Abstract
Multidrug-resistant (MDR) Acinetobacter spp. have emerged as a threat to public health. We investigated the various genes involved in resistance to fluoroquinolones, aminoglycosides, cephalosporins, and carbapenems in 75 clinical Acinetobacter isolates from a Taiwanese hospital. All isolates were tested for the gyrA mutations, the presence of integrons, blaAmpC, and carbapenem resistance genes. The Ser83Leu mutation in GyrA accounted for fluoroquinolone resistance. The presence of integrons containing aminoglycoside-modifying enzymes was associated with resistance to gentamicin and tobramycin but not with resistance to amikacin. The presence of an ISAba1 element upstream of blaAmpC was correlated with cephalosporin resistance. Although most Acinetobacter baumannii isolates with ISAba1-blaOXA-51-like were resistant to carbapenems, several isolates remained susceptible to carbapenems. Transformation by the introduction of ISAba1-blaOXA-23 or ISAba1-blaOXA-66 into A. baumannii ATCC 15151 (CIP 70.10), resulting in the overexpression of OXA-23 or OXA-66, respectively, suggested the role of the ISAba1 element as a strong promoter. The two transformants showed significantly increased resistance to piperacillin-tazobactam, imipenem, and meropenem. The cefepime resistance conferred by ISAba1-blaOXA-23 and the impact of ISAba1-blaOXA-66 on carbapenem resistance in A. baumannii are reported here for the first time. Continuous surveillance of antibiotic resistance genes in MDR Acinetobacter spp. and elucidation of their antibiotic resistance mechanisms are crucial for the development of therapy regimens and for the prevention of further dissemination of these antibiotic resistance genes.
In the past 2 decades, Acinetobacter spp. have become important opportunistic pathogens responsible for nosocomial infections, especially among patients in intensive care units (ICUs) (27). Infections with nosocomial Acinetobacter spp. have created a challenge for concordant therapy due to their acquisition of multidrug-resistant (MDR) phenotypes, such as resistance to fluoroquinolones, aminoglycosides, cephalosporins, and carbapenems (7).
Fluoroquinolone-resistant Acinetobacter spp. have emerged rapidly following an increase in the consumption of fluoroquinolones (primarily ciprofloxacin) (22). A major change from Ser83 to Leu83 (Ser83Leu) in the quinolone resistance-determining region (QRDR) of DNA gyrase subunit A (GyrA) is highly correlated with resistance to ciprofloxacin in Acinetobacter baumannii (36, 40). In addition, MDR Acinetobacter spp. frequently contain integrons, which provide bacteria with a gene capture system perfectly adapted to circumvent the challenges of multiple-antibiotic treatment regimens (24). Currently, a total of five classes of mobile integrons are known to play roles in the dissemination of antibiotic resistance genes (24). Class 1 integrons are the predominant class of integrons found in A. baumannii and contain the vast majority of gene cassettes encoding various aminoglycoside-modifying enzymes (13). The presence of such integrons may be associated with aminoglycoside resistance in Acinetobacter spp.
Ambler class C β-lactamases, known as AmpC cephalosporinases, are chromosomally inherited in A. baumannii. The major regulator of blaAmpC is an upstream insertion sequence (IS) element, ISAba1 (5, 35). The presence of ISAba1-blaAmpC in A. baumannii is highly correlated with overexpression of blaAmpC and resistance to extended-spectrum cephalosporins but not to carbapenems (5, 11, 35). Carbapenems are the broadest-spectrum β-lactams and are considered a “last resort” for serious infections with Gram-negative bacteria (21). However, carbapenem-resistant Acinetobacter spp. have emerged worldwide and are often associated with the presence of Ambler class B metallo-β-lactamases (MBLs) and carbapenem-hydrolyzing class D β-lactamases (CHDLs) (28). Two main MBL genes (blaIMP and blaVIM), and four main CHDL genes (the blaOXA-23, blaOXA-24-like, blaOXA-51-like, and blaOXA-58 genes) have been identified in carbapenem-resistant Acinetobacter spp. (30, 33). The blaOXA-51-like gene is chromosomally intrinsic only to A. baumannii (9). Furthermore, the ISAba1 element is often found upstream of blaOXA-23 and blaOXA-51-like, and it may provide a promoter that allows the overexpression of the two genes (12, 26).
In this study, we investigate the various genes of Acinetobacter spp. from a Taiwanese hospital that are involved in fluoroquinolone, aminoglycoside, cephalosporin, and carbapenem resistance. The roles of ISAba1-blaOXA-23 and ISAba1-blaOXA-66 in A. baumannii are also demonstrated.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A total of 75 nonduplicated Acinetobacter clinical isolates were collected from the National Taiwan University Hospital (Taipei, Taiwan) in 2006 and were identified by the API 20NE system. Genospecies were identified according to the 16S-23S rRNA gene intergenic spacer (ITS) region as described previously (20). Escherichia coli TOP10 and A. baumannii ATCC 15151 (CIP 70.10) were used as hosts for the cloning and expression experiments, respectively. Plasmid pCR2.1-TOPO (Invitrogen) was used as the cloning vector, and shuttle plasmid pAT801, with a PvuII fragment harboring the oriC of Acinetobacter from pW1277 (15) in pUc18, was obtained from P. Courvalin of Institut Pasteur and was used as the expression vector. E. coli and A. baumannii were cultured on Luria-Bertani broth (LB) and tryptic soy broth (TSB) plates, respectively, at 37°C for 20 h.
Antibiotic susceptibility testing.
