Abstract
Class D β-lactamases represent a heterogeneous group of active-site serine β-lactamases that show an extraordinary panel of functional features and substrate profiles, thus representing relevant models for biochemical and structural studies. OXA-46 is a narrow-spectrum enzyme belonging to the OXA-2 subgroup which was found in a Pseudomonas aeruginosa clinical isolate from northern Italy. In this work, we obtained the three-dimensional structure of OXA-46, which shows the overall fold of active serine β-lactamases and a dimeric quaternary structure. Significant differences with currently available structures of class D β-lactamases were found in the loops located close to the active site, which differ in length and conformation. Interestingly, the three subunits present in the asymmetric unit showed some structural heterogeneity, only one of which presented a carbamylated lysine recognized as an important functional feature of class D enzymes. The carbamylation state of residue Lys75 appeared to be associated with different shapes and dimensions of the active site. Moreover, a tartrate molecule from the crystallization buffer was found in the active site of the noncarbamylated subunits, which interacts with catalytically relevant residues. The OXA-46 crystal asymmetric units thus interestingly present the structures of the free carbamylated active site and of the ligand-bound uncarbamylated active site, offering the structural basis for investigating the potential of new scaffolds of β-lactamase inhibitors.
Class D β-lactamases are a group of bacterial enzymes involved in resistance to β-lactam antibiotics that show an extraordinary structural and functional diversity. Indeed, these enzymes share only nine strictly conserved residues and may behave as narrow-spectrum oxacillinases (e.g., OXA-1, OXA-2, and FUS-1 [OXA-85]) or exhibit an extended spectrum of activity that includes oxyimino-cephalosporins (e.g., OXA-32 and OXA-11, which are OXA-2- and OXA-10-derivatives, respectively, and OXA-18, an OXA-type enzyme with intrinsic extended-spectrum β-lactamase [ESBL] properties) or even hydrolyze the most recent carbapenem antibiotics (e.g., OXA-23, OXA-24, OXA-48, and OXA-58) (20, 24). The emergence of class D β-lactamases that show extended-spectrum or carbapenemase activity has contributed to the increased clinical relevance of these resistance determinants (15, 20, 24, 26).
OXA-46 was identified in a multidrug-resistant Pseudomonas aeruginosa clinical isolate from Pavia (northern Italy), which also produced a metallo-β-lactamase, and is encoded by an integron-borne gene cassette (9). On the basis of sequence similarity, OXA-46 belongs to the OXA-2 subgroup and shares 77.8% protein sequence identity with the latter, showed a narrow-spectrum substrate profile, and is inhibited by carbapenems, tazobactam and, to lesser extent, clavulanate (9). Notably, OXA-46 is poorly inhibited by NaCl, in comparison with other OXA-type enzymes, suggesting structural differences in the active-site region, as chloride ions have been hypothesized to bind in the vicinity of the active-site Lys of the SxxK motif (motif 1) and thus displace the water molecule involved in the deacylation step of the catalyzed reaction (21). Moreover, as already observed for some but not all class D β-lactamases, the native form of the enzyme adopts a dimeric quaternary structure whose stability does not depend on the presence of metal ions (9). These features make OXA-46 an alternative model of narrow-spectrum OXA-type β-lactamases, with a sequence rather divergent from OXA-1, whose three-dimensional structure is available.
Various crystal structures for class D β-lactamase are now available, including those for OXA-1 (29), OXA-10 (and its close variant, OXA-13) (16, 21, 22), OXA-24 (28), and OXA-48 (7). However, no structures of enzymes of the OXA-2 subgroup have been thus far published, although the structure of OXA-2 is present in the Protein Data Bank (PDB) database. In this work, we determined the crystal structure of OXA-46, taken as a model of OXA-2 subgroup narrow-spectrum enzymes, and compared it to the available class D β-lactamase structures.
MATERIALS AND METHODS
Crystallization of OXA-46.
OXA-46 was purified as previously reported (9). Preliminary crystallization trials were performed with a 10-mg/ml protein solution in 100 mM Tris-H2SO4 buffer at pH 7.0 using commercial screens (Crystal Screen 1 and 2; Hampton Research) and provided spherulites and very small and ill-formed crystals with two different precipitant solutions, 0.8 M Na,K l-tartrate tetrahydrate in 0.1 M HEPES, pH 7.5 (solution 1), and 2.0 M (NH4)2SO4 plus 2% (vol/vol) polyethylene glycol 400 (PEG 400) in 0.1 M HEPES, pH 7.5 (solution 2) (in all cases, 2-μl drops of the protein solution were mixed with an equal amount of the precipitant solution). Diffraction-quality crystals were obtained by repeated cycles of micro- and macroseeding in a sitting drop setup (2) by using the crystals obtained from solution 1 seeded in a drop made by 1 μl of a 10-mg/ml OXA-46 solution in ultrapure water and 1 μl of a 1 M Na,K l-tartrate tetrahydrate solution in 50 mM HEPES at pH 7.5 and 2 to 4% (vol/vol) PEG 400 as precipitants. The crystallization plates were stored at 20°C. Crystal growth was completed within 3 to 4 weeks from seeding.
