Abstract
β-Defensins are known to be both antimicrobial and able to chemoattract various immune cells. Although the sequences of paralogous genes are not highly conserved, the core defensin structure is retained. Defb14-1CV has bactericidal activity similar to that of its parent peptide (murine β-defensin Defb14) despite all but one of the canonical six cysteines being replaced with alanines. The 23-amino-acid N-terminal half of Defb14-1CV is a potent antimicrobial while the C-terminal half is not. Here, we use a library of peptide derivatives to demonstrate that the antimicrobial activity can be localized to a particular region. Overlapping fragments of the N-terminal region were tested for their ability to kill Gram-positive and Gram-negative bacteria. We demonstrate that the most N-terminal fragments (amino acids 1 to 10 and 6 to 17) are potent antimicrobials against Gram-negative bacteria whereas fragments based on sequence more C terminal than amino acid 13 have very poor activity against both Gram-positive and -negative types. We further test a series of N-terminal deletion peptides in both their monomeric and dimeric forms. We find that bactericidal activity is lost against both Gram types as the deletion region increases, with the point at which this occurs varying between bacterial strains. The dimeric form of the peptides is more resistant to the peptide deletions, but this is not due just to increased charge. Our results indicate that the primary sequence, together with structure, is essential in the bactericidal action of this β-defensin derivative peptide and importantly identifies a short fragment from the peptide that is a potent bactericide.
β-Defensins are believed to be important components in innate immunity. They are expressed predominantly at mucosal surfaces and the reproductive tract. They are a multigene family which has ∼40 members spread over five genomic loci in humans (18). They possess a strong, broad-spectrum antimicrobial action in vitro and also have been shown to be chemoattractants for various immune cells (23, 24). Some β-defensins are rapidly induced by exposure to lipopolysaccharide or Th1 cytokines, while human β-defensin 1 (hBD1) expression is mostly constitutive (14). Recently, increased copy number of the β-defensins at the chromosome 8 locus has been associated with an increased risk of psoriasis (7). Conversely, decreased copy number of this cluster has been associated with an increased risk of inflammatory bowel disease (5).
Defensins have a canonical six-cysteine motif, but the remaining sequence of these short peptides is not highly conserved. The consensus is X2-10CX5-7(G/A)XCX3-4CX9-13CX4-7CCXn, where X is any residue, G/A is glycine or alanine, and C is cysteine. The spacing and connectivity of the six cysteines separate α-defensins from β-defensins, and although highly conserved throughout evolution, the canonical six-cysteine motifs in either defensin class are not required for antimicrobial activity (2, 11, 20, 23). The canonical disulfide bridges may be important for protection of the peptide in vivo, as has been demonstrated for α-defensins (11).
Evolutionary studies of the mouse and human genomes have revealed that Defb14 is the mouse orthologue of human β-defensin 3 (DEFB103) (19). The peptide encoded by Defb14 has salt-resistant, wide-range antimicrobial activity very similar to that of the peptide encoded by DEFB103 (human β-defensin 3-hBD3) (6, 16, 20). Like all defensins, hBD3 has an antiparallel β-sheet scaffold with a short N-terminal α-helix (17), and this structure is conserved throughout evolution.
Given the promiscuity of the endogenous peptides, understanding the structure-activity relationships of β-defensins is essential. Work in this area aims to find key information for designing novel synthetic antibiotics and for clarifying their function in immunity. There is some controversy as to the mode of action of defensins with membrane disruption by some mechanism seeming to occur. However, studies with hBD3 in Staphylococcus aureus have revealed the occurrence of rapid killing and at the same time blockage of all biosynthetic pathways, although significant depolarization of the bacterial membranes was not observed and permeabilization was incomplete (1).
Wu et al. (23) elegantly used directional disulfide bonding to demonstrate that the disulfide connectivities influenced the chemoattractant potency of hBD3 but not the antibacterial effect. We have also shown that hBD3 or Defb14 peptides with all but one cysteine being replaced by alanine were still active as both a chemoattractant and an antimicrobial. The remaining cysteine was at position V of the canonical six-cysteine motif (-CI-CII-CIII-CIV-CV-CVI-), where the cysteines are interspersed with a variable number of amino acid residues. Hoover et al. (8) demonstrated that peptides from the C-terminal region of hBD3 had potent activity against Escherichia coli and Pseudomonas aeruginosa but not S. aureus. The shortest peptide was a decapeptide based on sequence from the C-terminal end of the molecule (RGRKSSRRKK). The two adjacent cysteines in the native peptide were replaced with serines to eliminate the potential to form disulfide bonds (8). Further work using modifications of this decapeptide sequence has shown that its antimicrobial activity is more complex than just being a function of charge (13). In addition, work on an hBD3 derivative where the cysteines of the full-length mature peptide are changed to alanines has revealed that this peptide is as active as the parent peptide against Gram-positive and -negative bacteria but is now sensitive to the ionic strength of the medium (3). As the cysteine content of the peptide does not affect the antimicrobial activity of the peptide, we have focused on the Defb14-1CV analogue (20). This peptide has all the cysteines replaced with alanines except Cys40, which resides at position V of the six-cysteine motif. In addition, it can easily be controlled to form monomer or dimer species. We have previously shown that this peptide analogue has the same salt-insensitive bactericidal activity as the parent Defb14 peptide (20). In addition, we divided the active Defb14-1CV peptide into an N-terminal and a C-terminal fragment and found that the C-terminal half (residues 24 to 45) was a very poor antimicrobial compared to the N-terminal fragment (residues 1 to 23), which was potent. In order to further dissect the residues important for the antimicrobial activity of Defb14-1CV, we created a library of peptide fragments. We show that the antimicrobial activity of the most N-terminal residues is most potent, and deletion of these residues reveals that the points at which the activity is most significantly lost vary for different bacterial strains. We conclude that both the sequence and structure that a given sequence imposes are important for antimicrobial activity.
MATERIALS AND METHODS
Peptide synthesis and purification.
