Abstract
The current standard of care for hepatitis C virus (HCV) infection, pegylated alpha interferon in combination with ribavirin, has a limited response rate and adverse side effects. Drugs targeting viral proteins are in clinical development, but they suffer from the development of high viral resistance. The inhibition of cellular proteins that are essential for viral amplification is thought to have a higher barrier to the emergence of resistance. Three cyclophilin inhibitors, the cyclosporine analogs DEBIO-025, SCY635, and NIM811, have shown promising results for the treatment of HCV infection in early clinical trials. In this study, we investigated the frequency and mechanism of resistance to cyclosporine (CsA), NIM811, and a structurally unrelated cyclophilin inhibitor, SFA-1, in replicon-containing Huh7 cells. Cross-resistance between all clones was observed. NIM811-resistant clones were selected only after obtaining initial resistance to either CsA or SFA-1. The time required to select resistance against cyclophilin inhibitors was significantly longer than that required for resistance selection against viral protein inhibitors, and the achievable resistance level was substantially lower. Resistance to cyclophilin inhibitors was mediated by amino acid substitutions in NS3, NS5A, and NS5B, with NS5A mutations conferring the majority of resistance. Mutation D320E in NS5A mediated most of the resistance conferred by NS5A. Taken together, the results indicate that there is a very low frequency and level of resistance to cyclophilin-binding drugs mediated by amino acid substitutions in three viral proteins. The interaction of cyclophilin with NS5A seems to be the most critical, since the NS5A mutations have the largest impact on resistance.
Hepatitis C virus (HCV) poses a serious medical problem, with more than 170 million people infected worldwide (27). Chronic HCV infection increases the risk of hepatocellular carcinoma and results in progressive liver disease and liver failure in approximately 30% of infected individuals (2, 13). HCV infection is the leading indication for liver transplantation in the United States, and HCV reinfection occurs in nearly all cases of chronically infected HCV patients receiving liver transplants. The effectiveness of the current standard therapy (pegylated alpha interferon [PEG-IFN-α] and ribavirin) is genotype dependent. The response rate in genotype 1 patients, the most prevalent genotype in North America, Europe, and Japan, is only 48%, whereas in genotype 2 and 3 patients there is an 88% response rate (4). In view of these limitations to the current standard of care, the development of alternative, more effective treatment regimens is urgently needed.
HCV is a positive-, single-stranded RNA virus with a genome approximately 9.6 kb in length that encodes a single polyprotein, which subsequently is cleaved into 10 distinct viral proteins. The NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase are the major foci of current anti-HCV drug discovery efforts. Both enzymes are essential to HCV viral replication and thus are attractive drug targets. However, like most RNA viruses, HCV has a high mutation rate as well as significant population heterogeneity due to the error-prone nature of the viral RNA-dependent RNA polymerase. This high mutation rate can result in the rapid emergence of drug-resistant viruses during treatment with compounds that target viral genes, such as protease and polymerase inhibitors (3). HCV therapy targeting host proteins rather than viral proteins are thought to reduce the emergence of drug-resistant viruses (7, 11). Furthermore, inhibitors of viral and cellular proteins could be used in combination to provide the more effective treatment of hepatitis C infection. One example of an HCV inhibitor that targets a cellular protein is cyclosporine (CsA) and its derivatives, DEBIO-025, SCY635, and NIM811, all of which exhibit anti-HCV effects by binding to the cyclophilin family of host factors (6, 7, 9, 12, 14, 15, 19, 22, 23, 28, 29).
We reported previously that the level of resistance and the resistance frequency to NIM811 is low compared to those of drugs inhibiting viral proteins (20). The underlying resistance mechanism, however, was not known. Here, we report the selection of two NIM811-resistant clones and the identification of viral mutations conferring resistance. NIM811-resistant clones were obtained after preselection with either cyclosporine A (designated CsA/NIMr) or a chemically distinct cyclophilin binder, the sanglifehrin A analog SFA-1 (SFA/NIMr) (30). CsA/NIMr and SFA/NIMr are cross-resistant, and viral RNA from both contained numerous mutations. Interestingly, both resistant clones contained the mutation D320E in NS5A, which alone conferred the majority of resistance observed for the resistant clones. Our results indicate that cyclophilins interact with several viral proteins. The binding of NS5A and NS5B to cyclophilins A and B had been shown in vitro recently (11). Our data obtained in the cellular environment support this interaction and suggest an additional interaction with the NS3 protease.
MATERIALS AND METHODS
Compounds.
NIM811, CsA, the sanglifehrin A analog SFA-1, and BILN2061 were prepared or isolated at Novartis (Basel, Switzerland).
Cells.
The subgenomic genotype 1b (Con1) HCV replicon cell line, clone A, was obtained from Charles Rice and Apath LLC (St. Louis, MO) (1). Cured Huh7 cells (Huh-cure cells) and the subgenomic genotype 1b replicon cell line with the firefly luciferase reporter, Huh-luc-neo-ET, were obtained from Ralf Bartenschlager/ReBLikon GmbH. Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 2 mM l-glutamine, 0.1 mM nonessential amino acids, and 10% fetal bovine serum (FBS). Replicon cells were cultured additionally in 1 mg/ml G418.
