Abstract
Quinolone resistance is an emerging problem in Streptococcus pyogenes, and recombination with Streptococcus dysgalactiae DNA has been implicated as a frequent mechanism leading to resistance. We have characterized a collection of S. dysgalactiae subsp. equisimilis isolates responsible for infections in humans (n = 314) and found a high proportion of levofloxacin-resistant isolates (12%). Resistance was associated with multiple emm types and genetic lineages, as determined by pulsed-field gel electrophoretic profiling. Since we could not find evidence for a role of efflux pumps in resistance, we sequenced the quinolone resistance-determining regions of the gyrA and parC genes of representative resistant and susceptible isolates. We found much greater diversity among the parC genes (19 alleles) than among the gyrA genes (5 alleles). While single mutations in either GyrA or ParC were sufficient to raise the MIC so that the strains were classified as intermediately resistant, higher-level resistance was associated with mutations in both GyrA and ParC. Evidence for recombination with S. pyogenes DNA was found in some parC alleles, but this was not exclusively associated with resistance. Our data support the existence of a common reservoir of genes conferring quinolone resistance shared between S. dysgalactiae subsp. equisimilis and S. pyogenes, while no recombination with the animal pathogen S. dysgalactiae subsp. dysgalactiae could be found.
Streptococcus dysgalactiae subsp. equisimilis is a beta-hemolytic, large-colony-forming species (diameter, >0.5 mm) presenting either the Lancefield group C or the Lancefield group G antigen (15). Among the members of this group, this species is the one the most commonly reported to be a cause of human infections worldwide (27, 33, 41, 45) and was the only one previously detected among Lancefield group C streptococci (GCS) and Lancefield group G streptococci (GGS) causing infections in humans in Portugal (37). Although S. dysgalactiae subsp. equisimilis may be part of the normal human flora (8), it has also been identified to be the causative agent of infections of the respiratory tract, infections of the skin and soft tissue, as well as life-threatening infections, such as endocarditis and bacteremia (37, 41, 45). More recently, an increasing number of reports have described the association of these organisms with streptococcal syndromes typically caused by Streptococcus pyogenes (a Lancefield group A streptococcus [GAS]), such as streptococcal toxic shock syndrome (22) and acute rheumatic fever (21). Several studies point to a close relationship between S. dysgalactiae subsp. equisimilis and GAS, with multiple common virulence (40) and antibiotic resistance (17) determinants being found in both species.
Fluoroquinolones (FQs) are nowadays valid therapeutic options for the treatment of streptococcal infections (24). However, the emergence of resistance to this antimicrobial class is a global concern and resistance is increasingly being reported among several streptococcal species (13, 26, 30), including S. dysgalactiae subsp. equisimilis (5). FQs act by inhibiting the bacterial DNA gyrase and DNA topoisomerase IV (24). Alteration of the target enzymes is the more widespread mechanism of resistance in streptococci and the only one that confers high-level resistance to FQs (24), but active efflux was also shown to confer low-level resistance in Streptococcus pneumoniae (4, 9) and in other viridans group streptococci (20), but this mechanism was not detected in GAS (30).
Both DNA gyrase and topoisomerase IV are tetramers of two different subunits, subunits GyrA and GyrB and subunits ParC and ParE, respectively (24). Previous studies have shown that FQ resistance arises from amino acid changes in the structurally similar proteins GyrA and ParC, while alterations in GyrB and ParE seem to be less relevant (4, 24). Mutations in the DNA sequences of gyrA and parC that confer resistance to FQs are found in the so-called quinolone resistance-determining regions (QRDRs) of these genes (34). Mutations in the gyrA and parC QRDRs are thought to occur spontaneously in bacterial populations, and this is considered the main mechanism of the emergence of FQ resistance (2, 24).
Evidence for the horizontal transfer of the QRDR between streptococci has been found, and this mechanism may also play a role in the rise in the incidence of FQ resistance (3, 14, 16, 38). One study suggested that S. dysgalactiae could act as the donor species in the emergence of FQ-resistant GAS (38), and more recently, a considerable proportion of the GAS isolates from Belgium presented characteristics suggestive of the acquisition of their QRDR of parC from Streptococcus dysgalactiae subsp. equisimilis (14). However, such studies have been hampered by the small number of S. dysgalactiae parC and gyrA sequences available, which limits the ability to assess the contribution of FQ resistance in this species to a putative QRDR gene pool common among beta-hemolytic streptococci.
We characterized the molecular mechanisms of levofloxacin resistance in isolates of S. dysgalactiae subsp. equisimilis responsible for infections in humans in Portugal. The clonal structure of the isolates was evaluated by using two well-established typing schemes: emm typing and SmaI macrorestriction profiling by pulsed-field gel electrophoresis (PFGE) (37). This approach allowed the identification of putative susceptible ancestors of resistant isolates and evaluation of the diversity of the genetic backgrounds of FQ-resistant isolates. We found that FQ resistance emerged by alteration of the target genes in diverse genetic lineages of S. dysgalactiae subsp. equisimilis, as previously shown for other streptococci (2), suggesting that mutation and selection are the primary drivers of resistance. The analysis of the QRDRs of a large number of genetically unrelated isolates allowed us to identify evidence of the frequent ongoing horizontal transfer of the QRDR of parC between S. dysgalactiae subsp. equisimilis and GAS, indicating the existence of a gene pool common between these streptococcal species.
MATERIALS AND METHODS
Bacterial isolates.
A collection of 314 beta-hemolytic, large-colony-forming (diameter, >0.5 mm) isolates of GGS (n = 249) and GCS (n = 65) responsible for infections in humans were collected from June 1998 to December 2005. The distribution of isolates over the study period was as follows: 11 in 1998, 28 in 1999, 20 in 2000, 28 in 2001, 29 in 2002, 70 in 2003, 50 in 2004, and 78 in 2005. The sources of the isolates were the following: blood (n = 34), other normally sterile sites (n = 11), pharyngeal exudate (n = 67), sputum (n = 37), skin and soft tissue (n = 155), vaginal exudate (n = 5), and urine (n = 5). The strains were recovered in 13 laboratories, located throughout Portugal, that were asked to submit all nonduplicate GCS and GGS isolates associated with human infections. Partial descriptions of some of these isolates were published previously (37).
Species identification.