Data collection and processing.
The data collection on OXA-46 crystals was carried out at the Deutsches Elektronen Synchrotron (DESY; Hamburg, Germany) on the BW7A beamline of EMBL. The data collection statistics are reported in Table 1. The diffraction data set was collected at a temperature of 100 K. The precipitating solution was added with 15% (vol/vol) ethylene glycol and used as the crystal cryoprotectant. The data set was processed using the program MOSFLM (14) and scaled with the program SCALA (8) from the CCP4 suite (4).
TABLE 1.
Data collection and refinement statisticsa
| Characteristic | Result |
|---|---|
| X-ray source | EMBL BW7A (DESY) |
| Wavelength (Å) | 0.92 Å |
| Data collection temp (K) | 100 |
| Space group | H32 (no. 155) |
| Cell dimensions (Å) | a = b = 123.84; c = 327.92 |
| No. of subunits/asymmetric unit | 3 |
| Matthews coefficient (Å3 Da−1) | 2.85 |
| Solvent content (%) | 56.83 |
| Resolution limits (Å) | 31.40-2.40 (2.57-2.40) |
| No. of reflections measured | 197,273 (31,943) |
| No. of unique reflections | 37,716 (6,058) |
| Completeness (%) | 99.5 (99.4) |
| Rmergeb (%) | 7.8 (43.0) |
| Multiplicity | 6.2 (5.3) |
| I/σ(I) | 6.2 (1.6) |
| Rcrystb (%) | 21.1 (26.0) |
| Rfreeb (%) | 28.5 (39.1) |
| No. of protein atoms | 5,825 |
| No. of ligand atoms | 63 |
| No. of water molecules | 398 |
| Average B factor (Å2) | 40.9 |
| RMSD bond length (Å) | 0.020 |
| RMSD bond angle (°) | 1.891 |
| Ramachandran plot residues in: | |
| Most favored regions (%) | 89.4 |
| Additionally allowed regions (%) | 9.6 |
| Disallowed regions (%) | 0 |
Data in parentheses refer to results for the highest-resolution shell.
Rmerge = ∑h∑i|li,h − l̄h|/∑h∑ili,h × 100. Rcryst(Rfree) = ∑h||Fh,obs| − |Fh,calc||/∑h|Fh,obs| × 100.
Structure determination and refinement.
The OXA-46 structure was solved by molecular replacement using the structure of OXA-48 (43% sequence homology) as a model (PDB code 3HBR), with all the water molecules and ions omitted. The correct orientation and translation of the molecule within the crystallographic unit cell were determined with standard Patterson search techniques (5, 27), as implemented in the software MOLREP (4, 30-32). The program provided an evident solution for the positioning of three independent OXA-46 subunits in the asymmetric unit (subunits A to C). The refinement was carried out by using REFMAC5 (4, 19). Between the refinement cycles, the model was subjected to manual rebuilding using XtalView (18). The same program has been used to model the various ligands (tartrate, PEG, and ethylene glycol). Water molecules have been added in all cases by using the standard procedure within the ARP/wARP suite (12, 23). The stereochemical quality of the refined model was assessed using the program PROCHECK (13). Figures were prepared with the program PyMol (http://www.pymol.org/) or with CCP4 mg from the CCP4 suite (4, 25).
Protein structure accession number.
The coordinates and structure factors of OXA-46 have been deposited at the Protein Data Bank under code 3IF6.
RESULTS
Overall fold and comparison with class D β-lactamase structures.
The OXA-46 crystals provided a complete data set to 2.4-Å resolution with maximum diffraction up to 2.0 Å and with data indexed in the rhombohedral space group H32 (no. 155; hexagonal axes) with cell parameters of a = b = 123.84 Å and c = 327.92 Å (Table 1). The crystal asymmetric unit contains three independent OXA-46 subunits and 56.83% solvent (Matthews coefficient, 2.85 Å3/Da), one PEG 400 molecule, and two tartrate anions from the crystallization buffer. After refinement, the final OXA-46 model consists of the three independent OXA-46 chains (A to C) starting from His24 (subunit B) or Val25 (subunits A and C) and ending with Asn266 (the amino acid numbering of OXA-46 SwissProt entry Q8GRH0 is used throughout) (Table 1). The residues at positions 97 to 108 and 152 to 155 in subunit A and residues 148 to 156 in subunit C are not visible in the electron density and are missing from the final model. On the other hand, subunit B is complete, with good electron density in the whole chain.