Generation of resistant replicon cells.
Clone A cells were serially passaged for 8 weeks in the presence of 250 μg/ml G418 and increasing concentrations of CsA, SFA-1, and NIM811. The starting concentration of the compounds was 1 μM and increased to 2 μM, and then it was increased to 5 μM at approximately 2-week intervals. Cells did not survive culture in the presence of 1 μM NIM811, regardless of how gradually the drug level was increased. Attempts at increasing CsA concentration above 5 μM and SFA-1 above 10 μM resulted in impaired cell growth. After the selection of CsA-resistant pools of cells in 5 μM CsA, cells were switched to continuous culture in 5 μM NIM811 for an additional 5 weeks. The SFA-1-resistant cells in 5 μM SFA-1 first were cultured for 2 weeks in 2 μM NIM811 followed by further selection in 5 μM NIM811. Colonies were pooled and assayed for resistance. To control the effect of the compound solvent, clone A cells were passaged twice weekly in medium containing 0.2% dimethylsulfoxide (DMSO) and 250 μg/ml G418 for the same length of time as it took to select resistant cell lines (DMSO-resistant [DMSOr] or DMSO control cell line).
RNA transfection.
Huh-cure cells were transfected with replicon RNA by electroporation. Plasmid DNA was linearized by digestion with ScaI, phenol-chloroform extracted, ethanol precipitated, washed, and dissolved in water. RNA transcription reactions were carried out using a T7 MEGAscript kit (Ambion) according to the manufacturer's protocol. For the electroporation, Huh-cure cells were harvested by trypsinization, washed in 50 ml phosphate-buffered saline (PBS), and resuspended in PBS at a concentration of 1 × 107 cells per ml. Four hundred microliters of the cell suspension was mixed with 100 ng viral RNA and 10 μg calf liver tRNA as a carrier. The cell-RNA mixture was electroporated in 0.4-cm-gap-width cuvettes using a Bio-Rad GenePulser set to 950 μF, 270 V, and maximum resistance. Cells were resuspended immediately in complete medium and transferred to an appropriate dish for culture. After 24 h, the medium was exchanged and G418 was added at a concentration of 250 μg/ml. For total RNA transfection, the following variation to the protocol described above were made: RNA was isolated from cell pellets using an RNeasy kit (Qiagen). Ten micrograms of total RNA was mixed with 200 μl of the cell suspension and electroporated in a 0.2-cm-gap-width cuvette set at 1,000 μF, 200 V, and 75 Ω.
qRT-PCR-based HCV replicon assay.
The antiviral activity and cytotoxicity of compounds were determined with various HCV replicon cell lines in a quantitative RT-PCR (qRT-PCR)-based assay as described previously (19). Briefly, 10,000 replicon cells were seeded in each well of a 96-well tissue culture plate and allowed to attach in complete culture medium without G418 overnight. The next day, the culture medium was replaced with medium containing serially diluted compounds in the presence of 2% FBS and 0.5% DMSO. After the cells were treated for 48 h, total RNA was extracted using an RNeasy 96 kit (Qiagen, Valencia, CA). The level of HCV RNA was measured by real-time quantitative PCR (TaqMan assay; Applied Biosystems, Foster City, CA) using HCV-specific primers (5′-TCT TCA CGC AGA AAG CGT CTA-3′ and 5′-CTG GCA ATT CCG GTG TAC T-3′) and probe (5′-6-carboxyfluorescein [FAM]-TCC TGG AGG CTG CAC GAC ACT CAT A-6-carboxytetramethylrhodamine [TAMRA]-3′). The absolute copy numbers of HCV RNA were determined using a standard curve that was established with known quantities of in vitro-transcribed RNA. The level of HCV RNA was normalized for each sample against the amount of total RNA extracted, which was determined using a Quant-iT RNA assay kit (Molecular Probes, Invitrogen). Each data point represents the average for six replicates in cell culture in a single experiment. The percentage of inhibition was calculated as follows: percent inhibition = 1 − (average of compound-treated cells)/(average of control cells). The EC50 is the concentration of compound at which the HCV RNA level in the replicon cells is reduced by 50%. To monitor cytotoxicity, the viability of the replicon cells following 48 h of compound treatment was determined using a tetrazolium compound (MTS)-based assay (CellTiter 96 Aqueous One Solution Cell Proliferation Assay; Promega, Madison, WI). The percentage of cytotoxicity was calculated as follows: percent cytotoxicity = 1 − (average of compound-treated cells)/(average of control cells). Each data point represents the average of three replicates in cell culture.
Plasmid construction.
Total RNA was isolated from NIM811-resistant clone A replicon cells using the RNeasy Mini kit (Qiagen). A reverse transcription reaction was carried out using the Transcriptor Reverse Transcriptase kit (Roche). The NS3- or NS5A-containing regions of the HCV replicon cDNA were amplified by PCR using the Expand Long Template PCR System (Roche) and cloned back into the wild-type pCon1 plasmid. The NS3-containing region was cloned into the DraI/MluI sites, while the NS5A-containing region was cloned into the MluI/MfeI sites. The single NS5B C575G mutation was introduced into the pCon1 plasmid using the QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. All constructs were sequenced to verify the correct manipulation.