Strains were identified to the genus level by the submitting laboratory. Upon receipt, the Lancefield group was confirmed by a commercial latex agglutination technique (Slidex Strepto kit; bioMérieux, Marcy l'Etoile, France). All further studies were carried out at the Instituto de Microbiologia, Faculdade de Medicina, Universidade de Lisboa. Beta-hemolysis and colony size were confirmed after overnight incubation at 37°C in tryptic soy agar (Oxoid, Hampshire, England) supplemented with 5% (vol/vol) defibrinated sheep blood. Further identification to the species level was done for all non-levofloxacin-susceptible isolates (levofloxacin MICs > 2 μg/ml [11]) and selected susceptible isolates used for comparative analysis by using the API 20 Strep system (bioMérieux). We had previously confirmed the efficacy of this system for the identification of S. dysgalactiae subsp. equisimilis strains by using 16S rRNA gene sequencing as the “gold standard” (37).
Molecular typing.
Isolates were characterized by emm typing and SmaI PFGE macrorestriction profiling, as described previously (37). The PFGE patterns were compared by using Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium) to create dendrograms by the unweighted-pair group method with arithmetic averages (UPGMA). The Dice similarity coefficient was used with optimization and position tolerance settings of 1.0 and 1.5, respectively. Clusters were defined as groups of isolates (n ≥ 2) presenting profiles with ≥80% similarity in the dendrogram (37).
Simpson's index of diversity (SID) and 95% confidence intervals (CIs) were calculated as described elsewhere (10).
Antimicrobial susceptibility testing and efflux screening.
Testing for susceptibility to levofloxacin was done for all 314 isolates by disk diffusion, according to the guidelines of the CLSI (11). Testing of the levofloxacin MIC for nonsusceptible isolates and selected susceptible isolates was performed with Etest strips (AB Biodisk, Solna, Sweden) and by use of the interpretative criteria of the CLSI (11). Nonsusceptible isolates were also tested for active efflux by the agar dilution method on Mueller-Hinton agar (Oxoid, Hampshire, England) supplemented with 5% sheep blood in the presence of levofloxacin or ciprofloxacin with or without 10 μg/ml of reserpine (Sigma-Aldrich, Steinheim, Germany), as described previously (9).
Analysis of DNA sequence conferring levofloxacin resistance.
The segments of the gyrA and parC genes containing the QRDRs were amplified and sequenced by using the primers previously proposed for use for this purpose (26). The nucleotide sequences and the deduced amino acid sequences of the gyrA and parC QRDRs were determined for 56 S. dysgalactiae subsp. equisimilis isolates which exhibited different levels of resistance to levofloxacin and which represented the main PFGE clusters and emm types identified in this work.
Phylogenetic analysis of gyrA and parC.
Sequence alignments were performed manually. MEGA (version 4) software (42) was used to construct phylogenetic trees by using the neighbor-joining algorithm and the Kimura two-parameter substitution model. RDP3 software (31) was used to test for recombination by the following four methods: an exact nonparametric method (7), sister scanning (18), the maximum χ2 method (32), and a likelihood-assisted recombination detection method (23).
Nucleotide sequence accession numbers.
The sequences of the QRDRs of the isolates studied here were submitted to GenBank under accession numbers GU002022 to GU002045.
RESULTS
Characterization of the non-levofloxacin-susceptible population.
Initial screening by disk diffusion identified 47 isolates that were not susceptible to levofloxacin (14.9%), and the MICs for these isolates were determined by Etest. The MICs determined were not in full agreement with the results of the disk diffusion test. Discrepancies were observed for one isolate that was resistant by disk diffusion and that had an MIC of 4 μg/ml (intermediate by use of the CLSI criteria) and for an additional set of five isolates that were intermediate by the disk diffusion test and that had MICs of 2 μg/ml (susceptible by use of the CLSI criteria). The results of MIC testing were considered definitive for analysis in this work.
A total of 38 isolates (12.1%) were resistant to levofloxacin (MIC50, >32 μg/ml; MIC range, 6 to >32 μg/ml) and 4 isolates (1.3%) expressed intermediate resistance to this antibiotic (MIC range, 3 to 4 μg/ml), resulting in a proportion of nonsusceptible isolates (MICs > 2 μg/ml) of 13.3%. Among the 30 susceptible isolates chosen for sequence analysis, the MIC range was 0.19 to 2 μg/ml.
Levofloxacin-resistant isolates were recovered from skin and soft tissue infections (n = 33), blood (n = 4), and urine (n = 1), while none was found among the 106 isolates recovered from the respiratory tract (P < 0.0001, Fisher's exact test). The recovery of small numbers of isolates in the first years of the study precluded the calculation of annual resistance rates. However, if we consider the data for the time period between 2003 and 2005, when more than 50 isolates were recovered each year, the rates of levofloxacin resistance were 11.4% in 2003 and 8.0% in 2004, and the rate reached its highest level (20.2%) in the last year of the study.
No difference in levofloxacin or ciprofloxacin MICs was observed for the nonsusceptible isolates in the presence of reserpine, excluding a role for active efflux in resistance in Streptococcus dysgalactiae subsp. equisimilis.
Analysis of the PFGE patterns generated after digestion with SmaI of the 314 isolates revealed 12 clusters and 10 unique profiles (SID ± 95% CI, 0.817 ± 0.021). More than 80% of the isolates were distributed in four large clusters: clusters H88, J77, C56, and E35 (Table 1). Levofloxacin-resistant isolates were present in nine of these PFGE clusters, while two isolates had unique profiles (SID ± 95% CI, 0.818 ± 0.101). PFGE cluster I12 was the only cluster with more than 10 isolates that did not include any levofloxacin-resistant isolates. Of the 31 distinct emm types that were found in the overall population (SID ± 95% CI, 0.924 ± 0.009), 10 were represented by levofloxacin-resistant isolates (SID ± 95% CI, 0.844 ± 0.071) (Table 1). stG166b (n = 13) and stG6792 (n = 8) accounted for more than half of the nonsusceptible isolates, followed by stG6 (n = 6) and stGLP3 (n = 4). stG166b was distributed into two main clusters: one comprised only levofloxacin-resistant isolates (grouped in cluster C56 [n = 11]) and the other mostly comprised susceptible ones (grouped in cluster J77 [n = 10], which included an isolate with intermediate susceptibility to levofloxacin) (Table 1). stGLP3 was the only emm type exclusively found in nonsusceptible isolates and, together with the stG166b resistant isolates, constituted the most significant presence of levofloxacin-resistant isolates in a single PFGE cluster (PFGE cluster C56; Table 1). The other resistant isolates were found to be dispersed in the population and were not concentrated in a specific PFGE cluster, as illustrated by stG6792 and stG6.