All peptides were chemically synthesized by standard solid-phase methodology. Defb14 and Defb14-1CV were obtained from Chemical Synthesis Services-Albachem Ltd. (Gladsmuir, United Kingdom). Disulfide connectivities were determined by proteolysis and peptide mass mapping by following the procedures outlined by Campopiano et al. (2). The sequences are shown in Table 1. The defensin 14-inspired peptides, including additional synthesis of Defb14-1CV, were made in-house using automated peptide synthesis. This was carried out on an Applied Biosystems model 433A peptide synthesizer using Rink amide-AM resin for peptide amides, preloaded NovaSyn TGT resin for peptide acids, and 9-fluorenylmethoxy carbonyl (Fmoc) amino acids, all from Novabiochem. All truncated Defb14-1CV peptides were synthesized with an acetyl group at the N terminus, rather than the free amino group, to best represent the conformation of the terminal amino acid. Liquid chromatography (LC) mass spectra confirming identity and purity were obtained on a Micromass Quattro LC mass spectrometer. Semipreparative high-performance liquid chromatography (HPLC) was performed using a Phenomenex Luna C18 column and a gradient of 5 to 95% acetonitrile (containing 0.1% trifluoroacetic acid) over 45 min (flow rate of 3.0 ml/min). All other chemical reagents were obtained from Aldrich. Automated solid-phase peptide synthesis was carried out on a 0.05-mmol scale using 0.5 mmol of each Fmoc amino acid per coupling reaction and 2-(1H-benzotriazol-1-yl)-1,1,3,3,-tetramethyluronium hexafluorophosphate/1-hydroxybenzotriazole as coupling reagents. Coupling time was 0.5 h. Peptide products were cleaved from the resin with 95% trifluoroacetic acid, 2.5% ethanedithiol, and 2.5% water for 3 h, the resin was filtered off and washed with trifluoroacetic acid, and filtrate was poured into diethyl ether (10 volumes). Following centrifugation (3,000 rpm at 15 min), the precipitate was resuspended in ether (5 volumes) and recentrifuged (3,000 rpm at 15 min). The crude peptides were dissolved in water and loaded directly onto a semipreparative HPLC column. Peptide fractions were identified by mass spectrometry (MS) and lyophilized.
TABLE 1.
Sequences of Defb14-derived peptides
| Peptide | Sequencea |
|---|---|
| Defb14 | FLPKTLRKFFCRIRGGRCAVLNCLGKEEQIGRCSNSGRKCCRKKK |
| Defb14-1CV | FLPKTLRKFFARIRGGRAAVLNALGKEEQIGRASNSGRKCARKKK |
| Defb14-1CVΔ(1) | LPKTLRKFFARIRGGRAAVLNALGKEEQIGRASNSGRKCARKKK |
| Defb14-1CVΔ(1-2) | PKTLRKFFARIRGGRAAVLNALGKEEQIGRASNSGRKCARKKK |
| Defb14-1CVΔ(1-5) | LRKFFARIRGGRAAVLNALGKEEQIGRASNSGRKCARKKK |
| Defb14-1CVΔ(1-8) | FFARIRGGRAAVLNALGKEEQIGRASNSGRKCARKKK |
| Defb14-1CVΔ(1-11) | RIRGGRAAVLNALGKEEQIGRASNSGRKCARKKK |
| Defb14-1CVΔ(1-14) | GGRAAVLNALGKEEQIGRASNSGRKCARKKK |
| Defb14-1CVΔ(1-17) | AAVLNALGKEEQIGRASNSGRKCARKKK |
| Defb14-1CVΔ(1-23) | LGKEEQIGRASNSGRKCARKKK |
| Defb14-1CV(1-23) | FLPKTLRKFFARIRGGRAAVLNA |
| Defb14-1CV(1-10) | FLPKTLRKFF |
| Defb14-1CV(6-17) | LRKFFARIRGGR |
| Defb14-1CV(18-23) | AAVLNA |
| Defb14-1CV(14-23) | RGGRAAVLNA |
| Defb14-1CV(12-23) | RIRGGRAAVLNA |
| Defb14-1CV(13-34) | IRGGRAAVLNALGKEEQIGRAS |
The sequences are given using single-letter abbreviations for amino acid residues; cysteine residues are in boldface. Deletion peptides are based on the Defb14-1CV active peptide used previously (20).
Bactericidal assays.
Bactericidal assays were carried out as previously described (20). Briefly, test organisms were grown to mid-logarithmic phase in Iso-Sensitest broth (Oxoid) growth medium and then diluted to between 1 × 106 CFU/ml and 5 × 106 CFU/ml in 10 mM potassium phosphate containing 1% (vol/vol) Iso-Sensitest broth, pH 7.4. Different concentrations of test peptide were incubated in 100 μl of cells (1 × 105 to 5 × 105 CFU) at 37°C for 3 h. Reduction of the peptides, where performed, was done by adding 10 mM dithiothreitol (DTT) and incubating the peptides at room temperature overnight. The oxidation state of each peptide was determined by mass spectrometry. Tenfold serial dilutions of the incubation mixture were spread on Iso-Sensitest plates and incubated at 37°C, and the numbers of CFU were determined the following day. The minimum bactericidal concentration (MBC) is the concentration of peptide at which we observed >99.99% killing of the initial inoculum. All assays were performed in duplicate and repeated on three independent occasions. The minimum bactericidal concentration was obtained by taking the mean of all of the results, and experimental errors were within one doubling dilution. Significant differences between MBCs were determined using a Mann-Whitney U test.
Molecular hydrophobicity analysis.
