Abstract
Expression of ampC, oprD, mexA, mexC, mexE, and mexX was studied in 25 Pseudomonas aeruginosa isolates from cystic fibrosis patients, including 14 isolates of the Liverpool epidemic strain. Overexpressed mexA or ampC and reduced oprD were associated with β-lactam resistance. A specific combination of mexR, nalC, and nalD mutations occurred in 11 Liverpool strain isolates, including 7 with upregulated mexA.
Once established in the respiratory tree of the cystic fibrosis (CF) patient, Pseudomonas aeruginosa survives aggressive chemotherapy, which may select for resistance-conferring mutations (28). Several mutations compromise β-lactams, including those that (i) upregulate AmpC β-lactamase (13, 24), (ii) inactivate or downregulate the carbapenem-specific OprD porin (13, 14, 22), or (iii) upregulate efflux by MexAB-OprM (8, 11, 22) and other resistance-nodulation-cell division (RND) pumps (1, 19-21, 30). Most of these pumps also excrete quinolones and one, MexXY-OprM, acts against aminoglycosides. While one can usually infer the resistance mechanisms of non-CF P. aeruginosa isolates by interpretive reading of antibiograms (17), this fails for CF isolates, which often have pan-resistance or antibiogram profiles discordant with known mechanisms.
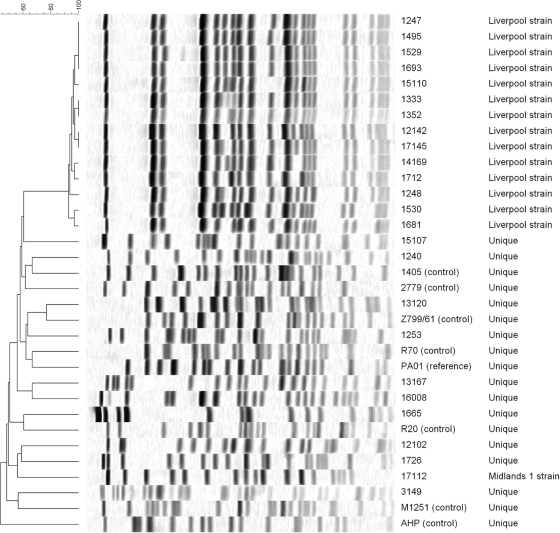
We investigated the combinations of resistance mechanisms in 25 CF patient P. aeruginosa isolates, including 14 representatives of the Liverpool epidemic strain (2) (Fig. 1), which is disseminated among United Kingdom CF patients. Selection was based on high or low meropenem MICs among isolates referred to the Health Protection Agency's Antibiotic Resistance Monitoring and Reference Laboratory (ARMRL) during 2007 and 2008. Susceptibility was determined using the British Society for Antimicrobial Chemotherapy agar dilution methodology or Etest and categorized using (i) European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoints, with a biological breakpoint of 64 μg/ml for cefotaxime to distinguish normal insensitivity (MICs of 8 to 32 μg/ml) from substantial resistance or (ii) a biological cutoff MIC >4× the mode for wild-type P. aeruginosa (http://www.srga.org/eucastwt/MICTAB/). We used P. aeruginosa PAO1 as a control along with seven strains with varied expression levels of resistance determinants: strains M1251 and R70 had upregulated efflux (15, 16); R20 (29) had normal efflux activity; AHP and Z799/61 (31) had reduced efflux; strains 2779-con and 1405-con D2− overexpressed AmpC β-lactamase (13), and the latter also lacked OprD porin.
FIG. 1.
Pulsed-field gel electrophoresis banding patterns of SpeI-digested genomic DNA from control strains and clinical isolates of P. aeruginosa.
Amplification of β-lactamase genes used primers for blaTEM (T-1, 5′-ATG AGT ATT CAA CAT TTC CG-3′; T-2, 5′-CCA ATG CTT AAT CAG TGA CG-3′), blaSHV (S-1, 5′-TCA GCG AAA AAC ACC TTG-3′; S-2, 5′-TCC CGC AGA TAA ATC ACC A-3′), or those described previously for IMP, VIM, SPM-1, GIM-1, and SIM-1 metallo-β-lactamase genes (5). Carbapenemase phenotypes were sought by (i) agar dilution imipenem-EDTA (320 μg/ml) synergy tests and (ii) a modified Hodge test (3, 10) with imipenem and meropenem disks (10 μg) on Mueller-Hinton (MH) agar inoculated with Escherichia coli ATCC 25922.
