Abstract
NlpE, an outer membrane lipoprotein, functions during envelope stress responses in Gram-negative bacteria. In this study, we report that overproduction of NlpE increases multidrug and copper resistance through activation of the genes encoding the AcrD and MdtABC multidrug efflux pumps in Escherichia coli.
Multidrug efflux pumps cause serious problems in cancer chemotherapy and treatment of bacterial infections. Bacterial drug resistance is often associated with multidrug efflux pumps that decrease drug accumulation in the cell (15). Bacterial multidrug efflux pumps are classified into five families based on sequence similarity: major facilitator, resistance-nodulation-cell division (RND), small multidrug resistance, multidrug and toxic compound extrusion, and the ATP-binding cassette (2, 24). Of these, the RND family efflux pumps play major roles in both intrinsic and elevated resistance of Gram-negative bacteria to a wide range of compounds including β-lactams (15, 18). RND efflux pumps require two other proteins to function: a membrane fusion protein and an outer membrane protein. Many drug efflux pumps in Escherichia coli need TolC to function (7, 18). Bacterial genome sequencing enables us to trace drug resistance genes (22). There are many putative and proven drug efflux pumps in E. coli, and we have previously identified 20 such functional drug efflux pumps (19). Because many such efflux pumps have overlapping substrate spectra (19), it is intriguing that bacteria, with their economically organized genomes, harbor such large sets of multidrug efflux genes.
The key to understanding how bacteria utilize these multiple efflux pumps lies in the regulation of pump expression. The currently available data show that multidrug efflux pumps are often expressed under precise and elaborate transcriptional control (8, 17). Expression of acrAB, which encodes the major AcrAB efflux pump, is subject to multiple levels of regulation. In E. coli, it is locally modulated by the repressor AcrR (13) and AcrS (11). At a more global level, it is modulated by stress conditions and global regulators such as MarA, SoxS, and Rob (14, 30). This example illustrates the complexity and diversity of the mechanisms regulating bacterial multidrug efflux pumps.
Stress responses in the bacterial cell envelope are transmitted to the cytoplasm in which gene expression is regulated in order to maintain the quality of the envelope. NlpE is anchored to the outer membrane through the lipid attached to its N-terminal cysteine and functions during envelope stress responses in Gram-negative bacteria. Adhesion to abiotic surfaces has been reported as an NlpE-dependent activation cue of the Cpx pathway (21). The Cpx envelope stress response is a two-component signal transduction pathway consisting of CpxA, an inner membrane histidine kinase, and CpxR, a cytoplasmic response regulator (5). Envelope stress responses play important roles in infection by many Gram-negative bacterial pathogens (25, 27). The expression level of NlpE is higher in a clinical, multidrug-resistant strain of the bacterium than that in a reference strain (28). In addition, overproduction of NlpE activates the Cpx pathway (29) and has been frequently utilized as a Cpx-specific activation signal in studies of the Cpx pathway (3, 6). However, the precise roles of NlpE in drug resistance are not clearly understood. In this study, we report that NlpE affects the multidrug resistance of E. coli by inducing the expression of drug efflux genes.
The expression of multidrug efflux genes is often regulated in a complex manner as described above. We therefore screened the genomic library of E. coli for genes that increased multidrug resistance levels in this organism. We screened a host strain (NKE96) lacking a functional acrB gene for identifying the regulatory elements involved in the expression of other multidrug resistance systems. A library was developed from the chromosomal DNA of the MG1655 strain (1), and then the recombinant plasmids were transformed into the ΔacrB strain NKE96. In one experiment, we found an 8-fold increase in oxacillin MIC against the transformant (data not shown). Introduction of the plasmid isolated from this strain into fresh ΔacrB cells resulted in the same oxacillin resistance phenotype; MIC was increased 8-fold over the recipient strain (data not shown).
The sequencing of the plasmid revealed an insertion containing the complete coding sequence of nlpE and a partial sequence of yaeJ. It seemed likely that in cells carrying this plasmid, overexpressed NlpE caused the transcriptional activation of genes involved in oxacillin resistance. Full-length wild-type nlpE was cloned in the pHSG398 vector to obtain pnlpE (Table 1). Oxacillin MICs for NKE96 cells harboring pnlpE were eight times higher (8 versus 1 μg/ml) than for cells harboring the pHSG398 vector (ΔacrB-vector) (Table 1). This suggests that the NlpE produced by this plasmid conferred oxacillin resistance on E. coli. During the screening, we also yielded the already known drug resistance activator BaeR (data not shown). We investigated the effect of nlpE overexpression on the susceptibility of E. coli to other toxic compounds. Various drugs were tested, including common substrates of multidrug efflux pumps, and we found that nlpE overexpression increased the resistance of the NKE96 strain to cloxacillin, nafcillin, cefamandole, aztreonam, carbenicillin, sulbenicillin, carumonam, kanamycin, novobiocin, and deoxycholate (Table 1). These results indicate that the overexpression of NlpE induces multidrug resistance in E. coli.