TABLE 1.
emm types and distribution of non-levofloxacin-susceptible isolates among PFGE clusters
| emm type | No. of nonsusceptible isolates/no. of susceptible isolates in PFGE clustera: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C56 | H88 | J77 | B11 | F4 | E35 | K8 | L3 | D2 | Otherb | Total | |
| stG166b | 11/0 | 0/1 | 1d/9 | 1/0 | 13/10 | ||||||
| stG6792 | 0/2 | 3/28 | 3/0 | 0/1 | 1/0 | 1/1 | 8/32 | ||||
| stG6 | 1d/2 | 1/4 | 2d/20 | 1/1 | 1/2 | 0/4 | 6e/33 | ||||
| stGLP3 | 4/0 | 4/0 | |||||||||
| stG485 | 0/1 | 0/2 | 1d/15 | 0/6 | 0/6 | 1/0 | 1/1 | 3/31 | |||
| stG2078 | 0/3 | 0/3 | 2/6 | 0/1 | 2/13 | ||||||
| stG245 | 2/3 | 0/1 | 0/1 | 0/2 | 2/7 | ||||||
| stG480 | 0/2 | 0/2 | 1/0 | 1/0 | 0/15 | 0/1 | 0/4 | 2/24 | |||
| stG643 | 0/1 | 0/3 | 1/0 | 0/5 | 1/9 | ||||||
| stC6979 | 0/1 | 1/0 | 0/3 | 1/4 | |||||||
| Otherc | 0/30 | 0/46 | 0/17 | 0/8 | 0/8 | 0/109 | |||||
| Total | 16/40 | 4/84 | 7/70 | 4/7 | 3/1 | 2/33 | 2/6 | 1/2 | 1/1 | 2/28 | 42/272 |
PFGE clusters were arbitrarily designated by the use of capital letters and a subscript number, which indicates the number of isolates included in the cluster.
Other included 3 PFGE clusters that did not contain resistant isolates and 10 unique PFGE profiles (i.e., the sharing of less than 80% similarity in the SmaI PFGE profile with that of any other isolate in the collection). This is the case for the two resistant isolates listed in this column.
Other included 21 emm types that were not found in resistant isolates. Among these, only stC839 (n = 30), stG10 (n = 27), and stG62647 (n = 11) comprised more than 10 isolates.
One isolate presenting intermediate resistance is included.
Two isolates presenting intermediate resistance are included.
Phylogenetic analysis of gyrA and parC.
The gyrA and parC QRDR sequences were determined for 56 S. dysgalactiae subsp. equisimilis isolates, including 22 isolates that were resistant to levofloxacin (representatives of 10 emm types and 11 PFGE clusters), the 4 intermediate isolates, and 30 susceptible isolates, including isolates that had the same emm types and that grouped together with resistant isolates in the same PFGE clusters.
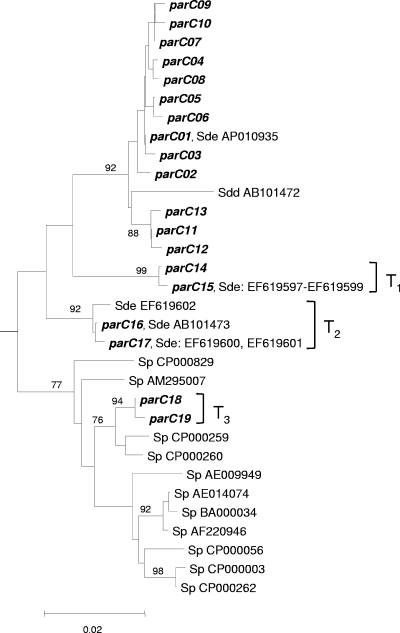
The gyrA sequences of the S. dysgalactiae subsp. equisimilis isolates exhibited minimal variation. Only 4 nucleotide changes were observed across the 399 bp sequenced, resulting in five distinct alleles. As these changes occurred at three specific codons and all resulted in amino acid replacements, they are all represented in Table 2. In contrast, multiple point mutations (mostly silent mutations) could be observed in the parC sequences of both susceptible and resistant isolates, resulting in 19 distinct alleles, which are indicated in Table 3. Surprisingly, the parC sequences of the majority of the isolates analyzed (33/56, parC01 to parC13 alleles) were more similar to the published sequence of S. dysgalactiae subsp. dysgalactiae (GenBank accession number AB101472) than to the majority of the S. dysgalactiae subsp. equisimilis parC sequences available to date in the GenBank database (GenBank accession numbers AB101473 and EF619597 to EF619601) (Fig. 1), with the exception of the sequence retrieved from the recently released genomic project (GenBank accession number AP010935).
TABLE 2.
Amino acid substitutions in ParC and GyrA of S. dysgalactiae subsp. equisimilis isolates with different levels of resistance to levofloxacina
| Allelesb | Amino acid coding change at the indicated position |
MIC range (μg/ml) | No. of isolates | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GyrA |
ParC |
|||||||||
| 81 | 85 | 142 | 79 | 83 | 91 | 140 | 166 | |||
| gyrA04, parCc | S81F | S79F | >32 | 5 | ||||||
| gyrA04, parC15 | S81F | S79F | N91D | >32 | 5 | |||||
| gyrA04, parC17 | S81F | S79F | N91D | P140S | >32 | 4 | ||||
| gyrA02, parC17 | S81Y | S79F | N91D | P140S | >32 | 1 | ||||
| gyrA03, parC08 | E85K | S79F | >32 | 1 | ||||||
| gyrA03, parC06 | E85K | S79Y | >32 | 1 | ||||||
| gyrA04, parC09 | S81F | D83N | 8, 24, >32 | 3 | ||||||
| gyrA04, parC13 | S81F | S79A | 12 | 1 | ||||||
| gyrA03, parC10 | E85K | D83G | 8 | 1 | ||||||
| gyrA04, parC14 | S81F | N91D | 4 | 1 | ||||||
| gyrA01, parC04 | S79F | 3 | 1 | |||||||
| gyrA01, parC17 | S79F | N91D | P140S | 1.5-3 | 3 | |||||
| gyrA01, parC19 | S79A | N91D | P140S | 2 | 1 | |||||
| gyrA01, parC12 | G166E | 1 | 1 | |||||||
| gyrA01, parC03 | S79P | 0.75 | 1 | |||||||
| gyrA01, parCd | N91D | P140S | 0.19-0.5 | 5 | ||||||
| gyrA01, parC14 | N91D | 0.5-0.75 | 3 | |||||||
| gyrA05, parC05 | D142N | 0.38-0.5 | 4 | |||||||
| gyrA01, parCe | 0.38-0.75 | 14 | ||||||||
A total of 56 isolates with different levels of resistance to levofloxacin were tested.