Antibiotic susceptibility testing was performed using the Vitek 2 automatic system (bioMérieux), with the AST-GN09 card for the identification of Gram-negative bacilli. The MIC of imipenem was further determined by Etest (AB Biodisk, Solna, Sweden). The criteria used were in accordance with the guidelines established by the Clinical and Laboratory Standards Institute (CLSI) (4).
Detection of antibiotic resistance genes.
Genomic DNA was extracted as described previously (20). All primers used in this study are listed in Table 1. The QRDR of gyrA was amplified and sequenced by procedures similar to those used in our previous study (36). The gyrA QRDR was defined as gyrA nucleotide positions 241 to 243 in Acinetobacter spp., and it is equivalent to Ser83 of E. coli. Integrons were amplified and sequenced with primers derived from the 5′ and 3′ conserved segments (18), and additional internal primers were designed to ensure complete identification. The carbapenem resistance genes, including blaIMP, blaVIM, blaOXA-23, blaOXA-24-like, blaOXA-51-like, and blaOXA-58, were detected by using primers and PCR conditions described previously (29, 41). The presence of the ISAba1 element was detected by PCR in order to determine the proximity of this element to blaOXA-23, blaOXA-51-like, and blaAmpC (5, 12, 39). The insertion sequence preceding the blaOXA-58 gene was detected by PCR using a reverse primer of the bla gene and a forward primer of the IS (ISAba1, ISAba2, or ISAba3) (31).
TABLE 1.
Oligonucleotide primers used in this study
| Name | Nucleotide sequence (5′-3′) | Location | Reference |
|---|---|---|---|
| Ab-GF | ACAAGAAATCTGCTCGT | gyrA | 36 |
| Ab-GR | CGAAGTTACCCTGACCATC | gyrA | 36 |
| 5′-CS | GGCATCCAAGCAGCAAG | Integron | 18 |
| 3′-CS | AAGCAGACTTGACCTGA | Integron | 18 |
| ACI10 | GCTGAACGCGATAAACTTC | ISAba1 of blaAmpC | 5 |
| ACI2 | TAGTACTGCTATTTACGGCT | blaAmpC | 5 |
| IMP-A | GAAGGYGTTTATGTTCATAC | blaIMP | 29 |
| IMP-B | GTAMGTTTCAAGAGTGATGC | blaIMP | 29 |
| VIM2004A | GTTTGGTCGCATATCGCAAC | blaVIM | 29 |
| VIM2004B | AATGCGCAGCACCAGGATAG | blaVIM | 29 |
| OXA23-F | GATCGGATTGGAGAACCAGA | blaOXA-23 | 40 |
| OXA23-R | ATTTCTGACCGCATTTCCAT | blaOXA-23 | 40 |
| OXA24-F | GGTTAGTTGGCCCCCTTAAA | blaOXA-24-like | 40 |
| OXA24-R | AGTTGAGCGAAAAGGGGATT | blaOXA-24-like | 40 |
| OXA51-F | TAATGCTTTGATCGGCCTTG | blaOXA-51-like | 40 |
| OXA51-R | TGGATTGCACTTCATCTTGG | blaOXA-51-like | 40 |
| OXA-58-F | AAGTATTGGGGCTTGTGCTG | blaOXA-58 | 40 |
| OXA-58-R | CCCCTCTGCGCTCTACATAC | blaOXA-58 | 40 |
| OXA-58A | CGATCAGAATGTTCAAGCGC | blaOXA-58 | 31 |
| OXA-58B | ACGATTCTCCCCTCTGCGC | blaOXA-58 | 31 |
| ISAba3B | CGTTTACCCCAAACATAAGC | tnpA of ISAba3 (but not in ISAba3-like) | 31 |
| ISAba3C | AGCAATATCTCGTATACCGC | tnpAof ISAba3-like and ISAba3 | 31 |
| ISAba1-F | GGATCCCTCTGTACACGAYAAATTTC | ISAba1 | This study |
| OXA23-R1 | GAATTCTTAAATAATATTCAGCTGTTTTAATG | blaOXA-23 | This study |
| OXA51-R1 | GAATTCCTATAAAATACCTAATTGTTCTAAAC | blaOXA-51-like | This study |
Cloning of ISAba1-blaOXA-23 and ISAba1-blaOXA-66.
For cloning and verification of the nucleotide sequences of ISAba1-blaOXA-23 and ISAba1-blaOXA-51-like, primers ISAba1-F and OXA23-R1 were used to amplify ISAba1-blaOXA-23 from strain 6AB15, and primers ISAba1-F and OXA51-R1 were used to amplify ISAba1-blaOXA-51-like from strain 6AB11. The PCR conditions for ISAba1-blaOXA-23 and ISAba1-blaOXA-51-like were as follows: preheating at 95°C for 2 min; 35 cycles consisting of 95°C for 10 s, 50°C for 10 s, and 72°C for 130 s; and a final extension at 72°C for 60 s. The two PCR products were separately ligated into the pCR2.1-TOPO vector, generating pCR2.1-OXA23 and pCR2.1-OXA51. The inserts of pCR2.1-OXA23 and pCR2.1-OXA51 were sequenced with an Applied Biosystems sequencer (ABI 3730). The nucleotide and deduced protein sequences were analyzed with software available on the website of the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov).
Expression of blaOXA-23 and blaOXA-66.
The sequence analysis identified blaOXA-51-like from 6AB11 as blaOXA-66. Fragments containing ISAba1-blaOXA-23 and ISAba1-blaOXA-66 were obtained from pCR2.1-OXA23 and pCR2.1-OXA51, respectively, by digestion with BamHI/EcoRI. BamHI/EcoRI-restricted ISAba1-blaOXA-23 and BamHI/EcoRI-restricted ISAba1-blaOXA-66 were ligated into BamHI/EcoRI-restricted pAT801, generating pOXA23 and pOXA66, respectively. pOXA23 and pOXA66 were separately transformed into A. baumannii ATCC 15151 by electroporation. The transformants were selected on LB plates containing 100 mg/liter ampicillin.