The relative molecular hydrophobicity of each monomeric peptide was evaluated by reverse-phase HPLC-MS using an Ultimate 3000 LC system equipped with a Famos autosampler (Dionex) and a VG Platform II mass spectrometer. A total of 30 μl of each sample, containing 50 μM peptide in 10 mM dithiothreitol (DTT) and 50 μM melittin (Sigma) as an internal standard, was injected onto a Waters Symmetry C18 column (3.9 by 150 mm). DTT was used to prevent dimerization of any of the peptides via cysteine oxidation; this was confirmed by mass analysis following the LC separation. Initial conditions were as follows: 100% eluent A (96% water, 3% acetonitrile, 1% formic acid); 0% eluent B (4% water, 95% acetonitrile, 1% formic acid); and flow rate, 1 ml min−1. Samples were eluted from the column by using a linear gradient of 0 to 40% eluent B over 40 min. The flow was split 1/50 prior to introduction to the electrospray ionization source of the mass spectrometer. In order to assess the relative molecular hydrophobicities, we compared the retention time of each peptide to that of melittin.
ΔG, the hydrophobicity score (kcal/mol) of monomeric peptides in water, was calculated using the scale of Wimley and White (22), where greater hydrophobicity is indicated by a less negative value.
Helical wheel projections.
Helical wheel projections were constructed with the aid of the program at http://rzlab.ucr.edu/scripts/wheel/wheel.cgi.
RESULTS
Bactericidal activity of peptide fragments.
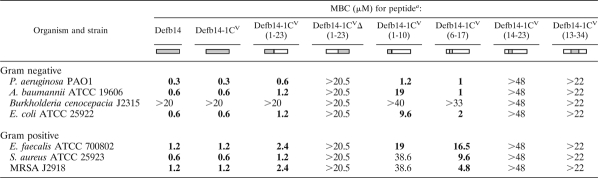
In order to narrow the antibacterial activity of Defb14-1CV to a particular region of the molecule, we synthesized three overlapping fragments (1 to 10 [1-10], 6 to 17 [6-17], and 14 to 23 [14-23]) of the antimicrobially active N-terminal Defb14-1CV. These fragments (see Table 1 for sequences) were tested against a panel of Gram-positive and Gram-negative bacteria. We determined the minimum bactericidal concentration (MBC) values for these peptide fragments (Table 2), i.e., the minimum concentration at which less than 99.99% of the initial inoculum is viable.
TABLE 2.
MBCs of Defb14-1CV derivatives against a range of Gram-negative and Gram-positive bacteria
Bactericidal activity is located in the N-terminal sequence of the β-defensin peptide. The region of the molecule that the peptide sequence is derived from is indicated beneath its name by gray shading. Boldface indicates MBC of less than 20 μM.
The 23-amino-acid N-terminal fragment Defb14-1CV(1-23) has bactericidal activity very similar to that of Defb14-1CV (as previously reported under the name D14ip 1 [20]) against Pseudomonas aeruginosa PAO1 and S. aureus ATCC 25923. The peptide fragment with the first 10 amino acids of Defb14-1CV [Defb14-1CV(1-10)] shows good activity against P. aeruginosa PAO1 (MBC of 1.0 μM), Acinetobacter baumannii (MBC of 0.58 μM), and E. coli (MBC of 2.3 μM), and these are within one doubling dilution of the Defb14-1CV(1-23) fragment and comparable to the parental peptide activity against these strains. The Defb14-1CV peptide fragment with the 12 internal amino acids [Defb14-1CV(6-17)] also has a strong bactericidal activity against the three Gram-negative strains tested. The peptide fragments containing amino acids 14 to 23 and 13 to 34 do not achieve an MBC against the bacteria at the concentrations tested here. Interestingly, only one of the two most N-terminal peptides has good activity against the Gram-positive strains tested. Only the fragment with residues 6 to 17 has the ability to kill all the Gram-positive strains at a concentration less than 19 μM (∼30 μg/ml). Defb14-1CV(6-17) had good activity against methicillin-resistant S. aureus (MRSA) strain J2918 (MBC of 4.8 μM), comparable with that of the Defb14-1CV(1-23) fragment, and its MBC against S. aureus was 8.2 μM. We previously reported that Defb14-1CV and the active N-terminal fragment Defb14-1CV(1-23) had salt-insensitive activity against P. aeruginosa (20). Interestingly, the bactericidal activities of both Defb14-1CV(1-10) and Defb14-1CV(6-17) against P. aeruginosa are unchanged with increasing salt concentrations of up to 200 mM NaCl (data not shown).
Bactericidal activity of N-terminal deletions.
In order to further dissect the antimicrobial sequence within the N-terminal region of Defb14-1CV and in the context of the molecule with the C-terminal region included, we made a series of N-terminal deletions (see Table 1 for sequences). As the deletions are based on Defb14-1CV, they all have a single cysteine residue (CysV40). This enabled us to examine both monomeric and covalent dimeric forms, with the latter occurring via a disulfide bridge. Following oxidation, the peptides spontaneously form a homodimer through the cysteine-cysteine disulfide bridge and reduction with DTT leaves the peptide in the reduced form, as verified by mass spectrometry, and this state is maintained throughout the assay (data not shown). We determined the MBCs of both the dimeric and monomeric molecules against three Gram-negative and three Gram-positive bacterial strains for all the deletion peptides (Table 3) .
TABLE 3.