Real-time reverse transcription-PCR (RT-PCR), with the primers detailed in Table 1, was used to quantify transcription of genes encoding efflux pump proteins MexA, MexC, MexE, and MexX, outer membrane protein OprD, and AmpC β-lactamase. Total RNA was isolated from late-log-phase cultures with RNeasy kits (Qiagen Inc., Crawley, United Kingdom) and treated with DNase (1 U/μl). RT-PCR was performed in triplicate using different RNA extractions (50 ng RNA per reaction mixture), LightCycler FastStrand DNA Master SYBR green I (Roche, Mannheim, Germany), and HotStarTaq Plus DNA polymerase (Qiagen). Gene expression was normalized versus that of rpoD in the same strain and then calibrated relative to P. aeruginosa PAO1 (27), which was assigned a value of 1.0. Increases or decreases in gene expression of ≥2-fold and ≤0.5-fold were taken as significant, and Student's t test was used to compare the occurrence of these changes between susceptible and resistant groups. Isolates with intermediate susceptibility were regarded as susceptible. Variables associated significantly (P ≤ 0.05) with resistance in this univariate analysis were used as covariates in multivariate logistic regression (SPSS v.16.0; Analytical Software, St. Paul, MN).
TABLE 1.
PCR primers used for real-time RT-PCR and to sequence regulatory genes
| Analysis method and gene | Primer | DNA sequence | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| RT-PCR | ||||
| rpoD | For | 5′-CGCAACAGCAATCTCGTCTGAAA-3′ | 130 | This study |
| Rev | 5′-GCGGATGATGTCTTCCACCTGTT-3′ | This study | ||
| ampC | For | 5′-GGTGCAGAAGGACCAGGCACAGAT-3′ | 97 | This study |
| Rev | 5′-CGATGCTCGGGTTGGAATAGAGGC-3′ | This study | ||
| oprD | For | 5′-CGGCGACATCAGCAACACC-3′ | 194 | This study |
| Rev | 5′-GGGCCGTTGAAGTCGGAGTA-3′ | This study | ||
| mexA | For | 5′-GGCGACAACGCGGCGAAGG-3′ | 203 | This study |
| Rev | 5′-CCTTCTGCTTGACGCCTTCCTGC-3′ | This study | ||
| mexC | For | 5′-GCAATAGGAAGGATCGGGGCGTTGG-3′ | 102 | This study |
| Rev | 5′-CCTCCACCGGCAACACCATTTCG-3′ | This study | ||
| mexE | For | 5′-TCATCCCACTTCTCCTGGCGCTACC-3′ | 150 | This study |
| Rev | 5′-CGTCCCACTCGTTCAGCGGTTGTTCGATG-3′ | This study | ||
| mexX | For | 5′-AATCGAGGGACACCCATGCACATCC-3′ | 82 | This study |
| Rev | 5′-CCCAGCAGGAATAGGGCGACCAG-3′ | This study | ||
| Regulatory gene sequencing | ||||
| mexR | For | 5′-TGTTCTTAAATATCCTCAAGCGG-3′ | 729 | 2 |
| Rev | 5′-GTTGCATAGCGTTGTCCTCA-3′ | 2 | ||
| Int1 | 5′-GCGCAACCCCAGCGACCA-3′ | This study | ||
| nalC | For | 5′-TCAACCCTAACGAGAAACGCT-3′ | 813 | 2 |
| Rev | 5′-TCCACCTCACCGAACTGC-3′ | 2 | ||
| Int 1 | 5′-CCGGCGATCGGCAAGTCC-3′ | This study | ||
| Int 2 | 5′-GATCCCCGCTGCCAGAGCC-3′ | This study | ||
| nalD | For | 5′-GCGGCTAAAATCGGTACACT-3′ | 788 | 2 |
| Rev | 5′-ACGTCCAGGTGGATCTTGG-3′ | 2 |
The mexR, nalC, and nalD genes were amplified using the primers listed in Table 1 and previously described conditions (23, 24, 26); sequences were determined with a CEQ 8000 XL genetic analysis system (Beckman Coulter, High Wycombe, United Kingdom) and compared with those of P. aeruginosa PAO1 (www.ncbi.nlm.gov/BLAST/).