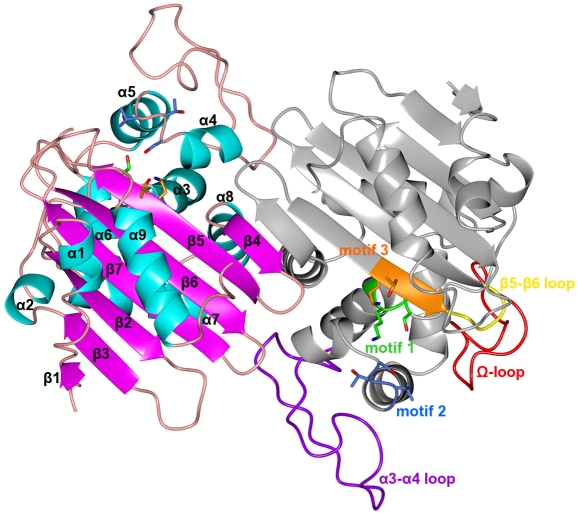
Each OXA-46 subunit adopts a tertiary structure (Fig. 1) consisting of a seven-stranded antiparallel β-sheet (β1, β3, β2, β7, β6, β5, and β4) sandwiched between the N- and C-terminal α-helices (α1 and α9) on one side and helices α2 and α7 on the other. Helices α2 and α7 are part of an all-α domain made of seven α-helices (α2 to α8). Helix α6 is linked to helix α7 by the large loop known as the Ω-loop (residues 143 to 168). Another large loop (residues 85 to 113) links α3 to α4 (α3-α4 loop). These latter helices define part of the active-site channel, and α3 bears the active-site motif 1 Ser72-Trp73-Phe74-Lys75.
FIG. 1.
Cartoon representation of the functional OXA-46 dimer, consisting of subunits B (right) and C (left). Subunit C shows the secondary structure elements of OXA-46, where helices appear in cyan, strands in magenta, and turns in pink. Subunit B shows the location of the amino acids constituting the conserved motifs 1 to 3 (visible as green, blue, and orange sticks, respectively) and of the functionally relevant loops, which are labeled. The different domains of each subunit are visible in the figure, which shows the N- and C-terminal helices in the foreground, separated by the seven-stranded β-sheet from the all-α domain in the background. The two subunits form an intermolecular β-sheet through their respective β4 strands.
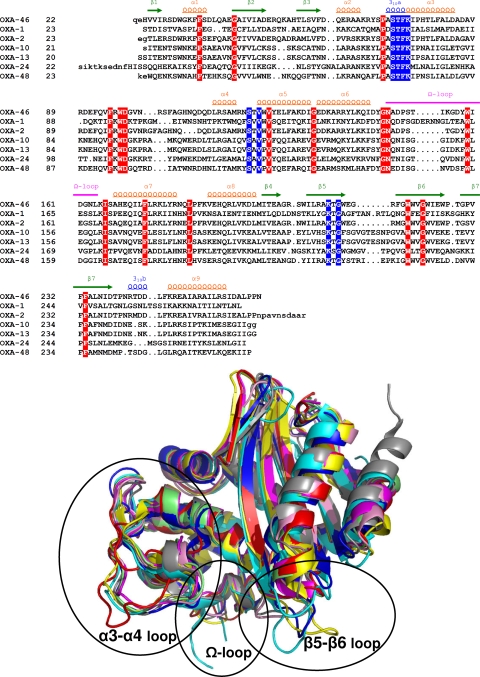
The overall structure of OXA-46 is close to that of the other class D β-lactamases so far determined by X-ray crystallography, including OXA-1 (29), OXA-2 (PDB code 1K38), OXA-10 (16, 21), OXA-13 (22), OXA-24 (28), and OXA-48 (7), whose amino acid sequences are shown in a structure-based alignment in Fig. 2, with a root mean square deviation (RMSD) on the backbone Cα atoms of the common residues ranging from 0.49 Å for OXA-2 to 2.20 Å for OXA-1 (Table 2). However, significant differences are located in the Ω-loop, the α3-α4 loop, and the loop connecting strands β5 and β6 (β5-β6 loop), where different conformations and lengths are observed (Fig. 2). Noteworthy, the β5-β6 loop, located close to the active site, is notably shorter in OXA-46 (and OXA-2) than in other class D β-lactamases and adopts a unique conformation compared to currently available structures (Fig. 2). Indeed, this loop consists of only 3 residues (by contrast with the 7 to 10 residues found in OXA-48 and OXA-10, respectively), which form a turn stabilized by a salt bridge between the side chains of residues Glu214 and Arg216. However, the orientation of this loop does not significantly alter active-site accessibility, as the dimensions of the OXA-46 active-site cavity are similar to those of other class D β-lactamases (see below). It remains unclear whether this structural feature could have an impact on the functional properties of OXA-46. Thus, the narrow spectrum exhibited by OXA-46 could not be explained by a simple active-site hindrance of larger molecules, such as expanded-spectrum cephalosporins.
FIG. 2.
(Top) Structure-based sequence alignment of OXA-46 with other class D β-lactamases whose structures have been determined. Amino acids of conserved motifs 1 to 3 are boxed in blue. Residues that are conserved in all sequences are boxed in red. Secondary structure elements of OXA-46 are shown on the top of the sequence. Lowercase amino acids refer to those missing in the crystal structure. (Bottom) Least-squares superimposition of representative class D β-lactamase structures and the β-lactam sensor domain of Staphylococcus aureus BlaR1. Color legends and PDB codes are as follows: OXA-46 (PDB code 3IF6), red; OXA-1 (PDB code 1M6K), cyan; OXA-2 (PDB code 1K38), magenta; OXA-10 (PDB code 1K55), yellow; OXA-24 (PDB code 2JC7), gray; OXA-48 (PDB code 3HBR), pink; BlaR1 (PDB code 1NRF), blue. The loops where the largest conformational differences occur are framed.