TABLE 1.
Susceptibility of E. coli strains to toxic compounds
| Strain | Genotype | MIC (μg/ml)a |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OXA | CLOX | NAF | FAM | ATM | CAR | SBPC | CRMN | KAN | NOV | DOC | ||
| NKE96 | ΔacrB | 1 | 1 | 2 | 0.063 | 0.063 | 1 | 2 | 0.031 | 2 | 4 | 5,000 |
| NKE154 | ΔacrB-vector | 1 | 1 | 2 | 0.063 | 0.063 | 1 | 2 | 0.031 | 2 | 4 | 5,000 |
| NKE155 | ΔacrB-pnlpE | 8 | 16 | 16 | 0.25 | 0.25 | 8 | 16 | 0.25 | 8 | 16 | 40,000 |
| NKE127 | ΔacrB cpxAR | 0.5 | 1 | 1 | 0.063 | 0.063 | 1 | 1 | 0.031 | 1 | 4 | 2,500 |
| NKE158 | ΔacrB cpxAR-vector | 0.5 | 1 | 1 | 0.063 | 0.063 | 1 | 1 | 0.031 | 1 | 4 | 2,500 |
| NKE159 | ΔacrB cpxAR-pnlpE | 0.5 | 1 | 1 | 0.063 | 0.063 | 1 | 1 | 0.031 | 1 | 4 | 2,500 |
| NKE128 | ΔacrB tolC | 0.5 | 0.25 | 1 | 0.063 | 0.063 | 1 | 1 | 0.031 | 1 | 2 | 156 |
| NKE160 | ΔacrB tolC-vector | 0.5 | 0.25 | 1 | 0.063 | 0.063 | 1 | 1 | 0.031 | 1 | 2 | 156 |
| NKE161 | ΔacrB tolC-pnlpE | 0.5 | 0.25 | 1 | 0.063 | 0.063 | 1 | 1 | 0.031 | 2 | 2 | 156 |
| NKE126 | ΔacrB acrD | 0.5 | 1 | 1 | 0.063 | 0.063 | 1 | 1 | 0.031 | 2 | 4 | 5,000 |
| NKE156 | ΔacrB acrD-vector | 0.5 | 1 | 1 | 0.063 | 0.063 | 1 | 1 | 0.031 | 2 | 4 | 5,000 |
| NKE157 | ΔacrB acrD-pnlpE | 2 | 2 | 4 | 0.13 | 0.063 | 2 | 2 | 0.063 | 4 | 8 | 10,000 |
| NKE1365 | ΔacrB mdtABC | 1 | 1 | 1 | 0.063 | 0.063 | 1 | 2 | 0.031 | 2 | 4 | 5,000 |
| NKE1366 | ΔacrB mdtABC-vector | 1 | 1 | 1 | 0.063 | 0.063 | 1 | 2 | 0.031 | 2 | 4 | 5,000 |
| NKE1367 | ΔacrB mdtABC-pnlpE | 4 | 4 | 4 | 0.13 | 0.25 | 4 | 8 | 0.13 | 8 | 8 | 10,000 |
| NKE1316 | ΔacrB acrD mdtABC | 0.5 | 0.5 | 1 | 0.063 | 0.063 | 1 | 1 | 0.031 | 2 | 2 | 2,500 |
| NKE1368 | ΔacrB acrD mdtABC-vector | 0.5 | 0.5 | 1 | 0.063 | 0.063 | 1 | 1 | 0.031 | 2 | 2 | 2,500 |
| NKE1369 | ΔacrB acrD mdtABC-pnlpE | 0.5 | 0.5 | 1 | 0.063 | 0.063 | 1 | 1 | 0.031 | 4 | 2 | 2,500 |
MIC determinations were repeated at least three times. Values in boldface are larger than those of a corresponding parental strain harboring the pHSG398 vector. OXA, oxacillin; CLOX, cloxacillin; NAF, nafcillin; FAM, cefamandole; ATM, aztreonam; CAR, carbenicillin; SBPC, sulbenicillin; CRMN, carumonam; KAN, kanamycin; NOV, novobiocin; DOC, deoxycholate.