Site-directed mutagenesis.
Individual NS5A mutations were introduced into pFKi398LucUbiNeo NS3-3′ (ReBLikon) to take advantage of the luciferase reporter readout for replication activity. The MulI/MfeI fragments of wild-type and resistant NS5A were PCR amplified with the primer pair 5′-CCCAAGCTTCATTCCCCATTAACGCGTACAC (forward) and 5′-TGCTCTAGAGGTGTCAATTGGTGTCTCAGTG (reverse) and cloned into the pCR2.1-TOPO vector (Invitrogen). Mutations were introduced using the QuikChange II XL site-directed mutagenesis kit (Stratagene) and the primers summarized in Table 1. The identified NS5A resistance mutations, D320E and R356Q, were introduced into the wild-type replicon, and the wild-type NS5A amino acid residues, E320D and Q356R, were reintroduced into the resistant replicon. These two NS5A constructs then were cloned into pFKi398LucUbiNeo NS3-3′ using the MulI/BclI restriction sites, and all constructs were confirmed by sequencing.
TABLE 1.
Primers used for site-directed mutagenesis
| Purpose | Sequence |
|---|---|
| Mutation D320E in NS5A | 5′-CATATGGGCACGCCCGGAGTACAACCCTCCACTGTTAG |
| 5′-CTAACAGTGGAGGGTTGTACTCCGGGCGTGCCCATATG | |
| Mutation R356Q in NS5A | 5′-CCTCCGATACCACCTCCACAGAGGAAGAGGACGGTTGTC |
| 5′-GACAACCGTCCTCTTCCTCTGTGGAGGTGGTATCGGAGG | |
| E320D reversal to WT NS5A | 5′-CATATGGGCACGCCCGGATTACAACCCTCCACTGTTAG |
| 5′-CTAACAGTGGAGGGTTGTAATCCGGGCGTGCCCATATG | |
| Q356R reversal to WT NS5A | 5′-CCTCCGATACCACCTCCACGGAGGAAGAGGCCGGTTGTC |
| 5′-GACAACCGGCCTCTTCCTCCGTGGAGGTGGTATCGGAGG |
RESULTS
Generation of CsA-, SFA-1-, CsA/NIM811-, and SFA-1/NIM811-resistant replicon cell lines.
The replicon clones resistant to CsA (CsAr) and SFA-1 (SFA-1r) were generated by the serial passage of clone A cells in the presence of increasing concentrations of drug to a final concentration of 5 μM. Attempts to generate NIM811-resistant replicon cells in the presence of increasing concentrations of NIM811 (starting concentration, 1 μM) failed. Therefore, CsAr and SFA-1r clones subsequently were passaged on increasing concentrations of NIM811 to a final concentration of 5 μM.
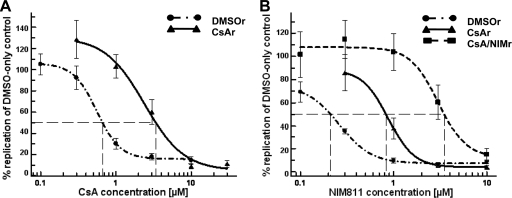
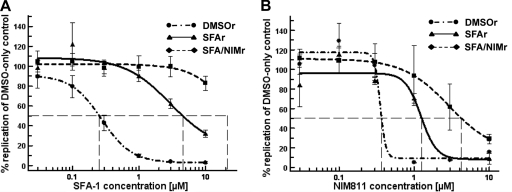
The replicon-containing cells that survived culture on 5 μM CsA or NIM811 were analyzed for susceptibility to CsA and NIM811. The EC50 curves shifted, indicating a gain of resistance to the drugs (Fig. 1A and B). After continuous culture on CsA, the CsAr clone showed a 4.8-fold increase of the EC50 for CsA and a 3.9-fold increase for NIM811. After culture on increasing concentrations of NIM811, the EC50 for NIM811 increased further to a 15-fold difference relative to that of the DMSO-only control cells (Fig. 1). A similar trend was observed for the SFA-1r and SFA/NIMr clones (Fig. 2). After continuous culture on SFA-1, the EC50s of the SFA-1r clone shifted 22.7- and 3.4-fold higher for SFA-1 and NIM811, respectively. Subsequent culture on NIM811 yielded an SFA/NIMr clone with a >40-fold increase in EC50 for SFA-1 and 12.5-fold increase for NIM811 (Fig. 2). Both the CsAr and SFA-1r clones exhibited some cross resistance to NIM811, a compound they were not selected with.
FIG. 1.
Susceptibility of wild-type, CsA-resistant, and CsA/NIM-resistant cells to CsA and NIM811. Wild-type (DMSOr), CsAr, and CsA/NIMr clones were treated for 48 h with CsA (A) or NIM811 (B). Viral RNA was quantified by qRT-PCR and normalized to total cellular RNA. The susceptibility curves of a representative experiment (A and B) show the level of viral RNA after drug treatment as the percentage of DMSO control cells. The standard deviation of six replicas is included. The experiment has been repeated several times, and the same trend has been observed. The EC50 for CsA was 0.66, 3.39, and >10 μM in DMSOr, CsAr, and CsA/NIMr replicon-containing cells, respectively. The EC50 for NIM811 was 0.21, 0.83, and 3.5 μM in DMSOr, CsAr, and CsA/NIMr replicon-containing cells, respectively.