TABLE 2.
Sequence and structure comparisons of OXA-46 with other class D β-lactamases whose structures are currently available
| β-Lactamase | Sequence identity (%) | RMSD (Å)a |
|---|---|---|
| OXA-1 | 23.3 | 2.20 (192) |
| OXA-2 | 77.8 | 0.49 (205) |
| OXA-10 | 33.3 | 0.98 (204) |
| OXA-24 | 27.0 | 1.15 (171) |
| OXA-48 | 38.5 | 1.01 (202) |
Data were computed using the coordinates of the OXA-46 subunit B. The number of Cα atoms used in the calculation is indicated in parentheses.
Surprisingly, considering the rather high sequence similarities between OXA-46 and OXA-2 (nearly 80% [Table 2]), structural differences in some of these loops are also found between the B subunits of OXA-46 and OXA-2 structures. The α3-α4 loop of OXA-46 is up to ∼5 Å away from the corresponding OXA-2 residues and is more distant from the protein core, underlining its greater conformational variability. Similarly, the Ω-loop, only partially visible in the OXA-2 structure, also seems to adopt a different orientation at the level of residues Asp150-Gly162. Overall, these data show the extreme variability and flexibility exhibited by these loops.
Quaternary structure and subunit interactions.
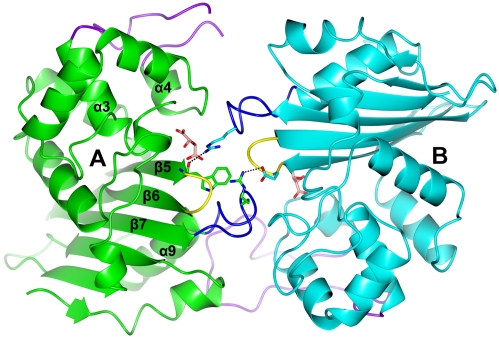
The crystal asymmetric unit contained three OXA-46 monomers and revealed the presence of OXA-46 homodimers (Fig. 1), in agreement with the dimeric nature of OXA-46 in solution, as shown by size exclusion chromatography (9). The three OXA-46 subunits establish different intermolecular contacts in the crystal. The OXA-46 dimer is formed by subunits B and C (and also by two symmetry-related A subunits belonging to adjacent asymmetric units) and is stabilized by an intermolecular β-sheet spanning the two subunits, similar to OXA-10, OXA-13, and OXA-48 (7, 16, 22). Additional contacts, which reflect crystal packing, are found between two symmetry-related B subunits through their β1 stands (data not shown) and between subunits A and B, involving structural elements that are close to the active site and the tartrate binding site (Fig. 3).
FIG. 3.
Intermolecular interface involving subunits A (green) and B (cyan). The interactions entail the β5-β6 loop (yellow), some residues from the α3-α4 loop (purple), and Arg242 (represented as sticks) located in the β7-α9 loop (blue), which reaches the active-site groove of the facing subunit. Tartrate molecules bound in the active site are shown as pink sticks. Arg242 of subunit A forms a salt bridge with the side chain of Glu214 of subunit B, while Arg242 of subunit B, which adopts a different conformation and is close to the tartrate molecule bound to subunit A, is H-bonded to the backbone carbonyl of residue Trp213 of subunit A.
The least-squares superimposition of OXA-46 subunit B with subunits A and C gives RMSDs of 0.33 Å and 0.51 Å, respectively. However, the conformation and orientation of loop α3-α4 differ greatly in subunits B and C, while they are not visible in subunit A, with observed displacements of up to 5.2 Å between corresponding residues. In subunit B, the α3-α4 loop lies at about 8.0 Å and 12.0 Å from the α4 and α5 helices, respectively, allowing sufficient room for PEG 400 from the crystallization buffer to bind in between. The PEG 400 molecule is almost completely shown by the electron density map and is located at the interface between loop α3-α4 and the loop α4-α5 characterized by the presence of several hydrophobic residues as well as that of motif 2 (Ser120-Val122).
Conserved structural motifs and the active-site cavity.
The three conserved motifs of class D β-lactamases (Fig. 1 and 2) were expectedly located as follows: (i) motif 1 (Ser72-Thr73-Phe74-Lys75) is at the beginning of helix α3, where Ser72 is the catalytic serine residue and Lys75 is the lysine residue whose role in deacylation has previously been demonstrated (10); (ii) motif 2 (Ser120-Thr121-Val122) is in the loop connecting helix α4 to helix α5; (iii) motif 3 (Lys210-Thr211-Gly212) is located in front of the catalytic site on the β5 strand.