Antibiotic susceptibilities and gene expression data are shown in Table 2. Control strains M1251 and R70, with upregulated efflux (11), had corresponding phenotypes, although R70 was additionally resistant to imipenem. The present analysis found overexpression of mexA (7.28-fold and 5.94-fold, respectively) and mexX (2.70- and 7.94-fold) in these strains. In addition, R70 had reduced OprD expression, correlating with imipenem resistance; also, a 5- to 6-fold increase in AmpC expression was observed, which was far less than the >300-fold increase in the two derepressed controls, 2779-con and 1405-con D2− (13). Strain 1405-con D2−, which showed imipenem resistance (MIC, 16 μg/ml) and, inconsistent with previous results (11), increased carbenicillin resistance (512 μg/ml), also had reduced oprD and increased mexA expression levels, explaining deviations from a straightforward AmpC phenotype. Strains AHP and Z799/61, with highly susceptible phenotypes and weak or absent efflux in direct assays (11, 12), showed weak transcription of all mex genes studied and of ampC; Z799/61 also had weak oprD expression but, since the organism lacks AmpC expression, this should not confer imipenem resistance (13), thus agreeing with the organism's phenotype. R20 had similar expression of most genes as PAO1, which it resembles in MIC profile, although with increased oprD expression.
TABLE 2.
Susceptibility and mRNA expression among control strains and clinical isolates of P. aeruginosa
| Isolate | MIC (μg/ml)a |
Relative gene expressionb |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPM | MEM | PTZ | CAZ | CTX | ATM | CIP | CAR | GEN | TOB | AMK | oprD* | ampC | mexA | mexC | mexE | mexX | |
| Controls | |||||||||||||||||
| M1251 | 4 | 8 | 32 | 4 | 128 | 32 | 0.5 | 512 | 0.5 | 0.25 | 1 | 0.29 | 1.78 | 7.28 | 1.24 | 2.50 | 2.70 |
| R70 | 16 | >32 | 32 | 8 | 256 | 64 | 0.5 | >512 | 1 | 0.5 | 1 | 0.05 | 5.72 | 5.94 | 1.72 | 1.32 | 7.94 |
| R20 | 2 | 0.5 | 8 | 1 | 4 | 4 | ≤0.12 | 64 | ≤0.25 | 0.5 | ≤0.5 | 3.27 | 0.73 | 0.75 | 0.77 | 0.30 | 1.79 |
| AHP | 0.12 | 0.25 | 4 | 0.5 | 4 | 0.5 | ≤0.12 | 32 | ≤0.12 | ≤0.12 | ≤0.5 | 1.23 | 0.004 | 0.12 | 0.02 | 0.01 | 0.02 |
| Z799/61 | 0.5 | ≤0.06 | ≤1 | ≤0.12 | 0.5 | ≤0.12 | ≤0.12 | ≤16 | ≤0.12 | 0.25 | 1 | 0.15 | 0.12 | 0.12 | 0.24 | 0.01 | 0.08 |
| 2779-con | 2 | 1 | >64 | 32 | >256 | 32 | ≤0.12 | 128 | 0.5 | 1 | 1 | 5.34 | 313 | 1.56 | 0.99 | 4.90 | 3.71 |
| 1405-con D2− | 16 | 8 | >64 | 64 | >256 | 64 | 0.25 | 512 | 2 | 2 | 4 | 0.35 | 599 | 6.92 | 2.62 | 2.26 | 1.22 |
| Clinical isolates | |||||||||||||||||
| 1530c | 8 | 0.5 | ≤1 | 4 | 32 | 2 | 8 | 64 | 32 | 4 | 32 | 0.06 | 1.13 | 0.65 | 0.69 | 1.23 | 11.6 |
| 1693c | 8 | 2 | ≤1 | 8 | 128 | 1 | 1 | 128 | 16 | 2 | 1 | 0.08 | 0.42 | 2.60 | 0.38 | 0.24 | 0.29 |