FIG. 2.
Susceptibility of wild-type, SFA-resistant, and SFA/NIM-resistant cells to SFA-1 and NIM811. Wild-type (DMSOr), SFA-1r, and SFA/NIMr clones were treated for 48 h with SFA-1 (A) or NIM811 (B). Viral RNA was quantified by qRT-PCR and normalized to total cellular RNA. The susceptibility curves of a representative experiment (A and B) show the level of viral RNA after drug treatment as the percentage of DMSO control cells. The standard deviations from six replicas are included. The experiment was repeated several times, and the same trend was observed. The EC50 for SFA-1 was 0.25, 4.55, and >10 μM in DMSOr, SFA-1r, and SFA/NIMr replicon-containing cells, respectively. The EC50 for NIM811 was 0.37, 1.26, and 4.26 μM in DMSOr, SFA-1r, and SFA/NIMr replicon-containing cells, respectively.
NIM811 resistance is not mediated by cellular changes.
We sought to determine whether NIM811 resistance was due to cellular or a viral mutation(s). To do this, we cured the CsA/NIMr and DMSO control cells of the HCV replicon by removing G418 selection and culturing the cells in the presence of 1 μM BILN2061 for 2 weeks and then increasing the compound concentration to 5 μM BILN2061 for two additional weeks. After treatment, the cells were assayed for HCV RNA content by qRT-PCR, which confirmed that the level of HCV RNA was below the limit of detection (data not shown). Wild-type replicon (pCon1) RNA then was reintroduced into the cured NIM811-resistant or DMSO control cells by electroporation and selected in the presence of 250 μg/ml G418. Colonies were pooled and assayed for NIM811 EC50 by qRT-PCR after at least 2 weeks of selection. There was no significant difference in the EC50s of NIM811 between the CsA/NIMr and DMSO control cells observed, suggesting that resistance was not mediated by cellular changes (Table 2).
TABLE 2.
Resistance to NIM811 is mediated by viral RNA
| Origin of total RNA | NIM811 EC50 (μM)a | BILN2061 EC50 (nM) |
|---|---|---|
| RNA transfected into cured CsA/NIMr cells | ||
| WT replicon | 0.12 | |
| DMSOr | 0.17 | |
| RNA transfected into Huh-cure cells | ||
| CsA/NIM811r | 1.5 (17.9) | 1.5 |
| DMSOr | 0.084 (1.0) | 1.4 |
Values in parentheses are the fold changes.
Viral mutations mediate resistance to NIM811.
To determine if viral mutations conferred NIM811 resistance, we extracted total RNA from the CsA/NIMr or DMSO-only control clone A replicon cells and transfected this RNA by electroporation into Huh-cure cells. These cells then were selected in medium with 250 μg/ml G418, and resistant colonies were pooled for further culture. Cells were assayed for the NIM811 EC50 by qRT-PCR. Cells transfected with total RNA from the CsA/NIMr cells had an EC50 for NIM811 of 1.5 μM, which is approximately 18-fold higher than the EC50 of DMSO control cells (Table 2). EC50s for BILN2061 were not significantly different between the two cell types (Table 2). These results indicate that changes in viral RNA were responsible for conferring resistance to NIM811.
NIM811 resistance is conferred mainly by mutations in NS5A.
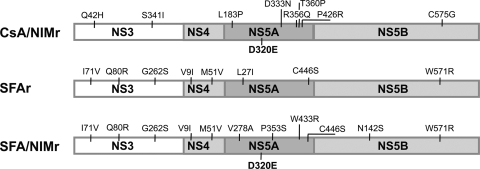
To identify the mutations in the viral RNA associated with NIM811 resistance, we sequenced the entire nonstructural coding region of HCV replicons (NS3 to NS5B) of the CsA/NIMr and SFA/NIMr clones and compared them to sequences of the control cells treated for the same amount of time with DMSO. A total of 9 and 12 mutations were identified in the CsA/NIMr and SFA/NIMr replicons, respectively (Fig. 3). The majority of the mutations mapped to the NS5A region (Fig. 3).
FIG. 3.
Resistant replicon-containing cell lines contain several mutations. RNA of the resistant cell lines and of control cells grown for the same time period with DMSO, the solvent of the drugs, was sequenced. Shown are the mutations in the replicon that were detected only in the resistant cells.