The allele numbers were attributed on the basis of the DNA sequence of the QRDR of each gene. When more than one allele that resulted in the same amino acid sequence was found, no number is indicated and the alleles found and the number of isolates presenting them are indicated in the subsequent footnotes.
For parC04, n = 3; for parC08, n = 2.
For parC16, n = 4; for parC18, n = 1.
For parC01, n = 5; for parC02, n = 3; for parC07, n = 5; for parC11, n = 1.
TABLE 3.
parC alleles of S. dysgalactiae subsp. equisimilis isolates
| Allele | No. of isolatesa | Base at nucleotide position/amino acid no.b: |
||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 51/17 | 54/18 | 60/20 | 69/23 | 78/26 | 100/34 | 108/36 | 111/37 | 117/39 | 141/47 | 156/52 | 207/69 | 225/75 | 235/79 | 236/79 | 237/79 | 247/83 | 248/83 | 271/91 | 279/93 | 330/110 | 339/113 | 405/135 | 417/139 | 418/140 | 420/140 | 426/142 | 432/144 | 456/152 | 459/153 | 497/166 | 513/171 | 519/173 | 540/180 | 552/184 | ||
| parC01 | 5 | T | A | C | C | A | A | G | A | T | A | T | T | A | T | C | C | G | A | A | A | T | T | A | T | C | C | T | C | G | G | G | C | T | A | T |
| parC02 | 3 | G | T | |||||||||||||||||||||||||||||||||
| parC03 | 1 | C* | ||||||||||||||||||||||||||||||||||
| parC04 | 4 | T* | ||||||||||||||||||||||||||||||||||
| parC05 | 4 | A | ||||||||||||||||||||||||||||||||||
| parC06 | 1 | A* | A | |||||||||||||||||||||||||||||||||
| parC07 | 5 | A | ||||||||||||||||||||||||||||||||||
| parC08 | 3 | T* | A | |||||||||||||||||||||||||||||||||
| parC09 | 3 | A* | A | |||||||||||||||||||||||||||||||||
| parC10 | 1 | G* | A | |||||||||||||||||||||||||||||||||
| parC11 | 1 | C | G | C | C | |||||||||||||||||||||||||||||||
| parC12 | 1 | C | G | C | C | A* | ||||||||||||||||||||||||||||||
| parC13 | 1 | C | G | C | C | G* | ||||||||||||||||||||||||||||||
| parC14 | 4 | C | C | T | T | G | C | A | G | C | G | C | G | G* | G | C | G | |||||||||||||||||||
| parC15 | 5 | C | C | T | T | G | C | A | G | C | G | C | G | T* | G* | G | C | G | ||||||||||||||||||
| parC16 | 4 | G* | G | C | C | G | C | T* | T* | G | C | C | G | A | ||||||||||||||||||||||
| parC17 | 8 | T* | G* | G | C | C | G | C | T* | T* | G | C | C | G | A | |||||||||||||||||||||
| parC18 | 1 | C | C | T | T | G | C | A | G | C | G | C | G | G* | G | C | G | C | T* | T* | G | T | C | C | C | G | A | |||||||||
| parC19 | 1 | C | C | T | T | G | C | A | G | C | G | C | G | G* | G* | G | C | G | C | T* | T* | G | T | C | C | C | G | A | ||||||||
A total of 56 isolates with different levels of resistance to levofloxacin were tested.
Base substitutions that result in amino acid coding changes are indicated by asterisks. Nucleotide and amino acid positions are numbered according to the S. pneumoniae numbering system (36).
FIG. 1.
Neighbor-joining tree for the parC QRDR. The Streptococcus agalactiae type strain NCTC 8181 parC sequence (GenBank accession number AB101464) was used to root the tree. If the percentage of replicate trees in which the associated sequences clustered together in the bootstrap test (1,000 replicates) was greater than 75%, the values are shown next to the branches. The tree is drawn to scale, with the branch lengths being in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed by the Kimura two-parameter method and are in the units of the number of base substitutions per site. Sp, Streptococcus pyogenes; Sde, Streptococcus dysgalactiae subsp. equisimilis; Sdd, Streptococcus dysgalactiae subsp. dysgalactiae. Sets of numbers and letters identify the GenBank entries whose sequences are identical to the sequences of the alleles found in our collection or the sequences of S. pyogenes used to construct the tree.
The parC18 and parC19 alleles were most similar to the parC sequences of S. pyogenes strains MGAS9429 (GenBank accession number CP000259) and MGAS10270 (GenBank accession number CP000260). These alleles were found in two Lancefield group C isolates of the same emm type (emm type stG62647) and with identical PFGE profiles (PFGE cluster J77) and differed only in the Ser-79-Ala (TCC-to-GCC) change in parC. The gyrA sequences of both isolates were indistinguishable and represented the most frequent gyrA allele (gyrA01) found in the S. dysgalactiae subsp. equisimilis isolates in our collection, in agreement with the species identification.
Phylogenetic trees were built by the neighbor-joining method (Fig. 1 and 2). The tree for parC shows a single branch that gave rise to the majority of S. dysgalactiae subsp. equisimilis parC alleles (parC01 to parC13), which were tightly clustered together with a high bootstrap support, but also included the more divergent sequence of S. dysgalactiae subsp. dysgalactiae (Fig. 1). Another branch groups the more diverse S. pyogenes alleles, although with a lower level of bootstrap support. As expected, the parC18 and parC19 alleles (group T3 in Fig. 1) of S. dysgalactiae subsp. equisimilis were found within the S. pyogenes branch. Two additional branches supported by high bootstrap levels were identified (transformant group 1 [T1] and T2 in Fig. 1, including parC14 to parC17), but they were not clearly grouped with either the S. dysgalactiae subsp. equisimilis or the S. pyogenes branches. An analysis of the alignment with RDP3 software (31) identified two recombination events that could explain the existence of these branches. A possible recombination event (T1) between the parC03 and parC18 alleles identified by an exact nonparametric method (P = 0.0346) and also supported by the sister-scanning method (P = 0.0024) could give rise to both the parC14 and the parC15 alleles, with the latter allele corresponding to multiple sequences deposited in the GenBank database (GenBank accession numbers EF619597 to EF619599). The parC14 and parC15 alleles differed at a single nucleotide, with one being more prevalent among resistant isolates (parC15, S79F amino acid substitution) and the other being more prevalent among susceptible isolates (Table 3). Another putative recombination event (T2) was detected by the maximum χ2 method (P = 0.0299) and was confirmed by the likelihood-assisted recombination detection method (P = 0.0400) between an S. pyogenes allele (GenBank accession number CP000260) and parC09. This could explain the origins of both parC16 and parC17. Such a recombination event could also explain the parC sequences of a number of S. dysgalactiae subsp. equisimilis strains currently deposited in the GenBank database (GenBank accession numbers EF619600 to EF619602 and AB101473). Similar to the previously presented putative recombination event, the parC16 allele was not found among resistant isolates, while the parC17 allele was found to be associated with resistance (S79F amino acid substitution; see below).