| 17145c | 32 | 4 | 2 | 64 | 128 | 0.5 | 4 | ≤16 | 32 | 16 | 64 | 0.0004 | 8.40 | 0.39 | 0.11 | 0.33 | 2.55 |
| 1529c | 16 | 4 | 2 | 4 | 32 | 0.5 | 8 | 8 | >32 | 16 | 64 | 0.12 | 3.29 | 0.56 | 0.18 | 1.18 | 12.4 |
| 14169c | 16 | 4 | 8 | 8 | 128 | 64 | 1 | 512 | 32 | 4 | 32 | 0.07 | 35.9 | 2.52 | 1.05 | 0.42 | 4.39 |
| 1247c | 16 | 4 | 4 | 8 | 128 | 4 | 4 | 128 | >32 | 8 | >64 | 0.018 | 36.4 | 1.81 | 0.75 | 0.44 | 2.96 |
| 1681c | 32 | 8 | >64 | 256 | >256 | 32 | 2 | 512 | 32 | 16 | 64 | 0.006 | 11.5 | 4.90 | 0.31 | 0.34 | 5.85 |
| 1352c | >32 | 16 | 64 | 64 | >256 | 32 | 2 | 512 | >32 | 16 | >64 | 0.004 | 10.8 | 1.82 | 0.49 | 0.89 | 6.41 |
| 1333c | 32 | 16 | >64 | 128 | >256 | >64 | 2 | >512 | >32 | 8 | 64 | 0.0007 | 368 | 2.66 | 1.77 | 9.01 | 33.9 |
| 1248c | 16 | 16 | 32 | 256 | >256 | >64 | 4 | >512 | >32 | 32 | >64 | 0.012 | 5.89 | 4.11 | 0.70 | 1.31 | 11.7 |
| 15110c | >32 | 16 | >64 | 256 | >256 | >64 | 2 | >512 | >32 | >32 | >64 | 0.05 | 474 | 5.61 | 1.00 | 0.7 | 2.37 |
| 12142c | 32 | 32 | >64 | 256 | >256 | >64 | 4 | >512 | >32 | >32 | >64 | 0.0004 | 262 | 15.9 | 3.1 | 3.7 | 6.52 |
| 1712c | 32 | 32 | >64 | >256 | >256 | >64 | 4 | >512 | >32 | 16 | >64 | 0.015 | 160 | 1.83 | 1.2 | 2.98 | 18.9 |
| 1495c | >32 | >32 | >64 | >256 | >256 | >64 | 4 | >512 | >64 | 8 | >64 | 0.06 | 284 | 3.49 | 2.92 | 6.91 | 78.3 |
| 1726 | 4 | 0.12 | ≤1 | 2 | 16 | 0.5 | 2 | ≤16 | 2 | 0.5 | 2 | 0.25 | 1.50 | 1.08 | 0.21 | 0.10 | 1.80 |
| 15107 | 8 | 2 | 32 | 8 | 256 | 2 | 0.25 | 128 | >32 | 2 | >64 | 0.03 | 1.36 | 1.87 | 1.06 | 0.26 | 10.5 |
| 16008 | 64 | 4 | ≤1 | 1 | 64 | 0.5 | 4 | ≤16 | 16 | 4 | 32 | 0.003 | 0.09 | 0.56 | 0.09 | 0.03 | 27.1 |
| 13167 | 4 | 4 | 4 | 64 | >256 | 16 | 8 | >512 | 32 | 4 | 64 | 1.53 | 7.22 | 3.51 | 0.76 | 0.4 | 1.17 |
| 1240 | 16 | 8 | 8 | 256 | 256 | 4 | 2 | 128 | 2 | 0.5 | 4 | 0.08 | 110 | 1.30 | 1 | 1.82 | 2.10 |
| 3149 | 32 | 8 | 16 | 8 | 64 | 64 | >8 | >512 | >32 | >32 | >64 | 0.007 | 10.5 | 4.87 | 1.35 | 0.37 | 0.3 |
| 12102 | 32 | 16 | 16 | 4 | 64 | 16 | 2 | 256 | 4 | 1 | 4 | 0.0006 | 0.79 | 2.46 | 1.51 | 1.02 | 11.8 |
| 1253 | >32 | 32 | 64 | 8 | 128 | 16 | 2 | 256 | 16 | 4 | 32 | 0.008 | 4.17 | 1.96 | 5.2 | 7.8 | 13.3 |
| 13120 | 16 | 32 | 16 | 4 | 128 | 16 | 0.5 | 256 | 0.5 | 0.25 | 1 | 0.9 | 2.04 | 2.30 | 0.87 | 0.4 | 15.0 |
| 1665 | 64 | >32 | >64 | 256 | >256 | >64 | 4 | >512 | 16 | 32 | >64 | 0.0001 | 150 | 5.89 | 1.65 | 1.14 | 0.51 |
| 17112 | 128 | >32 | 64 | 64 | >256 | >64 | 2 | 512 | 1 | 0.5 | 1 | 0.03 | 44.1 | 3.12 | 0.17 | 0.08 | 0.11 |
IPM, imipenem; MEM, meropenem; PTZ, piperacillin-tazobactam; CAZ, ceftazidime; CTX, cefotaxime; ATM, aztreonam; CIP, ciprofloxacin; CAR, carbenicillin; GEN, gentamicin; TOB, tobramycin; AMK, amikacin.