It is interesting that subunits A and B clearly show the presence of one tartrate anion from the crystallization solution bound in the active site (see below for the description of the interaction). On the contrary, the subunit C active site is free from exogenous ligands. The presence of the anions in the active site appears to be correlated to the chemical state of Lys75. Indeed, at variance with subunits A and B, subunit C has an active-site carbamylated lysine (Lys75). Furthermore, and contrary to subunit B, the conformation of the α3-α4 loop of subunit C is identical to that occurring in OXA-2 (PDB code 1K38), whose structure is that of the free enzyme (without ligand) and also bears a carbamylated lysine (motif 1). In other words, the crystal asymmetric unit thus offers the interesting feature of presenting the structure of the free carbamylated enzyme (subunit C) and of the uncarbamylated ligand-bound enzyme (subunits A and B) as subunit A and B active sites both occupied by an l-(R,R)-tartrate anion held in place by several H-bond and salt links (see below for details on the interaction).
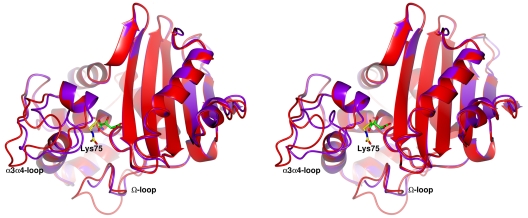
The active site is a narrow (∼10-Å) and shallow (∼7-Å) crevice defined by residues of motifs 2 and 3 but shows some flexibility, as the superimposition of the three subunits indicates that its width varies from about 9.5 Å in subunits A and B to 7.7 Å in subunit C (distance from Cα of Ser72 to Cα of Val122). The enlargement of the crevice appears to be correlated to the Lys75 carbamylation and in turn to the occupancy of the active-site cavity by the tartrate molecule. Indeed, the comparison with the crystal structures of OXA-10 determined at different pHs and with OXA-48 highlights this relevant difference present in the structure of the OXA-46 A and B subunits, namely, the different conformation presented by the α4-α5 loop when the tartrate molecule is bound. In these subunits, this loop adopts a different conformation, compared to subunit C or that observed in OXA-10 and OXA-48, causing a widening of the active-site channel by about 2.0 Å that provides room for the tartrate molecule (Fig. 4).
FIG. 4.
Stereo view of the least-squares superimposition of OXA-46 subunit B (red), showing a bound tartrate molecule (green), with subunit C (purple), in which Lys75 is carbamylated (yellow). The active-site width appears larger in subunit B where tartrate is bound. Significant differences are also present in the Ω-loop and in the α3-α4 loop.
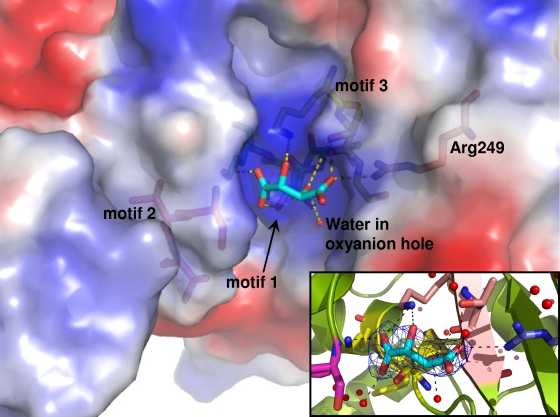
Figure 5 shows the electrostatic surface of OXA-46 subunit B, highlighting the overall positive charge present in the active site in correspondence to motifs 1 and 3. An additional positive charge is due to the presence of Arg249 (corresponding to Arg250 of OXA-10), which is known to interact with the β-lactam carboxylate and is present in many class D β-lactamases (17, 22).
FIG. 5.
Active-site cavity of OXA-46 subunit B. The surface is colored based on electrostatic potential as calculated by using PyMol (blue, positive; red, negative). Under the surface are visible the residues of motif 1 (Ser72 to Lys75; green), motif 2 (Ser120 to Val122; magenta), motif 3 (Lys210 to Gly212; yellow), and the conserved Arg249 (pink). The tartrate molecule (cyan) is shown docked in the cavity together with its H-bond network (yellow dotted lines). The water molecule bound into the “oxyanion hole” is represented as a red sphere of arbitrary radius. (Inset) Omit map contoured at 4σ calculated with phases computed from the refined model without ligands. Hydrogen bonds between tartrate and the active-site residues and water molecules are shown as black dashed lines.
Interaction with tartrate.