FIG. 2.
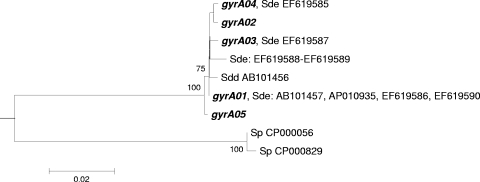
Neighbor-joining tree for the gyrA QRDR. The Streptococcus agalactiae type strain NCTC 8181 gyrA sequence (GenBank accession number AB101448) was used to root the tree. For details on the methods used to construct the tree, see the legend to Fig. 1. Sp, Streptococcus pyogenes; Sde, Streptococcus dysgalactiae subsp. equisimilis; Sdd, Streptococcus dysgalactiae subsp. dysgalactiae. Sets of numbers and letters identify the GenBank entries whose sequences are identical to the sequences of the alleles found in our collection or the sequences of S. pyogenes used to construct the tree.
In the phylogenetic tree for gyrA, all S. dysgalactiae subsp. equisimilis alleles clustered together with high bootstrap support (Fig. 2). The only gyrA sequence of S. dysgalactiae subsp. dysgalactiae available is included in this branch and represents more than 99% sequence identity to the S. dysgalactiae subsp. equisimilis alleles. In contrast to the case for parC, the S. pyogenes gyrA alleles are more distantly related to those of S. dysgalactiae subsp. equisimilis.
Contribution of mutations to levofloxacin resistance.
Not all the amino acid substitutions observed in the regions of gyrA and parC sequenced (4 and 9 amino acid changes, respectively) were exclusively detected in resistant isolates, as summarized in Table 2.
All the resistant isolates had amino acid changes within the QRDRs of both gyrA and parC compared to the consensus sequence of the susceptible isolates. The Ser-81 change to Phe (TCT to TTT) in GyrA and the Ser-79 change to Phe (TCC to TTC) in ParC were the most common combinations observed in resistant isolates (n = 14). These could also be accompanied by other changes in ParC and invariably resulted in an MIC of >32 μg/ml. These two amino acid changes were also the most widespread in nonsusceptible isolates, and both appeared in combination with other amino acid changes (Table 2). Ser-81-Tyr and Glu-85-Lys in GyrA and Ser-79-Tyr, Ser-79-Ala, Asp-83-Asn, and Asp-83-Gly in ParC were other amino acid substitutions found among resistant isolates and thought to contribute to resistance. The level of levofloxacin resistance achieved was dependent on the combination of the GyrA and ParC amino acid changes present in each isolate (Table 2). A single set of three isolates that shared the Ser-81-Phe and Asp-83-Asn substitutions in GyrA and ParC, respectively, displayed significantly variable MIC results (range, 8 to >32 μg/ml).
In contrast to the double mutations described above, a single amino acid change in ParC resulted in intermediate resistance to levofloxacin or in slightly increased MICs in susceptible strains (0.5 to 3 μg/ml). An intermediately resistant isolate with an MIC of 4 μg/ml was also the only one to have a mutation in the GyrA QRDR but no mutations in ParC. Common to both susceptible and resistant isolates were the Asn-91-Asp (AAT to GAT) and Pro-140-Ser (CCC to TCT) substitutions in ParC, present in 23 and 14 isolates, respectively.
DISCUSSION
A surprising finding in our work was the high overall rate of resistance to levofloxacin (12%). The only previous study that specifically addressed this problem (5) found that the rate of resistance to levofloxacin was less than 1% among GGS and GCS in a collection of isolates recovered in both Europe and North America. Portugal is the European country where FQs are more widely used (19), and their sustained use has been implicated in selection for resistance (24). Differences in consumption could thus be a major factor conditioning geographic differences in FQ resistance.
Several mechanisms have been implicated in bacterial resistance to quinolones. The existence of plasmids carrying specific genes that confer increased resistance to this class of antimicrobials was recently described in the Enterobacteriaceae family (39). However, this resistance mechanism has thus far not been found in Gram-positive cocci, in which the increased expression of multidrug resistance pumps and alterations of the fluoroquinolone targets are the major mechanisms of quinolone resistance (24, 30). Similar to what was described in other streptococci, in vitro studies with S. pyogenes, a closely related species, have implicated alterations in the QRDRs of gyrA and parC as the primary mechanism of the loss of susceptibility to fluoroquinolones (6). Studies with Streptococcus pneumoniae showed that the transformation of a susceptible strain with the QRDRs of these genes from a resistant strain was sufficient to confer resistance (43). Taken together, these findings indicate that alterations of the QRDRs of gyrA and parC are necessary and sufficient for the development of quinolone resistance in streptococci. We found no evidence of efflux-mediated resistance to levofloxacin in S. dysgalactiae subsp. equisimilis, similar to what was described in GAS (30, 38), indicating that this resistance mechanism is not widespread in beta-hemolytic streptococci and leaving the alterations in the quinolone targets as the sole and most probable mechanism of resistance.
The levofloxacin-resistant isolates presented a variety of emm types, comparable to that observed for the entire collection, and they were distributed in several PFGE clusters, indicating a polyclonal origin of resistance. It is known from epidemiological studies of GAS that some resistant lineages predominate, although mutational alterations can also be observed in diverse genetic backgrounds (1, 29). For instance, the ciprofloxacin-resistant lineage of S. pyogenes carrying the emm6 allele is disseminated in distinct geographic regions, such as Spain (1), Belgium (29, 30), and the United States (35). In our collection, we could also identify a PFGE cluster (PFGE cluster C56) that was significantly associated with resistance to levofloxacin (P = 0.0009, Fisher's exact test), and only two of the emm types found in this cluster (emm types stG166b and stGLP3) presented resistant isolates. This suggests that stable levofloxacin-resistant lineages also exist in S. dysgalactiae subsp. equisimilis. However, the latter did not dominate among the levofloxacin-resistant S. dysgalactiae subsp. equisimilis isolates recovered in Portugal. When we tested if the high rate of resistance found in the last year of the study could have been caused by an outbreak of a single clone, we verified that this was not so, since the isolates recovered in 2005 were a heterogeneous population (seven emm types and nine PFGE clusters).