Increased or decreased gene expression relative to P. aeruginosa PAO1 (which was assigned a value of 1.0) was defined by expression values of ≥2 and ≤0.5, respectively, and is indicated in bold.
Isolate belonging to the Liverpool epidemic strain.
Twelve CF isolates gave a weak positive Hodge test but with imipenem only, and two (12102 and 15110) were weakly positive in the EDTA synergy test; PCR did not detect genes encoding known metallo-β-lactamases or TEM or SHV enzymes. Although other β-lactamases, such as OXA-50 (6, 9), may have contributed to these phenotypes, we noted that 9/12 Hodge test-positive CF isolates overexpressed ampC. Similarly, controls R70, 2779-con, and 1405-con D2−, also with increased ampC expression, gave weak positive Hodge tests. None of the control strains was positive in the EDTA synergy test.
The CF isolates variously had altered expression of one to six genes implicated in resistance (Table 2) and, based on univariate analysis (data not shown), their antibiograms broadly agreed with the known spectra of these mechanisms. Thus, upregulation of mexAB-oprM was associated with resistance to β-lactams except imipenem, whereas upregulation of ampC was associated with resistance to cephalosporins and piperacillin-tazobactam but not to carbenicillin or imipenem. Among β-lactams, only carbapenem resistance was associated with reduced expression of oprD. However, there were anomalies, including (i) an association of OprD deficiency with aminoglycoside resistance and (ii) no association between upregulation of efflux components and ciprofloxacin resistance.
Multivariate analysis (results not shown) identified increased mexA expression as the main mechanism of resistance to aztreonam (P = 0.015), carbenicillin (P = 0.022), and piperacillin-tazobactam (P = 0.023), and also to meropenem but only when resistance was defined biologically as a MIC >4 times the mode for the species (i.e., MIC of >2 μg/ml; P = 0.027), not at the clinical breakpoint (P = 0.196). Imipenem was not included in the multivariate analysis, because downregulation of oprD was the sole factor associated with resistance by univariate analysis. Surprisingly, no associations with upregulated ampC remained significant in the multivariate analysis, and no mechanisms remained associated with aminoglycoside resistance.
Mutations in at least three genes (mexR, nalC, and nalD) reportedly lead to increased expression of MexAB-OprM (4, 7, 18, 24, 26). We found numerous mutations in these genes in both clinical isolates and controls (Table 3), but no individual lesion was overrepresented among isolates with upregulated mexA; rather, 17 different combinations of mexR, nalC, and nalD mutations were seen. MexR Arg83Cys, NalC Gly71Glu, and NalD Asp187His were found in 11 of 14 representatives of the Liverpool strain, but these varied widely in resistance, including an up to 40-fold change in mexA expression. Arg83Cys in MexR is known to have an impact on efflux activity (25), whereas Gly71Glu in NalC is considered insignificant (18); to the best of our knowledge, Asp187His in NalD has not been reported before. The significance of this combination for mexA expression merits further investigation.
TABLE 3.