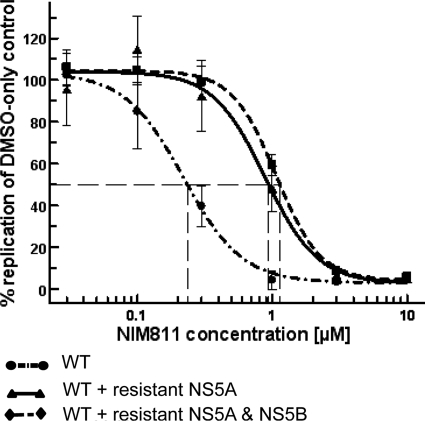
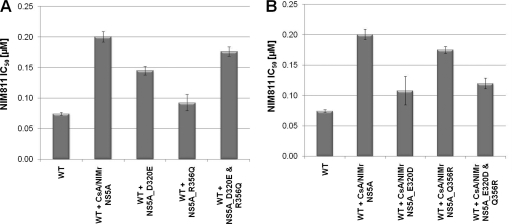
We sought to identify which mutation(s) conferred resistance in the CsA/NIMr clone by swapping the NS5A gene from the resistant replicon into the wild-type replicon. When assayed by qRT-PCR after treatment with NIM811, the EC50 of the wild-type replicon with the NIMr NS5A gene increased from 0.24 to 0.94 μM (Fig. 4). The addition of the C575G point mutation in NS5B increased the EC50 only modestly, to 1.14 μM (Fig. 4). This result was confirmed by the reverse experiment. Replacing NS5A in the CsA/NIMr clone with the wild-type NS5A reduced the resistance by >50%. The additional reversion of the NS5B residue at position 575 to wild type (G575C) yielded cells with 63% resistance compared to that of CsA/NIMr cells (NIM811 EC50 [μM]: CsA/NIMr, 8.4; CsA/NIMr plus WT NS5A, 4; CsA/NIMr plus WT NS5A and NS5B, 5.3). The mutations in NS3 most likely account for the remaining 37% of resistance. We analyzed the effects of the D320E and R356Q mutations in NS5A individually and in combination, since they had been identified in two independent approaches. D320E was identified in both the CsA/NIMr and the SFA/NIMr clones, and the R356Q mutation had been reported previously to confer resistance to CsA (26). For this experiment we chose to use the Huh/Luc-neo-ET replicon, since it allows monitoring replication via luciferase activity. We introduced both mutations by site-directed mutagenesis into the wild-type Huh/Luc-neo-ET replicon and reversed the mutations to wild type in the CsA/NIMr clone. After the selection of stable cell lines, we tested the resulting replicons and found that the R356Q mutation only marginally increased the EC50s, but the D320E mutation caused resistance almost to the level of the six NS5A mutations together (Fig. 5A). The reverse experiment matched this observation. The introduction of the wild-type amino acid at position 320 in the NS5A protein of the CsA/NIMr clone made the cells almost as susceptible to NIM811 as the wild-type clone. Changing amino acid 356 alone had marginal effects. Cross-resistance to SFA-1 remained constant in these experiments, while there was no resistance observed to BILN2061 (data not shown). These results suggest that the D320 residue in the NS5A protein is responsible for conferring a large degree of HCV viral resistance to CsA, NIM811, and SFA-1.
FIG. 4.
Contribution of NS5A and NS5B mutations to resistance against NIM811. The NS5A gene of the CsA/NIMr clone was engineered into the wild-type replicon, and the NS5B mutation was added by site-directed mutagenesis. NIM811 susceptibility curves were established. The graph shows the results of two experiments with six replicas each, with standard deviations indicated by error bars. The EC50 for NIM811 was 0.23, 0.94, and 1.14 μM in WT, WT + resistant NS5A, and WT + resistant NS5A & NS5B replicon-containing cells, respectively.
FIG. 5.
Contribution of the NS5A mutations D320E and R356Q to resistance against NIM811. The mutations were introduced separately or together into the wild-type replicon (A), and the mutations were reversed in the CsA/NIMr clone (B). Susceptibility to NIM811 was analyzed, and the EC50s are shown in comparison to those of the wild-type and mutated NS5A gene containing all six mutations. The graphs are the results from two experiments, with standard deviations indicated by error bars.
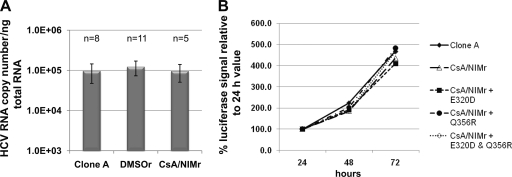
To test whether the viral mutations change the fitness of the virus, we calculated the viral RNA copy number per nanogram of total RNA in clone A, DMSOr, and CsA/NIMr cells. There were no drastic differences observed, especially in comparisons of clone A to CsA/NIMr cells (Fig. 6A). Also, the introduction of the wild-type NS5A gene or the reversion of the 320 and/or 356 mutations did not change luciferase reporter gene activity (Fig. 6B).
FIG. 6.
Mutations of the CsA/NIMr replicon do not affect the replication rate. Equal numbers of clone A, DMSOr, and CsA/NIMr cells were seeded, and the HCV RNA copy number/ng total RNA was analyzed by qRT-PCR after 48 h of growth. The number of experimental repeats and standard deviations are included for each cell type (A). RNA of the indicated replicons (B) was transfected into Huh-cure cells, and the luciferase signal was analyzed after 24, 48, and 72 h. The graphs show the percent increase of the luciferase signal relative to the 24-h value. A repeat of this experiment showed the same trend.