MBCs for monomer and dimer forms of the N-terminal deletion peptides
| Peptide | MBC (μM)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Pseudomonas aeruginosa PAO1 |
Escherichia coli ATCC 25922 |
Acinetobacter baumannii ATCC 19606 |
Staphylococcus aureus ATCC 25923 |
MRSA J2918 |
Enterococcus faecalis ATCC 700802 |
|||||||
| Monomer | Dimer | Monomer | Dimer | Monomer | Dimer | Monomer | Dimer | Monomer | Dimer | Dimer | Monomer | |
| Defb14-1CV | 0.3 (±0.0) | 0.2 (±0.0) | 0.6 (±0.0) | 0.3 (±0.0) | 0.6 (±0.0) | 0.3 (±0.0) | 1.4 (±0.5) | 0.5 (±0.0) | 1.2 (±0.0) | 0.6 (±0.0) | 1.2 (±0.2) | 0.6 (±0.2) |
| Defb14-1CVΔ(1) | ND | 0.3 (±0.0) | ND | 1.0 (±0.6) | ND | 0.6 (±0.0) | ND | 0.7 (±0.0) | ND | 0.7 (±0.0) | ND | 0.8 (±0.2) |
| Defb14-1CVΔ(1-2) | 0.7 (±0.0) | 0.5 (±0.2) | 3.3 (±0.8) | 0.6 (±0.2) | 1.7 (±0.4) | 0.6 (±0.0) | 1.0 (±0.3) | 0.8 (±0.2) | 2.1 (±0.4) | 0.8 (±0.6) | 4.2 (±0.8) | 1.0 (±0.2) |
| Defb14-1CVΔ(1-5) | 0.7 (±0.0) | 0.6 (±0.1) | 2.5 (±0.0) | 1.1 (±0.4) | ND | 1.0 (±0.2) | 1.4 (±0.0) | 1.1 (±0.4) | 2.5 (±0.0) | 1.7 (±0.8) | ND | 1.25 (±0.0) |
| Defb14-1CVΔ(1-8) | 1.1 (±0.4) | 0.7 (±0.4) | 10 (±0.0) | 2.5 (±0.0) | 2.9 (±1.1) | 1.3 (±0.6) | 1.5 (±0.0) | 1.1 (±0.3) | 2.2 (±0.4) | 1.3 (±0.0) | >5 | 1.9 (±0.6) |
| Defb14-1CVΔ(1-11) | 1.7 (±0.0) | 0.7 (±0.2) | >10 | >5 | 2.5 (±0.0) | 1.0 (±0.2) | 5.9 (±0.5) | 0.9 (±0.3) | >10 | 1.5 (±1.1) | >5 | >5 |
| Defb14-1CVΔ(1-14) | >15.2 | 1.4 (±0.3) | >10 | >5 | >10 | 2.5 (±0.0) | >15.2 | >7.6 | >10 | 6.7 (±3.3) | >10 | >5 |
| Defb14-1CVΔ(1-17) | >16.5 | 2.6 (±1.0) | >10 | >5 | >10 | 3.3 (±0.8) | >16.5 | >8.3 | >10 | >5 | >10 | >5 |
| Defb14-1CVΔ(1-23) | ND | >10 | ND | >10 | ND | >10 | ND | >10 | ND | >10 | ND | >10 |
| Defb14-1CV(18-23) | >83.5 | NA | ND | ND | ND | ND | >83.5 | NA | ND | NA | ND | ND |
Loss of bactericidal activity upon peptide N-terminal deletion is bacterial strain dependent. Sequences of peptides are given in Table 1. Peptides are in either the monomer (DTT treated) or dimer form. Values in parentheses are standard errors of the means. Values in boldface indicate that the MBC for the monomeric form is significantly different (P < 0.01) from that of the Defb14-1CV monomer. ND, not done; NA, not applicable.
As the deletions progress, the MBC increases in all cases, indicating loss of bactericidal activity. The points at which the bactericidal activity significantly reduces are not the same for all the bacterial strains. Deletion of 14 amino acids results in the monomeric form of the peptide being unable to kill any of the six bacterial strains at the concentrations tested. The critical point for activity against P. aeruginosa and A. baumannii ATCC 19606 is between Defb14-1CVΔ(1-11) and Defb14-1CVΔ(1-14). However, against another Gram-negative strain (E. coli), deletion of 8 amino acids results in the bactericidal activity falling 4-fold to 10 μM (37.3 μg/ml). Against Gram-positive S. aureus ATCC 25923 and MRSA J2918, the bactericidal activities drop significantly after deletion of 11 amino acids. However, deletion of only 5 amino acids results in the monomeric peptide significantly losing activity against Enterococcus faecalis.
The dimeric peptides are more robust than the monomers in their ability to kill the bacteria when the N-terminal amino acids are progressively deleted. All four strains tested show a significant increase in the MBC of the monomeric species before the dimeric species' MBC is affected. P. aeruginosa is killed at only 1.4 μM with the Defb14-1CVΔ(1-14) dimeric species, whereas the monomeric form is not able to kill the bacterium at 16.5 μM. The same effect was observed with E. coli and A. baumannii, where the monomer species loses bactericidal activity before the dimer forms. The Gram-positive strains also display this effect. For Defb14-1CVΔ(1-11), there is a 6-fold increase in the amount of monomer required to kill S. aureus ATCC 25923 compared to the amount of dimer (5.9 μM versus 0.9 μM), and with the same peptide, the monomer MBC against MRSA is >10 μM, compared to 1.5 μM for the dimer MBC.
The dimer deletion peptide activities against P. aeruginosa in increasing sodium chloride concentrations of up to 200 mM were tested. Like the parent peptide and peptide fragments 1 to 10 and 6 to 17, the bactericidal activities of deletion peptides up to and including Defb14-1CVΔ(1-5) were not significantly affected by addition of sodium chloride at concentrations of up to 200 mM. However, deletion peptide Defb14-1CVΔ(1-8) had a significantly (P < 0.01) raised MBC of 10 (±0.0) μM in 10 mM sodium chloride compared to 1.7 (±0.0) μM in 0 mM NaCl. The subsequent deletions also showed significantly (P < 0.01) increased MBCs of >10 mM in 100 mM NaCl, indicating that loss of the N-terminal region of Defb14-1CV rendered these peptides salt sensitive.
In order to check whether degradation of the peptides by bacterium-specific proteases could explain the loss of bactericidal activity, the integrity of the inactive peptide Defb14-1CVΔ(1-14) was evaluated by LC-MS following incubation with S. aureus ATCC 25923. After 4 h, the majority of the peptide remained intact (peak area of 66% relative to that of sample prior to incubation; data not shown), indicating that proteolysis of the peptide by the bacterial strain does not explain its inactivity.
Charge versus bactericidal activity.
