. Kupfer, and V. Khiani. 2006. Treatment of chronic hepatitis B. Nat. Clin. Pract. Gastroenterol. Hepatol. 3:446-458. [DOI] [PubMed] [Google Scholar]
- 15.Morshauser, R. C., W. Hu, H. Wang, Y. Pang, G. C. Flynn, and E. R. Zuiderweg. 1999. High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70. J. Mol. Biol. 289:1387-1403. [DOI] [PubMed] [Google Scholar]
- 16.Pérez-Vargas, J., P. Romero, S. López, and C. F. Arias. 2006. The peptide-binding and ATPase domains of recombinant hsc70 are required to interact with rotavirus and reduce its infectivity. J. Virol. 80:3322-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prange, R., M. Werr, and H. Löffler-Mary. 1999. Chaperones involved in hepatitis B virus morphogenesis. Biol. Chem. 380:305-314. [DOI] [PubMed] [Google Scholar]
- 18.Preston, B. D., B. J. Poiesz, and L. A. Loeb. 1988. Fidelity of HIV-1 reverse transcriptase. Science 242:1168-1171. [DOI] [PubMed] [Google Scholar]
- 19.Price, P. M., R. Banerjee, and G. Acs. 1989. Inhibition of the replication of hepatitis B virus by the carbocyclic analogue of 2′-deoxyguanosine. Proc. Natl. Acad. Sci. U. S. A. 86:8541-8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts, J. D., K. Bebenek, and T. A. Kunkel. 1988. The accuracy of reverse transcriptase from HIV-1. Science 42:1171-1173. [DOI] [PubMed] [Google Scholar]
- 21.Shafer, R. W. 2002. Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin. Microbiol. Res. 15:247-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukahara, F., and Y. Maru. 2004. Identification of novel nuclear export and nuclear localization-related signals in human heat shock cognate protein 70. J. Biol. Chem. 279:8867-8872. [DOI] [PubMed] [Google Scholar]
- 23.Vassilaki, N., P. Friebe, P. Meuleman, S. Kallis, A. Kaul, G. Paranhos-Baccalà, G. Leroux-Roels, P. Mavromara, and R. Bartenschlager. 2008. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J. Virol. 82:11503-11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, B. E. 2000. Treatment of chronic liver diseases with traditional Chinese medicine. J. Gastroenterol. Hepatol. 15(Suppl.):67-70. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., D. Menggen, W. Y. Zheng, H. U. Wu, and S. D. Liu. 2003. Pharmacokinetics of oxymatrine in patients with chronic hepatitis B. Chin. J. Clin. Pharm. 12:228-230. [Google Scholar]
- 26.Yao, G. B., Y. Y. Ji, Q. H. Wang, X. Q. Zhou, D. Z. Xu, and X. Y. Chen. 2002. A randomized double-blind controlled trial of bicyclol in treatment of chronic hepatitis B. Chin. J. New Drugs Clin. Rem. 21:457-462. [Google Scholar]
- 27.Yuan, M. F., J. Fung, D. K. H. Wong, and C. L. Lai. 2009. Prevention and management of drug resistance for antihepatitis B treatment. Lancet Infect. Dis. 9:256-264. [DOI] [PubMed] [Google Scholar]
- 28.Yuen, M. F., and C. L. Lai. 2001. Treatment of chronic hepatitis B. Lancet Infect. Dis. 1:232-241. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z., U. Protzer, Z. Hu, J. Jacob, and T. J. Liang. 2004. Inhibition of cellular proteasome activities enhances hepadnavirus replication in an HBX-dependent manner. J. Virol. 78:4566-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zoulim, F. 2006. Antiviral therapy of chronic hepatitis B. Antiviral Res. 71:206-215. [DOI] [PubMed] [Google Scholar]
- 31.Zoulim, F., D. Durantel, and P. Deny. 2009. Management and prevention of drug resistance in chronic hepatitis B. Liver Int. 29:108-115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.