As described above, it has been reported that overproduction of NlpE activates the Cpx pathway (29). In order to determine whether NlpE-mediated multidrug resistance depends on the CpxAR two-component signal transduction pathway, we constructed a deletion mutant of the cpxAR genes. The deletion was made by using the lambda Red system (4). In the ΔacrB cpxAR strain, overexpression of nlpE conferred no drug resistance (Table 1), indicating that NlpE conferred multidrug resistance of E. coli in a Cpx-dependent manner.
A major mechanism of bacterial multidrug resistance is active drug efflux. The results described above indicate that the expression of a multidrug efflux pump may be induced by nlpE overexpression. RND efflux pumps need two other proteins for their function: a membrane fusion protein and an outer membrane channel. It has been reported that many drug efflux systems need the membrane channel TolC for their function (7, 18). In order to determine whether NlpE-mediated multidrug resistance also depends on the TolC-dependent drug efflux pump(s), we investigated the effect of tolC deletion on the drug resistance in cells overexpressing nlpE. Deletion of tolC from the ΔacrB strain increased susceptibility to cloxacillin and deoxycholate (Table 1), which is in good agreement with a previous report (18). The tolC deletion inhibited NlpE-mediated multidrug resistance (Table 1). This result indicates that NlpE-mediated multidrug resistance is attributable to increased functioning of a TolC-dependent drug efflux pump.
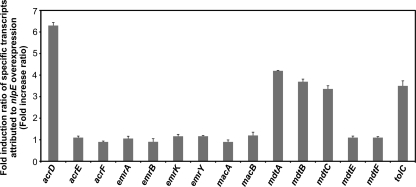
In order to determine which drug efflux pump shows increased expression when nlpE is overexpressed, we used quantitative reverse transcription-PCR (qRT-PCR) to investigate changes in the levels of drug efflux gene mRNAs dependent on nlpE overexpression. Total RNAs were isolated from exponential-phase cultures of NKE154 (ΔacrB-vector) and NKE155 (ΔacrB-pnlpE) strains, and cDNA samples were synthesized using TaqMan reverse transcription reagents (PE Applied Biosystems) with random hexamers as primers. Real-time PCR of cDNAs was performed with each specific primer pair using SYBR green PCR master mix (PE Applied Biosystems) as described previously (20). The changes in the expression levels of seven TolC-dependent-type drug efflux pump systems, including acrD, acrEF, emrAB, emrKY, macAB, mdtABC, mdtEF, and the outer membrane channel tolC genes were measured. The expression levels of acrD, mdtABC, and tolC were significantly increased by nlpE overexpression (Fig. 1). This increased expression was not observed in the ΔcpxAR strain (data not shown), indicating that NlpE stimulates these genes via the Cpx pathway.
FIG. 1.
Effect of NlpE on the expression levels of drug efflux and outer membrane channel genes. The level of the mRNA transcript was determined by qRT-PCR. The fold change ratio was calculated by dividing the expression level of the gene in the NKE155 strain by that in the NKE154 strain. The data correspond to mean values of three independent experiments. Error bars represent standard deviation.
In order to determine whether multidrug resistance mediated by nlpE overexpression is due to increased expression of acrD and/or mdtABC, we investigated the effects of deleting these genes on NlpE-mediated multidrug resistance (Table 1). In the ΔacrB acrD strain, overexpression of nlpE slightly conferred E. coli resistance to oxacillin, cloxacillin, nafcillin, cefamandole, carbenicillin, sulbenicillin, carumonam, kanamycin, novobiocin, and deoxycholate; however, these drug resistance levels were lower than those in the ΔacrB-pnlpE strain. This result indicates that AcrD is partially responsible for the NlpE-modulated multidrug resistance. AcrD was also fully responsible for aztreonam resistance (Table 1). In the ΔacrB mdtABC strain, overexpression of nlpE conferred resistance to oxacillin, cloxacillin, nafcillin, cefamandole, aztreonam, carbenicillin, sulbenicillin, carumonam, kanamycin, novobiocin, and deoxycholate. These resistance levels were lower than in the ΔacrB-pnlpE strain except for aztreonam and kanamycin, indicating that MdtABC partially contributes to the NlpE-mediated multidrug resistance. In the ΔacrB acrD mdtABC strain, overexpression of nlpE conferred no drug resistance (Table 1) except for slight increased resistance to kanamycin (2-fold increase). Together, these data indicate that the multidrug resistance conferred by NlpE is due to increased expression of both the acrD and mdtABC multidrug efflux genes.