Among the streptococci, horizontal gene transfer involving the QRDRs of the FQ target genes was first described for S. pneumoniae and members of the viridans group streptococci (3, 16). More recently, the parC QRDRs of GAS were also shown to present sequence polymorphisms (35), and these observations have been the basis for the suggestion of the occurrence of horizontal gene transfer events in this species (14, 38), with S. dysgalactiae acting as the donor. S. dysgalactiae subsp. dysgalactiae was suggested to be the most likely source of DNA by Duesberg et al. (14). However, such conclusions have been hampered by the lack of S. dysgalactiae subsp. equisimilis parC sequences, as only one parC sequence of an S. dysgalactiae subsp. equisimilis isolate was publicly available and was found in the GenBank database (GenBank accession number AB101473) at the time of these studies. S. dysgalactiae subsp. dysgalactiae is mainly a pathogen of animals and, contrary to the case for S. dysgalactiae subsp. equisimilis, does not usually infect humans (8, 15). No other reservoir for S. pyogenes besides the human host is known, making it unlikely that situations favoring gene exchange with S. dysgalactiae subsp. dysgalactiae would arise. Thus, S. dysgalactiae subsp. equisimilis would seem to be a more suitable candidate that engages in genetic exchange with GAS, as they can share ecological niches in the human host. Previous studies have documented the existence of bacteriophages capable of the in vitro transduction of chromosomal markers from GAS to GCS (12), as well as from GCS to GAS (44), providing a mechanism that could mediate genetic exchange between these two species. In agreement with the findings of previous studies, our results support the acquisition of S. pyogenes DNA by S. dysgalactiae subsp. equisimilis (25). The S. dysgalactiae subsp. equisimilis parC18 and parC19 alleles cluster within the S. pyogenes branch, but the gyrA alleles of the isolates in which the parC alleles were found were characteristic of those of S. dysgalactiae subsp. equisimilis. Taken together, these data strongly suggest that substitution of the entire QRDR of parC occurred in these isolates, through recombination with S. pyogenes DNA. The parC14 to parC17 alleles resulted from recombination within the QRDR with S. pyogenes alleles, with DNA stretches characteristic of both species being retained (Fig. 3). Interestingly, the QRDR of the parC sequence first available in the GenBank database for S. dysgalactiae subsp. equisimilis seems to have also resulted from recombination, a fact that may have confounded previous analyses that led to the suggestion that S. dysgalactiae subsp. dysgalactiae could have been the donor species for recombination with GAS. Our data support the ecologically more plausible hypothesis that gene exchange between S. dysgalactiae subsp. equisimilis and GAS is more frequent than that between S. dysgalactiae subsp. dysgalactiae and GAS. Horizontal transfer within S. dysgalactiae subsp. equisimilis may also be ongoing, since the same QRDR polymorphisms were found among distinct lineages, i.e., isolates belonging to different PFGE clusters and with distinct emm types (data not shown). No recombination events between S. dysgalactiae subsp. equisimilis and GAS were detected in the gyrA QRDR. A greater divergence between these species at this locus could explain this observation, since sequence divergence is a major barrier to gene exchange (28).
FIG. 3.
Diagram of the QRDRs of gyrA and parC in isolates showing recombination events. The box on the left represents the gyrA QRDR, while that on the right represents the parC QRDR. White boxes represent regions that are characteristic of S. dysgalactiae subsp. equisimilis, while black boxes represent regions characteristic of S. pyogenes. Each allele combination represents the sequences resulting from the putative recombination events indicated in Fig. 1 (T1 to T3). Recombination breakpoints were determined by the maximum χ2 method implemented in RDP3 software, refined by visual inspection of the sequences (Table 3).
All recombination events detected included both susceptible and resistant recombinants, indicating that there was not a strict relationship between recombination with foreign DNA and resistance and raising the possibility that the mutations conferring levofloxacin resistance occurred subsequent to DNA acquisition. In fact, the quinolone resistance-defining mutations have probably arisen independently in multiple strains sharing the same mosaic parC genes. If this scenario proves to be true, the expansion of levofloxacin resistance among S. dysgalactiae subsp. equisimilis isolates occurs mostly through vertical transmission and not through horizontal dissemination. Still, although our study was not designed to evaluate the frequency of recombination, we did identify a high proportion of recombinant QRDRs (n = 23/56 [41%] isolates), suggesting that recombination is frequent and ongoing.
Among our collection we could identify several pairs of levofloxacin-resistant and -susceptible isolates that shared the same emm type or SmaI profile, allowing us to investigate more closely the origin of levofloxacin resistance in these isolates. Only when mutations were concomitantly present in gyrA and parC were the isolates levofloxacin resistant. Changes to phenylalanine in Ser-81 in GyrA and Ser-79 in ParC were the most important in the acquisition of levofloxacin resistance in terms of both the frequency of occurrence and the level of resistance achieved, while mutations of only one of these amino acids predominated in isolates with reduced susceptibility. Our data support the hypothesis that the target for first-step mutations in S. dysgalactiae subsp. equisimilis is mainly ParC, as previously observed in other streptococci (34). A few groups of isolates with the same QRDR alleles showed some MIC variations, but most values were within a 1-dilution interval.
Isolates presenting high-level resistance to fluoroquinolones were described among different species of beta-hemolytic streptococci, but only one previous work described the mutations present in S. dysgalactiae subsp. equisimilis leading to fluoroquinolone resistance (5). The amino acid substitutions found to be present in the FQ-resistant isolates among our isolates recovered in Portugal resulted from changes in the same relative positions in GyrA (Ser-81, Glu-85) and ParC (Ser-79, Asp-83), as was the case with the changes described in other beta-hemolytic streptococcal species (5, 26). The Ser-79-Ala, Ser-79-Tyr, Asp-83-Gly, and Asp-83-Asn modifications, all in ParC, were not previously described in S. dysgalactiae subsp. equisimilis.
Quinolone resistance in S. dysgalactiae subsp. equisimilis was mediated by alterations in ParC and GyrA and not by efflux, similar to the situation in GAS. Sequence polymorphisms not related to FQ resistance are common in the parC QRDR of S. dysgalactiae subsp equisimilis; in contrast, the QRDR sequence of gyrA is highly conserved. Our data question the previously proposed role of S. dysgalactiae subsp. dysgalactiae as a donor for the mosaic structures found in GAS (14) and support the more plausible explanation that the exchanges occur with S. dysgalactiae subsp. equisimilis isolates that share ecological niches with S. pyogenes. A close proximity between strains of the two species would afford bacteriophages the opportunity to transduce the genetic information between them. The high proportion of recombinant sequences identified supports the occurrence of frequent genetic exchanges between the two species that do not seem to be driven exclusively by antimicrobial usage.