SDS-PAGE analysis.
Bacterial cells were grown overnight at 37°C in LB, harvested by centrifugation at 8,000 × g for 5 min, and washed once with iced phosphate-buffered saline (PBS). The cells were resuspended in fresh PBS with 1 mM dithiothreitol (DTT) and 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and were disrupted by sonication. The crude cell extracts were clarified by centrifugation at 12,000 × g for 10 min at 4°C, and the supernatants were collected. The protein concentration of the crude cell extract was determined using a commercial bicinchoninic acid (BCA) protein assay kit (Pierce, Rockford, IL) with bovine serum albumin (BSA) (0.05 to 2 mg/ml) as the standard. The crude cell extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a 4- to-20% gradient polyacrylamide gel, and protein patterns were detected by staining with Coomassie blue.
Statistical analysis.
Statistical analysis was performed using SPSS, version 10.0.7. Pearson's correlation coefficient was used to determine the relationships between the MIC distributions of antibiotic agents and antibiotic resistance genes. An r value of >0.7 (or <−0.7) and a P value of <0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
The nucleotide sequences of ISAba1-blaOXA-23 and ISAba1-blaOXA-66, reported in this paper, have been submitted to NCBI GenBank under accession numbers GQ849192 and GQ849191, respectively.
RESULTS
The 75 Acinetobacter clinical isolates were assigned to three distinct Acinetobacter genospecies based on their ITS sequences: A. baumannii (n = 53), Acinetobacter genospecies 13TU (n = 20), and Acinetobacter genospecies 3 (n = 2). The antibiotic susceptibilities and corresponding antibiotic resistance genes of these Acinetobacter clinical isolates are shown in Tables 2 and 3, respectively.
TABLE 2.
Antibiotic susceptibilities of 75 Acinetobacter clinical isolates
| Antibiotic and MIC (mg/liter)a | No. of isolatesb |
||
|---|---|---|---|
| A. baumannii (n = 53) | Acinetobacter genospecies 13TU (n = 20) | Acinetobacter genospecies 3 (n = 2) | |
| Ciprofloxacin | |||
| ≥4 (R) | 45 (44)a | 2 (2) | 1 (1) |
| 2 (I) | |||
| Levofloxacin | |||
| ≥8 (R) | 24 (24) | 1 (2) | 1 (1) |
| 4 (I) | 19 (19) | ||
| Amikacin | |||
| ≥64 (R) | 15 (15)b | 2 (2) | |
| 32 (I) | 2 (0) | ||
| Gentamicin | |||
| ≥16 (R) | 43 (40) | 2 (1) | |
| 8 (I) | 4 (2) | ||
| Tobramycin | |||
| ≥16 (R) | 40 (39) | 1 (1) | |
| 8 (I) | 2 (0) | 1 (1) | |
| Ceftazidime | |||
| ≥32 (R) | 44 (44)c | 5 (1) | |
| 16 (I) | 1 (0) | 2 (1) | 1 (0) |
| Cefepime | |||
| ≥32 (R) | 34 (34) | 6 (2) | 1 (0) |
| 16 (I) | 8 (8) | 1 (0) | |
| Imipenem | |||
| ≥16 (R) | 19 (17)d | 2 | |
| 8 (I) | 5 (4) | 2 | |
| Meropenem | |||
| ≥16 (R) | 20 (18) | 2 | |
| 8 (I) | 4 (3) | 1 | |
MICs followed by “(R)” indicate resistance; MICs followed by “(I)” indicate intermediate resistance.
The numbers of isolates with the following genetic features are given in parentheses: GyrA mutations (for fluoroquinolones [ciprofloxacin and levofloxacin]), integrons (for aminoglycosides [amikacin, gentamicin, and tobramycin]), ISAba1-blaAmpC structures (for cephalosporins [ceftazidime and cefepime]), and ISAba1-blaOXA-51-like structures (for carbapenems [imipenem and meropenem]).
TABLE 3.
Antibiotic resistance genes of 75 Acinetobacter clinical isolates
| Antibiotic resistance gene(s) | No. of isolates |
||
|---|---|---|---|
| A. baumannii (n = 53) | Acinetobactergenospecies 13TU (n = 20) | Acinetobacter genospecies 3 (n = 2) | |
| QRDR of GyrA | |||
| Ser83 (TCA) | 9 | 18 | 1 |
| Ser83Leu (TTA) | 44 | 1 | 1 |
| Ser83Phe (TTT) | 1 | ||
| Integron | |||
| Not present | 13 | 15 | 2 |
| aacA4-catB8-aadA1 | 35 | ||
| aacC1-orfX-orfX′-aadA1 | 3 | ||
| arr-3-aacA4 | 2 | ||
| arr-3-aacA4 + blaVIM-11 | 1 | ||
| dhfrXII-orfF-aadA2 | 2 | ||
| blaIMP-1-aac(6′)-II-aadA4 | 2 | ||
| AmpC | |||
| Not present | 1 | 18 | 2 |
| blaAmpC | 8 | ||
| ISAba1-blaAmpC | 44 | 2 | |
| CHDLs and MBLs | |||
| Not present | 15 | 2 | |
| ISAba1-blaOXA-23 + ISAba3-blaOXA-58 | 1 | ||
| ISAba3-blaOXA-58 only | 1 | ||
| ISAba3-blaOXA-58 + blaIMP-1 | 2 | ||
| ISAba3-blaOXA-58 + blaVIM-11 | 1 | ||
| ISAba1-blaOXA-23 + blaOXA-51-like | 1 | ||
| blaOXA-24-like + blaOXA-51-like | 1 | ||
| blaOXA-51-like only | 27 | ||
| ISAba1-blaOXA-51-like only | 24 | ||
Fluoroquinolones.