DISCUSSION
The rapid development of resistance to drugs targeting viral proteins prompted us to explore cellular proteins involved in HCV maintenance as possible drug targets. The cyclophilin inhibitor NIM811, as well as two other cyclosporine analogs, DEBIO-025 and SCY635, are currently in clinical development for the treatment of HCV. Targeting the host cell cyclophilins established a higher barrier in vitro to the development of resistance compared to that of viral targets (20). Here, we report how the virus gained resistance to NIM811 and other cyclophilin inhibitors.
The selection of resistant clones required several months, and it was impossible to raise NIMr clones directly via selection with NIM811 under the conditions employed in our experiments, which worked well for viral targets. However, starting with CsAr or SFA-1r clones enabled a stepwise increase in the concentration of NIM811 and selection of resistant clones with a 10- to 20-fold increase in NIM811 EC50 compared to that for wild-type replicon cells. This result was surprising for SFA-1 and NIM811, given that these compounds are distinct chemical molecules. Cross-resistance was observed for the CsAr and SFA-1r clones (see the CsAr or SFA-1r replicon tested with NIM811 in Fig. 1 and 2), which prompted us to use these clones for the further selection of NIM811-resistant clones. The three compounds bind to the target cyclophilins in a similar fashion and likely have a similar mechanism of inhibition (16). There was no cross-resistance observed to compounds that have a different target and mode of inhibition, such as the viral protease inhibitor BILN2061, indicating that the resistance profile of these cells is specific to cyclophilin inhibitors (Table 2).
Transfection of the RNA from the CsA/NIMr clone into Huh-cure cells made these cells as resistant to NIM811 as the original CsA/NIMr clone, indicating that resistance was obtained by mutations in the viral genome rather than in the target host factor. This was confirmed by the lack of an EC50 change when wild-type replicon RNA was transfected into cells cured of the CsA/NIMr replicon.
Sequencing the CsA/NIMr replicon RNA (NS3 through NS5B) identified several mutations, with the largest number of mutations located in the NS5A gene. The identified mutations are not known adaptive mutations that increase replication, and only the L183P mutation has been identified previously in quasispecies naturally occurring in patients (24). We searched the European HCV database (euHCVdb; http://euhcvdb.ibcp.fr/euHCVdb/) to determine the level of conservation of amino acids in NS5A that are mutated in clone A cells treated with compound (Table 3). The degree of conservation of the amino acids varies from 51.92% (L183P) to 99.93% (P426R). Position D320 is quite conserved, as only 17 out of 2,764 sequences contain a different amino acid at this position. It will be interesting to determine whether these 17 viruses are indeed resistant to cyclophilin inhibitors, since additional amino acid substitutions might counterbalance a D320 change.
TABLE 3.
Conservation of the mutated NS5A amino acids in HCV sequences of the European HCV database
| Mutation and treatment | No. of sequences containing a different amino acid than clone A |
|---|---|
| CsA/NIMr | |
| L183P | 1,330 |
| D320E | 17 |
| D333E | 229 |
| R356Q | 5 |
| T360P | 108 |
| P426R | 2 |
| SFA/NIMr | |
| L27I | 336 |
| D320E | 17 |
| W433R | 10 |
| C446S | 144 |
We sought to identify which mutations were responsible for the increase of resistance, and we used the CsA/NIMr clone for this analysis. The introduction of the NS5A gene containing all CsA/NIMr mutations into the wild-type replicon caused a large EC50 shift toward resistance. The addition of the NS5B mutation further increased resistance. However, introducing the NS5B single mutation alone did not change the resistance (data not shown). The fact that full resistance cannot be achieved without mutation(s) in NS3 indicates that NS3 also interacts with cyclophilins. Work is ongoing to identify the impact of each mutation on resistance.
We next investigated the NS5A mutation more closely. The D320E mutation in NS5A was identified in both resistant replicons, CsA/NIMr and SFA/NIMr. Another NS5A mutation at position 356 in the CsA/NIMr clone previously had been reported to confer resistance to CsA (5). Therefore, we chose to test the impact of these two mutations on resistance. We introduced both single mutations and the combination of them by site-directed mutagenesis into the wild-type replicon. In addition, we reverted each mutation in the CsA/NIMr clone, alone or in combination, to the corresponding wild-type NS5A amino acid. The level of resistance to NIM811 achieved by the single D320E mutation was nearly the same as that seen with the NS5A gene containing all six mutations. Reciprocally, changing the amino acid in the CsA/NIMr clone back to wild-type NS5A (E320D) resulted in a replicon that is susceptible to NIM811, which is similar to wild-type replicons. Thus, a single NS5A mutation mediated a large part of resistance, while additional mutations in other genes further increased resistance. The resistance obtained by viral mutations is not due to increased fitness, as we did not observe differences in replication between WT and CsA/NIMr cells or in replicons containing all six or individual mutations in the NS5A gene. It should be noted that the magnitude of resistance (<20-fold) observed here is significantly smaller than that for inhibitors targeting viral proteins. Single mutations can generate several-hundredfold resistances to polymerase or protease inhibitors (17, 18). Several attempts to further increase the resistance to NIM811 failed, indicating that resistance to a cellular target is more difficult to obtain, a characteristic that may make cellular targets more attractive candidates for anti-HCV therapeutics.