Acknowledgments
This work was partly supported by the Fundação para a Ciência e a Tecnologia (grant PTDC/SAU-ESA/72321/2006); the Fundação Calouste Gulbenkian, Portugal; and an unrestricted grant from GlaxoSmithKline, Portugal.
Members of the Portuguese Group for the Study of Streptococcal Infections are as follows: Luís Lito, and Lurdes Monteiro, Hospital de Santa Maria, Lisbon; Filomena Martins, Maria Ana Pessanha, Elsa Gonçalves, and Teresa Morais, Hospital de São Francisco Xavier, Lisbon; José Diogo, Ana Rodrigues, and Isabel Nascimento, Hospital Garcia de Orta, Almada; Manuela Ribeiro, Fernanda Cotta, and Dolores Pinheiro, Hospital de São João, Porto; Paulo Lopes, Ismália Calheiros, Luísa Felício, and Angelina Lameirão, Centro Hospitalar de Vila Nova de Gaia, Vila Nova de Gaia; Valquíria Alves, Antónia Read, and Margarida Monteiro, Hospital Pedro Hispano, Matosinhos; Ana Paula Castro, Hospital de Vila Real, Vila Real; Ana Paula M. Vieira, Hospital Senhora da Oliveira, Guimarães; Elmano Ramalheira, António Bastos Marques, and Fernanda Bessa, Hospital Infante Dom Pedro, Aveiro; Rui Tomé, Graça Ribeiro, Celeste Pontes, and Fátima Martins, Hospitais da Universidade de Coimbra, Coimbra; Helena Ramos, Ana Paula Castro, Susana Ferreira, and Paulo Pinto, Hospital de Santo António, Porto; Maria Alberta Faustino and Adelaide Alves, Hospital de São Marcos, Braga; and M. F. Menezes, Instituto Bacteriológico Câmara Pestana, Lisbon.
Footnotes
Published ahead of print on 9 February 2010.
REFERENCES
- 1.Albertí, S., G. Cortes, C. Garcia-Rey, C. Rubio, F. Baquero, J. A. Garcia-Rodriguez, E. Bouza, and L. Aguilar. 2005. Streptococcus pyogenes pharyngeal isolates with reduced susceptibility to ciprofloxacin in Spain: mechanisms of resistance and clonal diversity. Antimicrob. Agents Chemother. 49:418-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alou, L., M. Ramirez, C. Garcia-Rey, J. Prieto, and H. de Lencastre. 2001. Streptococcus pneumoniae isolates with reduced susceptibility to ciprofloxacin in Spain: clonal diversity and appearance of ciprofloxacin-resistant epidemic clones. Antimicrob. Agents Chemother. 45:2955-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balsalobre, L., M. J. Ferrandiz, J. Linares, F. Tubau, and A. G. de la Campa. 2003. Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob. Agents Chemother. 47:2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bast, D. J., D. E. Low, C. L. Duncan, L. Kilburn, L. A. Mandell, R. J. Davidson, and J. C. de Azavedo. 2000. Fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae: contributions of type II topoisomerase mutations and efflux to levels of resistance. Antimicrob. Agents Chemother. 44:3049-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biedenbach, D. J., M. A. Toleman, T. R. Walsh, and R. N. Jones. 2006. Characterization of fluoroquinolone-resistant beta-hemolytic Streptococcus spp. isolated in North America and Europe including the first report of fluoroquinolone-resistant Streptococcus dysgalactiae subspecies equisimilis: report from the SENTRY Antimicrobial Surveillance Program (1997-2004). Diagn. Microbiol. Infect. Dis. 55:119-127. [DOI] [PubMed] [Google Scholar]
- 6.Billal, D. S., D. P. Fedorko, S. S. Yan, M. Hotomi, K. Fujihara, N. Nelson, and N. Yamanaka. 2007. In vitro induction and selection of fluoroquinolone-resistant mutants of Streptococcus pyogenes strains with multiple emm types. J. Antimicrob. Chemother. 59:28-34. [DOI] [PubMed] [Google Scholar]
- 7.Boni, M. F., D. Posada, and M. W. Feldman. 2007. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandt, C. M., and B. Spellerberg. 2009. Human infections due to Streptococcus dysgalactiae subspecies equisimilis. Clin. Infect. Dis. 49:766-772. [DOI] [PubMed] [Google Scholar]
- 9.Brenwald, N. P., M. J. Gill, and R. Wise. 1998. Prevalence of a putative efflux mechanism among fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2032-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carriço, J. A., C. Silva-Costa, J. Melo-Cristino, F. R. Pinto, H. de Lencastre, J. S. Almeida, and M. Ramirez. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44:2524-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing, 16th informational supplement. CLSI document M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 12.Colón, A. E., R. M. Cole, and C. G. Leonard. 1972. Intergroup lysis and transduction by streptococcal bacteriophages. J. Virol. 9:551-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Campa, A. G., C. Ardanuy, L. Balsalobre, E. Perez-Trallero, J. M. Marimon, A. Fenoll, and J. Liñares. 2009. Changes in fluoroquinolone-resistant Streptococcus pneumoniae after 7-valent conjugate vaccination, Spain. Emerg. Infect. Dis. 15:905-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duesberg, C. B., S. Malhotra-Kumar, H. Goossens, L. McGee, K. P. Klugman, T. Welte, and M. W. Pletz. 2008. Interspecies recombination occurs frequently in quinolone resistance-determining regions of clinical isolates of Streptococcus pyogenes. Antimicrob. Agents Chemother. 52:4191-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Facklam, R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15:613-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrandiz, M. J., A. Fenoll, J. Linares, and A. G. De La Campa. 2000. Horizontal transfer of parC and gyrA in fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figueiredo, T. A., S. I. Aguiar, J. Melo-Cristino, and M. Ramirez. 2006. DNA methylase activity as a marker for the presence of a family of phage-like elements conferring efflux-mediated macrolide resistance in streptococci. Antimicrob. Agents Chemother. 50:3689-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibbs, M. J., J. S. Armstrong, and A. J. Gibbs. 2000. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573-582. [DOI] [PubMed] [Google Scholar]