Eighty-five percent of A. baumannii isolates (45/53) were resistant to fluoroquinolones, and all of these contained a Ser83Leu mutation in GyrA, except for one ciprofloxacin-resistant isolate with Ser83 in GyrA that was susceptible to levofloxacin. Only 55% (24/44) of the A. baumannii isolates described above with a Ser83Leu mutation in GyrA were also resistant to levofloxacin, and 43% (19/44) were intermediate to levofloxacin. Most of the Acinetobacter genospecies 13TU isolates (90% [18/20]) were susceptible to ciprofloxacin, with Ser83 in GyrA. Two Acinetobacter genospecies 13TU isolates were resistant to ciprofloxacin; one had a Ser83Leu mutation and the other had a Ser83Phe mutation in GyrA, and the isolate with Ser83Leu in GyrA was also resistant to levofloxacin. Of the two Acinetobacter genospecies 3 isolates, the one with Ser83Leu in GyrA was resistant to ciprofloxacin and levofloxacin. The GyrA mutations of Acinetobacter spp. were significantly correlated with susceptibility to ciprofloxacin (r, 0.972; P, <0.001) and levofloxacin (r, 0.847; P, <0.001).
Aminoglycosides.
Among the 75 Acinetobacter isolates, six distinct integrons were found, and their gene cassettes contained various aminoglycoside-modifying genes, including aacA4, aacC1, aac(6′)-II, aadA1, aadA2, and aadA4. Three integrons were detected in A. baumannii, including aacA4-catB8-aadA1 (2,381 bp), aacC1-orfX-orfX′-aadA1 (2,542 bp), and dhrfXII-orfF-aadA2 (1,873 bp), while three other integrons, including arr-3-aacA4 (1,395 bp), blaVIM-11 (1,062 bp), and blaIMP-1-aac(6′)-II-aadA4 (2,507 bp), were detected only in Acinetobacter genospecies 13TU. Twenty-eight percent (15/53) of A. baumannii isolates were resistant to amikacin and contained integrons. Furthermore, 81% (43/53) and 75% (40/53) of the A. baumannii isolates were resistant to gentamicin and tobramycin, respectively, and most of them contained integrons. In Acinetobacter genospecies 13TU, five isolates contained integrons, but some of them remained susceptible to aminoglycosides. No integrons were detected in the two Acinetobacter genospecies 3 isolates, and they were also susceptible to the three aminoglycosides analyzed. Consequently, the presence of integrons in Acinetobacter spp. was significantly correlated with susceptibility to gentamicin (r, 0.8; P, <0.001) and tobramycin (r, 0.841; P, <0.001) but not to amikacin (r, 0.403; P, <0.001).
Cephalosporins.
Eighty-three percent (44/53) of A. baumannii isolates were resistant to ceftazidime, and all of them contained the ISAba1-blaAmpC structure. The eight A. baumannii isolates lacking ISAba1 upstream of blaAmpC were susceptible to ceftazidime, and the sole isolate intermediate to ceftazidime did not contain blaAmpC. Seventy-nine percent (42/53) of A. baumannii isolates were resistant to cefepime, and they contained the ISAba1-blaAmpC structure. In Acinetobacter genospecies 13TU, seven isolates were resistant or intermediate to ceftazidime, and two of these contained the ISAba1-blaAmpC structure. Furthermore, these isolates were also not susceptible to cefepime and one ceftazidime-intermediate isolate harboring the ISAba1-blaAmpC structure showed resistance to cefepime. In addition, one Acinetobacter genospecies 3 isolates without blaAmpC was intermediate to ceftazidime and resistant to cefepime. The presence of ISAba1 upstream of blaAmpC in Acinetobacter spp. was significantly correlated with susceptibility to ceftazidime (r, 0.861; P, <0.001) and cefepime (r, 0.725; P, <0.001).
Carbapenems.
Forty-five percent (24/53) of A. baumannii isolates were resistant or intermediate to imipenem and meropenem; most (88% [21/24]) of them contained an ISAba1-blaOXA-51-like structure. Among the remaining isolates without ISAba1 upstream of blaOXA-51-like, one isolate containing ISAba1-blaOXA-23 and one containing blaOXA-24-like were resistant to carbapenems, while one isolate, intermediate to carbapenems, had no detectable carbapenemase genes. The presence of ISAba1 upstream of blaOXA-51-like in A. baumannii was highly correlated with susceptibility to imipenem (r, 0.753; P, <0.001) and meropenem (r, 0.764; P, <0.001). In addition, among the five Acinetobacter genospecies 13TU isolates harboring ISAba3-blaOXA-58, one isolate containing only ISAba3-blaOXA-58 was susceptible to carbapenems, while of the other four isolates, containing additional carbapenemase genes, two isolates with blaIMP-1 were resistant to carbapenems, one isolate with ISAba1-blaOXA-23 was intermediate to carbapenems, and one isolate with blaVIM-11 was intermediate to imipenem but susceptible to meropenem. Furthermore, all of the five Acinetobacter genospecies 13TU isolates harboring ISAba3-blaOXA-58 were also resistant to ceftazidime and cefepime, and only one of them, harboring blaIMP-1, contained ISAba1-blaAmpC. A total of 15 Acinetobacter genospecies 13TU and 2 Acinetobacter genospecies 3 isolates harbored no carbapenem resistance genes that we detected, and all of them were susceptible to carbapenems.