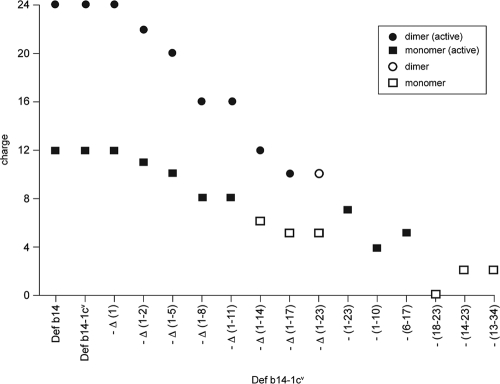
In an effort to determine whether charge rather than the primary sequence of the peptides had an effect on the ability to kill bacteria, we considered the net charge of each sequence in the peptide library we had synthesized. Figure 1 shows the charge of monomer molecules (and dimer molecules where relevant) and is annotated according to which peptides are active against P. aeruginosa PAO1 with an MBC of less than 5.0 μM (shown as filled dots or squares in the figure). All the inactive peptides (shown with open dots or squares) possess a charge of less than +11; however, six active peptides also have a charge of less than +11, with Defb14-1CV(1-10) having a charge of only +5 and an MBC of 1.2 μM. The Defb14-1CVΔ(1-17) dimer has a charge of +10 and is a potent antimicrobial agent with a low MBC, but in contrast, the Defb14-1CVΔ(1-23) dimer has an equal charge but is an inactive antimicrobial with an MBC in excess of 10 μM.
FIG. 1.
Charge and structure are important for antimicrobial activity of β-defensin derivatives. The Defb14-1CV derivative library was plotted according to the net charge (protonation state) of the synthesized peptides at a neutral pH. This takes into account the number of basic versus acidic amino acids and also the termini. The key indicates whether peptides are active bactericides and dimers or monomers.
Hydrophobicity versus bactericidal activity.
We also assessed the hydrophobicity of the deletion peptides both according to their primary sequences via the Wimley-White scale for proteins at a membrane interface (Fig. 2a) (22) and experimentally by measuring their retention times by reverse-phase HPLC (Fig. 2b). This second method is sensitive to the conformation of the free peptide in solution. However, we observe little difference between the hydrophobicities of these peptides, as gauged by either method (Fig. 2). The hydrophobicity via HPLC does decrease as the length of the peptide decreases, with the exception of Defb14-1CVΔ(1-17), which shows a slight increase relative to that of Defb14-1CVΔ(1-14). The observed differences in bactericidal activity with these peptides cannot be attributed to any dramatic difference in the hydrophobicity of the free peptide.
FIG. 2.
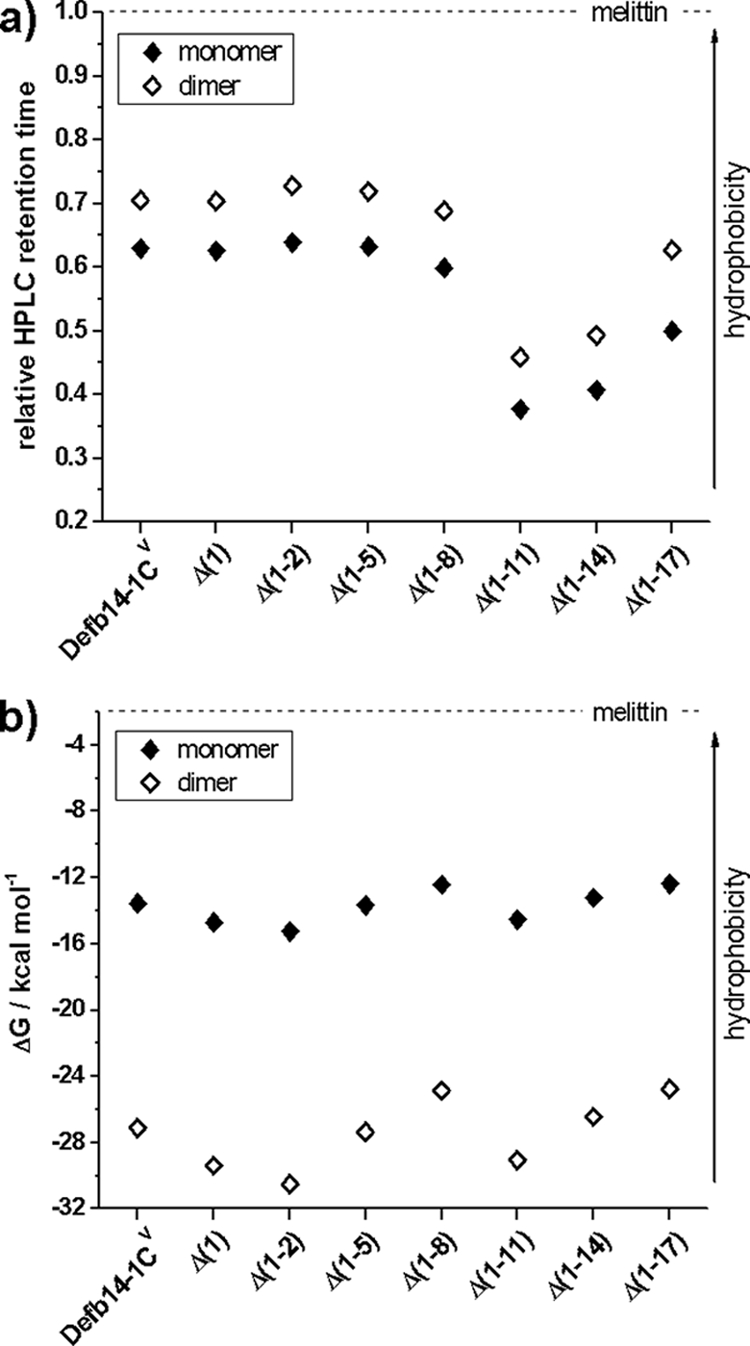
Assessment of the relative hydrophobicities of the Defb14-1CV N-terminal deletion series. (a) The retention times of each N-terminal deletion peptide, relative to that of melittin, are measured for both the monomer and dimer under reducing and nonreducing conditions, respectively. Increased relative retention time is indicative of more hydrophobic character, since under a reversed-phase gradient hydrophobic peptides will be retained longer on the C18 column. For comparison, panel b shows the theoretical hydrophobicity score (in kcal mol−1) of each peptide in water, calculated using the scale of Wimley and White (22). Greater hydrophobicity is indicated by a less negative value.