- 19.Goossens, H., M. Ferech, S. Coenen, and P. Stephens. 2007. Comparison of outpatient systemic antibacterial use in 2004 in the United States and 27 European countries. Clin. Infect. Dis. 44:1091-1095. [DOI] [PubMed] [Google Scholar]
- 20.Guerin, F., E. Varon, A. B. Hoi, L. Gutmann, and I. Podglajen. 2000. Fluoroquinolone resistance associated with target mutations and active efflux in oropharyngeal colonizing isolates of viridans group streptococci. Antimicrob. Agents Chemother. 44:2197-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haidan, A., S. R. Talay, M. Rohde, K. S. Sriprakash, B. J. Currie, and G. S. Chhatwal. 2000. Pharyngeal carriage of group C and group G streptococci and acute rheumatic fever in an Aboriginal population. Lancet 356:1167-1169. [DOI] [PubMed] [Google Scholar]
- 22.Hashikawa, S., Y. Iinuma, M. Furushita, T. Ohkura, T. Nada, K. Torii, T. Hasegawa, and M. Ohta. 2004. Characterization of group C and G streptococcal strains that cause streptococcal toxic shock syndrome. J. Clin. Microbiol. 42:186-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes, E. C., M. Worobey, and A. Rambaut. 1999. Phylogenetic evidence for recombination in dengue virus. Mol. Biol. Evol. 16:405-409. [DOI] [PubMed] [Google Scholar]
- 24.Hooper, D. C. 2002. Fluoroquinolone resistance among Gram-positive cocci. Lancet Infect. Dis. 2:530-538. [DOI] [PubMed] [Google Scholar]
- 25.Kalia, A., and D. E. Bessen. 2003. Presence of streptococcal pyrogenic exotoxin A and C genes in human isolates of group G streptococci. FEMS Microbiol. Lett. 219:291-295. [DOI] [PubMed] [Google Scholar]
- 26.Kawamura, Y., H. Fujiwara, N. Mishima, Y. Tanaka, A. Tanimoto, S. Ikawa, Y. Itoh, and T. Ezaki. 2003. First Streptococcus agalactiae isolates highly resistant to quinolones, with point mutations in gyrA and parC. Antimicrob. Agents Chemother. 47:3605-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopardo, H. A., P. Vidal, M. Sparo, P. Jeric, D. Centron, R. R. Facklam, H. Paganini, N. G. Pagniez, M. Lovgren, and B. Beall. 2005. Six-month multicenter study on invasive infections due to Streptococcus pyogenes and Streptococcus dysgalactiae subsp. equisimilis in Argentina. J. Clin. Microbiol. 43:802-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majewski, J. 2001. Sexual isolation in bacteria. FEMS Microbiol. Lett. 199:161-169. [DOI] [PubMed] [Google Scholar]
- 29.Malhotra-Kumar, S., C. Lammens, S. Chapelle, C. Mallentjer, J. Weyler, and H. Goossens. 2005. Clonal spread of fluoroquinolone non-susceptible Streptococcus pyogenes. J. Antimicrob. Chemother. 55:320-325. [DOI] [PubMed] [Google Scholar]
- 30.Malhotra-Kumar, S., L. Van Heirstraeten, C. Lammens, S. Chapelle, and H. Goossens. 2009. Emergence of high-level fluoroquinolone resistance in emm6 Streptococcus pyogenes and in vitro resistance selection with ciprofloxacin, levofloxacin and moxifloxacin. J. Antimicrob. Chemother. 63:886-894. [DOI] [PubMed] [Google Scholar]
- 31.Martin, D. P. 2009. Recombination detection and analysis using RDP3. Methods Mol. Biol. 537:185-205. [DOI] [PubMed] [Google Scholar]
- 32.Maynard Smith, J. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126-129. [DOI] [PubMed] [Google Scholar]
- 33.McDonald, M., R. J. Towers, R. M. Andrews, J. R. Carapetis, and B. J. Currie. 2007. Epidemiology of Streptococcus dysgalactiae subsp. equisimilis in tropical communities, Northern Australia. Emerg. Infect. Dis. 13:1694-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muñoz, R., and A. G. De La Campa. 1996. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 40:2252-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orscheln, R. C., D. R. Johnson, S. M. Olson, R. M. Presti, J. M. Martin, E. L. Kaplan, and G. A. Storch. 2005. Intrinsic reduced susceptibility of serotype 6 Streptococcus pyogenes to fluoroquinolone antibiotics. J. Infect. Dis. 191:1272-1279. [DOI] [PubMed] [Google Scholar]
- 36.Pan, X. S., J. Ambler, S. Mehtar, and L. M. Fisher. 1996. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2321-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinho, M. D., J. Melo-Cristino, and M. Ramirez. 2006. Clonal relationships between invasive and noninvasive Lancefield group C and G streptococci and emm-specific differences in invasiveness. J. Clin. Microbiol. 44:841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pletz, M. W., L. McGee, C. A. Van Beneden, S. Petit, M. Bardsley, M. Barlow, and K. P. Klugman. 2006. Fluoroquinolone resistance in invasive Streptococcus pyogenes isolates due to spontaneous mutation and horizontal gene transfer. Antimicrob. Agents Chemother. 50:943-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6:629-640. [DOI] [PubMed] [Google Scholar]
- 40.Sachse, S., P. Seidel, D. Gerlach, E. Gunther, J. Rodel, E. Straube, and K. H. Schmidt. 2002. Superantigen-like gene(s) in human pathogenic Streptococcus dysgalactiae, subsp. equisimilis: genomic localisation of the gene encoding streptococcal pyrogenic exotoxin G (speGdys). FEMS Immunol. Med. Microbiol. 34:159-167. [DOI] [PubMed] [Google Scholar]
- 41.Siljander, T., M. Karppelin, S. Vahakuopus, J. Syrjanen, M. Toropainen, J. Kere, R. Vuento, T. Jussila, and J. Vuopio-Varkila. 2008. Acute bacterial, nonnecrotizing cellulitis in Finland: microbiological findings. Clin. Infect. Dis. 46:855-861. [DOI] [PubMed] [Google Scholar]
- 42.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 43.Varon, E., C. Janoir, M. D. Kitzis, and L. Gutmann. 1999. ParC and GyrA may be interchangeable initial targets of some fluoroquinolones in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wannamaker, L. W., S. Almquist, and S. Skjold. 1973. Intergroup phage reactions and transduction between group C and group A streptococci. J. Exp. Med. 137:1338-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woo, P. C., A. M. Fung, S. K. Lau, S. S. Wong, and K. Y. Yuen. 2001. Group G beta-hemolytic streptococcal bacteremia characterized by 16S ribosomal RNA gene sequencing. J. Clin. Microbiol. 39:3147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]