We have shown recently that the three compounds used in this study can bind and inhibit several cyclophilins and regulate pathways in which these cyclophilins are involved (30). Recently, it has been demonstrated in vitro that cyclophilin B interacts with NS5B (29) and cyclophilin A with NS5A (10). Our data indicate an additional interaction of the viral NS3 protein with cyclophilins. The virus may need to mutate all three genes and perhaps as-of-yet unidentified cyclophilin-interacting proteins to become resistant to cyclophilin inhibitors. Also, it is possible that the disruption of the interaction of a viral protein with one cyclophilin promotes an interaction with another cyclophilin. Similar to results published previously on cyclosporine A resistance mutations (5), we found that changes in the NS5A protein have the strongest impact on resistance to CsA, NIM811, and SFA-1. NS5A is a very versatile protein that interacts with numerous cellular proteins, thereby fulfilling several functions. It contains two proline-rich domains, which mediate protein-protein interactions. These domains obtain their mature conformation through the peptidyl-prolyl cis/trans isomerase function of cyclophilins. We hypothesize that the change of an amino acid next to a proline (D320, D333, and R356) or a proline itself (P426) in NS5A influences the cis/trans isomerization of the peptide bond. In addition to this transient interaction, cyclophilins act as chaperones, guiding other molecules through the secretory pathway (25). All viral proteins need to traffic from their site of translation in the rough endoplasmic reticulum to the site where replication occurs (lipid raft/membranous web [8, 21]). Additional trafficking to lipid droplets was observed with a genomic replicon or infectious virus (26). We previously identified protein trafficking as one of the virus-essential pathways affected by NIM811 (30). Cyclophilins may assist viral proteins in this process. The fact that several viral proteins depend on cyclophilins and/or that several cyclophilins are exploited by the virus might explain the rather low resistance (20-fold, whereas it is 5,000-fold for a protease inhibitor [20]) and the need for the stepwise increase of drug concentration during a long time period, enabling mutations at different sites in the viral genome. Ongoing clinical studies will show whether this in vitro observation transfers to the situation in patients and targeting cyclophilins indeed raises a higher barrier for the development of resistant HCV.
Footnotes
Published ahead of print on 22 February 2010.
REFERENCES
- 1.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 2.Chisari, F. V. 2005. Unscrambling hepatitis C virus-host interactions. Nature 436:930-932. [DOI] [PubMed] [Google Scholar]
- 3.DeFrancesco, R., and G. Migliaccio. 2005. Challenges and successes in developing new therapies for hepatitis C. Nature 436:953-960. [DOI] [PubMed] [Google Scholar]
- 4.Feld, J. J., and J. H. Hoofnagle. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436:967-972. [DOI] [PubMed] [Google Scholar]
- 5.Fernandes, F., D. S. Poole, S. Hoover, R. Middleton, A.-C. Andrei, and J. Gerstner. 2007. Sensitivity of hepatitis C virus to cyclosporine A depends on nonstructural proteins NS5A and NS5B. Hepatology 46:1026-1033. [DOI] [PubMed] [Google Scholar]
- 6.Firpi, R. J., H. Zhu, G. Morelli, M. F. Abdelmalek, C. Soldevila-Pico, V. I. Machicao, R. Cabrera, A. I. Reed, C. Liu, and D. R. Nelson. 2006. Cyclosporine suppresses hepatitis C virus in vitro and increases the chance of a sustained virological response after liver transplantation. Liver Transpl. 12:51-57. [DOI] [PubMed] [Google Scholar]
- 7.Flisiak, R., J.-M. Dumont, and R. Crabbe. 2007. Cyclophilin inhibitors in hepatitis C viral infection. Exp. Opin. Investig. Drugs 16:1345-1354. [DOI] [PubMed] [Google Scholar]
- 8.Gao, L., H. Aizaki, J.-W. He, and M. M. C. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goto, K., K. Watashi, T. Murata, T. Hishiki, M. Hijikata, and K. Shimotohno. 2006. Evaluation of the anti-hepatitis C virus effects of cyclophilin inhibitors, cyclosporin A, and NIM811. Biochem. Biophys. Res. Commun. 343(3):879-884. [DOI] [PubMed] [Google Scholar]
- 10.Hanoulle, X., A. Badillo, J.-M. Wieruszenski, I. Verdegem, R. Landrieu, R. Bartenschlager, F. Penin, and G. Lippens. 2009. Hepatitis C virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. J. Biol. Chem. 284:13589-13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He, Y., W. Duan, and S.-L. Tan. 2007. Emerging host cell targets for hepatitis C therapy. Drug Discov. Today 12:209-217. [DOI] [PubMed] [Google Scholar]
- 12.Hopkins, S., D. Heuman, E. Gavis, J. Lalezari, E. Glutzer, B. Dimassimo, P. Rusnak, S. Wring, C. Smitley, and Y. Ribeill. 2009. Safety, plasma pharmacokinetics, and anti-viral activity of SCY-635 in adult patients with chronic hepatitis C virus infection. Proceedings of the 44th Annual Meeting of the European Association for the Study of the Liver. EASL, Geneva, Switzerland.