Structural change as determined by helical wheel projection.
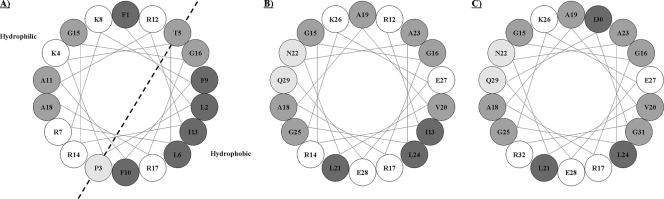
Figure 3 shows a helical wheel projection of Defb14-1CV (residues 1 to 18) which predicts that the N-terminal region of the full-length peptide has the potential to form an α-helix with charged residues concentrated on one face of the helix and hydrophobic residues on the other. The distinction between hydrophilic and hydrophobic faces is less clear in helical wheel projections of residues 12 to 29 and 15 to 32, which correspond to the N-terminal regions of the Defb14-1CVΔ(1-11) and Defb14-1CVΔ(1-14) peptides, respectively. This suggests a lesser propensity to form an α-helix, which we have also shown by circular dichroism (CD) (21), and may account, in part, for the diminished activity of the truncated peptides. There is little difference in these projections for residues 12 to 29 and 15 to 32.
FIG. 3.
Helical wheel projections of N-terminal regions of Defb14-1CV and inactive/active derivatives. Schematic helical wheel projections of Defb14-1CV (residues 1 to 18) (A), Defb14-1CVΔ(1-11) (residues 12 to 29) (B), and Defb14-1CVΔ(1-14) (residues 15 to 32) (C). Charged residues are denoted by white circles. Increasing hydrophobicity is denoted by increasing shading. The positions of the side chains are shown along a “regular” α-helix. For a right-handed helix with 3.6 residues per turn, rotation is clockwise as the polypeptide chain is followed from N to C, and the 5th residue ends up in a position exactly 40° clockwise relative to that of residue 1.
DISCUSSION
In order to further dissect the basis of the potent bactericidal activity of the N-terminal region of the Defb14-1CV derivative, we synthesized several overlapping fragments of the 23-amino-acid region and a series of Defb14-1CV-derived peptides with progressive N-terminal deletions. We determined their MBCs against a panel of Gram-positive and -negative bacterial strains. Although the Defb14-1CV(6-17) fragment is a strong bactericide, the deletion series reveals that there does not appear to be one short sequence responsible for activity against all strains.
Primary sequence determines bactericidal activity against different bacteria.
We show that the more C-terminal fragment Defb14-1CV(14-23) has very poor activity against both Gram-positive and -negative strains, whereas Defb14-1CV(1-10) and Defb14-1CV(6-17) retain some aspects of the antibacterial activity of the parent molecule. Both fragments are potent bactericides against the three Gram-negative strains tested; however, only Defb14-1CV(6-17) is robust against the Gram-positive S. aureus strains. The deletion series allowed us to determine whether these fragments influence bactericidal activity within the context of the full-length molecule. We find that as the N-terminal deletions progress, the monomeric peptide MBC increases (indicating loss of activity), but the points at which significant MBC increase occurs vary between strains. The deletion series indicates that the critical amino acid residues required for bactericidal activity are located between residues 5 and 14 (concordant with the activity of fragments 1 to 10 and 6 to 17). Gram-positive strains MRSA J2918 and S. aureus ATCC 25923 are effectively killed by Defb14-1CVΔ(6-17) but not Defb14-1CVΔ(1-10). In agreement with this, the ability of the truncated peptides to kill S. aureus ATCC 25923 and MRSA J2918 reduces significantly with the deletion of residues 9, 10, and 11 (FFA). The charge of active Defb14-1CVΔ(1-8) is the same (+8) as that of poorly active Defb141CVΔ(1-11), with little change in hydrophobicity, suggesting that the difference in activity is indeed primarily sequence/structure based. This contrasts with work on hBD3, where charge and hydrophobicity have been found to influence antimicrobial activity (9).
Although amino acid residues 9 to 11 appear to be important in the killing of Gram-positive S. aureus strains, deletion of these residues results in little change to the monomeric MBC against Gram-negative P. aeruginosa PAO1 and A. baumannii. However, the subsequent deletion of RIR in Defb14-1CV(1-14) results in a significant loss of activity. This decrease in sensitivity is not wholly dependent on the Gram status of the bacteria, since Gram-negative E. coli ATCC 25922 shows sensitivity to Defb14-1CVΔ(1-11) in a pattern similar to that of Gram-positive S. aureus species.
These data again suggest that the primary amino acid sequence is important for bactericidal activity. This notion is supported by a similar study with hBD1 which revealed that antimicrobial activity against E. coli was affected mostly by amino acid substitutions of charged residues in the positively charged C-terminal region (15). These substitutions did not affect peptide topology, and the study concluded that it was the side chain changes that affected the antibacterial activity. Recently, it was reported that the bactericidal activity of α-defensin cryptdin 4 was attenuated when all the arginines were mutated to lysine. Llenado et al. (10) postulate that the guanidine group in arginine may allow stronger electrostatic charges and hydrogen bonding interactions, which may be important for electronegative bacterial cell membrane interactions. However, we have tested whether inclusion of the RIR sequence results in a peptide with bactericidal activity, but peptide Defb14-1CV(12-23) with sequence RIRGGRAAVLNA is inactive as a bactericide at 20 μM against both P. aeruginosa and S. aureus (data not shown).