Cloning of ISAba1-blaOXA-23 and ISAba1-blaOXA-66.
DNA sequence analysis indicated that the insert of pCR2.1-OXA23, with a 2,036-bp fragment size, was 99% identical to ISAba1-blaOXA-23 in A. baumannii (GenBank accession number EF127491) and the insert of pCR2.1-OXA51 revealed a 2,012-bp fragment 100% identical to ISAba1-blaOXA-66 in A. baumannii (GenBank accession number DQ923479).
Expression of blaOXA-23 and blaOXA-66 in A. baumannii ATCC 15151.
The crude cell extracts of two transformants, A. baumannii ATCC 15151(pOXA23) and A. baumannii ATCC 15151(pOXA66), were analyzed by SDS-PAGE (Fig. 1). The protein patterns of clinical isolates 6AB11 and 6AB15 (Fig. 1, lanes 1 and 2) were significantly different from that of A. baumannii ATCC 15151 (lane 3), while the protein pattern of the A. baumannii transformant ATCC 15151(pAT801) (lane 4) was identical to that of A. baumannii ATCC 15151. Furthermore, the protein patterns of the two transformants A. baumannii ATCC 15151(pOXA23) and A. baumannii ATCC 15151(pOXA66) were very similar to that of A. baumannii ATCC 15151(pAT801) except for the conspicuous expression of two proteins of 30 and 32 kDa (Fig. 1, lanes 5 and 6).
FIG. 1.
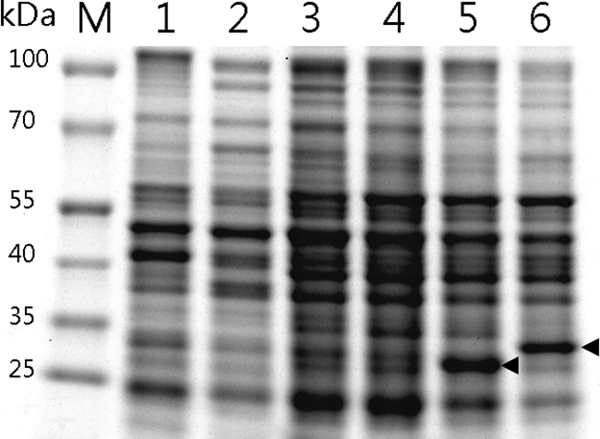
Expression of blaOXA-23 and blaOXA-66 from A. baumannii ATCC 15151. Lane 1, A. baumannii 6AB11 (ISAba1-blaOXA-66); lane 2, A. baumannii 6AB15 (ISAba1-blaOXA-23 and blaOXA-66); lane 3, A. baumannii ATCC 15151; lane 4, A. baumannii ATCC 15151(pAT801); lane 5, A. baumannii ATCC 15151(pOXA23); lane 6, A. baumannii ATCC 15151(pOXA66). The arrows in lanes 5 and 6 indicate the increased expression of 30-kDa and 32-kDa proteins in two transformants. The molecular masses of the size markers (lane M) are given on the left.
Antibiotic susceptibilities of two transformants.
We chose the wild-type A. baumannii strain ATCC 15151 as the host for investigation of the possible roles of ISAba1-blaOXA-23 and ISAba1-blaOXA-66. The susceptibilities of the isolates to β-lactams are shown in Table 4. Clinical strains 6AB11 and 6AB15 were resistant to all antibiotics tested. A. baumannii ATCC 15151 was susceptible to most of the antibiotic agents, and only the MICs of ampicillin, ampicillin-sulbactam, and piperacillin were increased for the transformant A. baumannii ATCC 15151(pAT801), harboring the AmpR-selectable marker from pAT801. The imipenem and meropenem MICs for transformants A. baumannii ATCC 15151(pOXA23) and ATCC 15151(pOXA66) were significantly increased (64-fold) over that for A. baumannii ATCC 15151(pAT801). Furthermore, the piperacillin-tazobactam MICs for those two transformants were increased 4- to 32-fold over that for A. baumannii ATCC 15151(pAT801). Only for ATCC 15151(pOXA23) was the cefepime MIC increased (16-fold) over that for the reference strain. ATCC 15151(pOXA23) and ATCC 15151(pOXA66) were susceptible to ciprofloxacin, levofloxacin, amikacin, gentamicin, and tobramycin (data not shown).
TABLE 4.
Antibiotic susceptibilities of A. baumannii clinical isolates and of A. baumannii reference strain ATCC 15151 alone or harboring a recombinant plasmid
| β-Lactam(s) | MICa (mg/liter) for: |
|||||
|---|---|---|---|---|---|---|
| A. baumannii 6AB11 (ISAba1-blaOXA-66) | A. baumannii 6AB15 (ISAba1-blaOXA-23 + blaOXA-66) | A. baumannii ATCC 15151 | A. baumannii ATCC 15151(pAT801) | A. baumannii ATCC 15151(pOXA23) (ISAba1-blaOXA-23) | A. baumannii ATCC 15151(pOXA66) (ISAba1-blaOXA-66) | |
| Ampicillin | ≥32 | ≥32 | 16 | 32 | ≥32 | ≥32 |
| Ampicillin-sulbactam | ≥32 | ≥32 | ≤2 | 16 | ≥32 | 16 |
| Piperacillin | ≥128 | ≥128 | 16 | ≥128 | ≥128 | ≥128 |
| Piperacillin-tazobactam | ≥128 | ≥128 | ≤4 | ≤4 | ≥128 | 16 |
| Ceftazidime | ≥64 | ≥64 | 4 | 4 | 8 | 4 |
| Cefepime | ≥64 | ≥64 | 4 | 4 | ≥64 | 4 |
| Imipenem | ≥32 | ≥32 | 0.25 | 0.25 | ≥16 | ≥32 |
| Meropenem | ≥16 | ≥16 | 0.25 | 0.25 | ≥16 | ≥16 |
The MICs of β-lactams were detected by Vitek 2 except for imipenem, for which the MIC was detected by Etest.