- 13.Inoue, K., K. Sekiyama, M. Yamada, T. Watanabe, H. Yasuda, and M. Yoshiba. 2003. Combined interferon alpha2b and cyclosporin A in the treatment of chronic hepatitis C: controlled trial. J. Gastroenterol. 38:567-572. [DOI] [PubMed] [Google Scholar]
- 14.Inoue, K., T. Umehara, U. T. Ruegg, F. Yasui, T. Watanabe, H. Yasuda, J.-M. Dumont, P. Scalfaro, M. Yoshiba, and M. Kohara. 2007. Evaluation of a cyclophilin inhibitor in hepatitis C virus-infected chimeric mice in vivo. Hepatology 45:921-928. [DOI] [PubMed] [Google Scholar]
- 15.Ishii, N., K. Watashi, T. Hishiki, K. Goto, D. Inoue, M. Hijikata, T. Wakita, and K. Shimotohno. 2006. Diverse effects of cyclosporine on hepatitis C virus strain replication. J. Virol. 80:4510-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kallen, J., R. Sedrani, G. Zenke, and J. Wagner. 2005. Structure of human cyclophilin A in complex with the novel immunosuppressant sanglifehrin A at 1.6 A resolution. J. Biol. Chem. 280:21965-21971. [DOI] [PubMed] [Google Scholar]
- 17.Lu, L., T. Dekhtyar, S. Masse, R. Pithawalla, P. Krishnan, W. He, T. Ng, G. Koev, D. Larson, T. Bosse, R. Wagner, T. Pilot-Matias, H. Mo, and A. Molla. 2007. Identification and characterization of mutations conferring resistance to an HCV RNA-dependent RNA polymerase inhibitor in vitro. Antiviral Res. 76:93-97. [DOI] [PubMed] [Google Scholar]
- 18.Lu, L., T. J. Pilot-Matias, K. D. Stewart, J. T. Randolph, R. Pithawalla, W. He, P. P. Huang, L. L. Klein, H. Mo, and A. Molla. 2004. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 48:2260-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, S., J. E. Boerner, C. Tiong Yip, B. Weidmann, N. S. Ryder, M. P. Cooreman, and K. Lin. 2006. NIM811, a cyclophilin inhibitor, exhibits potent in vitro activity against hepatitis C virus alone or in combination with alpha interferon. Antimicrob. Agents Chemother. 50:2976-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathy, J. E., M. Sue, T. Compton, and K. Lin. 2008. Combinations of cyclophilin inhibitor NIM811 with hepatitis C virus NS3-4A protease or NS5B polymerase inhibitors enhance antiviral activity and suppress the emergence of resistance. Antimicrob. Agents Chemother. 52:3267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, S., and J. Krijnse-Locker. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagawa, M., N. Sakamoto, N. Enomoto, Y. Tanabe, N. Kanazawa, T. Koyama, S. Maekawa, T. Yamashiro, C.-H. Chen, Y. Itsui, and S. Kakinuma. 2004. Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochem. Biophys. Res. Commun. 313:42-47. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa, M., N. Sakamoto, Y. Tanabe, T. Koyama, Y. Itsui, Y. Takeda, C. Chen, S. Oooka, S. Maekawa, N. Enomoto, and M. Watanabe. 2005. Suppression of hepatitis C virus replication by cyclosporin A is mediated by blockade of cyclophilins. Gastroenterology 129:1031-1041. [DOI] [PubMed] [Google Scholar]
- 24.Pellerin, M., Y. Lopez-Aguirre, F. Penin, D. Dhumeaux, and J.-M. Pawlotsky. 2004. Hepatitis C virus quasispecies variability modulates nonstructural protein 5A transcriptional activation, pointing to cellular compartmentalization of virus-host interactions. J. Virol. 78:4617-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, T., L. R. Ferreira, C. Hebert, K. Norris, and J. J. Sauk. 1995. Hsp47 and cyclophilin B traverse the endoplasmic reticulum with procollagen into pre-Golgi intermediate vesicles. A role for Hsp47 and cyclophilin B in the export of procollagen from the endoplasmic reticulum. J. Biol. Chem. 270:18323-18328. [DOI] [PubMed] [Google Scholar]
- 26.Targett-Adams, P., S. Boulant, and J. McLauchlan. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J. Virol. 82:2182-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 28.Watashi, K., M. Hijikata, M. Hosaka, M. Yamaji, and K. Shimotohno. 2003. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology 38:1282-1288. [DOI] [PubMed] [Google Scholar]
- 29.Watashi, K., N. Ishii, M. Hijikata, D. Inoue, T. Murata, Y. Miyanari, and K. Shimotohno. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19:111-122. [DOI] [PubMed] [Google Scholar]
- 30.Wiedmann, B., X. Puyang, K. Brown, A. Gaither, J. Baryza, J. Boerner, D. Poulin, S. Ma, X. Shen, J. Tao, P. Devay, T. Compton, and K. Lin. 2007. Roles of cyclophilins in HCV replication and mode of action of the cyclophilin inhibitor NIM811. Proceedings of the 14th International Symposium on Hepatitis C Virus and Related Viruses. EASL, Geneva, Switzerland.