Multiple sequences contribute to bactericidal activity.
E. coli, P. aeruginosa PAO1, and A. baumannii are killed effectively by peptide fragments Defb14-1CV(1-10) and Defb14-1CV(6-17). The deletion series monomer data support both of these fragments containing unique sequence that is important in the context of the peptide. Sequential deletion of amino acids 6, 7, and 8 and then 9, 10, and 11 (E. coli) and amino acids 9, 10, and 11 followed by 12, 13, and 14 (P. aeruginosa) results in an individual decrease in bactericidal potency. This suggests that there are two regions important to bactericidal activity, one in Defb14-1CV1-10 and one in Defb14-1CV6-17, so that only when both are deleted from the larger molecule is the activity very much reduced.
Supporting this is our previous finding that the sequence NTLQK in hBD3 (corresponding to KTLRK in Defb14) has been subjected to negative selection during primate evolution (19). This sequence is contained fully in Defb141CV(1-10) and partially in Defb141CV(6-17), suggesting that the region is important functionally. Interestingly, it is also this region (LRK) that appears to be critical in maintaining the salt insensitivity of the peptides against P. aeruginosa.
Dimeric structure retains activity.
The bactericidal activity of the dimeric deletion peptides is more resistant to the deletion of amino acids than that of the monomeric forms up until the Defb14-1CVΔ(1-14) fragment (against S. aureus) and deletion of RIR. This indicates that dimer structure, in addition to sequence, is important for bactericidal activity. The increased charge of dimeric molecules may implicate charge as a contributor to antibacterial activity; however, the inactivity of dimeric Defb14-1CVΔ(1-14) implies that sequence and structure are more important than charge for this strain. In order to consider this further, we compare the charges and bactericidal activities for the various peptides against P. aeruginosa PAO1 (Fig. 1). Comparison of bactericidal activities between Defb14-1CVΔ(1-23) and Defb14-1CVΔ(1-17) dimers with the same charge (+5) and similar low hydrophobicities but vastly different bactericidal activities supports that sequence and structure are more important than charge.
The importance of dimeric structure is further supported by evidence that covalent dimeric defensins are more active bactericides than monomeric or noncovalent dimers (2, 4) and by the recent report by Antcheva et al. (1) which demonstrated that covalent dimerization of an artificial defensin could improve its antimicrobial activity.
Helical structure may affect function.
We performed circular dichroism (CD) on an analogue of Defb14-1CVΔ(1-23), which has poor antimicrobial activity identical to that of Defb14-1CVΔ(1-23) but alanines at all the cysteine positions (20). When compared to the potent N-terminal peptide Defb14-1CV(1-23), it is shown to have reduced helical propensity (21; data not shown). We also show by helical wheel projection that the helix-forming propensity of the N-terminal region is likely to be lost as the peptide deletions increase. This may be important since the formation of helical oligomers is implicated in the pore model of antimicrobial action. However, there is little difference in these projections for residues 12 to 29 and 15 to 32 to rationalize the lack of bactericidal activity against P. aeruginosa of the Defb14-1CVΔ(1-14) monomer relative to that of Defb14-1CVΔ(1-11). In addition, the activities found in these deletion peptides and mode of action may not reflect those present in the full-length disulfide stabilize molecule.
Of the peptides tested here, Defb14-1CV(6-17) has the most potential as a short and effective bactericidal agent in vitro. We have, however, found that the bactericidal activities of the peptide fragments and parental β-defensin are inhibited by serum, although not by trypsin digestion (data not shown), which is also observed for hBD3 (12).
In conclusion, our results show that both structure and sequence are important for the antimicrobial activity of these β-defensin derivatives. We find that the activities of the N-terminal peptide fragments are supported by the N-terminal deletions, and we find that the precise sequences that confer bactericidal activity vary for different bacteria, showing selectivity at the amino acid level. We have shown that deletion of FFA and RIR is important for loss of activity against S. aureus, deletion of LRK for E. coli and E. faecalis, and deletion of RIR for P. aeruginosa and A. baumannii. We also show that, in all cases, covalent dimer peptides retain activity better than monomeric forms. Approaches aimed at determining the molecular basis of structure-activity relationships will greatly assist in the design of novel therapeutic agents.
Acknowledgments
This research was supported by the EPSRC, the Royal Society, Cystic Fibrosis Research Trust UK, MRC, EaStCHEM, and the University of Edinburgh.
Footnotes
Published ahead of print on 22 February 2010.