DISCUSSION
This study analyzed various genes of Acinetobacter clinical isolates that are responsible for resistance to fluoroquinolones, aminoglycosides, cephalosporins, and carbapenems. Although Acinetobacter genospecies 3 and Acinetobacter genospecies 13TU are genetically closely related to A. baumannii and are also associated with nosocomial infections (1), A. baumannii is still the predominant genospecies isolated and has a higher ratio of MDR than other Acinetobacter spp. Previous studies have found differences in antibiotic susceptibility and antibiotic resistance genes among distinct Acinetobacter genospecies (16, 19), but they were focused mainly on carbapenem resistance. We found differences in phenotypic and genetic characteristics between A. baumannii and Acinetobacter genospecies 13TU not only with regard to carbapenem resistance but also with regard to fluoroquinolone, aminoglycoside, and cephalosporin resistance. In our previous studies, ciprofloxacin was the most effective antibiotic against non-A. baumannii Acinetobacter isolates(20, 36). In this study, only three non-A. baumannii Acinetobacter isolates were ciprofloxacin resistant, and a mutation in Ser83Leu or Ser83Phe was identified, thus further supporting the idea that GyrA mutation was the major mechanism for the resistance of Acinetobacter spp. to fluoroquinolones. Although fluoroquinolone-resistant proteins (Qnr) in A. baumannii were reported in 2008 (37), this mechanism does not appear to be common in Acinetobacter spp. We did not find Qnr-positive isolates within our collections. But this is an interesting topic for further study.
Integrons play an important role in the horizontal spread of antibiotic resistance genes (24). In this study, aacA4-catB8-aadA1 was the most prevalent (88% [35/40]) integron-borne gene cassette in A. baumannii, a finding similar to that of one Taiwanese report (13). aacA4, which encoded an aminoglycoside 6′-N-acetyltransferase [AAC(6′)-Ib], was shown to confer resistance to amikacin, netilmicin, and tobramycin (34). Although the presence of aacA4 in Acinetobacter spp. was highly correlated with gentamicin and tobramycin resistance, 66% (25/38) of aacA4-harboring Acinetobacter isolates remained susceptible to amikacin (data not show). Similar findings were obtained in another study (13). In addition, some antibiotic resistance genes were identified on integrons even though their corresponding antibiotics, such as chloramphenicol (catB8), rifampin (arr-3), trimethoprim (dhfrXII), streptomycin, and spectinomycin (aadA1, aadA2, and aadA4), are no longer in use. Such integrons were frequently found in epidemic strains of A. baumannii and were associated with a high prevalence of multiple antibiotic resistance, especially aminoglycoside resistance (13, 38).
The basal level of expression of the chromosome-borne blaAmpC gene in A. baumannii seemed not to reduce susceptibility to expanded-spectrum cephalosporins (2). Our results also indicated that all blaAmpC-harboring A. baumannii isolates were susceptible to cephalosporins, whereas all ISAba1-blaAmpC-harboring A. baumannii isolates were resistant to ceftazidime, and most of them were not susceptible to cefepime. Although the presence of ISAba1-blaAmpC was correlated with cefepime resistance according to the statistical analysis, other studies showed that the inhibition or overproduction of AmpC had no significant effect on cefepime susceptibility in A. baumannii (6, 14). In addition, five ISAba3-blaOXA-58-harboring Acinetobacter genospecies 13TU isolates resistant to ceftazidime and cefepime without ISAba1-blaAmpC were observed, and it has been suggested that alternative mechanisms may be responsible for resistance to extended-spectrum cephalosporins (3, 6). Among carbapenem resistance genes, we found blaIMP-1, blaVIM-11, and blaOXA-58 only in Acinetobacter genospecies 13TU, while blaOXA-51-like was detected only in A. baumannii, suggesting that distinct Acinetobacter genospecies contained different carbapenemases (17). In other countries, the widespread dissemination of carbapenem-resistant Acinetobacter spp. with blaOXA-23 or blaOXA-24 has been reported (25, 32). However, we found only 1 blaOXA-23-harboring and 1 blaOXA-24-harboring A. baumannii isolate in this study, and similar observations have been reported in other Taiwanese studies, suggesting that the presence of ISAba1-blaOXA-51-like was the most common mechanism of carbapenem resistance in A. baumannii strains in Taiwan (17, 23). Although ISAba1-blaOXA-51-like was considered a major factor in carbapenem resistance in A. baumannii, several A. baumannii isolates with ISAba1-blaOXA-51-like remained susceptible to carbapenems in our study. It is likely that differences in transcriptional-level regulation of blaOXA-51-like could affect carbapenem susceptibility in different A. baumannii strains (12).