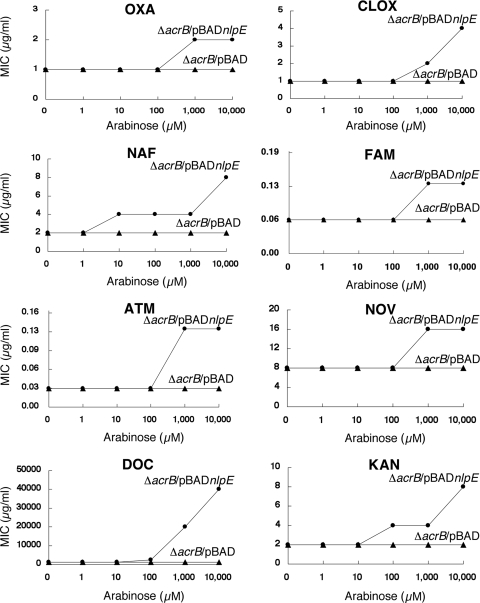
Recent characterization of the Cpx regulon in E. coli strain MC4100 by Price and Raivio (23) showed that overexpression of nlpE from the cloning vector pBR322 did not increase the expression levels of acrD and mdtABC; however, we found that pnlpE generated from the high-copy-number cloning vector pHSG398 clearly increases the expression levels of these genes and AcrD-MdtABC-dependent multidrug resistance in E. coli. These results suggest that high levels of NlpE overproduction may be required to induce the multidrug resistance phenotype. To test this hypothesis, the nlpE gene was cloned into pBAD33 (10) to produce the plasmid pBADnlpE. In this plasmid, nlpE is under the control of an arabinose-inducible PBAD promoter. The multidrug resistance phenotype of the ΔacrB-pBADnlpE strain was investigated under different concentrations of arabinose (from 1 to 10,000 μM) (Fig. 2). MICs of several drugs against the ΔacrB-pBADnlpE strain increased under a high concentration of arabinose (Fig. 2). Results presented in Fig. 2 support our hypothesis that NlpE enhances the drug resistance level of E. coli in a dose-dependent manner.
FIG. 2.
Multidrug resistance enhanced by the induction of NlpE using arabinose. MICs of oxacillin (OXA), cloxacillin (CLOX), nafcillin (NAF), cefamandole (FAM), aztreonam (ATM), novobiocin (NOV), deoxycholate (DOC), and kanamycin (KAN) to NKE1355 (ΔacrB-pBAD) and NKE1359 (ΔacrB-pBADnlpE) strains were measured under several concentrations (0 to 10,000 μM) of arabinose. MIC determinations were repeated at least three times.
In this study, we performed a genome-wide search for a regulator of multidrug resistance in E. coli by random shotgun cloning. We found NlpE, which increases resistance to oxacillin, cloxacillin, nafcilllin, cefamandole, aztreonam, carbenicillin, sulbenicillin, carumonam, kanamycin, novobiocin, and deoxycholate by upregulating acrD and mdtABC. NlpE-modulated kanamycin resistance was partially dependent on AcrD, but not on MdtABC, probably because aminoglycoside is the specific substrate for AcrD (19, 26). Slightly increased resistance to kanamycin (2-fold increase) was observed in ΔacrB tolC-pnlpE and ΔacrB acrD mdtABC-pnlpE strains, whereas it was not observed in the ΔacrB cpxAR-pnlpE strain. This suggests the possibility that NlpE may also affect other kanamycin resistance factors through the Cpx pathway in addition to AcrD. Because it was reported that NlpE is involved in copper tolerance (9), we tested effect of nlpE overexpression on the susceptibility of E. coli to CuSO4. NlpE slightly increased resistance to copper (MIC of 4 mM for NKE154 versus 6 mM for NKE155). This phenotype was not observed in NKE159, NKE161, and NKE1369, suggesting that NlpE-mediated copper resistance was due to increased expression of acrD and mdtABC via the Cpx pathway. This is reminiscent of the copper and zinc resistance mechanism by the AcrD and MdtABC multidrug efflux systems regulated by the BaeSR two-component signal transduction system in Salmonella enterica (16). In this study, it was also revealed that NlpE enhances multidrug resistance of E. coli in a dose-dependent manner. Structural study suggests that unfolded NlpE is plausibly related to activation of the Cpx pathway (12). Thus, it is possible that overproduction of NlpE may have resulted in an increased amount of unfolded protein and then it stimulated multidrug resistance via the Cpx pathway. Previously, a relationship between NlpE and biofilm formation was also highlighted by a proteome analysis of Acinetobacter baumannii (28). We found the importance of NlpE as a drug resistance factor through the induction of the multidrug efflux genes in this study. Further investigation of the regulation of multidrug efflux systems in several natural environments such as those inside hosts is needed to elucidate the biological significance of their regulatory networks.