The tartrate molecule strongly interacts with the cavity by making 9 and 10 H-bonds, in subunits A and B, respectively, with conserved active-site residues (Ser72, Lys75, Asn119, Lys210, Thr211, and Arg249) and three or four water molecules (Fig. 5, inset). The sets of H-bonds slightly differ between the two subunits, as do the orientations of the two ligand molecules. The Lys75 side chains in subunits A and B are not carbamylated and adopt a different conformation with respect to that observed in other OXA structures free from ligands or inhibitors, and they are not H-bonded to Ser72. It is worth noting that in subunit C, where no tartrate is bound, the side chain of Lys75 is clearly carbamylated and found in the usual conformation where one carboxylate oxygen is H-bonded to Ser72. The tartrate carboxylate O1 and O2 atoms are engaged in H-bonds with Arg249 and Thr211 and occupy a position similar to that of the meropenem carboxylate group, as observed in the active site of OXA-13 (PDB code 1H8Y) (22). In addition, the so-called oxyanion hole, composed of the backbone NH of Ser72 and NH and CO of Trp213, is occupied in all three subunits by a water molecule, which in the A and B subunits is H-bonded to the tartrate ligands (Fig. 5). A single other water molecule is structurally conserved in the active site of the three subunits and corresponds to the water that is H-bonded to His78, the carbamylated Lys75 (in subunit C), or other water molecules (subunits A and B). Finally, as a consequence of the interaction existing between subunits A and B, the side chain of Arg242 of subunit B comes in the vicinity of the tartrate anion bound in the facing subunit A and closes one side of the tartrate binding site (Fig. 3).
The comparison of subunits B and C highlights the widening of the active-site crevice observed when a tartrate molecule is bound (Fig. 4), and the distance between the active-site opposite walls in subunits A and B increases by about 2.2 Å compared to that in subunit C. Such a finding shows that the enzyme is able to accommodate molecules of slightly different sizes. The comparison with the crystal structures of the OXA-1 and OXA-13 complexes with imipenem and meropenem shows that tartrate has a unique binding mode, being able to link four conserved and catalytically relevant residues in the active site, namely, Ser72, Lys75, Lys210, and Arg249, besides the oxyanion hole water molecule. On the contrary, the interactions in the active site of OXA-13 of the inhibitors imipenem (1H5X) and meropenem (1H8Y) involve only Arg250 (corresponding to OXA46 Arg249) and the backbone carbonyl oxygen of Phe209 (corresponding to OXA-46 Trp213) (22), besides the covalent bond to the catalytic serine.
Inhibition of OXA β-lactamases by anions.
Chloride anions have little effect on the activity of OXA-46 (9). Inspection of the OXA-46 structure shows that OXA-46 interacts with anions (here, a tartrate molecule). In subunits A and B, where Lys75 is not carbamylated, a water molecule is observed in the same position of the putative water/chloride site found in the low-pH crystal structure of OXA-10 (PDB code 1FOF) (21). This water molecule is absent in subunit C due to the presence in its place of the carbamyl group on Lys75 (the same observation is made for the high-pH form of OXA-10, where Lys70 is carbamylated as well [10]). Paetzel et al. proposed that the inhibition by anions characteristic of some OXA enzymes was due to the replacement of the deacylating water by chloride (21). Later Golemi et al. proposed that anions might inhibit OXAs by promoting the decarbamylation of the active-site lysine, leading to enzyme inactivation, and they showed that carbamylation of motif 1 lysine is needed for enzyme activity (10). Indeed, an [Au(CN)2]− anion, which was used for phasing, was found in the active-site cavity of the first OXA-10 structure determined (11), whereas Lys70 (equivalent to OXA-46 Lys75) appeared to be not carbamylated.
The OXA-46 structure shows that (i) when Lys75 is not carbamylated, the deacylating water molecule is present in its usual site and a tartrate molecule is observed in the active site, indicating that anions like chloride ions could bind the enzyme, and that (ii) when Lys75 is carbamylated, this water is absent and tartrate cannot bind. Whatever case is present in solution for the OXA-46 molecules, the enzyme is only weakly inhibited by chloride ions (9). Our conclusion is that a possible mechanism of inhibition by anions involves both the active-site lysine decarbamylation (possibly promoted by the anion itself) and the subsequent binding of anions to the positive charge shown by the decarbamylated lysine side chain. Thus, the relatively high inhibition constant of OXA-46 for chloride ions (Ki, 230 ± 20 mM) (9) might reflect the incomplete decarbamylation of the enzyme by the anion, as shown by the crystal structure.
DISCUSSION
The determination of the structure of the narrow-spectrum OXA-46 β-lactamase demonstrates its dimeric quaternary structure and allows us to understand some of the properties of this enzyme. Indeed, both the OXA-10 and OXA-14 dimeric structures are stabilized by the binding of divalent metal ions (6, 21), while for the OXA-46 dimer, as with OXA-29, stability is only due to intersubunit (B and C) H-bonds, which involve not only the residues of the β7 strands responsible for the intermolecular β-sheet but also other interactions at the interface, like the H-bonds and salt links intervening between Glu91-Asn181, Glu91-Arg191, and Asp195-Arg198.
The comparison of the different class D β-lactamase structures reported in Fig. 2 shows that the length of the β5-β6 loop of OXA-46 is similar to that of OXA-2 and much shorter than in all the other class D β-lactamases, and it appears not to influence active-site accessibility. In other enzymes, like OXA-48, the residues present in this loop have been suggested to influence the enzyme catalytic profile toward the different classes of β-lactams (7).