In the cloning experiments, both A. baumannii ATCC 15151(pOXA23) and A. baumannii ATCC 15151(pOXA66) were resistant to carbapenems, indicating that the presence of ISAba1-blaOXA-23 or ISAba1-blaOXA-66 was sufficient to confer carbapenem resistance even without the assistance of other mechanisms. Figueiredo et al. (8) reported in vivo selection of reduced susceptibility to carbapenems in A. baumannii associated with the ISAba1-related overexpression of blaOXA-66. Furthermore, the inactivation of blaOXA-66 in A. baumannii resulted in higher susceptibility to carbapenems, indicating that blaOXA-66 was involved in reduced susceptibility to carbapenems even when weakly expressed (8). In addition, although OXA-23 was considered a CHDL, we found that the MIC of cefepime, but not that of ceftazidime, was also significantly increased for A. baumannii ATCC 15151(pOXA23) (Table 4). Similar results have been observed previously by introducing a natural blaOXA-23-harboring plasmid, pFER, into A. baumannii ATCC 15151 (CIP 70.10) (10), but the authors did not describe these results in detail. Here we introduced ISAba1-blaOXA-23 into A. baumannii ATCC 15151 (CIP 70.10), and its contribution to cefepime resistance is reported for the first time.
In this study, we showed that the Acinetobacter clinical isolates contained various antibiotic resistance genes to combat fluoroquinolones, aminoglycosides, cephalosporins, and carbapenems. Other mechanisms, such as overexpression of an efflux pump and loss of outer membrane protein, were not investigated in this study. Bratu et al. (3) suggested that efflux pumps seemed not to be important contributors to aminoglycoside or fluoroquinolone resistance. Furthermore, nonenzymatic mechanisms were considered not to be major factors contributing to carbapenem resistance (30). Hu et al. (12) determined the transcription level of ISAba1-blaOXA-66 in A. baumannii by reverse transcription (RT-PCR) and suggested that the expression of blaOXA-66 in E. coli was correlated with imipenem resistance. However, the direct impact of ISAba1-blaOXA-66 in carbapenem-susceptible A. baumannii isolates had not been determined. This is the first report directly demonstrating the impact of ISAba1-blaOXA-66 in transforming carbapenem-susceptible A. baumannii strains to carbapenem-resistant A. baumannii strains. In conclusion, A. baumannii has been the most successful Acinetobacter species in nosocomial infections and contains various antibiotic resistance genes. Continuous surveillance of MDR Acinetobacter spp. and elucidation of their antibiotic resistance mechanisms in the hospital are crucial to help develop effective therapy regimens and to prevent the further dissemination of these MDR species.
Acknowledgments
This work was supported by grants DOH96-DC-2012 and DOH98-DC-2005 from the Centers for Disease Control, Department of Health, Taiwan.
Footnotes
Published ahead of print on 1 March 2010.
REFERENCES
- 1.Bergogne-Bérézin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bou, G., and J. Martinez-Beltran. 2000. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 44:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bratu, S., D. Landman, D. A. Martin, C. Georgescu, and J. Quale. 2008. Correlation of antimicrobial resistance with beta-lactamases, the OmpA-like porin, and efflux pumps in clinical isolates of Acinetobacter baumannii endemic to New York City. Antimicrob. Agents Chemother. 52:2999-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing: fifteenth informational supplement, M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Corvec, S., N. Caroff, E. Espaze, C. Giraudeau, H. Drugeon, and A. Reynaud. 2003. AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J. Antimicrob. Chemother. 52:629-635. [DOI] [PubMed] [Google Scholar]
- 6.Danes, C., M. M. Navia, J. Ruiz, F. Marco, A. Jurado, M. T. Jimenez de Anta, and J. Vila. 2002. Distribution of beta-lactamases in Acinetobacter baumannii clinical isolates and the effect of Syn 2190 (AmpC inhibitor) on the MICs of different beta-lactam antibiotics. J. Antimicrob. Chemother. 50:261-264. [DOI] [PubMed] [Google Scholar]
- 7.Dijkshoorn, L., A. Nemec, and H. Seifert. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939-951. [DOI] [PubMed] [Google Scholar]
- 8.Figueiredo, S., L. Poirel, J. Croize, C. Recule, and P. Nordmann. 2009. In vivo selection of reduced susceptibility to carbapenems in Acinetobacter baumannii related to ISAba1-mediated overexpression of the natural bla(OXA-66) oxacillinase gene. Antimicrob. Agents Chemother. 53:2657-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Héritier, C., L. Poirel, P. E. Fournier, J. M. Claverie, D. Raoult, and P. Nordmann. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Héritier, C., L. Poirel, T. Lambert, and P. Nordmann. 2005. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:3198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Héritier, C., L. Poirel, and P. Nordmann. 2006. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 12:123-130. [DOI] [PubMed] [Google Scholar]
- 12.Hu, W. S., S. M. Yao, C. P. Fung, Y. P. Hsieh, C. P. Liu, and J. F. Lin. 2007. An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3844-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, L. Y., T. L. Chen, P. L. Lu, C. A. Tsai, W. L. Cho, F. Y. Chang, C. P. Fung, and L. K. Siu. 2008. Dissemination of multidrug-resistant, class 1 integron-carrying Acinetobacter baumannii isolates in Taiwan. Clin. Microbiol. Infect. 14:1010-1019. [DOI] [PubMed] [Google Scholar]