Acknowledgments
This research was supported by the Asahi Glass Foundation; Astellas Foundation for Research on Metabolic Disorders; the Nakajima Foundation; Senri Life Science Foundation; the Takeda Science Foundation; the Uehara Memorial Foundation; the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation; a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; a Grant-in-Aid for Young Scientists (S) from the Japan Society for the Promotion of Science; and PRESTO, Japan Science and Technology Agency.
Footnotes
Published ahead of print on 8 March 2010.
REFERENCES
- 1.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 2.Brown, M. H., I. T. Paulsen, and R. A. Skurray. 1999. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 31:394-395. [DOI] [PubMed] [Google Scholar]
- 3.Connolly, L., A. De Las Penas, B. M. Alba, and C. A. Gross. 1997. The response to extracytoplasmic stress in Escherichia coli is controlled by partially overlapping pathways. Genes Dev. 11:2012-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Wulf, P., A. M. McGuire, X. Liu, and E. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277:26652-26661. [DOI] [PubMed] [Google Scholar]
- 6.DiGiuseppe, P. A., and T. J. Silhavy. 2003. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grkovic, S., M. H. Brown, and R. A. Skurray. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta, S. D., H. C. Wu, and P. D. Rick. 1997. A Salmonella typhimurium genetic locus which confers copper tolerance on copper-sensitive mutants of Escherichia coli. J. Bacteriol. 179:4977-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirakawa, H., A. Takumi-Kobayashi, U. Theisen, T. Hirata, K. Nishino, and A. Yamaguchi. 2008. AcrS/EnvR represses expression of the acrAB multidrug efflux genes in Escherichia coli. J. Bacteriol. 190:6276-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano, Y., M. M. Hossain, K. Takeda, H. Tokuda, and K. Miki. 2007. Structural studies of the Cpx pathway activator NlpE on the outer membrane of Escherchia coli. Structure 15:963-976. [DOI] [PubMed] [Google Scholar]
- 13.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 14.Martin, R. G., and J. L. Rosner. 2003. Analysis of microarray data for the marA, soxS, and rob regulons of Escherichia coli. Methods Enzymol. 370:278-280. [DOI] [PubMed] [Google Scholar]
- 15.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishino, K., E. Nikaido, and A. Yamaguchi. 2007. Regulation of multidrug efflux systems involved in multidrug and metal resistance of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189:9066-9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishino, K., E. Nikaido, and A. Yamaguchi. 2009. Regulation and physiological function of multidrug efflux pumps in Escherichia coli and Salmonella. Biochim. Biophys. Acta 1794:834-843. [DOI] [PubMed] [Google Scholar]
- 18.Nishino, K., J. Yamada, H. Hirakawa, T. Hirata, and A. Yamaguchi. 2003. Roles of TolC-dependent multidrug transporters of Escherichia coli in resistance to β-lactams. Antimicrob. Agents Chemother. 47:3030-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishino, K., and A. Yamaguchi. 2002. EvgA of the two-component signal transduction system modulates production of the yhiUV multidrug transporter in Escherichia coli. J. Bacteriol. 184:2319-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulsen, I. T., L. Nguyen, M. K. Sliwinski, R. Rabus, and M. H. Saier, Jr. 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301:75-100. [DOI] [PubMed] [Google Scholar]
- 23.Price, N. L., and T. L. Raivio. 2009. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J. Bacteriol. 191:1798-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raivio, T. L., D. L. Popkin, and T. J. Silhavy. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J. Bacteriol. 181:5263-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg, E. Y., D. Ma, and H. Nikaido. 2000. AcrD of Escherichia coli is an aminoglycoside efflux pump. J. Bacteriol. 182:1754-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowley, G., M. Spector, J. Kormanec, and M. Roberts. 2006. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat. Rev. Microbiol. 4:383-394. [DOI] [PubMed] [Google Scholar]
- 28.Siroy, A., P. Cosette, D. Seyer, C. Lemaitre-Guillier, D. Vallenet, A. Van Dorsselaer, S. Boyer-Mariotte, T. Jouenne, and E. Dé. 2006. Global comparison of the membrane subproteomes between a multidrug-resistant Acinetobacter baumannii strain and a reference strain. J. Proteome Res. 5:3385-3398. [DOI] [PubMed] [Google Scholar]
- 29.Snyder, W. B., L. J. Davis, P. N. Danese, C. L. Cosma, and T. J. Silhavy. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 177:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]