The present crystal structure shows that (i) two out of the three independent subunits have a noncarbamylated Lys75 residue and are bound to a tartrate molecule and that (ii) the OXA-46 α4-α5 loop is flexible enough to allow the binding of the tricarboxylate molecule (in this case, a tartrate ligand). This observation raises the question about the chemistry underlying this asymmetric behavior of the OXA-46 subunits. Tartrate was added to the protein during the crystallization experiment, several days after protein purification. This time lag should have been sufficient for the complete carbamylation of the protein by the ambient carbon dioxide. On the other hand, it has been shown for OXA-10 that soaking with anions like chloride causes decarbamylation of the active-site lysine, leading to enzyme inactivation (10). It is then possible that the tartrate itself promotes its own binding by decarbamylating Lys75. However, the mechanism of this putative reaction is unknown, and it is puzzling that the reaction occurs for OXA-46 in the exact stoichiometry of 2:3 with respect to the OXA-46 subunits. Interestingly, the binding interaction of tartrate in the OXA-46 active site is apparently not similar to that of the β-lactam substrates, except for the carboxylate group, which interacts with Thr211 and Arg249. It is noteworthy that tartrate did not displace the so-called “acylation” water molecule located in the oxyanion hole, which was observed in all subunits independently of the presence of the ligand.
The observed binding of the tartrate molecule suggests that organic polycarboxylates might behave as serine β-lactamase competitive inhibitors, but only when the motif 1 Lys residue is not carbamylated either constitutively or after a decarbamylation reaction. This finding would suggest that such inhibitors should preferentially inhibit class A β-lactamases, rather than class D enzymes. Nevertheless, such polycarboxylates might represent promising scaffolds for the design of new β-lactamase inhibitors, as recently reported by Beck et al., who performed the screening of amino citrate and amino isocitrate derivatives toward the class D enzyme OXA-10 (1). The present crystal structure might provide a useful rationale for the inhibitory properties found for the above citrate derivatives (1). It also underlines the importance of the assay conditions when screening for class D β-lactamase inhibitors and of the question whether all or a portion of the enzyme is actually carbamylated under physiological conditions.
Finally, the heterogeneity exhibited by class D β-lactamases in terms of susceptibility to chloride ions might depend on their ability to promote the decarbamylation of the active-site lysine residue, a process that might exhibit very different kinetics depending on the enzyme. This hypothesis is in agreement with our present data (OXA-46 binds only tartrate when Lys75 is free) and the structure of OXA-24, obtained at low pH and thus with a free lysine, which shows the presence of a sulfate ion in the active site. The prediction of the class D β-lactamase spectrum of hydrolysis remains, at this stage, beyond the capabilities of structural analysis, as the active site of OXA-46 is apparently large enough to accommodate larger molecules, such as expanded-spectrum cephalosporins. Thus, the mutations conferring extended-spectrum properties, such as the Asn63Ser substitution in the OXA-17 ESBL variant of OXA-10, are unlikely to affect the enzyme active site but rather the reactivity of conserved water molecules toward substrate acylation and/or deacylation.
Acknowledgments
We acknowledge the EMBL Hamburg outstation for the use of their beamline for data collection.
The work at EMBL Hamburg was supported by grant HPRI-CT-1999-00017. This work was also funded in part by grants from the European Union (contract no. HPRN-CT-2002-00264) and by the Italian Ministero dell'Istruzione, Università e Ricerca (MIUR contract no. 2005061894_004).
Footnotes
Published ahead of print on 9 February 2010.
REFERENCES
- 1.Beck, J., L. Vercheval, C. Bebrone, A. Herteg-Fernea, P. Lassaux, and J. Marchand-Brynaert. 2009. Discovery of novel lipophilic inhibitors of OXA-10 enzyme (class D β-lactamase) by screening amino analogs and homologs of citrate and isocitrate. Bioorg. Med. Chem. Lett. 19:3593-3597. [DOI] [PubMed] [Google Scholar]
- 2.Benvenuti, M., and S. Mangani. 2007. Crystallization of soluble proteins in vapor diffusion for X-ray crystallography. Nat. Protoc. 2:1633-1651. [DOI] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Collaborative Computational Project, N4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 5.Crowther, R. A. 1972. The fast rotation function, p. 173-178. In M. J. Rossmann (ed.), The molecular replacement method. Gordon & Breach, New York, NY.