REFERENCES
- 1.Antcheva, N., F. Morgera, L. Creatti, L. Vaccari, U. Pag, S. Pacor, Y. Shai, H. G. Sahl, and A. Tossi. 2009. Artificial beta-defensin based on a minimal defensin template. Biochem. J. 421:435-447. [DOI] [PubMed] [Google Scholar]
- 2.Campopiano, D. J., D. J. Clarke, N. C. Polfer, P. E. Barran, R. J. Langley, J. R. Govan, A. Maxwell, and J. R. Dorin. 2004. Structure-activity relationships in defensin dimers: a novel link between beta-defensin tertiary structure and antimicrobial activity. J. Biol. Chem. 279:48671-48679. [DOI] [PubMed] [Google Scholar]
- 3.Chandrababu, K. B., B. Ho, and D. Yang. 2009. Structure, dynamics, and activity of an all-cysteine mutated human beta defensin-3 peptide analogue. Biochemistry 48:6052-6061. [DOI] [PubMed] [Google Scholar]
- 4.Circo, R., B. Skerlavaj, R. Gennaro, A. Amoroso, and M. Zanetti. 2002. Structural and functional characterization of hBD-1(Ser35), a peptide deduced from a DEFB1 polymorphism. Biochem. Biophys. Res. Commun. 293:586-592. [DOI] [PubMed] [Google Scholar]
- 5.Fellermann, K., D. E. Stange, E. Schaeffeler, H. Schmalzl, J. Wehkamp, C. L. Bevins, W. Reinisch, A. Teml, M. Schwab, P. Lichter, B. Radlwimmer, and E. F. Stange. 2006. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am. J. Hum. Genet. 79:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinrichsen, K., R. Podschun, S. Schubert, J. M. Schroder, J. Harder, and E. Proksch. 2008. Mouse beta-defensin-14, an antimicrobial ortholog of human beta-defensin-3. Antimicrob. Agents Chemother. 52:1876-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollox, E. J., U. Huffmeier, P. L. Zeeuwen, R. Palla, J. Lascorz, D. Rodijk-Olthuis, P. C. van de Kerkhof, H. Traupe, G. de Jongh, M. den Heijer, A. Reis, J. A. Armour, and J. Schalkwijk. 2008. Psoriasis is associated with increased beta-defensin genomic copy number. Nat. Genet. 40:23-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoover, D. M., Z. Wu, K. Tucker, W. Lu, and J. Lubkowski. 2003. Antimicrobial characterization of human beta-defensin 3 derivatives. Antimicrob. Agents Chemother. 47:2804-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kluver, E., S. Schulz-Maronde, S. Scheid, B. Meyer, W. G. Forssmann, and K. Adermann. 2005. Structure-activity relation of human beta-defensin 3: influence of disulfide bonds and cysteine substitution on antimicrobial activity and cytotoxicity. Biochemistry 44:9804-9816. [DOI] [PubMed] [Google Scholar]
- 10.Llenado, R. A., C. S. Weeks, M. J. Cocco, and A. J. Ouellette. 2009. Electropositive charge in alpha-defensin bactericidal activity: functional effects of Lys-for-Arg substitutions vary with the peptide primary structure. Infect. Immun. 77:5035-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maemoto, A., X. Qu, K. J. Rosengren, H. Tanabe, A. Henschen-Edman, D. J. Craik, and A. J. Ouellette. 2004. Functional analysis of the alpha-defensin disulfide array in mouse cryptdin-4. J. Biol. Chem. 279:44188-44196. [DOI] [PubMed] [Google Scholar]
- 12.Maisetta, G., L. M. Di, S. Esin, W. Florio, F. L. Brancatisano, D. Bottai, M. Campa, and G. Batoni. 2008. Evaluation of the inhibitory effects of human serum components on bactericidal activity of human beta defensin 3. Peptides 29:1-6. [DOI] [PubMed] [Google Scholar]
- 13.Papanastasiou, E. A., Q. Hua, A. Sandouk, U. H. Son, A. J. Christenson, M. L. Van Hoek, and B. M. Bishop. 2009. Role of acetylation and charge in antimicrobial peptides based on human beta-defensin-3. APMIS 117:492-499. [DOI] [PubMed] [Google Scholar]
- 14.Pazgier, M., D. M. Hoover, D. Yang, W. Lu, and J. Lubkowski. 2006. Human beta-defensins. Cell. Mol. Life Sci. 63:1294-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pazgier, M., A. Prahl, D. M. Hoover, and J. Lubkowski. 2007. Studies of the biological properties of human beta-defensin 1. J. Biol. Chem. 282:1819-1829. [DOI] [PubMed] [Google Scholar]
- 16.Rohrl, J., D. Yang, J. J. Oppenheim, and T. Hehlgans. 2008. Identification and biological characterization of mouse beta-defensin 14, the orthologue of human beta-defensin 3. J. Biol. Chem. 283:5414-5419. [DOI] [PubMed] [Google Scholar]
- 17.Schibli, D. J., H. N. Hunter, V. Aseyev, T. D. Starner, J. M. Wiencek, P. B. McCray, Jr., B. F. Tack, and H. J. Vogel. 2002. The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus. J. Biol. Chem. 277:8279-8289. [DOI] [PubMed] [Google Scholar]
- 18.Schutte, B. C., J. P. Mitros, J. A. Bartlett, J. D. Walters, H. P. Jia, M. J. Welsh, T. L. Casavant, and P. B. McCray, Jr. 2002. Discovery of five conserved beta-defensin gene clusters using a computational search strategy. Proc. Natl. Acad. Sci. U. S. A. 99:2129-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semple, C. A., A. Maxwell, P. Gautier, F. M. Kilanowski, H. Eastwood, P. E. Barran, and J. R. Dorin. 2005. The complexity of selection at the major primate beta-defensin locus. BMC Evol. Biol. 5:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor, K., D. J. Clarke, B. McCullough, W. Chin, E. Seo, D. Yang, J. Oppenheim, D. Uhrin, J. R. Govan, D. J. Campopiano, D. Macmillan, P. E. Barran, and J. R. Dorin. 2008. Analysis and separation of residues important for the chemoattractant and antimicrobial activities of beta-defensin 3. J. Biol. Chem. 283:6631-6639. [DOI] [PubMed] [Google Scholar]
- 21.Tyrrell, C., M. De Cecco, N. L. Reynolds, F. Kilanowski, D. Campopiano, P. Barran, D. Macmillan, and J. R. Dorin. 2010. Isoleucine/leucine(2) is essential for chemoattractant activity of beta-defensin Defb14 through chemokine receptor 6. Mol. Immunol. 47:1378-1382. [DOI] [PubMed] [Google Scholar]
- 22.Wimley, W. C., and S. H. White. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3:842-848. [DOI] [PubMed] [Google Scholar]
- 23.Wu, Z., D. M. Hoover, D. Yang, C. Boulegue, F. Santamaria, J. J. Oppenheim, J. Lubkowski, and W. Lu. 2003. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human beta-defensin 3. Proc. Natl. Acad. Sci. U. S. A. 100:8880-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang, D., O. Chertov, S. N. Bykovskaia, Q. Chen, M. J. Buffo, J. Shogan, M. Anderson, J. M. Schroder, J. M. Wang, O. M. Howard, and J. J. Oppenheim. 1999. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-528. [DOI] [PubMed] [Google Scholar]