- 14.Hujer, K. M., N. S. Hamza, A. M. Hujer, F. Perez, M. S. Helfand, C. R. Bethel, J. M. Thomson, V. E. Anderson, M. Barlow, L. B. Rice, F. C. Tenover, and R. A. Bonomo. 2005. Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 beta-lactamase: defining a unique family of class C enzymes. Antimicrob. Agents Chemother. 49:2941-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunger, M., R. Schmucker, V. Kishan, and W. Hillen. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45-51. [DOI] [PubMed] [Google Scholar]
- 16.Lee, J. H., C. H. Choi, H. Y. Kang, J. Y. Lee, J. Kim, Y. C. Lee, S. Y. Seol, D. T. Cho, K. W. Kim, D. Y. Song, and J. C. Lee. 2007. Differences in phenotypic and genotypic traits against antimicrobial agents between Acinetobacter baumannii and Acinetobacter genomic species 13TU. J. Antimicrob. Chemother. 59:633-639. [DOI] [PubMed] [Google Scholar]
- 17.Lee, Y. T., L. Y. Huang, D. H. Chiang, C. P. Chen, T. L. Chen, F. D. Wang, C. P. Fung, L. K. Siu, and W. L. Cho. 2009. Differences in phenotypic and genotypic characteristics among imipenem-non-susceptible Acinetobacter isolates belonging to different genomic species in Taiwan. Int. J. Antimicrob. Agents 34:580-584. [DOI] [PubMed] [Google Scholar]
- 18.Lévesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim, Y. M., K. S. Shin, and J. Kim. 2007. Distinct antimicrobial resistance patterns and antimicrobial resistance-harboring genes according to genomic species of Acinetobacter isolates. J. Clin. Microbiol. 45:902-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, Y. C., W. H. Sheng, S. C. Chang, J. T. Wang, Y. C. Chen, R. J. Wu, K. C. Hsia, and S. Y. Li. 2008. Application of a microsphere-based array for rapid identification of Acinetobacter spp. with distinct antimicrobial susceptibilities. J. Clin. Microbiol. 46:612-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 22.Lockhart, S. R., M. A. Abramson, S. E. Beekmann, G. Gallagher, S. Riedel, D. J. Diekema, J. P. Quinn, and G. V. Doern. 2007. Antimicrobial resistance among Gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J. Clin. Microbiol. 45:3352-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, P. L., M. Doumith, D. M. Livermore, T. P. Chen, and N. Woodford. 2009. Diversity of carbapenem resistance mechanisms in Acinetobacter baumannii from a Taiwan hospital: spread of plasmid-borne OXA-72 carbapenemase. J. Antimicrob. Chemother. 63:641-647. [DOI] [PubMed] [Google Scholar]
- 24.Mazel, D. 2006. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4:608-620. [DOI] [PubMed] [Google Scholar]
- 25.Mendes, R. E., J. M. Bell, J. D. Turnidge, M. Castanheira, and R. N. Jones. 2009. Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: report from the SENTRY Surveillance Program. J. Antimicrob. Chemother. 63:55-59. [DOI] [PubMed] [Google Scholar]
- 26.Mugnier, P. D., L. Poirel, and P. Nordmann. 2009. Functional analysis of insertion sequence ISAba1 responsible for genomic plasticity of Acinetobacter baumannii. J. Bacteriol. 191:2414-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munoz-Price, L. S., and R. A. Weinstein. 2008. Acinetobacter infection. N. Engl. J. Med. 358:1271-1281. [DOI] [PubMed] [Google Scholar]
- 28.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pitout, J. D., D. B. Gregson, L. Poirel, J. A. McClure, P. Le, and D. L. Church. 2005. Detection of Pseudomonas aeruginosa producing metallo-beta-lactamases in a large centralized laboratory. J. Clin. Microbiol. 43:3129-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poirel, L., and P. Nordmann. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826-836. [DOI] [PubMed] [Google Scholar]
- 31.Poirel, L., and P. Nordmann. 2006. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:1442-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi, C., M. Malczynski, M. Parker, and M. H. Scheetz. 2008. Characterization of genetic diversity of carbapenem-resistant Acinetobacter baumannii clinical strains collected from 2004 to 2007. J. Clin. Microbiol. 46:1106-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rather, P. N., H. Munayyer, P. A. Mann, R. S. Hare, G. H. Miller, and K. J. Shaw. 1992. Genetic analysis of bacterial acetyltransferases: identification of amino acids determining the specificities of the aminoglycoside 6′-N-acetyltransferase Ib and IIa proteins. J. Bacteriol. 174:3196-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segal, H., E. C. Nelson, and B. G. Elisha. 2004. Genetic environment and transcription of ampC in an Acinetobacter baumannii clinical isolate. Antimicrob. Agents Chemother. 48:612-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheng, W. H., Y. C. Lin, J. T. Wang, Y. C. Chen, S. C. Chang, K. C. Hsia, R. J. Wu, and S. Y. Li. 2009. Identification of distinct ciprofloxacin susceptibility in Acinetobacter spp. by detection of the gyrA gene mutation using real-time PCR. Mol. Cell. Probes 23:154-156. [DOI] [PubMed] [Google Scholar]
- 37.Touati, A., L. Brasme, S. Benallaoua, A. Gharout, J. Madoux, and C. De Champs. 2008. First report of qnrB-producing Enterobacter cloacae and qnrA-producing Acinetobacter baumannii recovered from Algerian hospitals. Diagn. Microbiol. Infect. Dis. 60:287-290. [DOI] [PubMed] [Google Scholar]
- 38.Turton, J. F., M. E. Kaufmann, J. Glover, J. M. Coelho, M. Warner, R. Pike, and T. L. Pitt. 2005. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J. Clin. Microbiol. 43:3074-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turton, J. F., M. E. Ward, N. Woodford, M. E. Kaufmann, R. Pike, D. M. Livermore, and T. L. Pitt. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]
- 40.Vila, J., J. Ruiz, P. Goni, A. Marcos, and T. Jimenez de Anta. 1995. Mutation in the gyrA gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 39:1201-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodford, N., M. J. Ellington, J. M. Coelho, J. F. Turton, M. E. Ward, S. Brown, S. G. Amyes, and D. M. Livermore. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351-353. [DOI] [PubMed] [Google Scholar]