Combinations of mutations in genes reported to affect regulation of mexA expression
| Mutation(s) detected |
No. of isolates with mutation |
|||
|---|---|---|---|---|
| mexR | nalC | nalD | Clinical isolates | Control strains |
| Arg83→Cys | Gly71→Glu | Asp187→His | 11a | —b |
| Arg83→Cys | Gly71→Glu | Asp187→His/Leu201→Pro | 2 | — |
| Arg83→Cys | Gly71→Glu | None | 1 | — |
| Arg83→Cys | Gly71→Glu/Ser209→Arg | Asp187→His/Leu201→Pro | 1 | — |
| Val126→Glu | Gly71→Glu | Asp187→His | 1 | — |
| Val126→Glu | Gly71→Glu | None | 1 | 1 |
| Val126→Glu | Gly71→Glu/Ser209→Arg | Ala145→Thr | — | 1 |
| Val126→Glu | Gly71→Glu/Ser209→Arg | None | — | 1 |
| Val126→Glu | Gly71→Glu/Ala186→Thr | Asp187→His | 1 | — |
| Val126→Glu | Gly71→Glu/Ser209→Arg/Ala78→Thr | None | — | 1 |
| None | Gly71→Glu | Asp187→His/Leu201→Pro | 1 | — |
| None | Gly71→Glu | None | 3 | — |
| None | Gly71→Glu/Ser209→Arg | Thr188→Ala | 1 | — |
| None | Gly71→Glu/Ser209→Arg | None | — | 1 |
| None | Gly71→Glu/Ala186→Thr | None | — | 1 |
| None | Gly71→Glu/Ser209→Arg/Ala145→Val | None | 1 | — |
| None | None | None | 1 | 1 |
All isolates were representatives of the Liverpool epidemic strain.
—, no isolate.
To conclude, we used RT-PCR to investigate complex combinations of resistance mechanisms in CF patient isolates of P. aeruginosa, many of them multiresistant. Despite many predicted statistical agreements between phenotype and gene up- or downregulation, the approach had limitations for analyzing resistance at the level of individual strains, with numerous anomalies, and showed some spurious statistical associations. Aside from distortions arising through multiple coresident mechanisms in the same strain, there were the following issues: (i) assay reproducibility, although use of HotStarTaq Plus DNA polymerase and assessment of expression using three separate RNA extractions resulted in values for all strains that clustered tightly around the means; (ii) RT-PCR provided only a snapshot of gene expression, which may vary through the growth cycle; (iii) P. aeruginosa has up to 13 efflux pump systems, and only the 4 best-characterized were studied here; (iv) the function of RND efflux pumps may be modulated not only by expression of the pump protein, as examined, but also by that of other components and by the architecture and energetics of the membrane within which it functions. Failure to associate resistance with an expected codeterminant, such as (i) between efflux-component overexpression and ciprofloxacin and (ii) only a weak association between overexpression of mexX and aminoglycoside resistance, suggests that other uninvestigated factors—mutations in DNA gyrase or topoisomerase IV and transmembrane aminoglycoside uptake or aminoglycoside-modifying enzymes, respectively—may be relatively more important than efflux as codeterminants of resistance. A more definitive evaluation would require isogenic strains, each differing by defined resistance mechanisms, and would include isolates with specific genes knocked out.
Acknowledgments
M. Tomás was financially supported by Program Río Hortega (CH Universitario A Coruña and ISCIII) and received a scholarship from the Spanish Society of Infections Diseases and Clinical Microbiology to stay in London. This work was partially funded by the Spanish Network for Research in Infectious Diseases (REIPI RD06/0008).
Footnotes
Published ahead of print on 1 March 2010.