- 6.Danel, F., J. M. Frere, and D. M. Livermore. 2001. Evidence of dimerisation among class D β-lactamases: kinetics of OXA-14 β-lactamase. Biochim. Biophys. Acta 1546:132-142. [DOI] [PubMed] [Google Scholar]
- 7.Docquier, J. D., V. Calderone, L. F. De, M. Benvenuti, F. Giuliani, L. Bellucci, A. Tafi, P. Nordmann, M. Botta, G. M. Rossolini, and S. Mangani. 2009. Crystal structure of the OXA-48 β-lactamase reveals mechanistic diversity among class D carbapenemases. Chem. Biol. 16:540-547. [DOI] [PubMed] [Google Scholar]
- 8.Evans, P. R. 1997. SCALA, continuous scaling program. Joint CCP4 ESF-EACBM Newslett. 33:22-24. [Google Scholar]
- 9.Giuliani, F., J. D. Docquier, M. L. Riccio, L. Pagani, and G. M. Rossolini. 2005. OXA-46, a new class D β-lactamase of narrow substrate specificity encoded by a blaVIM-1-containing integron from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 49:1973-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golemi, D., L. Maveyraud, S. Vakulenko, J. P. Samama, and S. Mobashery. 2001. Critical involvement of a carbamylated lysine in catalytic function of class D β-lactamases. Proc. Natl. Acad. Sci. U. S. A. 98:14280-14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golemi, D., L. Maveyraud, S. Vakulenko, S. Tranier, A. Ishiwata, L. P. Kotra, J. P. Samama, and S. Mobashery. 2000. The first structural and mechanistic insights for class D β-lactamases: evidence for a novel catalytic process for turnover of β-lactam antibiotics. J. Am. Chem. Soc. 122:6132-6133. [Google Scholar]
- 12.Lamzin, V. S., and K. S. Wilson. 1993. Automated refinement of protein models. Acta Crystallogr. D Biol. Crystallogr. 49:129-147. [DOI] [PubMed] [Google Scholar]
- 13.Laskowski, R. A., M. W. Macarthur, D. S. Moss, and J. M. Thornton. 1993. Procheck: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 14.Leslie, A. G. W. 1991. Molecular data processing, p. 50-61. In V. Moras, A. D. Podjarny, and J. P. Thierry (ed.), Crystallographic computing V. Oxford University Press, Oxford, England.
- 15.Livermore, D. M. 2007. Introduction: the challenge of multiresistance. Int. J. Antimicrob. Agents 29(Suppl. 3):S1-S7. [DOI] [PubMed] [Google Scholar]
- 16.Maveyraud, L., D. Golemi, L. P. Kotra, S. Tranier, S. Vakulenko, S. Mobashery, and J. P. Samama. 2000. Insights into class D β-lactamases are revealed by the crystal structure of the OXA10 enzyme from Pseudomonas aeruginosa. Structure 8:1289-1298. [DOI] [PubMed] [Google Scholar]
- 17.Maveyraud, L., D. Golemi-Kotra, A. Ishiwata, O. Meroueh, S. Mobashery, and J. P. Samama. 2002. High-resolution X-ray structure of an acyl-enzyme species for the class D OXA-10 β-lactamase. J. Am. Chem. Soc. 124:2461-2465. [DOI] [PubMed] [Google Scholar]
- 18.McRee, D. E. 1999. XtalView/Xfit: a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156-165. [DOI] [PubMed] [Google Scholar]
- 19.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 20.Naas, T., and P. Nordmann. 1999. OXA-type β-lactamases. Curr. Pharm. Des. 5:865-879. [PubMed] [Google Scholar]
- 21.Paetzel, M., F. Danel, L. De Castro, S. C. Mosimann, M. G. Page, and N. C. Strynadka. 2000. Crystal structure of the class D β-lactamase OXA-10. Nat. Struct. Biol. 7:918-925. [DOI] [PubMed] [Google Scholar]
- 22.Pernot, L., F. Frenois, T. Rybkine, G. L'Hermite, S. Petrella, J. Delettre, V. Jarlier, E. Collatz, and W. Sougakoff. 2001. Crystal structures of the class D β-lactamase OXA-13 in the native form and in complex with meropenem. J. Mol. Biol. 310:859-874. [DOI] [PubMed] [Google Scholar]
- 23.Perrakis, A., R. Morris, and V. S. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458-463. [DOI] [PubMed] [Google Scholar]
- 24.Poirel, L., and P. Nordmann. 2002. Acquired carbapenem-hydrolyzing β-lactamases and their genetic support. Curr. Pharm. Biotechnol. 3:117-127. [DOI] [PubMed] [Google Scholar]
- 25.Potterton, E., S. McNicholas, E. Krissinel, K. Cowtan, and M. Noble. 2002. The CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 58:1955-1957. [DOI] [PubMed] [Google Scholar]
- 26.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossmann, M. J., and D. M. Blow. 1962. The detection of sub-units within the crystallographic asymmetric unit. Acta Crystallogr. 15:24-31. [Google Scholar]
- 28.Santillana, E., A. Beceiro, G. Bou, and A. Romero. 2007. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc. Natl. Acad. Sci. U. S. A. 104:5354-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun, T., M. Nukaga, K. Mayama, E. H. Braswell, and J. R. Knox. 2003. Comparison of β-lactamases of classes A and D: 1.5-Å crystallographic structure of the class D OXA-1 oxacillinase. Protein Sci. 12:82-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vagin, A., and A. Teplyakov. 1998. A translation-function approach for heavy-atom location in macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 54:400-402. [DOI] [PubMed] [Google Scholar]
- 31.Vagin, A., and A. Teplyakov. 2000. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 56:1622-1624. [DOI] [PubMed] [Google Scholar]
- 32.Vagin, A., and A. Teplyakov. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022-1025. [Google Scholar]