REFERENCES
- 1.Aires, J. R., T. Kohler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Aloul, M., J. Crawley, C. Winstanley, C. A. Hart, M. J. Ledson, and M. J. Walshaw. 2004. Increased morbidity associated with chronic infection by an epidemic Pseudomonas aeruginosa strain in CF patients. Thorax 59:334-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, K. F., D. R. Lonsway, J. K. Rasheed, J. Biddle, B. Jensen, L. K. McDougal, R. B. Carey, A. Thompson, S. Stocker, B. Limbago, and J. B. Patel. 2007. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J. Clin. Microbiol. 45:2723-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, L., R. Srikumar, and K. Poole. 2004. MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 53:1423-1436. [DOI] [PubMed] [Google Scholar]
- 5.Ellington, M. J., J. Kistler, D. M. Livermore, and N. Woodford. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J. Antimicrob. Chemother. 59:321-322. [DOI] [PubMed] [Google Scholar]
- 6.Girlich, D., T. Naas, and P. Nordmann. 2004. Biochemical characterization of the naturally occurring oxacilinase OXA-50 of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48:2043-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hocquet, D., X. Bertrand, T. Kohler, D. Talon, and P. Plesiat. 2003. Genetic and phenotypic variations of a resistant Pseudomonas aeruginosa epidemic clone. Antimicrob. Agents Chemother. 47:1887-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Köhler, T., M. Michea-Hamzehpour, S. F. Epp, and J. C. Pechere. 1999. Carbapenem activities against Pseudomonas aeruginosa: respective contributions of OprD and efflux systems. Antimicrob. Agents Chemother. 43:424-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong, K. F., S. R. Jayawardena, A. Del Puerto, L. Wiehlmann, U. Laabs, B. Tummler, and K. Mathee. 2005. Characterization of poxB, a chromosomal-encoded Pseudomona aeruginosa oxacillinase. Gene 358:82-92. [DOI] [PubMed] [Google Scholar]
- 10.Lee, K., Y. Chong, H. B. Shin, Y. A. Kim, D. Yong, and J. H. Yum. 2001. Modified Hodge and EDTA-disk synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 7:88-91. [DOI] [PubMed] [Google Scholar]
- 11.Li, X. Z., D. Ma, D. M. Livermore, and H. Nikaido. 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to β-lactam resistance. Antimicrob. Agents Chemother. 38:1742-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, X. Z., D. M. Livermore, and H. Nikaido. 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob. Agents Chemother. 38:1732-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livermore, D. M. 1992. Interplay of impermeability and chromosomal β-lactamase activity in imipenem-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:2046-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 15.Livermore, D. M., and K. W. Davy. 1991. Invalidity for Pseudomonas aeruginosa of an accepted model of bacterial permeability to β-lactam antibiotics. Antimicrob. Agents Chemother. 35:916-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livermore, D. M., R. J. Williams, and J. D. Williams. 1981. In-vitro activity of MK0787 (N-formimidoyl thienamycin) against Pseudomonas aeruginosa and other gram-negative organisms and its stability to their β-lactamases. J. Antimicrob. Chemother. 8:355-362. [DOI] [PubMed] [Google Scholar]
- 17.Livermore, D. M., T. G. Winstanley, and K. P. Shannon. 2001. Interpretative reading: recognizing the unusual and inferring resistance mechanisms from resistance phenotypes. J. Antimicrob. Chemother. 48:87-102. [DOI] [PubMed] [Google Scholar]
- 18.Llanes, C., D. Hocquet, C. Vogne, D. Benali-Baitich, C. Neuwirth, and P. Plesiat. 2004. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 48:1797-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pai, H., J. Kim, J. H. Lee, K. W. Choe, and N. Gotoh. 2001. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 45:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pirnay, J. P., D. De Vos, D. Mossialos, A. Vanderkelen, P. Cornelis, and M. Zizi. 2002. Analysis of the Pseudomonas aeruginosa oprD gene from clinical and environmental isolates. Environ. Microbiol. 4:872-882. [DOI] [PubMed] [Google Scholar]
- 24.Quale, J., S. Bratu, J. Gupta, and D. Landman. 2006. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 50:1633-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito, K., H. Akama, E. Yoshihara, and T. Nakae. 2003. Mutations affecting DNA-binding activity of the MexR repressor of mexR-mexA-mexB-oprM operon expression. J. Bacteriol. 185:6195-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobel, M. L., D. Hocquet, L. Cao, P. Plesiat, and K. Poole. 2005. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:1782-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobel, M. L., G. A. McKay, and K. Poole. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, E. E., G. D. Buckley, Z. Wu, C. Saenphimmachak, R. L. Hoffman, D. A. D'Argenio, S. L. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul, and M. V. Olson. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams, R. J., M. A. Lindridge, A. A. Said, D. M. Livermore, and J. D. Williams. 1984. National survey of antibiotic resistance in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 14:9-16. [DOI] [PubMed] [Google Scholar]
- 30.Wolter, D. J., E. Smith-Moland, R. V. Goering, N. D. Hanson, and P. D. Lister. 2004. Multidrug resistance associated with mexXY expression in clinical isolates of Pseudomonas aeruginosa from a Texas hospital. Diagn. Microbiol. Infect. Dis. 50:43-50. [DOI] [PubMed] [Google Scholar]
- 31.Zimmermann, W. 1980. Penetration of β-lactam antibiotics into their target enzymes in Pseudomonas aeruginosa: comparison of a highly sensitive mutant with its parent strain. Antimicrob. Agents Chemother. 18:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]